Summary
Background
In preimplantation mouse embryos, the first cell fate specification to the trophectoderm or inner cell mass occurs by the early blastocyst stage. The cell fate is controlled by cell position-dependent Hippo signaling, although the mechanisms underlying position-dependent Hippo signaling are unknown.
Results
We showed that a combination of cell polarity and cell–cell adhesion establishes position-dependent Hippo signaling, where the outer and inner cells are polar and nonpolar, respectively. The junction-associated proteins angiomotin (Amot) and Amotl2 are essential for Hippo pathway activation and appropriate cell fate specification. In the nonpolar inner cells, Amot localizes to adherens junctions (AJs) and cell–cell adhesion activates the Hippo pathway. In the outer cells, the cell polarity sequesters Amot from basolateral AJs to apical domains, thereby suppressing Hippo signaling. The N-terminal domain of Amot is required for actin binding, Nf2/Merlin-mediated association with the E-cadherin complex, and interaction with Lats protein kinase. In AJs, Ser176 in the N-terminal domain of Amot is phosphorylated by Lats, which inhibits the actin-binding activity, thereby stabilizing the Amot–Lats interaction to activate the Hippo pathway.
Conclusion
We propose that the phosphorylation of S176 in Amot is a critical step for activation of the Hippo pathway in AJs and that cell polarity disconnects the Hippo pathway from cell–cell adhesion by sequestering Amot from AJ. This mechanism converts positional information into differential Hippo signaling, thereby leading to differential cell fates.
Introduction
During preimplantation development, mouse embryos form blastocysts that comprise two cell types: the outer epithelial trophectoderm (TE) layer and the inner cell mass (ICM). TE is required for implantation and later contributes to the placenta. ICM further differentiates into the pluripotent epiblast, which later forms the embryo proper and the primitive endoderm.
Historically, two models have been proposed for the first cell fate specification process: the Inside–Outside (or Positional) Model [1], in which the cell position within the embryo specifies the cell fate, and the Polarity Model [2], in which the acquisition of cell polarity at the eight-cell stage is a critical step in the establishment of differential cell fates. The Polarity Model was further developed to include the promotion of TE fate based on the presence of the apical domain [3, 4]. We recently found that Hippo signaling pathway components, i.e., the TEAD family transcription factor, Tead4 [5–7], its co-activator proteins, Yap (encoded by Yap1) and Taz (encoded by Wwtr1), and the protein kinases, Lats1/2, play critical roles in this cell fate specification process [6, 8]. In the inner cells, cell–cell adhesions activate Hippo signaling, which inactivates Tead4 by suppressing the nuclear accumulation of Yap. In the outer cells, weak Hippo signaling facilitates the nuclear accumulation of Yap. The resulting active Tead4–Yap complex induces the TE-specific transcription factors Cdx2 and Gata3, which promote differentiation into TE [6, 9]. Therefore, establishment of position-dependent Hippo signaling is a critical step during differential cell fate specification, which supports the Inside–Outside Model [6, 8]. We previously proposed that a possible mechanism for differential Hippo signaling may be differences in the degrees of cell–cell contacts between the inner and outer cells [6]. However, the exact mechanisms underlying position-dependent Hippo signaling remain largely unknown.
In support of the Polarity Model, several recent studies have suggested the importance of cell polarity during TE development. The Par-aPKC system plays central roles in the regulation of the apicobasal polarity of cells (see reviews [10–12]). Knockdown of Pard6b resulted in the reduced expression of Cdx2 and the failure of functional TE formation [13]. The complete absence of E-cadherin disrupted cell polarization, while the membrane localization of PKCζ correlated with the nuclear accumulation of Yap and the expression of Cdx2 [14]. These observations suggest that cell polarity is probably important for cell fate specification and the regulation of Hippo signaling in preimplantation embryos. Studies in Drosophila also suggest that the cell polarity regulators Crumbs and aPKC control Hippo signaling in epithelial cells [15–18], although the relationships between cell polarity and the Hippo activation status are opposite in fly epithelial cells and preimplantation embryos. Thus, the exact roles and mechanisms of cell polarity during the regulation of Hippo signaling in preimplantation embryos remain unknown.
The Hippo pathway is controlled by various stimuli (see reviews and references in [19–21]). Cell–cell adhesion is an important activation signal for the Hippo pathway, although the mechanisms that connect junctions to Hippo signaling remain largely unknown. Angiomotin (Amot) family proteins [Amot, Amot-like 1 (Amotl1)/JEAP and Amot-like 2 (Amotl2)/MASCOT [22]] are Hippo signaling components [23, 24] that bind to the tight junction proteins, MUPP1/Patj. [25, 26]. Amot proteins also bind to Yap/Taz and the Nf2 tumor suppressor protein Merlin [23, 24, 27, 28]. Therefore, Amot is a potentially important protein that may connect junctions and the Hippo pathway.
In this study, we analyzed the roles of cell polarity during regulation of Hippo signaling in preimplantation embryos. We found that a combination of cell polarity and cell–cell adhesion established position-dependent Hippo signaling. We also found that phosphorylation of Amot at adherens junctions (AJs) stabilized its interaction with Lats and activated the Hippo pathway. Thus, cell polarity control through the junctional localization of Amot is the molecular basis for establishment of cell position-dependent Hippo signaling and the regulation of cell fate.
Results
Combination of cell polarity and cell adhesion establishes position-dependent Hippo signaling in preimplantation embryos
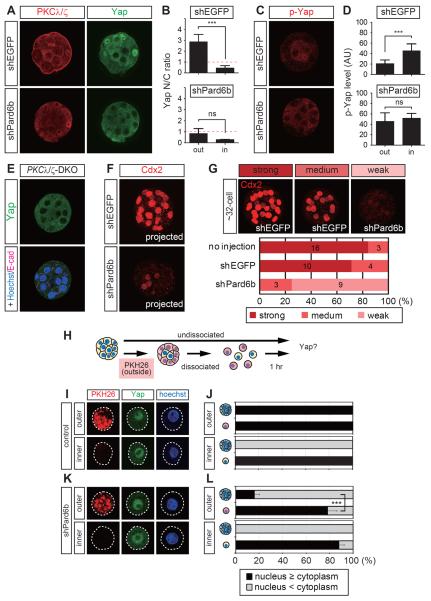
To examine the role of cell polarity during the regulation of Yap, we initially focused on the apical domain regulator, aPKC–Par6–Par3 complex, which we disrupted by knocking down Pard6b via pronuclear injection of short hairpin RNA (shRNA) expression plasmids. As demonstrated previously [13], shPard6b clearly reduced Pard6b proteins around the 32-cell stage, which disrupted the apical domain, as indicated by reduction in apically localized PKCλ/ζ (n = 5/5) and p-ERM (n = 6/6) (Figures 1A and S1A). In contrast, the distributions of the basolateral regulators Scribble (n = 8/8) and Lgl1 (n = 3/3) were expanded into the outside domains (Figure S1A, dots).
Figure 1. Combination of cell polarity and cell–cell-cell adhesion established position-dependent Hippo signaling in 32-cell stage preimplantation embryos.
(A) Effects of Pard6b knockdown (KD) on the apical domain marker PKCλ/ζ and Yap. Nuclear Yap was reduced in the outer cells of Pard6b KD embryos.
(B) Quantification of the ratio of nuclear (N) to cytoplasmic (C) Yap shown in A. ns, not significant. ***, P < 0.001.
(C) Increase in p-Yap in the outer cells of Pard6b KD embryos.
(D) Quantification of the signal intensity of p-Yap shown in C. ns, not significant. ***, P < 0.001.
(E) PKCλ−/−:PKCζ−/− embryos with reduced nuclear Yap in the outer cells.
(F, G) Expression of Cdx2 in 32-cell stage Pard6b KD embryos. (F) Representative Pard6b KD embryo with reduced Cdx2 expression. (G) Embryos were classified into three categories, depending on the Cdx2 expression level. A representative embryo for each category is shown in the upper panels. The numbers in the graphs indicate the numbers of embryos in each category.
(H–L) Effects of cell dissociation on the Yap distribution in normal and Pard6b KD embryos.
(H) Schematic representation of the cell dissociation experiments.
(I, J) Distribution of Yap in dissociated control (uninjected) embryos.
(K, L) Distribution of Yap in dissociated Pard6b KD embryos.
