Abstract
The molecular circuitry underlying innate immunity is constructed of multiple, evolutionarily conserved signaling modules with distinct regulatory targets. The MAP kinases and the IKK-NF-κB molecules play important roles in the initiation of immune effector responses. We have found that the Drosophila NF-κB protein Relish plays a crucial role in limiting the duration of JNK activation and output in response to Gram-negative infections. Relish activation is linked to proteasomal degradation of TAK1, the upstream MAP kinase kinase kinase required for JNK activation. Degradation of TAK1 leads to a rapid termination of JNK signaling, resulting in a transient JNK-dependent response that precedes the sustained induction of Relish-dependent innate immune loci. Because the IKK-NF-κB module also negatively regulates JNK activation in mammals, thereby controlling inflammation-induced apoptosis, the regulatory cross-talk between the JNK and NF-κB pathways appears to be broadly conserved.
Keywords: MAPKKK, signal transduction, rel protein, proteosome, Imd
Multicellular organisms defend themselves from microbial infections by mounting an innate immune response that serves to neutralize pathogenic invaders (Kimbrell and Beutler 2001; Janeway and Medzhitov 2002). Receptors that recognize pathogen-associated molecular patterns (PAMPs) trigger signaling cascades that culminate in expression of immune effector genes encoding antimicrobial peptides, cytokines, chemokines, and various inflammatory mediators. In both vertebrates and invertebrates, the NF-κB family of transcription factors serves a pivotal role in signal-induced gene activation during the innate immune response (Silverman and Maniatis 2001).
Owing to the broad evolutionary conservation of innate immunity, studies in genetically tractable invertebrates, especially Drosophila melanogaster, have contributed greatly to our understanding of the mechanisms underlying innate immunity. Drosophila, like other invertebrates, lacks an adaptive immune system, being entirely dependent on innate immunity for its resistance to microbial infection. Genetic screens for Drosophila mutants susceptible to pathogenic challenge have identified two distinct signaling cascades—the Toll and Imd pathways—that govern expression of antimicrobial peptides (Hoffmann and Reichhart 2002; Tzou et al. 2002; Hultmark 2003).
The Toll pathway, originally characterized for its role in establishing embryonic dorsoventral polarity, mediates resistance to fungal and Gram-positive infections by activating two NF-κB proteins: Dorsal and Dif (Wasserman 2000). The Imd pathway induces expression of a distinct set of genes coding for antimicrobial peptides, such as Diptericin and Attacin, that provide immunity to Gram-negative bacteria. Components of the Imd pathway include a peptidoglycan recognition protein (PGRP-LC; Choe et al. 2002; Gottar et al. 2002; Ramet et al. 2002), as well as several counterparts to signal transducers involved in NF-κB activation in mammalian cells. These include IMD itself, a homolog of the TNF signaling pathway kinase RIP (Georgel et al. 2001), as well as TAK1, a MAP kinase kinase kinase (MAPKKK), and two IκB kinase (IKK) complex components, IKKβ (ird5), and IKKγ (Kenny; Rutschmann et al. 2000; Silverman et al. 2000, 2003; Lu et al. 2001; Vidal et al. 2001). In conjunction with the IKK complex, the Drosophila caspase Dredd functions in proteolytic activation of a third NF-κB family protein, Relish, that directly activates antibacterial peptide genes (Leulier et al. 2000; Stöven et al. 2000, 2003). There is also evidence that the Drosophila FADD homolog acts downstream of IMD, independently of TAK1 and IKK, to control Dredd activation (Leulier et al. 2002; Naitza et al. 2002).
Although Relish, Dif, and Dorsal are critical for transcriptional induction of antimicrobial peptides, they are unlikely to be the sole mediators of PAMP-triggered signal transduction. In mammalian macrophages and dendritic cells, Toll-like receptors (TLRs) specific to particular PAMP ligands induce activation of ERK, JNK, and p38 MAP kinases (MAPKs) in addition to NF-κB (Akira et al. 2001). Furthermore, the nematode Caenorhabditis elegans and the plant Arabidopsis thaliana encode no orthologs of NF-κB, but rely instead on MAPK signaling cascades to mediate innate immune responses (Asai et al. 2002; Kim et al. 2002; Mallo et al. 2002).
