Abstract
Background & Aims
During tumorigenesis, loss of rapid messenger RNA (mRNA) decay allows for overexpression of cancer-associated genes. The RNA-binding proteins Hu antigen R (HuR) and tristetraprolin (TTP) bind AU-rich elements in the 3′ untranslated region of many cancer-associated mRNAs and target them for stabilization or rapid decay, respectively. We examined the functions of HuR and TTP during colon tumorigenesis and their ability to regulate cyclooxygenase (COX-2), a mediator of prostaglandin synthesis that increases in the colon tumor microenvironment.
Methods
We evaluated expression of HuR and TTP during colorectal tumorigenesis and in colon cancer cells and associated them with COX-2 expression. HuR and TTP-inducible cells were created to investigate HuR- and TTP-mediated regulation of COX-2.
Results
In normal colon tissues, low levels of nuclear HuR and higher levels of TTP were observed. By contrast, increased HuR expression and cytoplasmic localization were observed in 76% of adenomas and 94% of adenocarcinomas, and TTP expression was lost in >75% of adenomas and adenocarcinomas. Similar results were obtained for HuR and TTP mRNA levels in normal and staged tumor samples. In both adenomas and adenocarcinomas, COX-2 overexpression was associated with increased HuR and decreased TTP (P < .0001); similar associations were observed in colon cancer cells. HuR overexpression in cells up-regulated COX-2 expression, whereas overexpression of TTP inhibited it; limited TTP expression antagonized HuR-mediated COX-2 overexpression.
Conclusions
Increased expression of the mRNA stability factor HuR and loss of the decay factor TTP occurs during early stages of colorectal tumorigenesis. These changes promote COX-2 overexpression and could contribute to colon tumorigenesis.
Colorectal cancer is the third most common cancer among adult Americans and accounts for approximately 10% of all cancer-related deaths. In colorectal tumors, various genetic alterations have been identified that promote the initiation and progression of tumorigenesis. As a consequence of these defects, activation of multiple signaling pathways leads to enhanced expression of many growth- and inflammation-associated immediate-early response genes. A critical point in controlling the expression of these factors in intestinal epithelium occurs through posttranscriptional mechanisms that promote rapid mRNA decay, and a majority of immediate-early gene transcripts are inherently unstable because of the presence of 3′-untranslated region (3′UTR) adenylate- and uridylate (AU)-rich elements (AREs) that target the mRNA for rapid decay.1 However, dysregulation in ARE-mediated decay is observed in colon cancer cells and tumors,2,3 indicating the functional significance of posttranscriptional regulation in carcinogenesis.4
AREs mediate their regulatory function through association with multiple RNA-binding proteins that display high affinity for AREs.1 The best studied ARE-binding proteins can promote rapid mRNA decay, mRNA stabilization, and translational silencing.1 The Hu antigen R (HuR) protein is a ubiquitously expressed member of the ELAV-like family of RNA-binding proteins. HuR can function in an mRNA stabilizing capacity; when overexpressed in cells, HuR stabilizes ARE-containing transcripts and promotes their translation.5 Contrasting the effects of HuR, tristetraprolin ([TTP], ZFP36, TIS11) is a member of a small family of tandem Cys3His zinc finger proteins and promotes rapid decay of ARE-containing mRNAs.6 The binding of TTP to AREs targets the mRNA for rapid degradation through exosome recruitment and association with mRNA decay enzymes.7
Cyclooxygenases (COX) are key enzymes in the production of prostaglandins, and overexpression of the inducible isoform COX-2 has been shown to occur at multiple stages of colon carcinogenesis allowing for elevated prostaglandin synthesis to occur in the tumor microenvironment.8 In normal cells, COX-2 expression levels are potently regulated through AREs present in its mRNA,9 whereas, under conditions of neoplastic transformation, the ability of the COX-2 ARE to promote posttranscriptional regulation is compromised.10 Our prior work has demonstrated the ability of HuR to promote the stability of COX-2 and other ARE-containing mRNAs.2 These findings and others demonstrate increased expression of HuR to occur in a variety of human cancers, including colorectal tumors, and promote ARE-containing gene expression.2,3 However, the status of TTP expression and its ability to promote ARE-mediated mRNA decay in colorectal cancer is not known. In this report, we demonstrate that elevated HuR expression occurs concomitant with loss of TTP expression at an early stage of colorectal tumorigenesis that is associated with increased COX-2 expression and defines the role these opposing RNA-binding proteins have in controlling COX-2 expression. These findings offer what we believe are new insights into the loss of posttranscriptional regulation allowing for enhanced expression of COX-2 and other cancer- and inflammation-associated genes in colorectal cancer.
Materials and Methods
Colorectal Tissue Specimens
Immunohistochemical analysis was performed on paraffin-embedded human tissue array samples obtained from 2 sources. The colorectal carcinoma progression tissue array (CHTN2003CRCprog) from the Cooperative Human Tissue Network (National Cancer Institute [NCI], Rock-ville, MD) contained 42 normal colorectal tissue samples derived from 14 cases of nonneoplastic colonic mucosa, 42 adenoma tissue samples derived from 14 cases of adenomatous polyps, and 42 adenocarcinoma tissue samples derived from 14 cases of primary colorectal adeno-carcinomas classified by pTNM staging. The colon cancer tissue array CO801 (US Biomax, Rockville, MD) contained 40 tissue cores each of colorectal adenocarcinomas and matched or unmatched adjacent normal tissue graded by histology. TissueScan qPCR colon cancer arrays (HCRT501) classified by American Joint Committee on Cancer staging were obtained from Origene (Rockville, MD).
Immunohistochemistry
Detection of HuR, TTP, and COX-2 protein was performed on serial sectioned tissue arrays. The following antibodies were used for immunohistochemical staining: monoclonal 19F12 HuR antibody (Molecular Probes, Eugene, OR) at 160 ng/mL (1:1250), 2 polyclonal TTP antibodies (N-18 and G-20) used in combination (Santa Cruz Biotechnology, Santa Cruz, CA) each at 800 ng/mL (1:250), and polyclonal COX-2 antibody (160126; Cayman Chemical Company, Ann Arbor, MI) at 1250 ng/mL (1:400). Standard staining protocols were used. Briefly, slides were hydrated, and antigen retrieval was performed in citrate buffer in a steam bath for 30 minutes. All primary antibodies were incubated on slides for 18 hours at 4°C. After washing in TBST, slides were incubated in corresponding biotinylated secondary antibodies following the Vecta Stain ABC kit protocol (Vector Laboratories, Burlingame, CA). Immunohistochemistry was visualized using the DAB peroxidase substrate kit (Vector Laboratories) and counterstained with hematoxylin (Sigma–Aldrich, St. Louis, MO).
Immunoreactivity Scoring
Stained tissues were examined for intensity of staining using a method similar to that previously described.11 The intensity of staining in tumor sections was evaluated independently by 2 blinded investigators (L.E.Y. and D.A.D.). For each tissue section, the percentage of positive cells was scored on a scale of 0 to 4 for the percentage of tissue stained: 0 (0% positive cells), 1 (<10%), 2 (11% to 50%), 3 (51% to 80%), or 4 (>80%). Staining intensity was scored on a scale of 0 to 3: 0, negative staining; 1, weak staining; 2, moderate staining; or 3, strong staining. The 2 scores were multiplied resulting in an immunoreactivity score (IRS) value ranging from 0 to 12. These scores were then grouped together in 1 of 2 IRS categories: low (IRS, 0–6) and high (IRS, 7–12).