(J, L) Quantification of Yap distribution. ***, P < 0.001
See Figure S1 for related data.
In these apical domain-disrupted embryos, nuclear Yap signals were markedly reduced in the outer cells (n = 9/9; Figures 1A, 1B). In this paper, Yap is used to describe Yap and Taz, because the anti-Yap antibody detects both proteins. The activation of Hippo signaling promotes the phosphorylation of Yap (p-Yap), including the 112th serine residue (S112) by Lats1/2 [29]. In normal embryos, the p-Yap signal or Hippo signaling is strong in the inner cells and weak in the outer cells (Figures 1C, 1D) [6]. In Pard6b knockdown (KD) embryos, the levels of p-Yap signals in the outer cells were increased to a level similar to that in the inner cells (Figure 1C, 1D; n = 9/9). Consistent with the importance of Hippo signaling for cell fate control, Pard6b KD embryos had lower Cdx2 expression at the 32-cell stage (Figures 1F, 1G).
Essentially, the same results were obtained after suppressing PKCλ/ζ activity through overexpression of dnPKCλ (the kinase activity-negative form of PKCλ) [30] (Figure S1B) (Yap: n = 15/16, p-Yap: n = 8/9) and the genetic ablation of aPKC in PKCλ−/−:PKCζ−/− embryos (Figure 1E, n = 3/3; and Figures S1F, S1G) [31]. Overall, these results suggest that the presence of an apical domain suppresses Hippo signaling in the outer cells.
Disruption of the apical Par–aPKC complex activated the Hippo pathway. To understand whether this activation requires cell–cell adhesions, we dissociated 32-cell stage embryos and examined the distribution of Yap. To mark the original cell position within the embryos, the outer cells were labeled with the red fluorescent dye PKH26 before dissociation [32] (Figure 1H). The inner and outer cells of the normal undissociated embryos exhibited clear cytoplasmic and nuclear Yap, respectively (Figures 1A, 1J). When these embryos were dissociated, all the cells exhibited clear nuclear Yap, irrespective of the original cell position (Figures 1I, 1J). In the undissociated Pard6b KD embryos, 84% of the outer cells had cytoplasmic Yap (Figures 1A, 1L). After the Pard6b KD embryos were dissociated, the majority of the outer and inner cells exhibited nuclear Yap, or nuclear and cytoplasmic Yap (Figures 1K, 1L). These results suggest that cell adhesion is required for all blastomeres to exclude Yap from the nuclei. Therefore, in the outer cells of normal embryos, the presence of the apical domain or the operation of the Par–aPKC system inhibits the activation of the Hippo pathway via cell–cell adhesion.
The Hippo pathway component Amot has a cell position-dependent differential distribution
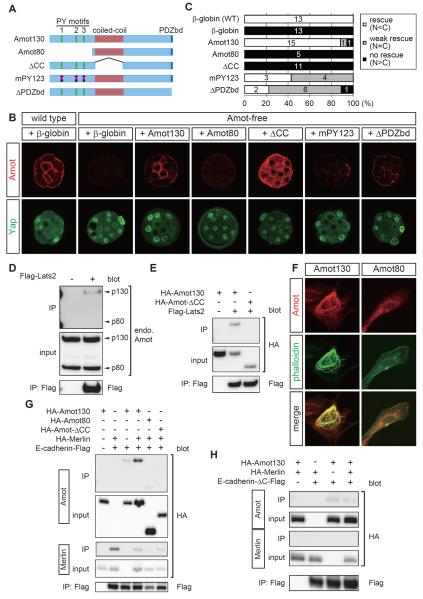
Amot is a junction-associated Hippo pathway component [23, 24]. There are two isoforms of Amot protein: p130 Amot (Amot130) and p80 Amot (Amot80), the latter lacking the N-terminal portion of Amot130 [33] (Figure 4A). We focused on Amot130, which we refer to as Amot unless stated otherwise. Yeast two-hybrid screening of Amot-interacting proteins identified Yap. Like other researchers, we found that multiple Hippo pathway components, including Merlin, Lats2, and Kibra, also bind to Amot [23, 24, 27, 28] (Figures 4D, 4E, and S4A–S4C).
Figure 4. The N-terminal domain of Amot is required for actin polymerization, association with AJs and Lats2, and Hippo pathway activation.
(A) Schematic representations of mutant Amot proteins.
(B) Representative results from the rescue experiments. mRNAs encoding the proteins indicated were injected into wild-type or Amot-free embryos. The upper panels show the distributions of injected Amot proteins (except the wild-type embryos). The lower panels show the distributions of Yap proteins.
(C) Graphs summarizing the rescue activities of each protein. The numbers in the graphs show the number of embryos in each category.
(D) Co-immunoprecipitation experiments showing the interaction between endogenous Amot130 and Lats2.
(E) Co-immunoprecipitation experiments showing the requirement for the coiled-coil domain of Amot for the interaction with Lats2.
(F) F-actin polymerization and actin-binding activities of Amot proteins in NIH3T3 cells. Exogenous Amot proteins were detected using the HA-tag.
(G) Co-immunoprecipitation experiments showing the interactions of Amot with E-cadherin and Merlin. Note that interaction between Merlin and E-cadherin was not enhanced by Amot.
(H) Co-immunoprecipitation experiments showing the requirement for the β-catenin-binding domain of E-cadherin in the Merlin-dependent interaction with Amot.
See Figure S4 for related data.
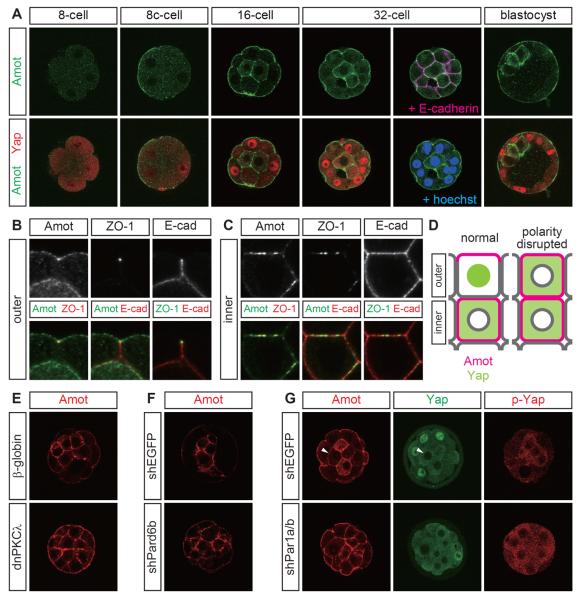
In the 32-cell stage, Amot proteins had different subcellular distributions in the inner and outer cells (Figure 2A). In the outer cells, Amot was localized to the apical membrane and was not present in the (baso)lateral membrane. In the inner cells, Amot was distributed throughout the plasma membrane and weakly in the cytoplasm. The polarized Amot distribution in the outer cells was established via cell polarization during compaction of the eight cell stage embryos (Figures 2A and S2A). The cell position-dependent differential distribution of Amot was established after formation of the inner cells during the 16-cell stage, and this difference was maintained until the early blastocyst stage (Figure 2A). The absence of Amot protein from the basal membrane of polarized cells was detected in the outer cells of the blastocysts (Figure 2A).
Figure 2. The junction-associated Hippo component angiomotin had different position- and polarity-dependent distributions in preimplantation embryos.
(A) Correlation between the position-dependent distributions of Amot and Yap in eight cell stage embryos and blastocysts. 8c-cell: compacted eight cell stage.
(B) Amot distribution in the outer cells relative to the distributions of ZO-1 and E-cadherin. Note that the ZO-1 signals in the inner cells were significantly weaker than those in the outer cells (tight junctions). Therefore, the ZO-1 signals in the inner cells are not visible in this panel.
(C) Comparison of the Amot distribution in the inner cells with those of ZO-1 and E-cadherin.
(D) Schematic representation showing the distributions of Amot and Yap proteins in the inner and outer cells of normal and polarity-disrupted embryos.
(E) Amot distribution in PKCλ/ζ-inhibited embryos.
(F) Amot distribution in Pard6b KD embryos.
(G) Par1a/b DKD disrupted the Amot distribution and Hippo signaling. The arrowhead indicates an inner cell with nuclear exclusion of Yap in the absence of a clear Amot signal in the cytoplasm. Therefore, cytoplasmic Amot is probably not important for the regulation of Yap.
See Figure S2 for related data.