JNK signaling is required for a variety of biological processes in flies and mammals, including developmental morphogenesis, apoptosis, and immunity (Stronach and Perrimon 1999; Chang and Karin 2001). In mammals, JNK activation is required for inflammation-induced apoptosis (Deng et al. 2003; Maeda et al. 2003). Curiously, JNK activation is negatively regulated by the IKK-NF-κB pathway (De Smaele et al. 2001; Tang et al. 2001). Although this cross-talk appears to be an important mediator of the antiapoptotic function of NF-κB (Karin and Lin 2002), the mechanism by which NF-κB negatively regulates JNK activation in mammals is not well understood (Kyriakis 2001). In Drosophila, lipopolysaccharide (LPS) has been reported to activate JNK (Sluss et al. 1996), which is required for induction of a subset of immune response genes (Boutros et al. 2002), as well as the NF-κB family member Relish, which directs transcription of a distinct group of immune response loci (Boutros et al. 2002). However, it has been difficult to assess the biological role of the JNK module in the innate immune response because loss-of-function mutations in this pathway are usually embryonic lethal (Glise et al. 1995; Riesgo-Escovar et al. 1996; Sluss et al. 1996).
We have found that activation of the Relish module in Drosophila results in attenuation of JNK activity after challenge with a commercial LPS preparation. Moreover, we demonstrate that Relish activation is mechanistically linked to switching off the JNK signaling module after LPS stimulation. In the absence of Relish activation, LPS treatment causes a sustained activation of JNK, which, in turn, leads to prolonged expression of JNK target genes. Biochemical experiments reveal that the negative regulation of JNK signaling involves proteasomal degradation of TAK1. Hence, it appears that the Relish-dependent genes not only serve their own immune effector roles, but also participate in shaping the temporal pattern of the JNK-mediated immune response. These findings reveal an unanticipated cross-talk between Drosophila innate immune pathways and hold promise for mechanistic insight into the mammalian counterpart of this regulatory interaction.
Results
An LPS preparation induces transient and sustained transcriptional responses via distinct signaling modules
Activation of innate immune signaling in whole flies can arise from the primary challenge with the infectious organism and from secondary responses to cytokines and inflammatory mediators produced during the primary response. To reduce the complexity of the systemic immune response, we chose to work with cultured S2 cells. These cells are known to be immune-responsive and are believed to be derived from phagocytes (Ramet et al. 2002). We first pretreated the cells with ecdysone, which enhances the innate immune response (Dimarcq et al. 1997). Then, 1 d after ecdysone treatment, we exposed the cells to a commercial LPS preparation; such preparations contain Gram-negative bacterial peptidoglycan that induces the Imd pathway (Leulier et al. 2003). We then isolated total RNA at different time points post-challenge and analyzed the mRNA expression profile using high-density oligonucleotide microarrays representing nearly all of the Drosophila transcriptome. To identify genes exhibiting the largest response, we screened for loci with an increase in mRNA expression of at least fourfold within 6 h of treatment. In all, 64 genes met this criterion. We then restricted our analysis to a subset of 27 genes that were induced at least twofold within 90 min of bacterial infection of adult flies, based on published data obtained with the same array platform and data analysis scheme (De Gregorio et al. 2001).
The responsive genes could be readily divided into two classes (Fig. 1A,B). Expression of transient response genes peaked at 2 h and declined two- to ninefold between the 2- and 4-h time points. In contrast, expression of the sustained response genes remained steady (less than two-fold variation) or increased from 2 h of LPS addition onward. The composition of the two response classes was markedly different. Most of the sustained response genes encoded either antimicrobial peptides or peptidoglycan recognition proteins (Fig. 1A) and belonged to the group of Relish-dependent genes (De Gregorio et al. 2002). Neither type of protein was found among the products of the transient response class, which, instead, contained several novel genes that had not previously been classified as immune response loci (Fig. 1B; data not shown). The fact that their expression very quickly returned to uninduced levels precluded their passing the screening criteria previously used in identifying immune response loci in whole flies (De Gregorio et al. 2002). Many of the same transient response genes were, however, identified in an analogous study carried out without steroid hormone treatment (Boutros et al. 2002).
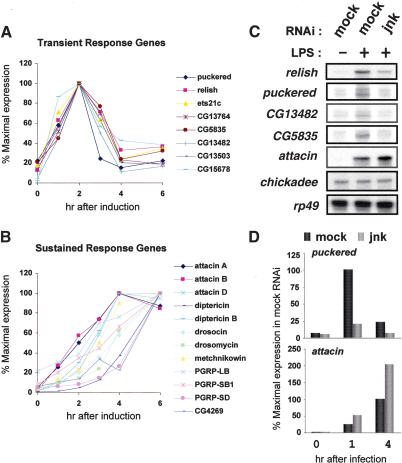
Figure 1.
Requirement for JNK in the transient response gene induction by LPS. (A) LPS-induced transient response gene expression in S2 cells. S2 cells were stimulated with LPS for the length of time indicated on the abscissa. Total RNA was prepared and analyzed using the Affymetrix Drosophila gene chip. Values on the ordinate indicate the relative mRNA level. (B) LPS-induced sustained response gene expression in S2 cells. (C) S2 cells were mock-treated or treated with jnk dsRNA for 3 d and then left unstimulated or stimulated with LPS for 1 h. Total RNA isolated from those cells was analyzed by RNase protection to assay the expression of the genes listed on the left. In this and subsequent panels, attacin refers to the attacin A gene. (D) After treating S2 cells with dsRNA and LPS as in C, the puckered and attacin mRNA levels in S2 cells were measured by real-time PCR analysis. The results shown are averages of three determinations.