Messenger RNA Analysis
Relative levels for HuR, TTP, and COX-2 messenger RNA (mRNA) in human colon cancer tissues were determined by real-time PCR (qPCR) using the TissueScan qPCR colon cancer array (Origene, Rockville, MD). qPCR was performed according to the manufacturer’s guidelines using Taqman probes for TTP (ZFP36) and HuR (ELAVL1) purchased from Applied Biosystems (Foster City, CA) using the 7300 PCR Assay System (Applied Biosystems). qPCR for COX-2 was performed using SYBR green PCR master mix (Applied Biosystems) and primers for COX-2 sense, 5′-GTCACAAGATGGCAAAATGCTG-3′ and antisense, 5′-TAAGATAACACTGCAGTGGCTC-3′. β-actin amplification was used as a loading control. Fold change in mRNA expression levels for each individual sample were normalized to the cycle threshold (Ct) using the first normal sample (N1). Total RNA extracted from HuR and TTP-inducible HeLa cells was used for complementary DNA (cDNA) synthesis as previously described,9 and COX-2 mRNA levels were detected using Taqman probes for COX-2 (PTGS2) and normalized to 18S ribosomal RNA (rRNA) levels.
Cell Culture, DNA Transfection, and Adenoviral Infection
Human colon cancer cell lines LoVo, HT-29, SKCO1, and CaCo2 were obtained from the American Type Culture Collection (ATCC; Manassas, VA); HCA7 and Moser cells were kindly provided by S. Kirkland (Imperial College, London, United Kingdom) and R. D. Beauchamp (Vanderbilt University Medical Center, Nashville, TN), respectively. COS7 cells were purchased from the ATCC, and HeLa tetracycline (Tet)-OFF cells were purchased from BD Clontech (Mountain View, CA). All cells were maintained in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS; Hyclone, Logan, UT) except SKCO1 and CaCo2 cells, which were maintained in Eagle’s medium containing 10% and 20% fetal bovine serum (FBS), respectively; HeLa Tet-OFF cell media were supplemented with 100 jug/mL G418 (Cellgro, Herndon, VA).
Transient transfections of COS7 cells with full-length COX-2 cDNA expression plasmid (pRC/cytomegalovirus immediate-early gene [CMV]-COX-2+3′UTR)9 along with Flag epitope-tagged HuR (pcDNA3-HuR-C-Flag)2 and TTP cDNAs (pcDNA3.1-TTP-Flag)12 were accomplished using Lipofectamine Plus (Invitrogen, Carlsbad, CA) according to the vendor’s protocol for 48 hours. Stable transfections of HeLa-Tet-OFF cells with Flag epitope-tagged HuR cDNA cloned into pTRE2hyg (Clontech, Mountain View, CA) to create pTRE2hyg-HuR-Flag or Flag epitope-tagged TTP to create pTRE2hyg-TTP-Flag were accomplished using Lipofectamine Plus (Invitrogen, Carlsbad, CA). Stably transfected cells were selected in normal growth medium containing 100 µg/mL G418, 200 ug/mL hygromycin B (Invitrogen, Carlsbad, CA), and 2 µg/mL doxycycline (Dox) (Clontech) for 2–3 weeks. Individual clones were isolated by limiting dilution in 96-well plates. Positive HeLa-Tet-OFF/HuR-Flag and HeLa-Tet-OFF/TTP-Flag clones were screened by growing cells in the presence or absence of Dox (2µg/ mL) for 48 hours to induce expression of HuR-Flag or TTP-Flag, respectively; the absence of Dox induces expression. For stable cell maintenance, the hygromycin B concentration was reduced to 100 µg/mL.
TTP-Flag expressing adenovirus was created by cloning TTP-Flag cDNA into the shuttle vector Dual-CCM-CMV-EGFP (Vector Biolabs, Philadelphia, PA), which contains dual CMV promoters to drive expression of TTP-Flag and GFP. Construction of TTP-expressing adenovirus (AdGFP/ TTP-Flag) was conducted by Vector Biolabs. Control GFP-expressing adenovirus (AdGFP) was purchased from Vector Biolabs. HCA7 and Moser cells were infected at the indicated multiplicity of infection (MOI) in serum-free DMEM for 2 hours, after which FBS was added to 10%. Forty-eight hours after infection, phase contrast images were obtained, and cells were harvested in SDS-PAGE lysis buffer for Western blot analysis.
Immunofluorescence
Cells grown on coverslips were fixed in 4% paraformaldehyde for 20 minutes and permeabilized with 0.2% Triton X-100 in phosphate-buffered saline (PBS) for 5 minutes. After blocking with 10% horse serum diluted in 1% bovine serum albumin (BSA)/PBS for 1.5 hours, cells were incubated for 1 hour with monoclonal 19F12 HuR antibody (800 ng/mL, 1:250) diluted in blocking serum. Fluorescent anti-mouse secondary antibody conjugated to fluorescein (5 µg/mL, 1:100; Vector Laboratories, Burlingame, CA) was used for secondary detection. Cells were counterstained with DAPI (300 nmol/L in PBS), and confocal images were obtained using a Zeiss LSM 510 META confocal microscope (Zeiss MicroImaging Inc, Thornwood, NY).
Immunoblot Analysis
Western blots were performed as previously described2 using a monoclonal anti-HuR antibody (clone 3A2; Santa Cruz Biotechnology, Santa Cruz, CA) at 1:12,500 for 1 hour at room temperature, a polyclonal anti-TTP antibody (ab36558; Abcam, Cambridge, MA) used at 324 ng/mL (1:10,000) for 16 hours at 4°C, a monoclonal anti-COX-2 antibody (160112; Cayman Chemical Company, Ann Arbor, MI) used at 1:1000 for 16 hours at 4°C, and a monoclonal anti-Flag antibody (F3165; Sigma–Aldrich) used at 330 ng/mL (1:15,000) for 1 hour at room temperature. Where indicated, blots were stripped and then probed with β-actin antibody (Clone C4; MP Biomedicals, Aurora, OH). Detection and quantitation of blots were performed as described.9
Statistical Analysis
χ2 Contingency table analysis was used to determine the significance of the differences in the number of tissue cores displaying low vs high IRS for HuR, TTP, or COX-2 in normal colonic epithelium, colon adenomas, and colon adenocarcinomas. Student t test was used to determine significant differences between normal cDNA samples and tumor cDNA samples for each tumor stage (I–IV). P values less than .05 were considered significant.
Results
HuR, TTP, and COX-2 Protein Expression in Human Colorectal Tumorigenesis
We and others have shown increased HuR expression to occur in human colorectal tumors2,3,11; however, the state of TTP expression in colon cancer is not known. Based on the opposing effects these RNA-binding proteins elicit on ARE-containing gene expression, we sought to determine whether corresponding changes in HuR and TTP expression exist during colorectal tumorigenesis. HuR and TTP immunoreactivity were evaluated on serial sections of human tissue arrays containing adenocarcinomas, adenomas, and specimens extracted from matched or unmatched normal tissue samples (Figure 1). In normal colon tissue, HuR was localized solely to the nucleus of epithelial cells in 89% of samples. HuR immunoreactivity was increased in adenomas and more intense in adenocarcinomas. The cytoplasmic abundance of HuR was increased in adenomas, with 76% of samples displaying cytoplasmic HuR and 94% of adenocarcinoma samples show this change in HuR subcellular localization compared with normal mucosa (Figure 1A and Supplementary Figure 1, see supplementary Figure 1 online at www.gastrojournal.org). By contrast, expression of TTP was highest in normal colonic epithelium and predominantly localized to the cytoplasm. In both adenomas and adenocarcinomas, TTP immunoreactivity is substantially decreased with heterogeneous TTP staining observed in nonneoplastic stromal cells (Figure 1B and Supplementary Figure 1).