Importantly, the presence of Amot proteins in the apical membrane or throughout the plasma membrane was always correlated with the presence of Yap in the nuclei or in the cytoplasm, respectively (Figure 2A, D). In the outer cells, Amot was present at tight junctions, where it overlapped with two major junction proteins, ZO-1 and E-cadherin (Figure 2B). In the inner cells, Amot colocalized with ZO-1 and E-cadherin (Figure 2C), which suggested that Amot was present in the vicinity of AJs. Of the junctional Hippo components, these correlations were probably specific to Amot because three other Hippo components, i.e., Merlin, Kibra, and α-catenin, were present throughout the plasma membrane (Figures S2B–S2D).
The Par–aPKC system controls the distribution of Amot
Amot localized to the apical domains in polarized cells, and thus, we examined the role of the apical regulator, Par–aPKC, during the regulation of Amot. In embryos expressing dnPKCλ, the apical restriction of Amot was absent from the outer cells. Amot was also present in the basolateral membrane, similar to the inner cells (n = 4/4) (Figure 2E). Pard6b KD embryos produced the same results (n = 17/18) (Figure 2F). Thus, the subcellular distribution of Amot was controlled by the apical Par–aPKC complex.
The activity of the apical Par–aPKC complex is suppressed by the basolateral regulator Par1 [10], and thus, we examined the effects of downregulating two Par1 proteins, Par1a and Par1b, by coinjecting shRNA plasmids for Par1a/b (Figure S2E). Par1a/b double knockdown (DKD) embryos exhibited no apparent changes in apically localized PKCλ/ζ (n = 6/7) and Pard6b (n = 6/6), or the basolateral localization of E-cadherin (n = 6/6) (Figure S2F). However, the Amot distribution was changed (n = 7/7), and Amot was present in the lateral membrane of the outer cells, thereby mimicking the inner cells (Figure 2G). The importance of the junctional localization of Amot in the Hippo pathway activation was supported further because Yap was excluded from the nuclei of the outer cells (n = 7/10) (Figure 2D, G) while the phosphorylation of Yap was increased in the outer cells (n = 3/3) (Figure 2G). These results suggest that the Par–aPKC system, including Par1, controls the junctional localization of Amot and Hippo signaling in the outer cells.
Amot family proteins are required for activation of the Hippo pathway
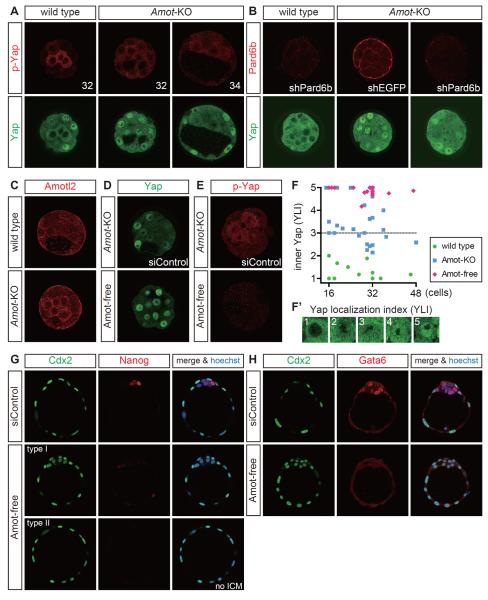
To examine the role of Amot, we used Amot mutant embryos [34]. Amot-null mutant embryos (Amot−/− or Amot−/Y; Amot is on the X chromosome) lacked Amot proteins (Figure S3A), and these embryos failed to exclude Yap from the nuclei of the inner cells until the 32-cell stage (Figures 3A, 3F, 3F'; the subcellular distribution of Yap in the inner cells was determined semiquantitatively based on the Yap localization index (YLI): 1 = cytoplasmic Yap, 5 = nuclear Yap). The strong p-Yap signals in the inner cells were also reduced (n = 14/14) (Figure 3A), which supports the importance of Amot in activating the Hippo pathway and regulating Yap. Pard6b KD in Amot mutant embryos led to nuclear Yap in all cells (n = 3/4) (Figure 3B) resembling the Amot mutants, which indicates that Amot functions downstream of cell polarity.
Figure 3. Amot family proteins are required for Hippo pathway activation in preimplantation embryos.
(A) Distribution of Yap and p-Yap proteins in Amot mutant (Amot-KO) embryos. The numbers in the upper panels indicate the number of nuclei in the embryos shown.
(B) Distribution of Yap in Pard6b KD Amot mutant embryos.
(C) Distribution of Amotl2 proteins in wild-type and Amot mutant embryos.
(D) Distribution of Yap in Amot mutant and Amot-free embryos. Amot-free indicates Amotl2 KD in Amot mutant embryos.
(E) Distribution of p-Yap in Amot mutant and Amot-free embryos. Note that virtually no p-Yap signal was observed in Amot-free embryos.
(F) Ontogenic change in the subcellular distribution of Yap in the inner cells. The subcellular distribution of Yap was determined semiquantitatively based on the Yap localization index (YLI) shown in F′. Each dot represents the mean index value of the inner cells in a single embryo.
(G) Expression of Cdx2 and Nanog in Amot-free embryos at E4.5.
(H) Expression of Cdx2 and Gata6 in Amot-free embryos at E4.5.
See Figure S3 for related data.
In the later stages, however, the inner cells of most Amot mutants exhibited moderate nuclear Yap (Figure 3A,F), and Amot mutants survived until the postimplantation stages [34]. There are two other Amot family genes, Amotl1 and Amotl2, and thus, we hypothesized that these proteins may also play roles in the regulation of Yap. Indeed, Amotl2 (but not Amotl1) was expressed (Figure 3C, and data not shown). In wild-type embryos, Amotl2 was localized only to the apical membrane of the outer cells (Figure 3C). However, in Amot mutant embryos, it was interesting that Amotl2 also localized to the plasma membrane of the inner cells, similar to Amot (Figure 3C).
To examine the role of Amotl2, we knocked down Amotl2 by siRNA injection. In Amotl2 KD embryos, Amotl2 protein was clearly reduced, but these embryos exhibited a normal Yap distribution and Hippo pathway activation, which was monitored based on p-Yap (Figures S3B, S3C). However, when Amotl2 KD was performed with Amot mutants, these embryos, which we describe as Amot-free embryos, exhibited strong nuclear localization of Yap in the inner cells, even in the later stages (n = 30/30) (Figures 3D, 3F), and the activation of the Hippo pathway was almost completely lost (n = 7/7) (Figure 3E). Therefore, Amot family proteins are essential for the activation of the Hippo pathway in preimplantation embryos, where Amot and Amotl2 play major and supplementary roles, respectively. Consistent with the idea that cell polarity acts upstream of Amot, Amot-free embryos had a normal distribution of PKCλ/ζ (n = 5/5) (Figure S3D).
At E4.5, the Amot-free embryos exhibited two morphological types: a blastocyst-like morphology (type I) and a cyst lacking inner cells (type II). In both types, all the cells strongly expressed the TE regulator Cdx2 (n = 27/27) (Figures 3G, 3H, and S3E). Two similar phenotypes were also observed with Lats1/2 double mutants, in which the Hippo pathway was inactive (Figure S3F) [6]. In Amot-free embryos, the expression of the epiblast marker Nanog was lost or significantly reduced (n = 2/2) (Figure 3G), while expression of the primitive endoderm marker Gata6 was lost (n = 3/3) (Figure 3H). Formation of blastocoels in Amot-free embryos suggested the formation of functional TE because Tead4−/− and Pard6b KD embryos, which lacked TE, failed to form blastocoels. Therefore, activation of the Hippo pathway by Amot family proteins is critically important for appropriate cell fate specification.
The N-terminal and coiled-coil domains are required for efficient activation of the Hippo pathway in vivo
To understand the mechanisms that allow Amot to activate the Hippo pathway in AJs, we performed in vivo domain analysis of Amot. We injected the RNAs of various Amot proteins (Figure 4A), together with siAmotl2, into both blastomeres of two-cell stage Amot mutant embryos and examined the rescue activities of the expressed Amot proteins. Injection with an appropriate dose of the Amot130 RNA clearly rescued the mutant phenotype, which was demonstrated by the nuclear exclusion of Yap in the inner cells (Figures 4B, 4C). The expressed Amot proteins had similar distribution patterns to the endogenous proteins (Figure 4B). Overexpression of Amot led to the formation of abnormal F-actin-mediated aggregates in the cytoplasm and nuclear exclusion of Yap in the outer cells (data not shown). Therefore, we used the same RNA dose for all constructs to ensure that they were expressed at physiological levels.