As induction of many transient response genes by LPS preparations, including puckered, requires JNK signaling (Boutros et al. 2002), we treated S2 cells with JNK-specific double-stranded RNA to deplete endogenous JNK by RNA interference (RNAi). The cells were then challenged with LPS, and their gene expression profile analyzed by an RNase protection assay (Fig. 1C) and by real-time PCR analysis (Fig. 1D). When we examined expression of four transient response genes (Relish, puckered, CG13482, and CG5835), we found that their induced expression was greatly diminished or abolished by JNK dsRNA treatment. In contrast, expression of the Relish target gene attacin was not abolished and, indeed, increased slightly, upon JNK inactivation by RNAi. This finding, together with previous results (Boutros et al. 2002), indicates that JNK signaling governs expression of transient response genes in Drosophila immunity. Furthermore, the JNK and Relish branches of the Imd pathway control induction of genes that differ with regard to their temporal patterns of expression and their function.
Defects in the IKK/Relish module result in prolonged expression of JNK target genes
In infected flies, cytokines (Agaisse et al. 2003) and other inflammatory mediators (Foley and O'Farrell 2003) mediate secondary responses that have the potential to contribute to JNK activation. Temporal expression patterns and requirements for particular signaling modules could therefore differ from those in S2 cells. To address this possibility, we analyzed expression of transient and sustained response genes in wild-type (wt) flies.
Infection of wild-type flies with the Gram-negative bacterium Enterobacter cloacae induced JNK target gene expression with a pattern similar to that observed in LPS-treated S2 cells (Fig. 2A). The puckered and CG13482 loci were induced immediately after infection, with expression peaking after 30 min and then declining rapidly to basal levels. Other transient response genes also exhibited similar patterns of induced expression in both cells and flies (data not shown). In contrast, diptericin expression in wild-type flies increased gradually and continued to increase until 12 h of infection. Thus, the two temporal patterns of induction seen in S2 cells also characterize the immune response in whole flies.
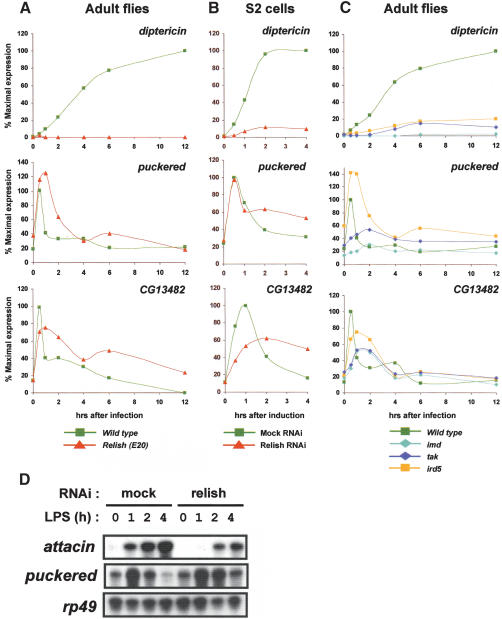
Figure 2.
Prolonged expression of JNK targets in flies and S2 cells that have a defect in Relish activation. (A) Wild-type and relish mutant adult flies were infected by wounding with a needle dipped in E. cloacae. Total RNA was prepared at the indicated time points after infection and subjected to real-time PCR analysis. (B) S2 cells were mock-treated or treated with relish dsRNAs for 3 d and then left unstimulated or stimulated with LPS. Total RNA was prepared and analyzed as in A. (C) Wild-type adult flies and imd, tak1, and ird5 mutant flies were analyzed as in A. (D) RNAi in S2 cells and LPS stimulation were carried out as in B. Total RNA was analyzed by RNase protection assay to measure the expression of the genes listed on the left.
In the Imd pathway, signaling branches downstream from TAK1 (Boutros et al. 2002; Chen et al. 2002). The JNK signaling cascade is required for expression of transient response genes, whereas the IKK/Relish module is essential for sustained gene induction (Boutros et al. 2002). To determine whether there is any cross-talk between these two branches of the Imd pathway, we repeated the infection experiments, using adult flies mutant for individual pathway components. Evidence for cross-talk was readily apparent in studies with relish mutant flies. Although Relish was not required for induction of JNK target genes, the relish E20 mutation increased the duration (puckered and CG13482) and in some cases the intensity (puckered) of target gene expression. These effects did not reflect residual Relish activity or any peculiarity of the E20 mutation, because diptericin expression was abolished (Fig. 2A) and the prolonged expression was also seen in a relish mutant line harboring a different mutation (E38 allele; Hedengren et al. 1999; data not shown). Furthermore, similar effects were observed in S2 cells treated with ds relish RNA. For example, LPS-induced attacin expression was greatly reduced, whereas both puckered and CG13482 displayed sustained expression (Fig. 2B,D). Thus, Relish acts as a negative regulator of JNK target gene expression in both flies and cultured cells.