Figure 1.
Immunohistochemical detection of HuR, TTP, and COX-2 expression in colon tissue. Representative serial sections from tissue samples of normal colon epithelium (left), tubular adenoma (center), and adenocarcinoma (stage T3; right) were examined for HuR, TTP, or COX-2 expression. (A) Nuclear HuR localization is observed in normal epithelium, and elevated HuR expression localized to the nucleus and cytoplasm occurs in adenoma and adenocarcinoma. (B) Cytoplasmic TTP expression is observed in normal epithelium, whereas TTP immunostaining of tumor tissue is low or negative. (C) COX-2 expression is negative in normal epithelium and elevated in adenoma and adenocarcinoma. Original magnification, 200×.
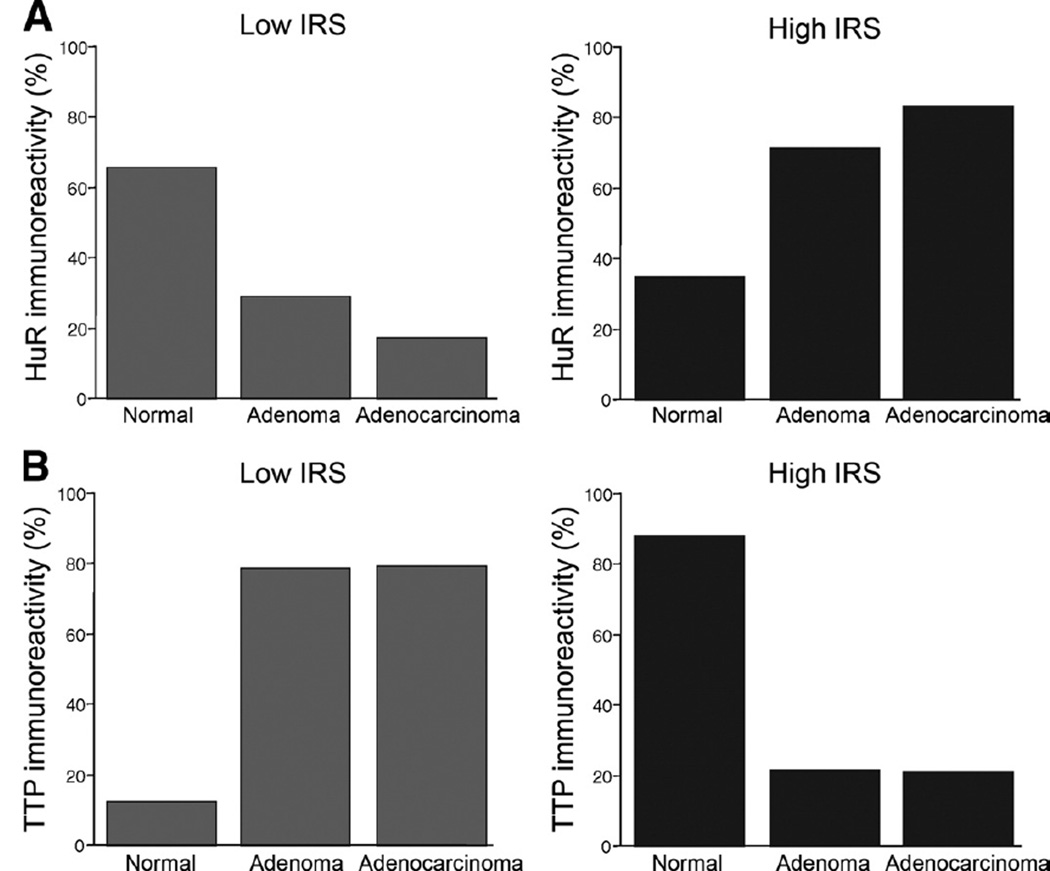
To evaluate expression patterns of HuR and TTP in normal colonic epithelium compared with adenomas and adenocarcinomas, tissue sections were grouped into low or high expression categories based on the IRS. As shown in Table 1 and Figure 2, low HuR staining was observed in 53 of the 81 normal epithelium tissues (65.4%). In adenomatous tissue, HuR expression displayed a shift toward high immunoreactivity in 71.1% of the samples (P < .0001). Adenocarcinomas exhibit the largest number of samples with high HuR immunoreactivity scores in 63 of 76 samples (82.9%, P < .0001), consistent with prior observations demonstrating increased HuR expression in colorectal adenocarcinoma.2,3,11 Interestingly, expression patterns for TTP provided opposite results of those observed with HuR. In normal tissue samples, TTP immunoreactivity was high in 72 of 82 (87.8%) samples, whereas expression was significantly lower in 33 of 42 adenoma samples (78.6%, P = .00019). Additionally, 65 of 72 (79.3%) adenocarcinoma tissue samples display low TTP expression (P < .0001).
Table 1.
Immunoreactivity Score Distribution for HuR, TTP, and COX-2 Protein Expression in Colon Tumor Samples
| Tissue type | Total (n) | Low immunoreactivity score, n (%)a | High immunoreactivity score, n (%)a | Significanceb |
|---|---|---|---|---|
| HuR | ||||
| Normal | 81 | 53 (65.4) | 28 (34.6) | |
| Adenoma | 38 | 11 (28.9) | 27 (71.1) | <.0001 |
| Adenocarcinoma | 76 | 13 (17.1) | 63 (82.9) | <.0001 |
| TTP | ||||
| Normal | 82 | 10 (12.2) | 72 (87.8) | |
| Adenoma | 42 | 33 (78.6) | 9 (21.4) | .00019 |
| Adenocarcinoma | 82 | 65 (79.3) | 17 (20.7) | <.0001 |
| COX-2 | ||||
| Normal | 81 | 70 (87.0) | 11 (13.0) | |
| Adenoma | 41 | 16 (39.0) | 25 (61.0) | <.0001 |
| Adenocarcinoma | 78 | 17 (21.8) | 61 (78.2) | <.0001 |
Immunoreactivity score for each tissue was on a scale of 0 to 12 as indicated in Materials and Methods section; low immunoreactivity score distribution ranged from 0 to 6, and high immunoreactivity score distribution ranged from 7 to 12.
χ2 Test.
Figure 2.
Immunoreactivity score (IRS) distribution of HuR (A) and TTP (B) protein expression in normal colon tissue, adenomas, and adenocarcinomas. Scores were calculated and plotted as percentage of samples displaying a low IRS from 0 to 6 (grey bars) and high IRS from 7–12 (black bars) for each tissue type.