Expression of Amot80, which lacks the N-terminal domain, failed to rescue the Amot-free phenotype (Figures 4A–4C). The Amot80 protein level was low and the localization of Amot80 to inner cell AJs was not seen clearly (Figure 4B). Expression of Amot-ΔCC, which lacks the central coiled-coil domain, also failed to rescue Amot-free embryos (Figures 4A–4C). In this case, however, Amot-ΔCC proteins were abundant, including AJs. These results suggest that the N-terminal and coiled-coil domains are both required for activation of the Hippo pathway, while the N-terminal domain is required for the localization of Amot proteins to AJs and probably to maintain their stability. It appears that Amot is stabilized in the AJs of Hippo-active cells. The role of the coiled-coil domain in the Hippo pathway activation is distinct from that in the N-terminal domain.
The N-terminal domain contains three PY motifs. The first two PY motifs interact with NEDD4-like E3 ubiquitin ligase [35] and Yap [23, 24, 27], respectively. We found that the third motif interacts with Kibra (Figures S4B,S4C). Amot also interacts with the junctional proteins, MUPP1/Patj, via the C-terminal PDZ domain-binding motif [25, 26]. To examine the importance of these motifs, we also expressed Amot-mPY123, which had mutations in all three PY motifs, and Amot-ΔPDZbd, which lacked the C-terminal PDZ domain-binding motif, in Amot-free embryos (Figure 4A). The rescue activities of Amot-mPY123 and Amot-ΔPDZbd appeared to be weaker than that of unmodified Amot, but they clearly rescued the mutant phenotype (Figures 4B,4C), thereby indicating that these motifs do not play critically important roles by themselves.
The N-terminal and coiled-coil domains are both required for efficient interaction with Lats2
The N-terminal domain of Amotl2 binds to and activates Lats2 [36]. Thus, we examined the requirements of the Amot domains for interaction with Lats2 by co-immunoprecipitation (co-IP) of tagged proteins expressed in HEK293T cells. In HEK293T cells, endogenous Amot130, but not Amot80, co-immunoprecipitated with exogenous Lats2 (Figure 4D). When overexpressed, Amot80 also interacted with Lats2, but this interaction was clearly weaker than that with Amot130 (Figure S4D). Amot-ΔCC did not interact with Lats2 (Figure 4E). Therefore, the functionally important N-terminal and coiled-coil domains are both required for the strong interaction between Amot and Lats2.
The N-terminal domain is also required for the actin-binding activity and the Merlin-mediated interaction with the E-cadherin complex
Amot80 did not exhibit strong signals in AJs (Figure 4B), and thus, we examined the molecular basis of this result. Consistent with a previous report that the N-terminal domain of Amot interacts with and promotes the formation of F-actin [37], Amot130-expressing HEK293T and NIH3T3 cells formed thick F-actin bundles where Amot130 co-localized, whereas Amot80 did not exhibit these activities (Figures 4F, S4E). Therefore, the localization to AJs may involve binding to cortical actin fibers.
To further elucidate the relationship between Amot and AJs, we examined the interaction between Amot and the E-cadherin complex. When E-cadherin was immunoprecipitated from cell lysates co-expressing E-cadherin and Amot, weak co-IP of Amot was observed (Figure 4G). Amot interacts with Merlin [28], which also interacts with α-catenin, a core component of cadherin complexes [38]. Thus, we hypothesized that the interaction between Amot and the E-cadherin complex involved Merlin. As expected, co-expression of Merlin enhanced co-IP of Amot and E-cadherin (Figure 4G). Amot interacts directly with Merlin via its coiled-coil domain [28]. In support of the importance of its interaction with Merlin, Amot-ΔCC did not interact with E-cadherin (Figure 4G). We also observed weak co-IP of Amot80 and E-cadherin, but no enhancement was observed by the co-expression of Merlin (Figure 4G and data not shown). Amot130 and Amot80 contain the coiled-coil domain, and thus, the N-terminal domain has accessory roles that stabilize its interaction with the E-cadherin complex.
Amot also had weak co-IP with E-cadherin-ΔC, which lacked the β-catenin interaction domain [39, 40] (Figure 4H). The interaction of α-catenin with cadherin requires β-catenin, and thus, this result suggested that a catenin-independent mechanism was also involved in the interaction between Amot and E-cadherin. Consistent with the hypothesis that Merlin cooperates with α-catenin, Merlin did not interact with E-cadherin-ΔC and the interaction between Amot and E-cadherin-ΔC was not enhanced by Merlin (Figure 4H). Overall, these results suggest that the N-terminal domain of Amot is essential for its strong interaction with Lats2, for actin binding, and for the strong Merlin-dependent interaction with the E-cadherin complex.
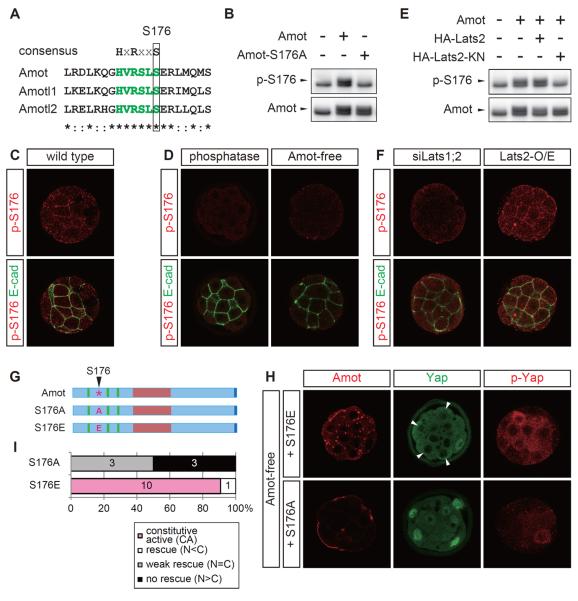
Phosphorylation of the Lats target site in Amot, S176, activates the Hippo pathway
The N-terminal domain of Amot family proteins possesses a sequence, HVRSLS, which matches the consensus motif for the Lats phosphorylation site HxRxxS [29, 41]. The 176th serine residue (S176) in this Amot motif is a possible Lats phosphorylation site (Figure 5A). We generated an antibody that recognized phosphorylated S176 specifically (p-S176). In the western blot analysis, this antibody detected Amot but not the nonphosphorylatable mutant Amot-S176A (Figure 5B). In preimplantation embryos, this antibody produced signals in the inner cell AJs (Figure 5C). These signals were not observed in the embryos treated with lambda phosphatase or Amot-free embryos (Figure 5D), which indicated that the antibody specifically detected phosphorylated Amot (p-S176-Amot).
Figure 5. Phosphorylation of S176 in Amot by Lats activates the Hippo pathway.
(A) Identification of the Lats phosphorylation consensus sequence in the N-terminal domain of Amot family proteins. The amino acid sequences of the mouse proteins are shown.
(B) Western blot analysis showing the specificity of anti-p-S176-Amot antibody. The anti-p-S176-Amot and anti-Amot antibodies detected endogenous (lower bands) and tagged-exogenous (upper bands indicated with arrowheads) Amot proteins.
(C) Localization of p-S176-Amot to AJs in the inner cells of 32-cell stage embryos.
(D) Anti-p-S176-Amot antibody specifically detected phosphorylated Amot proteins. Amot-free and phosphatase-treated embryos did not exhibit signals.
(E) Western blot analysis showing the Lats-dependent phosphorylation of S176-Amot.
(F) Distribution of p-S176-Amot in Lats1/2 DKD and Lats2-overexpressing embryos. The efficient knockdown of Lats1/2 in Lats1/2 DKD embryos was confirmed by the nuclear accumulation of Yap in all blastomeres, which was similar to that in Lats1−/−; Lats2−/− embryos (data not shown).
(G) Schematic representations of mutant Amot proteins.
(H) Distribution of Amot-S176A/E, Yap, and p-Yap proteins in an Amot-free embryo. The arrowheads indicate outer cells that exhibited the clear nuclear exclusion of Yap.
(I) Graphs summarizing the rescue activities of each protein. The numbers in the graphs show the number of embryos in each category.