To determine the point of intersection between the branches of the Imd pathway, we extended our studies to mutations in other pathway components. In ird5 flies, JNK target gene expression exhibited a broadened peak of induction, whereas diptericin expression was substantially decreased, a pattern very similar to that in the relish mutants (Fig. 2C). In contrast, flies mutant for either imd or tak1 were incapable of activating either class of genes, indicating that these loci are essential for triggering both the IKK/Relish and the JNK signaling cascades. We noted, however, that the residual induction of JNK targets in imd and tak1 mutant flies also followed a broadened time course.
Inhibition of Relish activation results in a sustained JNK activation after challenge
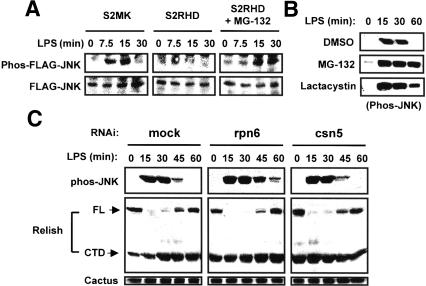
To explore the mechanism of Relish-mediated negative regulation of JNK target genes, we used RNAi to deplete S2 cells either of Relish or of Ird5 or Kenny, components of the IKK complex that directs proteolytic activation of Relish. We then monitored JNK activation in the treated cells by immunoblot analysis, using the phospho-JNK-specific antibody that detects the activated enzyme. In mock-treated cells, phosphorylated JNK was detected at 15 and 30 min after LPS stimulation, with its levels diminishing almost to that of the unstimulated state by 45 min (Fig. 3A). In contrast, RNAi against kenny, ird5, or relish resulted in JNK activation that persisted for at least 60 min after LPS stimulation. We conclude that Relish processing is required for transient JNK activation by the Imd pathway.
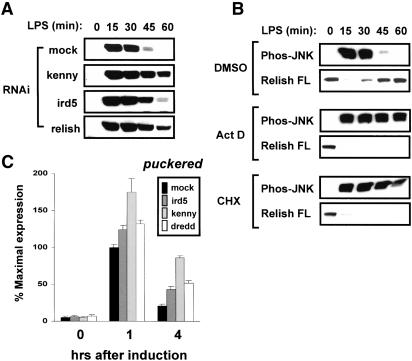
Figure 3.
Sustained activation of JNK signaling in S2 cells lacking Relish activation. (A) S2 cells were treated with dsRNAs specific to kenny, ird5, or relish for 3 d and then unstimulated or stimulated with LPS for the length of time indicated. Cell lysates were prepared and analyzed by immunoblotting with antibodies specific to the phosphorylated forms of JNK. (B) S2 cells were treated with 0.1% (v/v) dimethylsulfoxide (DMSO), 1 μg/mL actinomycin D (Act D), or 25 μg/mL cycloheximide (CHX) for 1 h and then unstimulated or stimulated with LPS for the length of time indicated. Cell lysates were prepared and analyzed as in A with antibodies specific to the phosphorylated forms of JNK (Phos-JNK) and Relish. For Relish, only the full-length (FL) form is shown. (C) S2 cells were mock-treated or were treated with ird5, kenny, or dredd dsRNAs and analyzed as in Figure 2B.
To determine whether Relish-mediated target gene expression is required for JNK regulation, we examined whether down-regulation of JNK in the LPS response requires ongoing RNA and protein synthesis. Pretreatment of S2 cells with either the transcriptional inhibitor actinomycin D or the translational inhibitor cycloheximide resulted in a sustained JNK activation upon LPS stimulation (Fig. 3B). These inhibitors also blocked the resynthesis of full-length Relish protein following proteolytic depletion of the pool present prior to LPS stimulation. These results suggest that activation of one or more target genes by processed Relish protein is required for rapid shutoff of JNK activity. The Relish-dependent negative regulation of JNK signaling is distinct from the previously described negative feedback loop involving the MAPK phosphatase Puckered (Martin-Blanco et al. 1998), because puckered expression is enhanced, not suppressed, in response to inhibition of Relish (see Fig. 2).
To examine whether a prolonged JNK activation in S2 cells lacking Relish activation affects JNK target expression, the level of puckered mRNA was measured after RNAi against ird5, kenny, and dredd. Like S2 cells devoid of Relish itself (Fig. 2B), S2 cells depleted of the components in either branch of the Imd pathway showed an increased and prolonged expression of puckered (Fig. 3C).