Based on the ability of both HuR and TTP to regulate COX-2 expression posttranscriptionally through the COX-2 ARE,2,10,12 we investigated the correlation between HuR or TTP expression and COX-2 immunoreactivity. As shown in Figure 1C and Table 1, normal tissue sections display little to no COX-2 expression, whereas there was significant COX-2 immunoreactivity in the tumor tissue sections. In adenomas, COX-2 overexpression was observed in approximately 61% of the samples (P < .0001), and 78.2% of adenocarcinomas had elevated COX-2 (P < .0001), consistent with previous results.8 There was a significant association among low COX-2 IRS, low HuR, and high TTP expression (P < .0001) in normal tissues. Furthermore, increased COX-2 immunoreactivity observed in tumor tissues was associated with high HuR levels and decreased TTP in both adenomas and adenocarcinomas (P < .0001). Taken together, these results demonstrate that distinct changes in HuR and TTP expression occur early during colorectal tumorigenesis and indicate that this alteration in ARE-binding protein expression is associated with increased COX-2 levels observed in tumors.
Altered Expression of HuR and TTP mRNA Occurs in Colorectal Tumors
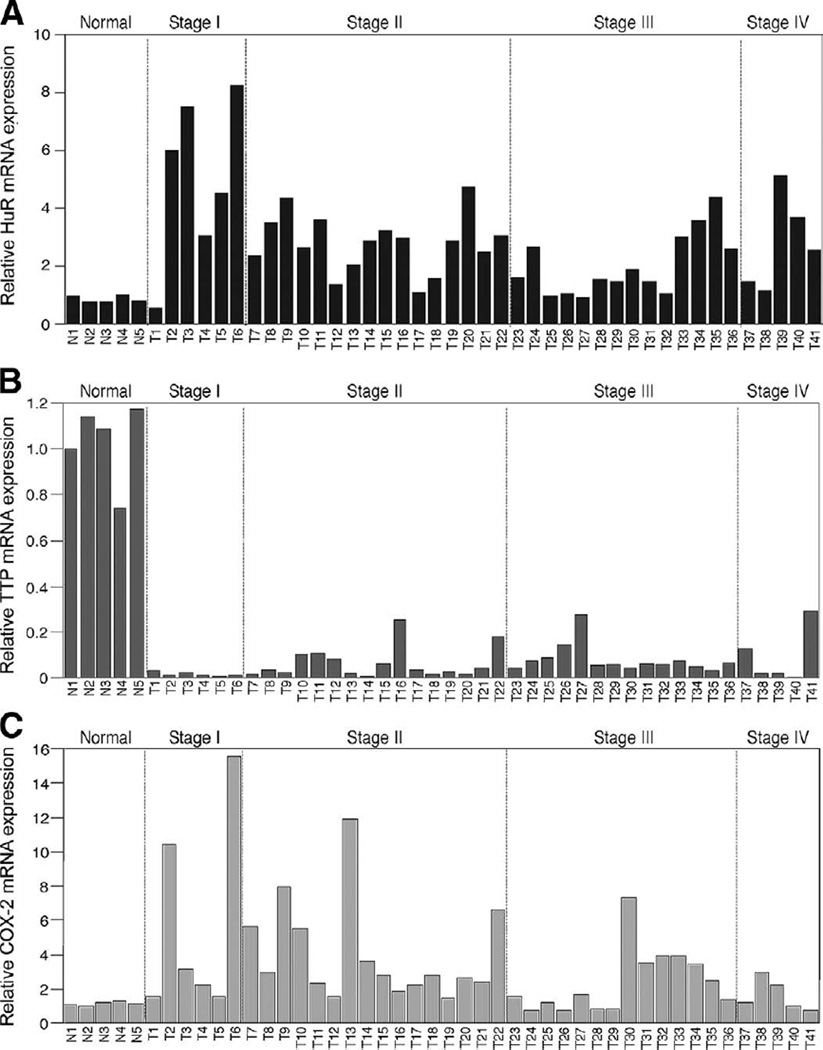
To determine whether changes in HuR and TTP mRNA expression correlate with those observed on the protein level, colorectal tumors of various stages were assayed for HuR and TTP mRNA by qPCR. HuR mRNA expression was increased in essentially all of the tumor samples compared with normal tissue (Figure 3A). The Student t test revealed that changes between normal samples and individual tumor stages were all significant (Table 2). Conversely, TTP mRNA was expressed at high levels in all normal samples, and low levels were detected in tumor samples (Figure 3B). The Student t test comparing normal samples to each tumor stage determined that changes in TTP mRNA levels were also significant (Table 2). Consistent with these observations, elevated COX-2 mRNA expression was detected in all stages I and II tumors, 71% of stage III tumors, and 60% of stage IV tumors as compared with normal tissues (Figure 3C). These results agree with those obtained via immunohistochemistry of normal/tumor tissue and imply that these changes arise through respective changes in HuR and TTP gene expression occurring at an early stage of tumorigenesis.
Figure 3.
HuR, TTP, and COX-2 mRNA expression are altered in colorectal cancer. Human colon tissue (N1-N5, normal; T1-T41, tumors) gene expression panels were analyzed for HuR (A), TTP (B), and COX-2 (C) expression by qPCR and standardized using β-actin. The data are represented as fold change in mRNA expression normalized to N1 for HuR, TTP, or COX-2.
Table 2.
Summary of Statistical Analysis for HuR and TTP mRNA Expression in Colon Tumor Samples
| Group | Total (n) | Meana | Standard error | Significanceb |
|---|---|---|---|---|
| HuR | ||||
| Normal | 5 | 0.870 | 0.054 | |
| Stage I | 5 | 4.332 | 1.197 | .0115 |
| Stage II | 16 | 3.128 | 0.423 | .0004 |
| Stage III | 14 | 2.053 | 0.293 | .0312 |
| Stage IV | 6 | 2.765 | 0.598 | .0303 |
| TTP | ||||
| Normal | 5 | 1.028 | 0.078 | |
| Stage I | 5 | 0.017 | 0.005 | <.0001 |
| Stage II | 16 | 0.053 | 0.016 | <.0001 |
| Stage III | 15 | 0.088 | 0.018 | <.0001 |
| Stage IV | 6 | 0.087 | 0.044 | <.0001 |
Average relative mRNA expression.
Student t test.