Western blot analysis of cell lysates expressing Lats2 or kinase activity-negative Lats2 (Lats2-KN) detected Lats activity-dependent phosphorylation of S176 (Figure 5E). In embryos, junction-localized p-S176-Amot was not detected in Lats1/2 DKD embryos but was ectopically observed in Lats2-overexpressing embryos (Figure 5F). These results suggest that Lats phosphorylated S176 in Amot in the AJs of the inner cells.
p-S176-Amot localized to the AJs of the Hippo-active inner cells, and thus, we examined the role of this phosphorylation in the Hippo pathway activation. RNA injection into Amot-free embryos showed that the nonphosphorylatable mutant Amot-S176A had the very weak rescue activity of the mutant phenotype (Figures 5G–5I). The inner cells exhibited nuclear Yap, and the p-Yap signal remained very low (Figure 5H). Similar to Amot80, the level of Amot-S176A was low and no clear signal was observed in the AJs (Figure 5H). In contrast, a phospho-mimetic mutant, Amot-S176E, excluded Yap from the inner cell nuclei (Figures 5G–5I). Importantly, Yap was also excluded from the outer cell nuclei, which suggests that Amot-S176E is a constitutively active protein, although the degree and frequency of nuclear exclusion in the outer cells were lower than those in the inner cells (Figures 5H, 5I). Ectopic activation of Hippo signaling was also confirmed by increased p-Yap signals (Figure 5H). The Amot-S176E proteins formed dense discrete dots on the plasma membranes (Figure 5H). These results suggest that the phosphorylation of S176 has critically important roles in controlling the functions of the N-terminal domain: localization/stabilization in the AJs and the activation of the Hippo pathway. Therefore, the phosphorylation of S176 in Amot is important for Hippo pathway activation in AJs.
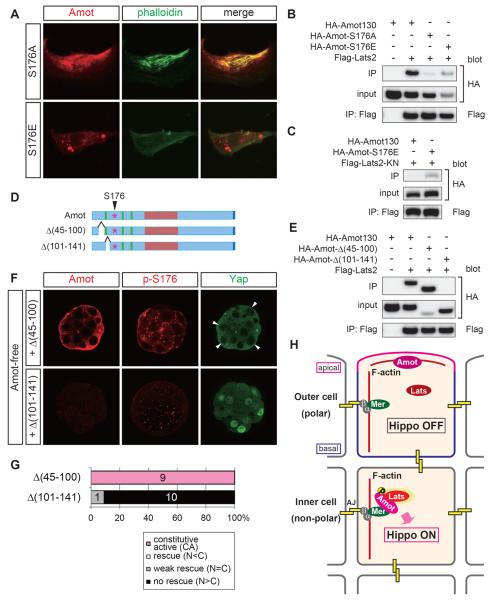
Phosphorylation of S176 suppresses the actin-binding activity of Amot
To determine the molecular basis of Hippo pathway activation via the phosphorylation of S176, we investigated how this phosphorylation modifies the molecular properties of Amot. First, we examined the actin-binding activity. In HEK293T and NIH3T3 cells, Amot-S176A had actin polymerization and binding activities similar to Amot, whereas Amot-S176E had neither activity (Figures 6A and S5A). Therefore, nonphosphorylated Amot binds to actin, and the phosphorylation of S176 suppresses its actin-binding activity.
Figure 6. Phosphorylation of S176 in Amot stabilizes the interaction with Lats2.
(A) The F-actin polymerization and actin-binding activities of Amot-S176A/E proteins in NIH3T3 cells. Exogenous Amot proteins were detected with the HA-tag.
(B) Co-immunoprecipitation experiments showing the interaction of Amot-S176A/E with Last2.
(C) Co-immunoprecipitation experiments showing the interaction of Amot-S176A/E with Lats2-KN.
(D) Schematic representations of the mutant Amot proteins.
(E) Co-immunoprecipitation experiments showing the interaction between Amot-Δ(45-100)/(101-141) and Lats2.
(F) Distribution of Amot, p-S176-Amot, and Yap proteins in Amot-Δ(45–100)/Δ(101–141)-expressing Amot-free embryos. The arrowheads indicate outer cells that exhibited clear nuclear exclusion of Yap.
(G) Graphs summarizing the rescue activities of each protein. The numbers in the graphs show the number of embryos in each category.
(H) Model of Amot-regulated differential Hippo activation in preimplantation embryos. α, α-catenin; β, β-catenin; Mer, Merlin; AJ, adherens junction.
See Discussion for details.
See Figure S5 for related data.
Phosphorylation of S176 does not affect the Merlin-dependent interaction between Amot and the E-cadherin complex
We examined the effects of S176 phosphorylation on the interaction between Amot and the E-cadherin complex. Amot-S176A and Amot-S176E had Merlin-dependent interactions with E-cadherin (Figure S5B). Therefore, the phosphorylation status of S176 did not have significant effects on the interaction between Amot and the E-cadherin complex.
Phosphorylation of S176 in Amot is required for its strong interaction with Lats
Finally, we examined the effects of S176 phosphorylation on the interaction between Amot and Lats2. In co-IP experiments, Amot-S176A and Amot-S176E had weak and strong interactions with Lats2, respectively (Figure 6B). Therefore, the phosphorylation of S176 is required for the strong interaction with Lats2. Consistent with the idea that S176 is a Lats target site, Amot did not have clear interaction based on co-IP from the lysates of cells co-transfected with Amot and Lats2-KN (Figure 6C). In this system, Amot-S176E had a strong interaction with Lats2-KN (Figure 6C). These results suggest that the phosphorylation of S176 by Lats enhances the interaction between Amot and Lats.
Strong correlations between Lats binding, the protein stability at AJs, and the Hippo pathway activation
Further support of the importance of S176 phosphorylation was provided by the correlations between Lats binding, the in vivo protein stability at AJs, and mutant rescue activity using two additional deletion mutants of Amot. One mutant, Amot-Δ(45–100), had a strong interaction with Lats2 (Figures 6D, 6E). In rescue experiments, Amot-Δ(45–100) protein was abundant, including those in AJs, where S176 was phosphorylated, and the embryos exhibited clear nuclear exclusion of Yap, which was an indication of the strong activation of the Hippo pathway, in all cells (Figures 6F, 6G). Amot-Δ(45–100) activated the Hippo pathway without disturbing the cell polarity, which was demonstrated by apically localized PKCλ/ζ (Figure S5C). The second mutant Amot-Δ(101–141), which had only a very weak interaction with Lats2 (Figures 6D,6E), was not detected in embryos and failed to rescue the mutant phenotype (Figures 6F, 6G). Therefore, the phosphorylation of S176 by Lats is an important step during Hippo pathway activation in AJs, which includes protein stabilization in the AJs and a stable interaction with Lats.
Discussion
Combination of cell adhesion and cell polarity converts positional information into cell fate via Hippo signaling ifate via Hippo signaling in preimplantation embryos
We previously showed that cell position-dependent Hippo signaling is the key mechanism underlying position-dependent cell fate specification in preimplantation embryos [6, 8]. In the present study, we demonstrated the involvement of cell polarity and cell–cell adhesion with the regulation of Hippo signaling. In the outer cells, the presence of cell polarity or the operation of the Par–aPKC system suppressed the activation of the Hippo pathway via cell–cell adhesion. In the inner cells, the absence of cell polarity facilitated Hippo pathway activation via cell–cell adhesion, thereby leading to “inside-ON, outside-OFF” differential Hippo signaling in the embryo. This mechanism implies that cells in preimplantation embryos interpret their positional information via a combination of two cellular processes, i.e., cell polarity and cell–cell adhesion, to control their cell fate. Furthermore, this mechanism is consistent with the two historical models of the initial cell fate specification process: the Inside–Outside Model [1] and the newer version of the Polarity Model [3, 4].
A model of Amot-dependent Hippo pathway regulation in preimplantation embryos
At the molecular level, the localization of Amot family proteins to AJs is essential for the activation of the Hippo pathway, while the cell polarity/Par–aPKC system controls the AJ localization of Amot. The roles of Amot in Hippo pathway activation and the establishment of position-dependent Hippo signaling are summarized in the following model (Figure 6H).