Relish activation prior to Imd pathway activation attenuates JNK activation
The notion that Relish is responsible for preventing sustained JNK activation implies that there is a step in the JNK signaling cascade that is sensitive to the inhibitory action of Relish or one of its targets. To test this hypothesis, we devised a protocol in which we temporally separated activation of Relish from that of JNK and then examined whether forced Relish activation prior to LPS stimulation suppressed JNK activation. To this end, we used a constitutively active Relish that contains the N-terminal 600 amino acids, including the Rel-homology domain (Dushay et al. 1996). This derivative, designated RHD, is similar to mature nuclear Relish, which is generated by endoproteolytic cleavage (Fig. 4A; Stöven et al. 2000). We generated an S2 cell line stably transfected with a copper-inducible RHD construct (S2RHD), as well as a control cell line with the backbone vector construct (S2MK). By immunoblot analysis, we confirmed high-level expression of RHD in induced S2RHD cells (Fig. 4B). We also confirmed that the expressed RHD protein was functional by examining Relish target gene expression. Induction of RHD expression directed attacin expression, albeit with a lower efficiency than LPS treatment (Fig. 4C). Other Relish targets were induced to a similar extent by RHD, whereas JNK targets did not show any noticeable induction (data not shown).
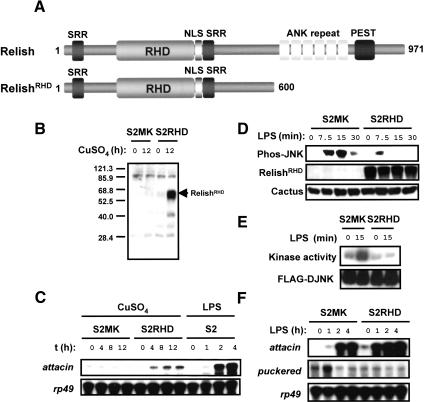
Figure 4.
Inhibition of the JNK signaling module by Relish activation prior to LPS stimulation. (A) Structure of Relish protein and its Rel-homology domain (RHD) derivative. (B) S2 cells stably transfected with this RHD construct (S2RHD) and control cells transfected with the backbone vector construct (S2MK) were untreated or treated with 0.7 mM CuSO4 for 12 h. Cell lysates were prepared and analyzed by immunoblotting with antibodies specific to the RHD of Relish (RelishRHD). (C) S2MK and S2RHD cells were unstimulated or stimulated with CuSO4. Total RNA was prepared after the length of time indicated and analyzed by RNase protection assay. (D) S2MK and S2RHD cells were treated with CuSO4 for 12 h and then unstimulated or stimulated with LPS for the length of time indicated. Cell lysates were prepared and analyzed by immunoblotting with antibodies specific to phos-JNK, RelishRHD, and Cactus. (E) S2MK and S2RHD cells were transfected with a plasmid that expresses Flag-DJNK and then treated with CuSO4 and stimulated with LPS as in D. Flag-DJNK proteins were immunoprecipitated from cell lysates and subjected to in vitro kinase assay using GST-DJun as a substrate. (F) S2MK and S2RHD cells were treated with CuSO4 and stimulated with LPS. Total RNA isolated at the indicated time point was analyzed by RNase protection assay.
Next we tested whether RHD expression prior to Imd pathway activation could inhibit JNK signaling. For this, 12 h after copper addition, we stimulated cells with LPS and then examined JNK activation by two methods. First, we analyzed the level of phosphorylated JNK in the protein extract by immunoblotting. JNK activation in S2RHD cells was greatly reduced in level and duration in comparison with the S2MK line (Fig. 4D). In a second set of experiments, we transfected S2MK and S2RHD with an expression vector for Flag-epitope-tagged JNK and then performed an immune complex kinase assay. We found that RHD expression almost completely inhibited activation of JNK in response to LPS (Fig. 4E). Importantly, this effect was not due to reduced JNK expression. Consistent with our model for Relish function, the inhibition of JNK signaling by RHD expression also altered the induction of target genes. In copper-treated S2RHD cells, puckered induction was abolished, whereas the onset of attacin induction was shifted toward an earlier time point (Fig. 4F).
Relish-mediated inactivation of JNK signaling requires target protein degradation
To further investigate the mechanism underlying Relish-mediated JNK inactivation, we treated S2 cells with an array of chemical agents that perturb inflammatory processes in mammalian immune cells, screening for any that relieved the RHD-mediated inhibition of JNK activation. The compounds analyzed included inhibitors to different classes of serine proteases, cathepsins and caspases, proteasomal inhibitors, scavengers of reactive oxygen and nitrogen intermediates, alkalinizing agents, nitric oxide synthase inhibitors, and NADPH oxidase inhibitors.