Overexpression of Cytoplasmic HuR and Loss of TTP Are Observed in Colon Cancer Cell Lines
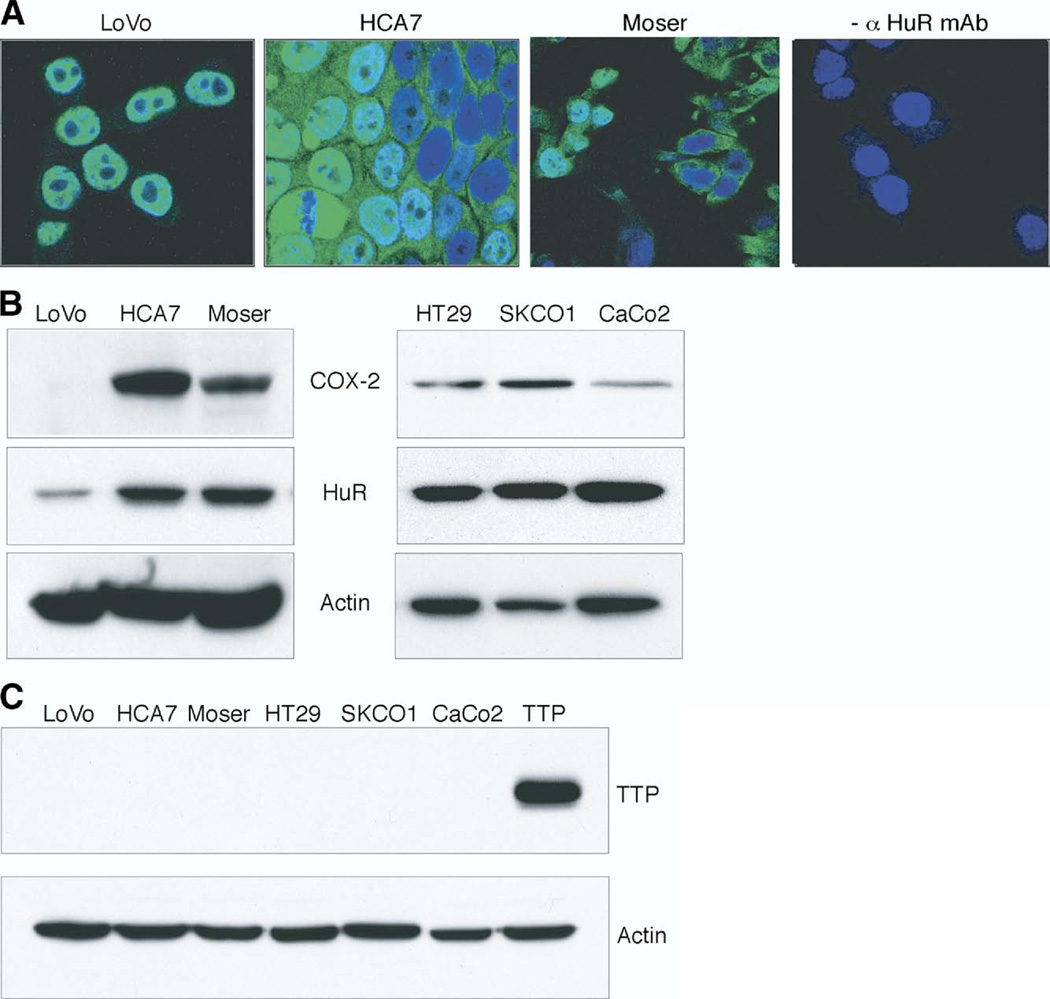
In normal cells, HuR is localized predominantly in the nucleus, and nuclear export of HuR to the cytoplasm is associated with ARE-containing mRNA stabilization.5 Based on this, we sought to determine whether increased cytoplasmic localization of HuR could contribute to COX-2 overexpression observed in tumor tissue. As shown in Figure 4A, HuR was almost exclusively localized to the nuclei of LoVo cells, which is consistent with rapid degradation of COX-2 mRNA in this cell line.2 In contrast, cytoplasmic HuR was robustly detected by immunostaining in HCA7 and Moser cells. To demonstrate the relationship between HuR and COX-2 expression, Western blot analysis of colon cancer cell lines was performed (Figure 4B). HuR expression was variable among the cell lines, with LoVo cells expressing approximately 2.5-fold less HuR than HCA7, Moser, HT29, SKCO1, and CaCo2 cells. Similarly, COX-2 expression correlated with HuR expression patterns in colon cancer cells. Concurrent with increased HuR levels, TTP protein was undetectable in colon cancer cell lines compared with control cells transfected with a human TTP expression vector (Figure 4C). These findings further support the observation that expression of TTP is deficient in colon cancer.
Figure 4.
HuR and TTP expression in colon cancer cell lines. (A) Immunofluorescent detection of HuR, shown in green, in LoVo, HCA7, and Moser colon cancer cell lines; nuclei are shown in blue. Moser cells treated without primary antibody are shown. (B and C) Western blot analysis of HuR, COX-2, and TTP expression in LoVo, HCA7, Moser, HT29, SKCO1, and CaCo2 cells. Total protein extracts were analyzed by SDS-PAGE and probed for HuR, COX-2, or TTP. β-actin served as a loading control. COS7 cells transfected with a human TTP expression vector served as a positive control for TTP.
HuR and TTP-Mediated Regulation of COX-2 Expression
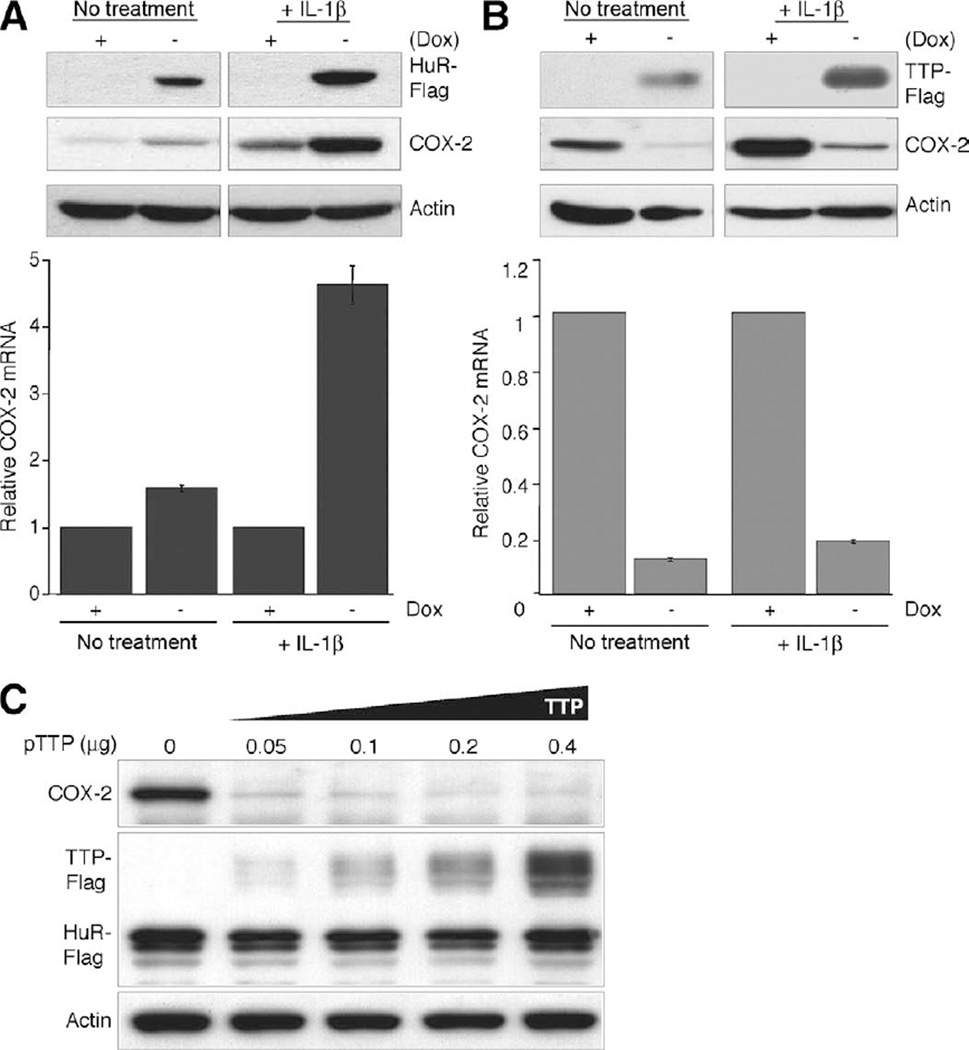
To demonstrate the ability of these ARE-binding proteins to specifically impact endogenous COX-2 expression, Tet-regulated HuR or TTP expression system in HeLa cells was established. HeLa Tet-OFF cells were stably transfected with Tet-regulated expression vectors containing either a Flag epitope-tagged HuR or TTP cDNA; cells grown in the absence of Dox allow for the overexpression of HuR or TTP. As shown in Figure 5A, in both actively growing cells or cells stimulated with interleukin (IL)-1β for 24 hours, overexpression of HuR promotes an increase in both COX-2 mRNA and protein expression. However, inducible expression of TTP strongly inhibited COX-2 mRNA and protein expression in both nontreated and IL-1β-stimulated cells (Figure 5B). Similar results were obtained from 3 other independent HuR or TTP HeLa Tet-OFF clones. Control HeLa Tet-OFF parental cells grown in the presence or absence of Dox did not show any differences in COX-2 expression (data not shown).