In the inner cells, (1) Amot localizes to AJs by binding to cortical F-actin and the E-cadherin complex by interacting with Merlin. E-cadherin and Amot are probably connected by a large complex of E-cadherin–β-catenin–α-catenin–Merlin–Amot because Merlin interacts with α-catenin [38] and Amot [28]. Nf2/Merlin mutant mice survive until the early postimplantation stage [42], and thus, other Amot interacting molecules may also be involved and/or maternal Merlin proteins/RNAs could support preimplantation development. In this state, Amot is fairly unstable and is probably degraded via ubiquitination [35]. (2) Lats phosphorylates S176 in the AJ-associated Amot. The dimerization of E-cadherin [43] may contribute to Amot phosphorylation by promoting the dimerization and/or cross-phosphorylation of Lats [44, 45]. (3) p-S176-Amot is detached from cortical F-actin and is stabilized in the AJs. p-S176-Amot also forms a stable complex with Lats, which activates the Hippo pathway, probably by activating Lats. This may not be the only mechanism, but the phosphorylation of S176-Amot is an important molecular switch that turns on the Hippo pathway in AJs.
In the outer cells, the operation of the Par–aPKC system sequesters Amot from the basolateral AJs to the apical membrane domain. AJs lack the essential switching protein, Amot, and thus, AJs do not activate Lats and the Hippo pathway remains inactive. Nonphosphorylated Amot probably binds to apical actin bundles. The mechanism by which cell polarity restricts Amot remains to be elucidated. The direct/indirect interaction of Amot with the polarity regulators Par3 and Crumbs and the junctional proteins Patj/MUPP1 [25, 26, 48] probably contributes to the regulation of the subcellular localization of Amot.
General roles of Amot in the Hippo pathway
How general are the mechanisms described above? An Amot-mediated mechanism does not operate in Drosophila because it lacks a homolog [22]. In mammals, the roles of Amot appear to differ in epithelial cells and preimplantation embryos. Amot is localized to the tight junctions of polarized epithelial cells and outer cells. However, the outer cells exhibit nuclear Yap while the epithelial cells suppress nuclear Yap by activating the Hippo pathway and tethering Yap to tight junctions [23, 27, 36]. Despite these differences, Amot activates the Hippo pathway via Lats at intercellular junctions in the inner and epithelial cells [36]. The relationship between the cell polarity and Hippo pathway activation is opposite in these cell types, but there must be common activation mechanisms, which probably involve the phosphorylation of S176.
An important feature of Amot is its actin-binding activity, and we determined correlations between S176 phosphorylation, actin binding, Lats binding, and Hippo pathway activation. F-actin suppresses Hippo signaling via Lats in cultured cells [49, 50], and thus, it is intriguing to speculate that Amot is involved with the actin-mediated regulation of Hippo signaling. It is also likely that Hippo signaling controls F-actin by modulating the actin-binding/polymerization activity of Amot via phosphorylation. In this context, it is interesting to note that the apical domain of the outer cells of the preimplantation embryo, in which the Hippo pathway is inactive, contained a large amount of cortical F-actin.
Roles of Hippo signaling during cell fate specification in preimplantation embryos
In the context of preimplantation embryos, two important questions need to be addressed in future. Cell polarization is the first important step during the establishment of position-dependent Hippo signaling. Therefore, the first question is what promotes the initial polarization of the outer cells, either in the early embryo or in isolated ICMs [51]? The second question is, what is the relationship between the Hippo-regulated mechanism and other mechanisms during initial cell fate specification? Several different mechanisms are known to operate, including very early biases among blastomeres and polarity-dependent asymmetric segregation of mRNAs (see the reviews and references in [52, 53]). Irrespective of the operation of these mechanisms, the experimental manipulation of 16-cell and 32-cell stage embryos demonstrated the complete and partial adjustment of cell fates, respectively, with new cell positions [32]. Therefore, the continuous operation of position-dependent Hippo signaling after the 16-cell stage probably has central roles in cell fate specification by generating, enhancing, and stabilizing position-dependent differences throughout preimplantation development. However, it is important to clarify the relationship among these different mechanisms during normal developmental processes.
Experimental Procedures
Mouse lines
Wild-type embryos were obtained by intercrossing [C57BL/6xDBA]F1 (BDF1) or by crossing ICR female with BDF1 male mice. The Amot mutant (Acc. No. CDB0089K; http://www.cdb.riken.jp/arg/mutant%20mice%20list.html) [34] and PKCλ mutant [31] mice were described previously. PKCζ mutant mice were generated via homologous recombination in ES cells. Detailed procedures for the production of PKCζ mutant mice and the housing of mice are provided in the Supplemental Information.
DNA/RNA injection into preimplantation embryos
Poly(A)-tailed RNA was injected into both blastomeres of two-cell stage embryos, as described previously [6]. An RNA concentration of 25 ng/μl was used in the rescue experiments. To achieve gene knockdown by the injection of siRNAs, Stealth RNAi siRNAs (Invitrogen) were injected into both blastomeres of two-cell stage embryos at a concentration of 8 μM. The purified and verified shRNA plasmid DNA solution (10 ng/μl) was injected into the male pronuclei using standard protocols [13, 54]. The embryos were cultured for 48 h and 80 h after injection to analyze 32-cell and late blastocyst stage embryos, respectively. Details of the plasmids and siRNAs used as well as other embryo manipulations are provided in the Supplemental Information.
Immunofluorescent staining and data acquisition
Immunofluorescent staining of preimplantation embryos and transfected cells was performed as described previously [6, 55], with slight modifications. Goat anti-mouse, anti-rabbit, and anti-rat IgG antibodies labeled with Alexa Fluor 488, Alexa Fluor 555 (Invitrogen), or Dylight 649 (Jackson ImmunoResearch) were used as the secondary antibodies. Hoechst 33258 stain (Dojindo, Japan) was used for nuclear staining. Images were acquired using confocal laser scanning microscopes, LSM510META (Zeiss) or A1 (Nikon). Details of the analyses of the acquired images are provided in the Supplemental Information.
Immunoprecipitation (IP)
IP was performed according to previously described procedures [56], with slight modifications. In brief, expression plasmids (0.8 μg) were transfected into 6 × 105 HEK293T cells plated onto six-well plates. One day after transfection, the cells were lysed with 0.5 ml of high salt RIPA buffer [50 mM Tris–HCl (pH 8.0), 500 mM NaCl, 1% NP-40, 0.5% deoxycholic acid, and 0.1% SDS] supplemented with complete protease inhibitor cocktail (Roche). Genomic DNA was disrupted by sonication. The lysate was centrifuged at 12,000 ×g for 10 min, and the supernatant was diluted with 0.5 ml of NaCl-free RIPA buffer. The resulting diluted lysates were used as samples for IP. In all of the IP experiments, Flag-tagged proteins were precipitated using anti-Flag M2-Agarose (Sigma). All the procedures were performed at 4°C or on ice.
Statistical analysis
All statistical analyses were performed using Prism5 (GraphPad). Student's t-test was used for comparisons between two groups.
Supplementary Material
Highlights
-
➢
Amot localizes to adherens junctions (AJs) in inner cells but not in outer cells.
-
➢
Phosphorylation of S176 in Amot by Lats in AJs activates the Hippo pathway.
-
➢
Polarity disconnects the Hippo pathway from adhesion by sequestering Amot from AJs.
-
➢
This mechanism converts positional information into differential Hippo signaling.