Of the compounds tested, only the proteasomal inhibitors MG-132 and lactacystin were effective in restoring LPS-induced JNK activation in copper-treated S2RHD cells (Fig. 5A; data not shown). To confirm that the effect of these inhibitors on JNK activation reflected a restoration of signaling rather than a stabilization of JNK, we transfected Flag-JNK into S2RHD cells and examined the levels of total and phosphorylated JNK. As shown in Figure 5A, MG-132 increased JNK activation without affecting the amount of JNK protein, indicating that MG-132 inhibits a Relish-dependent proteolytic event responsible for attenuation of JNK activation. We also confirmed that proteasomal degradation is essential for negative feedback regulation of JNK during the LPS response in untransfected S2 cells. Pretreatment with MG-132 or lactacystin resulted in sustained JNK activation (Fig. 5B) in much the same way as did RNAi against IKK or Relish or pretreatment with actinomycin D or cycloheximide (see Fig. 3A,B).
Figure 5.
Involvement of proteasomal degradation in Relish-dependent JNK inhibition. (A) S2MK and S2RHD cells were transfected with Flag-DJNK expression plasmid and then treated with CuSO4. S2RHD cells were untreated or further treated with 50 μM MG-132 for 2 h. After stimulating with LPS, cell lysates were prepared and analyzed by immunoblotting with antibodies specific to phos-JNK (upper panels) and Flag (lower panels). (B) S2 cells were treated with DMSO, 50 μM MG-132, or 5 μM lactacystin for 2 h and then unstimulated or stimulated with LPS for the length of time indicated. Phos-JNK was analyzed by immunoblotting. (C) S2 cells were treated with dsRNAs specific to rpn6 or csn5 for 3 d and then unstimulated or stimulated with LPS for the length of time indicated. Cell lysates were prepared and analyzed by immunoblotting with the antibodies indicated on the left.
To confirm a requirement for the proteasome in JNK inactivation during the LPS response, we used RNAi in S2 cells to inactivate Rpn6, a component of the 19S proteasomal regulatory complex. As a control, we also used RNAi against Csn5/Jab1, a component of the nonproteasomal COP9 protein degradation complex. RNAi against Rpn6, but not Csn5, promoted sustained JNK activation (Fig. 5C). Under the same conditions, processing of Relish, as assayed by levels of full-length protein, was unaffected. Because inactivation of SkpA and Slimb, components of the SCF-E3 ubiquitin ligase, has been shown to increase the level of both full-length and processed Relish protein (Khush et al. 2002), we also carried out RNAi against these factors. RNAi against SkpA or Slimb had no effect on JNK activation (data not shown). We conclude that a ubiquitin E3 ligase other than SCF-E3 mediates the negative regulation of JNK activation.
TAK1 is the target for Relish-mediated feedback regulation
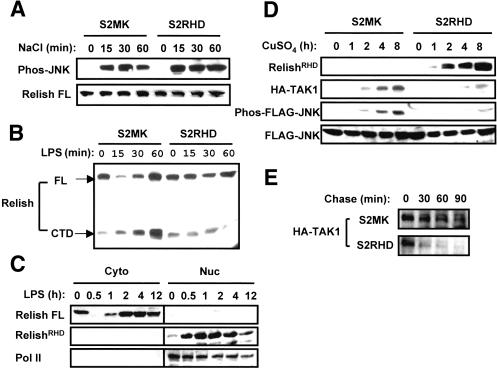
To identify the target for proteasome-mediated inhibition of JNK activation, we first examined whether such inhibition is specific to the Imd pathway. Among other stimuli known to activate JNK in Drosophila, we chose to examine hyperosmolarity. Multiple MAPKKKs, including TAK1, Slipper, Ask, and Mekk1, mediate JNK activation in S2 cells subjected to osmotic stress triggered by sorbitol or NaCl (Chen et al. 2002). In the case of MAPKKs, however, osmotic stress activates the same enzymes, Hep/Mkk7 and Mkk4, as does LPS. In control S2MK cells, we observed JNK activation, but not Relish processing, within 15 min after addition of NaCl to the culture medium; this activation persisted for at least 60 min (Fig. 6A). We detected similar activation of JNK by NaCl in copper-treated S2RHD cells, even when RHD was induced 12 h prior to osmotic stress. Because such RHD expression suppressed JNK activation by LPS (see Fig. 4D), these results suggest that Relish does not inhibit JNK directly, nor does it target the two MAPKKs common to the LPS and osmotic stress pathways.
Figure 6.