Figure 5.
Regulation of COX-2 by HuR and TTP. (A) COX-2 protein (top panel) and mRNA (bottom panel) expression in HeLa-Tet-OFF/HuR-Flag cells grown in the presence or absence of Dox (2 µg/mL) to induce HuR-Flag. After 48 hours of growth, cells were treated with IL-1β (10 ng/mL) or left untreated for 24 hours. (B) Identical treatments were performed using HeLa-Tet-OFF/TTP-Flag cells. COX-2, Flag-TTP, Flag-HuR, and β-actin were detected by Western blot. COX-2 mRNA was examined by qPCR and normalized to 18S rRNA levels. Values shown are based on COX-2 levels from uninduced cells (+ Dox). (C) TTP antagonizes the ability of HuR overexpression to promote COX-2 expression. Expression constructs containing full-length COX-2 (COX-2+3’ UTR), Flag-tagged HuR, and Flag-tagged TTP were trans-fected into COS7 cells. The amount of COX-2 and HuR constructs was kept constant at 0.2 and 0.4 µg, respectively. Increasing amounts of TTP construct were transfected using empty vector to keep total DNA transfected at 1 µg. COX-2, Flag-TTP, Flag-HuR, and β-actin were detected by Western blot.
The findings above demonstrate the ability of TTP to target COX-2 for down-regulation, whereas HuR overexpression promotes COX-2 overexpression. Because both HuR and TTP share overlapping ARE-binding specificity to the COX-2 mRNA, we sought to determine whether TTP could antagonize the ability of HuR to promote COX-2 expression when overexpressed. COS7 cells were transfected with expression vectors containing the full-length COX-2 (COX-2 + 3′UTR), HuR, and TTP cDNAs; COX-2 and HuR vector amounts were kept constant, and TTP vector amounts were progressively increased. As demonstrated in Figure 5C, TTP effectively down-regulates COX-2 protein expression even at the lowest concentration despite the presence of elevated levels of HuR. The ability of TTP to antagonize HuR-mediated COX-2 expression did not appear to involve their physical interaction because coimmunoprecipitation of TTP with HuR was not observed (Supplementary Figure 2).
TTP-Mediated Down-Regulation of COX-2 and Growth Inhibition in Colon Cancer Cells
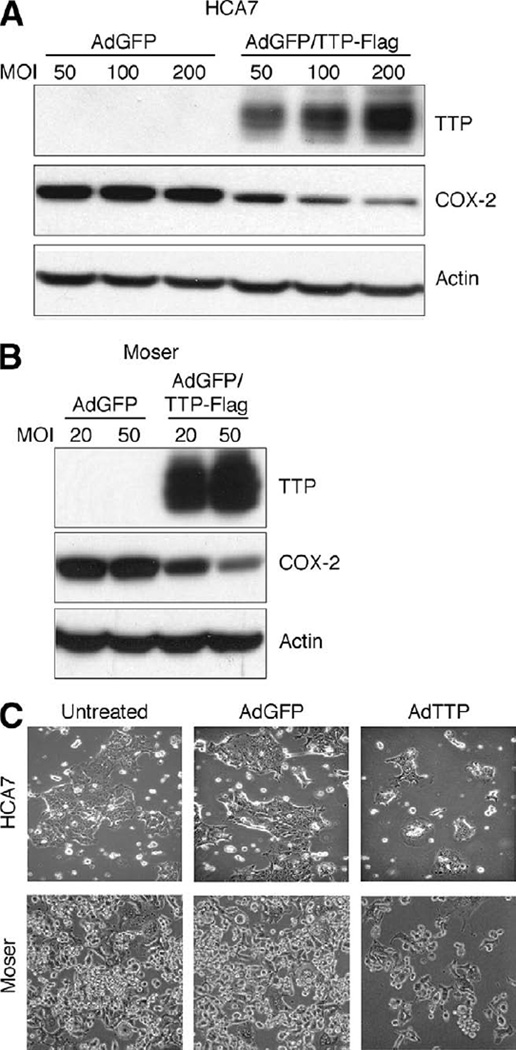
Based on our data demonstrating the ability of TTP to attenuate COX-2 expression in HeLa cells, adenoviral delivery of TTP to colon cancer cells was performed to determine its impact on COX-2 levels and cell growth. As shown in Figure 6A and B, both HCA7 and Moser cells showed a dose-dependent decrease in COX-2 expression when infected with increasing amounts of adenovirus expressing TTP compared with the control adenovirus expressing GFP. Consistent with this, adenoviral delivery of TTP to HCA7 and Moser cells dramatically inhibited cell growth (Figure 6C). These results demonstrate the ability of TTP to target COX-2 for down-regulation in the presence of elevated HuR and impact colon cancer cell growth similar to treatment of colon cancer cells with selective COX-2 inhibitors.13
Figure 6.
TTP expression in colon cancer cells inhibits COX-2 and attenuates cell growth. HCA7 (A) and Moser (B) cells infected with AdGFP or AdGFP/TTP-Flag virus at the indicated MOI for 48 hours were examined for TTP and COX-2 expression by Western blotting. β-actin was used as a loading control. (C) Phase contrast microscopy of HCA7 and Moser cells after 48 hours of infection with AdGFP or AdGFP/TTP-Flag virus using MOIs of 100 and 20, respectively. Uninfected cells are shown. Original magnification, 100×.
Discussion
The control of COX-2 expression is a complex regulatory process that requires input from multiple pathways impacting gene expression on both transcriptional and posttranscriptional levels.10 It is generally well accepted that transcriptional activation of the COX-2 gene PTGS2 is an early event in tumorigenesis because evidence demonstrates the presence of COX-2 mRNA in virtually all colorectal adenomas, adenocarcinomas, and colon cancer cells.2,10,14,15 However, the presence of COX-2 mRNA does not necessarily reflect respective protein levels because variable expression of COX-2 protein and prostaglandins is observed in colon cancer tissue and cells.2,8,15,16 These results imply that enhanced expression of COX-2 protein requires loss of posttranscriptional regulation to occur. This is consistent with observations demonstrating that tumors with increased size and invasiveness display elevated COX-2 mRNA and protein levels17–19 and indicate a link between tumor progression and defects in both the regulation of PTGS2 gene transcription and subsequent mRNA decay.