Acknowledgments
We thank T. Ideue, M. Shibata, M. Harano, A. Katsuyama, and N. Fujita for technical assistance; K. Yamagata, H. Niwa, J.-i. Miyazaki, H. Kondoh, Y. Kamachi, M. G. Halder, M. Takeichi, and A. Nagafuchi for plasmids; LARGE at RIKEN CDB for providing Amot mutants, collecting mouse embryos, and housing the mice; and CARD at Kumamoto Univ. for housing the mice. This work was supported by grants from RIKEN and the Uehara Memorial Foundation to H.S. and by Grants-in-Aid for Scientific Research (KAKENHI) from MEXT (21116003) and JSPS (23247036) to H.S. and from JSPS (23770265) to Y.H.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Tarkowski AK, Wroblewska J. Development of blastomeres of mouse eggs isolated at the 4- and 8-cell stage. Journal of embryology and experimental morphology. 1967;18:155–180. [PubMed] [Google Scholar]
- 2.Johnson MH, Ziomek CA. The foundation of two distinct cell lineages within the mouse morula. Cell. 1981;24:71–80. doi: 10.1016/0092-8674(81)90502-x. [DOI] [PubMed] [Google Scholar]
- 3.Yamanaka Y, Ralston A, Stephenson RO, Rossant J. Cell and molecular regulation of the mouse blastocyst. Dev Dyn. 2006;235:2301–2314. doi: 10.1002/dvdy.20844. [DOI] [PubMed] [Google Scholar]
- 4.Niwa H, Toyooka Y, Shimosato D, Strumpf D, Takahashi K, Yagi R, Rossant J. Interaction between Oct3/4 and Cdx2 determines trophectoderm differentiation. Cell. 2005;123:917–929. doi: 10.1016/j.cell.2005.08.040. [DOI] [PubMed] [Google Scholar]
- 5.Nishioka N, Yamamoto S, Kiyonari H, Sato H, Sawada A, Ota M, Nakao K, Sasaki H. Tead4 is required for specification of trophectoderm in pre-implantation mouse embryos. Mechanisms of development. 2008;125:270–283. doi: 10.1016/j.mod.2007.11.002. [DOI] [PubMed] [Google Scholar]
- 6.Nishioka N, Inoue K, Adachi K, Kiyonari H, Ota M, Ralston A, Yabuta N, Hirahara S, Stephenson RO, Ogonuki N, et al. The Hippo signaling pathway components Lats and Yap pattern Tead4 activity to distinguish mouse trophectoderm from inner cell mass. Developmental cell. 2009;16:398–410. doi: 10.1016/j.devcel.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 7.Yagi R, Kohn MJ, Karavanova I, Kaneko KJ, Vullhorst D, Depamphilis ML, Buonanno A. Transcription factor TEAD4 specifies the trophectoderm lineage at the beginning of mammalian development. Development (Cambridge, England) 2007;134:3827–3836. doi: 10.1242/dev.010223. [DOI] [PubMed] [Google Scholar]
- 8.Hirate Y, Cockburn K, Rossant J, Sasaki H. Tead4 is constitutively nuclear, while nuclear vs. cytoplasmic Yap distribution is regulated in preimplantation mouse embryos. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:E3389–E3390. doi: 10.1073/pnas.1211810109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ralston A, Cox BJ, Nishioka N, Sasaki H, Chea E, Rugg-Gunn P, Guo G, Robson P, Draper JS, Rossant J. Gata3 regulates trophoblast development downstream of Tead4 and in parallel to Cdx2. Development (Cambridge, England) 2010;137:395–403. doi: 10.1242/dev.038828. [DOI] [PubMed] [Google Scholar]
- 10.Suzuki A, Ohno S. The PAR-aPKC system: lessons in polarity. Journal of cell science. 2006;119:979–987. doi: 10.1242/jcs.02898. [DOI] [PubMed] [Google Scholar]
- 11.Assemat E, Bazellieres E, Pallesi-Pocachard E, Le Bivic A, Massey-Harroche D. Polarity complex proteins. Biochim Biophys Acta. 2008;1778:614–630. doi: 10.1016/j.bbamem.2007.08.029. [DOI] [PubMed] [Google Scholar]
- 12.St Johnston D, Ahringer J. Cell polarity in eggs and epithelia: parallels and diversity. Cell. 2010;141:757–774. doi: 10.1016/j.cell.2010.05.011. [DOI] [PubMed] [Google Scholar]
- 13.Alarcon VB. Cell polarity regulator PARD6B is essential for trophectoderm formation in the preimplantation mouse embryo. Biology of reproduction. 2010;83:347–358. doi: 10.1095/biolreprod.110.084400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stephenson RO, Yamanaka Y, Rossant J. Disorganized epithelial polarity and excess trophectoderm cell fate in preimplantation embryos lacking E-cadherin. Development (Cambridge, England) 2010;137:3383–3391. doi: 10.1242/dev.050195. [DOI] [PubMed] [Google Scholar]
- 15.Grzeschik NA, Parsons LM, Allott ML, Harvey KF, Richardson HE. Lgl, aPKC, and Crumbs regulate the Salvador/Warts/Hippo pathway through two distinct mechanisms. Curr Biol. 2010;20:573–581. doi: 10.1016/j.cub.2010.01.055. [DOI] [PubMed] [Google Scholar]
- 16.Ling C, Zheng Y, Yin F, Yu J, Huang J, Hong Y, Wu S, Pan D. The apical transmembrane protein Crumbs functions as a tumor suppressor that regulates Hippo signaling by binding to Expanded. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:10532–10537. doi: 10.1073/pnas.1004279107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen CL, Gajewski KM, Hamaratoglu F, Bossuyt W, Sansores-Garcia L, Tao C, Halder G. The apical-basal cell polarity determinant Crumbs regulates Hippo signaling in Drosophila. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:15810–15815. doi: 10.1073/pnas.1004060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robinson BS, Huang J, Hong Y, Moberg KH. Crumbs regulates Salvador/Warts/Hippo signaling in Drosophila via the FERM-domain protein Expanded. Curr Biol. 2010;20:582–590. doi: 10.1016/j.cub.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Halder G, Dupont S, Piccolo S. Transduction of mechanical and cytoskeletal cues by YAP and TAZ. Nature reviews. Molecular cell biology. 2012;13:591–600. doi: 10.1038/nrm3416. [DOI] [PubMed] [Google Scholar]
- 20.Schroeder MC, Halder G. Regulation of the Hippo pathway by cell architecture and mechanical signals. Seminars in cell & developmental biology. 2012;23:803–811. doi: 10.1016/j.semcdb.2012.06.001. [DOI] [PubMed] [Google Scholar]
- 21.Yu FX, Guan KL. The Hippo pathway: regulators and regulations. Genes & development. 2013;27:355–371. doi: 10.1101/gad.210773.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bratt A, Wilson WJ, Troyanovsky B, Aase K, Kessler R, Van Meir EG, Holmgren L. Angiomotin belongs to a novel protein family with conserved coiled-coil and PDZ binding domains. Gene. 2002;298:69–77. doi: 10.1016/s0378-1119(02)00928-9. [DOI] [PubMed] [Google Scholar]
- 23.Zhao B, Li L, Lu Q, Wang LH, Liu CY, Lei Q, Guan KL. Angiomotin is a novel Hippo pathway component that inhibits YAP oncoprotein. Genes & development. 2011;25:51–63. doi: 10.1101/gad.2000111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang W, Huang J, Chen J. Angiomotin-like proteins associate with and negatively regulate YAP1. The Journal of biological chemistry. 2011;286:4364–4370. doi: 10.1074/jbc.C110.205401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sugihara-Mizuno Y, Adachi M, Kobayashi Y, Hamazaki Y, Nishimura M, Imai T, Furuse M, Tsukita S. Molecular characterization of angiomotin/JEAP family proteins: interaction with MUPP1/Patj and their endogenous properties. Genes Cells. 2007;12:473–486. doi: 10.1111/j.1365-2443.2007.01066.x. [DOI] [PubMed] [Google Scholar]
- 26.Wells CD, Fawcett JP, Traweger A, Yamanaka Y, Goudreault M, Elder K, Kulkarni S, Gish G, Virag C, Lim C, et al. A Rich1/Amot complex regulates the Cdc42 GTPase and apical-polarity proteins in epithelial cells. Cell. 2006;125:535–548. doi: 10.1016/j.cell.2006.02.045. [DOI] [PubMed] [Google Scholar]