Destabilization of Tak1 protein by Relish activation. (A) S2MK and S2RHD cells were treated with CuSO4 and then unstimulated or stimulated with 0.4 M NaCl for the length of time indicated. Phospho-JNK and FL Relish were analyzed by immunoblotting. (B) S2MK and S2RHD cells treated with CuSO4 were unstimulated or stimulated with LPS. FL Relish and its C-terminal domain (CTD) fragment generated by endoproteolytic processing were analyzed by immunoblotting. (C) S2 cells were treated with LPS for the length of time indicated. Cytoplasmic and nuclear protein fractions were prepared and analyzed by immunoblotting. (D) S2MK and S2RHD cells were transfected with HA-TAK1 and Flag-DJNK expression plasmids and then treated with CuSO4. Protein lysates were prepared at the indicated time point and analyzed by immunoblotting. (E) S2MK and S2RHD cells were transfected with the HA-TAK1 expression plasmid and then treated with CuSO4 for 8 h. Cells were then pulse-labeled with [35S]methionine for 1 h and chased with nonradioactive medium for the time points indicated. HA-Tak1 protein was immunoprecipitated from protein lysates and analyzed by SDS-PAGE and autoradiography.
We next examined the effect of RHD overexpression on the LPS-mediated activation of the IKK branch of the Imd pathway. Expression of RHD prior to LPS treatment blocked efficient processing of Relish (Fig. 6B). Thus, Relish must inactivate at least one component that is shared by the JNK and IKK/Relish modules. Nevertheless, there is sustained expression of attacin, diptericin, and other Relish target genes (Fig. 1A). The pattern of Relish target expression may reflect a stabilization of RHD in the nucleus. To test this idea, we prepared cytoplasmic and nuclear fractions of S2 cells at different times after LPS treatment and monitored the level of full-length and processed Relish in the two subcellular fractions. The processed form was found to translocate to the nucleus as early as 30 min after stimulation. Moreover, the amount of processed Relish in the nucleus remained above the basal level for at least 12 h, indicating that a transcriptionally active pool of processed Relish can be maintained in the absence of detectable cytoplasmic processing event (Fig. 6C).
Of the components shared by the JNK and IKK/Relish modules, we chose to examine TAK1, the MAPKKK that is common to both pathways. To assay TAK1 activity, we used a vector expressing Tak1 protein fused to the flu haemagglutinin (HA) epitope tag from the copper-inducible MT promoter and examined its ability to activate JNK in transient transfected cells.
In control S2MK cells, copper-mediated induction of the HA-TAK1 construct resulted in gradually increasing levels of both Tak1 protein and activated JNK (Fig. 6D). In contrast, HA-TAK1 accumulated at only very low levels in S2RHD cells treated with copper, and there was a concomitant minor induction of JNK activation (Fig. 6D). To determine whether the lack of TAK1 accumulation in copper-treated S2RHD reflected increased degradation or decreased production, we carried out a pulse-chase experiment. As shown in Figure 6E, the turnover rate of TAK1, as determined after 8 h of copper treatment, was much higher in S2RHD cells than in S2MK cells. Taken together with our earlier findings, the shortened half-life of TAK1 and minimal JNK activation in cells expressing RHD provide strong support for the idea that Relish triggers negative feedback regulation of immediate early JNK signaling by directing the proteasome-mediated degradation of TAK1.
Discussion
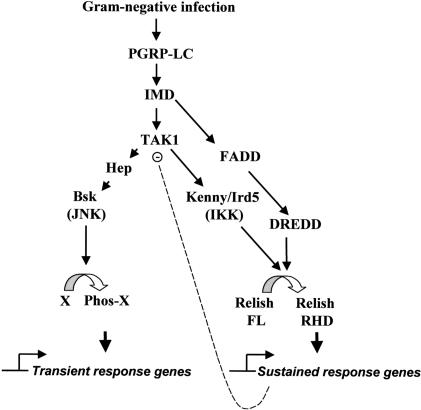
The JNK and IKK/Relish branches of the Imd pathway mediate distinct gene induction responses in Drosophila innate immunity (Fig. 7). After diverging downstream from TAK1, these two signaling cascades regulate two separate groups of target genes that are distinct in their induction kinetics and function. The IKK/Relish targets have been extensively characterized and most encode products whose role in innate immunity is relatively well established. On the other hand, the JNK-regulated, LPS-responsive genes represent a largely uncharacterized set of loci whose function in innate immunity is not clear. These genes exhibit transient induction kinetics, reaching a maximum ∼1 h after induction. We found that the transient kinetics of the JNK target genes is controlled by the transient kinetics of the JNK module of the Imd pathway and that the IKK/Relish branch plays an active role in turning off JNK activity. Hence, the two seemingly independent branches of the Imd pathway are wired in such a way as to coordinate the temporal order of individual responses.
Figure 7.
Model of branched Imd pathway.