This study set out to examine the basis for the loss of COX-2 posttranscriptional regulation occurring in colorectal tumors. The expression levels of the mRNA stability factor HuR and decay factor TTP were evaluated in normal colonic epithelium compared with colorectal adenomas, adenocarcinomas, and colon cancer cell lines. Normally expressed at low levels and located in the nucleus, HuR overexpression and cytoplasmic localization were observed in tissues obtained from colon adenomas, adenocarcinomas, and metastases (Figure 1 and Supplementary Figure 1, and data not shown). Furthermore, in cells displaying elevated HuR, we observed a concurrent loss of TTP expression. Consistent with these observations, overexpression of the ARE-containing gene COX-2 coincided with elevated HuR and loss of TTP expression. Several studies indicate that HuR overexpression and cytoplasmic localization are a marker for elevated cancer-associated gene expression that is correlated with advancing stages of malignancy and poor clinical outcome.11,20–22 The findings presented here are in agreement indicating a role for HuR overexpression in colon cancer development. It is not known whether HuR overexpression is an oncogenic event in colon or other cancers. Using an ubiquitously expressed HuR transgenic mouse model, Levadoux–Martin et al23 demonstrated impaired gametogenesis to occur; however, loss of transgene expression in somatic tissues limited determination of the oncogenic capacity of HuR in vivo. Similarly, an inducible HuR transgene restricted to myeloid cell lineages promoted stabilization of inflammatory mediator mRNAs but did not induce an observable neoplastic phenotype.24 Although these in vivo studies do not indicate a putative role for HuR overexpression as a tumor-initiating event, they suggest a possible function for HuR overexpression in tumorigenesis by serving as a tumor growth promoter or in a tumor maintenance capacity.
The results presented here imply that the accompanied loss of TTP expression is a critical factor for cancer-associated gene overexpression in tumors. The tumor-derived loss of TTP is of significance because low levels of TTP can efficiently suppress COX-2 expression in the presence of elevated HuR (Figure 5C) along with attenuating COX-2 overexpression and cell growth in colon cancer cells (Figure 6). This ability to antagonize HuR-mediated COX-2 mRNA stabilization implies that both loss-of-TTP and gain-of-HuR function are required events for COX-2 overexpression. Furthermore, the presence of TTP in normal colon epithelium suggests that it serves in a protective capacity by controlling inflammatory mediator expression levels. This is evident in TTP knock-out mice that develop multiple inflammatory syndromes resulting from increased COX-2 and inflammatory factors because of defects in their respective rapid mRNA turnover.25 These aspects, coupled with the observation that TTP can inhibit tumorigenesis of an H-ras-dependent mast cell model through degradation of ARE-containing IL-3 mRNA,26 suggest that TTP can serve in a tumor suppressor capacity by attenuating ARE-containing gene expression.
The mechanisms allowing for HuR overexpression and TTP loss in colon tumors are largely undefined. The data presented here in colon tumors (Figure 3) are consistent with prior results demonstrating increased HuR and decreased TTP mRNA levels to occur in colon cancer cell lines.2,27 The HuR (ELAVL1) and TTP (ZFP36) genes are located on 19p13.2 and 19q13.1, respectively, and do not appear to lie in regions of genomic alterations occurring in colorectal cancer.28 Pertaining to HuR overexpression, an alternative explanation suggests that increased HuR transcription contributes to overexpression in colon tumors. With regard to this, characterization of the human and murine HuR promoter has identified a number of responsive elements associated with cellular signaling pathways commonly altered in colon cancer.29,30 One explanation for the lack of TTP expression observed in tumor tissue may reside in epigenetic silencing of the TTP promoter. Examination of the human TTP promoter (accession No. AY771351) identified a putative CpG island present in the proximal 3′ region of the promoter, and, in colon cancer cell lines that were deficient in TTP expression, the presence of hypermethylation was observed (unpublished observations). Based on this, we hypothesize that epigenetic alterations governed by changes in DNA methylation or altered chromatin structure promote TTP gene silencing in colorectal tumors. Alternatively, TTP has been demonstrated to regulate its own expression through a negative feedback loop by binding an ARE present in its 3’UTR.31 However, we are unable to detect TTP protein in tumor tissue or colon cancer cells, suggesting that this mechanism of TTP autoregulation may be a means to limit TTP levels in nonneoplastic cells under inflammatory conditions.
There is a growing body of evidence suggesting that defects in ARE-mediated mRNA decay play a central role in chronic inflammation and tumorigenesis. It is estimated that 5%–8% of the human transcriptome is composed of ARE-containing mRNAs,32 and recent findings have demonstrated an enrichment of this subset of transcripts to occur during colon tumorigenesis. Gene expression profiling comparing adenomas to late-stage adenocarcinomas shows a 3- to 4-fold enrichment in ARE-containing genes compared with the genome as a whole, and a similar enrichment is observed as early as stage I tumors.33 The findings presented here provide a mechanistic basis for these results and indicate ARE-mediated posttranscriptional gene regulation to be an important regulatory mechanism involved during the early stages of colorectal tumorigenesis.
Supplementary Material
Acknowledgments
Funding
Supported by grants P20 RR017698 from the National Institutes of Health, and RSG-06-122-01-CNE from the American Cancer Society (to D.A.D.).
Abbreviations used in this paper
- 3′ UTR
3′-untranslated region
- ARE
adenylate- and uridylate-rich element
- COX-2
cyclooxygenase-2
- HuR
Hu antigen R
- IRS
immunoreactivity score
- TTP
tristetraprolin
Footnotes
Supplementary Data
Note: To access the supplementary material accompanying this article, visit the online version of Gastroenterology at www.gastrojournal.org, and at doi:10.1053/j.gastro.2009.01.010.
Conflicts of interest
The authors disclose no conflicts.