- 27.Chan SW, Lim CJ, Chong YF, Venkatesan Pobbati A, Huang C, Hong W. Hippo pathway-independent restriction of TAZ and YAP by angiomotin. The Journal of biological chemistry. 2011;286:7018–7026. doi: 10.1074/jbc.C110.212621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yi C, Troutman S, Fera D, Stemmer-Rachamimov A, Avila JL, Christian N, Luna Persson N, Shimono A, Speicher DW, Marmorstein R, et al. A Tight Junction-Associated Merlin-Angiomotin Complex Mediates Merlin's Regulation of Mitogenic Signaling and Tumor Suppressive Functions. Cancer cell. 2011;19:527–540. doi: 10.1016/j.ccr.2011.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim J, Xie J, Ikenoue T, Yu J, Li L, et al. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes & development. 2007;21:2747–2761. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suzuki A, Yamanaka T, Hirose T, Manabe N, Mizuno K, Shimizu M, Akimoto K, Izumi Y, Ohnishi T, Ohno S. Atypical protein kinase C is involved in the evolutionarily conserved par protein complex and plays a critical role in establishing epithelia-specific junctional structures. J Cell Biol. 2001;152:1183–1196. doi: 10.1083/jcb.152.6.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matsumoto M, Ogawa W, Akimoto K, Inoue H, Miyake K, Furukawa K, Hayashi Y, Iguchi H, Matsuki Y, Hiramatsu R, et al. PKClambda in liver mediates insulin induced SREBP-1c expression and determines both hepatic lipid content and overall insulin sensitivity. J Clin Invest. 2003;112:935–944. doi: 10.1172/JCI18816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suwinska A, Czolowska R, Ozdzenski W, Tarkowski AK. Blastomeres of the mouse embryo lose totipotency after the fifth cleavage division: expression of Cdx2 and Oct4 and developmental potential of inner and outer blastomeres of 16- and 32-cell embryos. Developmental biology. 2008;322:133–144. doi: 10.1016/j.ydbio.2008.07.019. [DOI] [PubMed] [Google Scholar]
- 33.Ernkvist M, Birot O, Sinha I, Veitonmaki N, Nystrom S, Aase K, Holmgren L. Differential roles of p80- and p130-angiomotin in the switch between migration and stabilization of endothelial cells. Biochim Biophys Acta. 2008;1783:429–437. doi: 10.1016/j.bbamcr.2007.11.018. [DOI] [PubMed] [Google Scholar]
- 34.Shimono A, Behringer RR. Angiomotin regulates visceral endoderm movements during mouse embryogenesis. Curr Biol. 2003;13:613–617. doi: 10.1016/s0960-9822(03)00204-5. [DOI] [PubMed] [Google Scholar]
- 35.Wang C, An J, Zhang P, Xu C, Gao K, Wu D, Wang D, Yu H, Liu JO, Yu L. The Nedd4 like ubiquitin E3 ligases target angiomotin/p130 to ubiquitin dependent degradation. The Biochemical journal. 2012;444:279–289. doi: 10.1042/BJ20111983. [DOI] [PubMed] [Google Scholar]
- 36.Paramasivam M, Sarkeshik A, Yates JR, 3rd, Fernandes MJ, McCollum D. Angiomotin family proteins are novel activators of the LATS2 kinase tumor suppressor. Molecular biology of the cell. 2011;22:3725–3733. doi: 10.1091/mbc.E11-04-0300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ernkvist M, Aase K, Ukomadu C, Wohlschlegel J, Blackman R, Veitonmaki N, Bratt A, Dutta A, Holmgren L. p130-angiomotin associates to actin and controls endothelial cell shape. The FEBS journal. 2006;273:2000–2011. doi: 10.1111/j.1742-4658.2006.05216.x. [DOI] [PubMed] [Google Scholar]
- 38.Gladden AB, Hebert AM, Schneeberger EE, McClatchey AI. The NF2 Tumor Suppressor, Merlin, Regulates Epidermal Development through the Establishment of a Junctional Polarity Complex. Developmental cell. 2010;19:727–739. doi: 10.1016/j.devcel.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nagafuchi A, Takeichi M, Tsukita S. The 102 kd cadherin-associated protein: similarity to vinculin and posttranscriptional regulation of expression. Cell. 1991;65:849–857. doi: 10.1016/0092-8674(91)90392-c. [DOI] [PubMed] [Google Scholar]
- 40.Ozawa M, Baribault H, Kemler R. The cytoplasmic domain of the cell adhesion molecule uvomorulin associates with three independent proteins structurally related in different species. Embo J. 1989;8:1711–1717. doi: 10.1002/j.1460-2075.1989.tb03563.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lei QY, Zhang H, Zhao B, Zha ZY, Bai F, Pei XH, Zhao S, Xiong Y, Guan KL. TAZ promotes cell proliferation and epithelial-mesenchymal transition and is inhibited by the hippo pathway. Molecular and cellular biology. 2008;28:2426–2436. doi: 10.1128/MCB.01874-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McClatchey AI, Saotome I, Ramesh V, Gusella JF, Jacks T. The Nf2 tumor suppressor gene product is essential for extraembryonic development immediately prior to gastrulation. Genes & development. 1997;11:1253–1265. doi: 10.1101/gad.11.10.1253. [DOI] [PubMed] [Google Scholar]
- 43.Gumbiner BM. Regulation of cadherin-mediated adhesion in morphogenesis. Nature reviews. Molecular cell biology. 2005;6:622–634. doi: 10.1038/nrm1699. [DOI] [PubMed] [Google Scholar]
- 44.Yabuta N, Okada N, Ito A, Hosomi T, Nishihara S, Sasayama Y, Fujimori A, Okuzaki D, Zhao H, Ikawa M, et al. Lats2 is an essential mitotic regulator required for the coordination of cell division. The Journal of biological chemistry. 2007;282:19259–19271. doi: 10.1074/jbc.M608562200. [DOI] [PubMed] [Google Scholar]
- 45.Visser S, Yang X. LATS tumor suppressor: a new governor of cellular homeostasis. Cell Cycle. 2010;9:3892–3903. doi: 10.4161/cc.9.19.13386. [DOI] [PubMed] [Google Scholar]
- 46.Song H, Mak KK, Topol L, Yun K, Hu J, Garrett L, Chen Y, Park O, Chang J, Simpson RM, et al. Mammalian Mst1 and Mst2 kinases play essential roles in organ size control and tumor suppression. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:1431–1436. doi: 10.1073/pnas.0911409107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yue T, Tian A, Jiang J. The cell adhesion molecule echinoid functions as a tumor suppressor and upstream regulator of the Hippo signaling pathway. Developmental cell. 2012;22:255–267. doi: 10.1016/j.devcel.2011.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Varelas X, Samavarchi-Tehrani P, Narimatsu M, Weiss A, Cockburn K, Larsen BG, Rossant J, Wrana JL. The Crumbs complex couples cell density sensing to Hippo-dependent control of the TGF-beta-SMAD pathway. Developmental cell. 2010;19:831–844. doi: 10.1016/j.devcel.2010.11.012. [DOI] [PubMed] [Google Scholar]
- 49.Wada K, Itoga K, Okano T, Yonemura S, Sasaki H. Hippo pathway regulation by cell morphology and stress fibers. Development (Cambridge, England) 2011;138:3907–3914. doi: 10.1242/dev.070987. [DOI] [PubMed] [Google Scholar]
- 50.Zhao B, Li L, Wang L, Wang CY, Yu J, Guan KL. Cell detachment activates the Hippo pathway via cytoskeleton reorganization to induce anoikis. Genes & development. 2012;26:54–68. doi: 10.1101/gad.173435.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Eckert JJ, McCallum A, Mears A, Rumsby MG, Cameron IT, Fleming TP. Relative contribution of cell contact pattern, specific PKC isoforms and gap junctional communication in tight junction assembly in the mouse early embryo. Developmental biology. 2005;288:234–247. doi: 10.1016/j.ydbio.2005.09.037. [DOI] [PubMed] [Google Scholar]
- 52.Zernicka-Goetz M. Development: do mouse embryos play dice? Curr Biol. 2013;23:R15–17. doi: 10.1016/j.cub.2012.10.032. [DOI] [PubMed] [Google Scholar]
- 53.Bruce AW, Zernicka-Goetz M. Developmental control of the early mammalian embryo: competition among heterogeneous cells that biases cell fate. Curr Opin Genet Dev. 2010;20:485–491. doi: 10.1016/j.gde.2010.05.006. [DOI] [PubMed] [Google Scholar]
- 54.Hogan B, Beddington R, Constantini F, Lacy E. Manipulating the Mouse Embryo: Laboratory Manual. 2nd edn Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- 55.Ota M, Sasaki H. Mammalian Tead proteins regulate cell proliferation and contact inhibition as a transcriptional mediator of Hippo signaling. Development (Cambridge, England) 2008;135:4059–4069. doi: 10.1242/dev.027151. [DOI] [PubMed] [Google Scholar]
- 56.Yamamoto S, Nishimura O, Misaki K, Nishita M, Minami Y, Yonemura S, Tarui H, Sasaki H. Cthrc1 selectively activates the planar cell polarity pathway of Wnt signaling by stabilizing the Wnt-receptor complex. Dev. Cell. 2008;15:23–36. doi: 10.1016/j.devcel.2008.05.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.