References
- 1.Garneau NL, Wilusz J, Wilusz CJ. The highways and byways of mRNA decay. Nat Rev Mol Cell Biol. 2007;8:113–126. doi: 10.1038/nrm2104. [DOI] [PubMed] [Google Scholar]
- 2.Dixon DA, Tolley ND, King PH, et al. Altered expression of the mRNA stability factor HuR promotes cyclooxygenase-2 expression in colon cancer cells. J Clin Invest. 2001;108:1657–1665. doi: 10.1172/JCI12973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lopez de Silanes I, Fan J, Yang X, et al. Role of the RNA-binding protein HuR in colon carcinogenesis. Oncogene. 2003;22:7146–7154. doi: 10.1038/sj.onc.1206862. [DOI] [PubMed] [Google Scholar]
- 4.Benjamin D, Moroni C. mRNA stability and cancer: an emerging link? Expert Opin Biol Ther. 2007;7:1515–1529. doi: 10.1517/14712598.7.10.1515. [DOI] [PubMed] [Google Scholar]
- 5.Fan XC, Steitz JA. Overexpression of HuR, a nuclear-cytoplasmic shuttling protein, increases the in vivo stability of ARE-containing mRNAs. EMBO J. 1998;17:3448–3460. doi: 10.1093/emboj/17.12.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lai WS, Carballo E, Thorn JM, et al. Interactions of CCCH zinc finger proteins with mRNA. Binding of tristetraprolin-related zinc finger proteins to Au-rich elements and destabilization of mRNA. J Biol Chem. 2000;275:17827–17837. doi: 10.1074/jbc.M001696200. [DOI] [PubMed] [Google Scholar]
- 7.Hau HH, Walsh RJ, Ogilvie RL, et al. Tristetraprolin recruits functional mRNA decay complexes to ARE sequences. J Cell Biochem. 2007;100:1477–1492. doi: 10.1002/jcb.21130. [DOI] [PubMed] [Google Scholar]
- 8.Wang D, Mann JR, DuBois RN. The role of prostaglandins and other eicosanoids in the gastrointestinal tract. Gastroenterology. 2005;128:1445–1461. doi: 10.1053/j.gastro.2004.09.080. [DOI] [PubMed] [Google Scholar]
- 9.Dixon DA, Kaplan CD, McIntyre TM, et al. Posttranscriptional control of cyclooxygenase-2 gene expression. The role of the 3′-untranslated region. J Biol Chem. 2000;275:11750–11757. doi: 10.1074/jbc.275.16.11750. [DOI] [PubMed] [Google Scholar]
- 10.Dixon DA. Regulation of COX-2 expression in human cancer. Prog Exp Tumor Res. 2003;37:52–71. doi: 10.1159/000071363. [DOI] [PubMed] [Google Scholar]
- 11.Denkert C, Koch I, von Keyserlingk N, et al. Expression of the ELAV-like protein HuR in human colon cancer: association with tumor stage and cyclooxygenase-2. Mod Pathol. 2006;9:1261–1269. doi: 10.1038/modpathol.3800645. [DOI] [PubMed] [Google Scholar]
- 12.Sawaoka H, Dixon DA, Oates JA, et al. Tristetrapolin binds to the 3′ untranslated region of cyclooxygenase-2 mRNA: a polyadenylation variant in a cancer cell line lacks the binding site. J Biol Chem. 2003;278:13928–13935. doi: 10.1074/jbc.M300016200. [DOI] [PubMed] [Google Scholar]
- 13.Williams CS, Watson AJ, Sheng H, et al. Celecoxib prevents tumor growth in vivo without toxicity to normal gut: lack of correlation between in vitro and in vivo models. Cancer Res. 2000;60:6045–6051. [PubMed] [Google Scholar]
- 14.Eberhart CE, Coffey RJ, Radhika A, et al. Up-regulation of cycloxygenase 2 gene expression in human colorectal adenomas and adenocarcinomas. Gastroenterology. 1994;107:1183–1188. doi: 10.1016/0016-5085(94)90246-1. [DOI] [PubMed] [Google Scholar]
- 15.Shao J, Sheng H, Inoue H, et al. Regulation of constitutive cyclooxygenase-2 expression in colon carcinoma cells. J Biol Chem. 2000;43:33951–33956. doi: 10.1074/jbc.M002324200. [DOI] [PubMed] [Google Scholar]
- 16.Kargman S, O’Neill G, Vickers P, et al. Expression of prostaglandin G/H synthase-1 and −2 protein in human colon cancer. Cancer Res. 1995;55:2556–2559. [PubMed] [Google Scholar]
- 17.Zhang H, Sun XF. Overexpression of cyclooxygenase-2 correlates with advanced stages of colorectal cancer. Am J Gastroenterol. 2002;97:1037–1041. doi: 10.1111/j.1572-0241.2002.05625.x. [DOI] [PubMed] [Google Scholar]
- 18.Hasegawa K, Ichikawa W, Fujita T, et al. Expression of cycloOxygenase-2 (COX-2) mRNA in human colorectal adenomas. Eur J Cancer. 2001;37:1469–1474. doi: 10.1016/s0959-8049(01)00137-x. [DOI] [PubMed] [Google Scholar]
- 19.Einspahr JG, Krouse RS, Yochim JM, et al. Association between cyclooxygenase expression and colorectal adenoma characteristics. Cancer Res. 2003;63:3891–3893. [PubMed] [Google Scholar]
- 20.Heinonen M, Bono P, Narko K, et al. Cytoplasmic HuR expression is a prognostic factor in invasive ductal breast carcinoma. Cancer Res. 2005;65:2157–2161. doi: 10.1158/0008-5472.CAN-04-3765. [DOI] [PubMed] [Google Scholar]
- 21.Erkinheimo TL, Lassus H, Sivula A, et al. Cytoplasmic HuR expression correlates with poor outcome and with cyclooxygenase 2 expression in serous ovarian carcinoma. Cancer Res. 2003;63:7591–7594. [PubMed] [Google Scholar]
- 22.Mrena J, Wiksten JP, Thiel A, et al. Cyclooxygenase-2 is an independent prognostic factor in gastric cancer and its expression is regulated by the messenger RNA stability factor HuR. Clin Cancer Res. 2005;11:7362–7368. doi: 10.1158/1078-0432.CCR-05-0764. [DOI] [PubMed] [Google Scholar]
- 23.Levadoux-Martin M, Gouble A, Jégou B, et al. Impaired gametogenesis in mice that overexpress the RNA-binding protein HuR. EMBO Rep. 2003;4:394–399. doi: 10.1038/sj.embor.embor803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Katsanou V, Papadaki O, Milatos S, et al. HuR as a negative posttranscriptional modulator in inflammation. Mol Cell. 2005;19:777–789. doi: 10.1016/j.molcel.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 25.Phillips K, Kedersha N, Shen L, et al. Arthritis suppressor genes TIA-1 and TTP dampen the expression of tumor necrosis factor α, cyclooxygenase 2, and inflammatory arthritis. Proc Natl Acad Sci USA. 2004;101:2011–2016. doi: 10.1073/pnas.0400148101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stoecklin G, Gross B, Ming XF, et al. A novel mechanism of tumor suppression by destabilizing AU-rich growth factor mRNA. Oncogene. 2003;22:3554–3561. doi: 10.1038/sj.onc.1206418. [DOI] [PubMed] [Google Scholar]
- 27.Carrick DM, Blackshear PJ. Comparative expression of tristetraprolin (TTP) family member transcripts in normal human tissues and cancer cell lines. Arch Biochem Biophys. 2007;462:278–285. doi: 10.1016/j.abb.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 28.Staub E, Grone J, Mennerich D, et al. A genome-wide map of aberrantly expressed chromosomal islands in colorectal cancer. Mol Cancer. 2006;5:37. doi: 10.1186/1476-4598-5-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kang MJ, Ryu BK, Lee MG, et al. NF-κB activates transcription of the RNA-binding factor HuR, via PI3K–AKT signaling, to promote gastric tumorigenesis. Gastroenterology. 2008;135:2030–2042. doi: 10.1053/j.gastro.2008.08.009. [DOI] [PubMed] [Google Scholar]
- 30.King PH, Fuller JJ, Nabors LB, et al. Analysis of the 5′ end of the mouse Elavl1 (mHuA) gene reveals a transcriptional regulatory element and evidence for conserved genomic organization. Gene. 2000;242:125–131. doi: 10.1016/s0378-1119(99)00537-5. [DOI] [PubMed] [Google Scholar]
- 31.Tchen CR, Brook M, Saklatvala J, et al. The stability of tristetraprolin mRNA is regulated by mitogen-activated protein kinase p38 and by tristetraprolin itself. J Biol Chem. 2004;279:32393–32400. doi: 10.1074/jbc.M402059200. [DOI] [PubMed] [Google Scholar]
- 32.Bakheet T, Williams BR, Khabar KS. ARED 3.0: the large and diverse AU-rich transcriptome. Nucleic Acids Res. 2006;34:D111–D114. doi: 10.1093/nar/gkj052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kanies CL, Smith JJ, Kis C, et al. Oncogenic Ras and transforming growth factor-β synergistically regulate AU-rich element-containing mRNAs during epithelial to mesenchymal transition. Mol Cancer Res. 2008;6:1124–1136. doi: 10.1158/1541-7786.MCR-07-2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.