Our evidence indicates that Relish-mediated JNK inhibition involves proteasomal degradation of TAK1, the MAPKKK responsible for JNK activation in response to LPS. Treatment with proteasomal inhibitors or RNAi against a component of the proteasome complex results in sustained JNK activation during the LPS response. Furthermore, in cells expressing constitutively active Relish, the stability of TAK1 is greatly decreased. Based on these findings, we suggest that certain targets of Relish that are induced during immune responses facilitate destruction of TAK1 and switch off the JNK cascade (Fig. 7). The fact that cycloheximide and actinomycin D also block the down-regulation of JNK activity indicates the involvement of targets of Relish rather than Relish itself. The Relish target involved in this cross-talk likely increases the susceptibility of TAK1 to proteasomal degradation by direct targeting of TAK1 or by antagonizing factors responsible for TAK1 stabilization.
In considering this model, it should be noted that TAK1 is critically required for activating both IKK and JNK. Elimination of TAK1 during the LPS response thus turns off both of the downstream signaling cascades. Yet IKK/Relish target genes do not show a transient expression pattern, whereas JNK targets do. One possible explanation for this discrepancy lies in the nature of JNK and Relish activation. JNK is activated through its phosphorylation, a modification that is highly reversible. Thus, termination of the input that contributes to JNK phosphorylation is sufficient to result in its rapid inactivation, especially when one of the JNK targets encode a JNK phosphatase, Puckered. Relish, on the other hand, is activated through proteolysis, an irreversible modification. Once activated Relish enters the nucleus, it may remain bound to its target genes for some time even after termination of the upstream signal.
Interestingly, the antagonism between NF-κB and JNK signaling is evolutionarily conserved. In mice, inactivation of either IKKβ or NF-κB RelA (p65) in cells leads to a sustained JNK activation in response to TNFα (Karin and Lin 2002). The sustained JNK activation by TNFα has been associated with TNFα-induced apoptosis. Several independent studies have proposed different molecules as mediators for the NF-κB inhibition of JNK signaling in the TNFα pathway including XIAP (Tang et al. 2001), GADD45β (De Smaele et al. 2001), and reactive oxygen species (Sakon et al. 2003). Nevertheless, the underlying mechanism remains largely unknown (Liu 2003). As the Drosophila Imd pathway and the signaling pathway downstream of mammalian TNF receptor share many conserved features (Hoffmann and Reichhart 2002), a similar mechanism may govern the antagonistic relationship between IKK/NF-κB and JNK in both flies and mammals. Thus, the mechanistic insights gained from our studies in Drosophila should be of relevance to elucidating the mechanism that connects NF-κB to JNK signaling in mammals.
Materials and methods
S2 cell culture and treatment
Immunocompetent S2 cells were grown in Schneider's Drosophila medium supplemented with 10% fetal bovine serum and 1% antibiotic–antimycotic solution (Invitrogen). Cells were maintained as described previously (Sun et al. 2002). A commercial LPS preparation (Sigma; 055:B5) was dissolved in H2O and applied to cells at a final concentration of 10 μg/mL. Chemical agents used in screening for an inhibitor of the RHD-mediated JNK inhibition were N-tosyl-L-phenylalanine, AEBSF, z-VAD-fmk, ac-YVAD-cmk, z-DEVD-fmk, z-FA-fmk, ac-LV-lysinal, diphenyleneiodonium chloride, bafilomycin A1, aminoguanidine, NG-monomethyl-L-arginine monoacetate, MG-132, and lactacystin (Calbiochem).
Fly immune challenge
All Drosophila stocks were raised at room temperature on standard cornmeal, agar, and yeast medium. The relE20 relE38, ird52, imd1, and tak11 mutations have been described previously (Hedengren et al. 1999; Georgel et al. 2001; Lu et al. 2001; Vidal et al. 2001). To study bacterial infection, adult flies were pricked with a fine glass needle dipped in a live culture of E. cloacae resuspended at high concentration in sterile Ringer's solution.
Microarray analysis
S2 cells were grown in Schnieder's Drosophila medium supplemented with 10% fetal bovine serum and 1% glutamine (Invitrogen). Ecdysone was added to a final concentration of 1 μM before 3 × 106 of these cells were plated in a 3-mL volume per well of a 6-well dish. Cells were induced with LPS 24 h later. At specified intervals, media was removed by aspiration and the cells harvested by scraping into 4°C PBS. Total RNA was isolated using the QIAGEN RNeasy kit and QIAshredder; final elution was done with water.
Affymetrix Drosophila gene chips were used for gene expression analysis. Briefly, double-stranded cDNA was synthesized from 5 μg of total RNA. Biotin-labeled cRNA was synthesized using the MessageAmp aRNA kit (Ambion); 15 μg of cRNA was fragmented and hybridized to each array. Each RNA sample was processed twice to obtain replicate biotin-labeled probes. Arrays were washed with the EukGW2 protocol on the GeneChip Fluidics Station (Affymetrix) and scanned using the Gene Array scanner (Affymetrix). Analysis of intensity data, including scaling, normalization, and treatment of replicates, were as described previously (De Gregorio et al. 2001).
RNAi experiments
Templates for RNA transcription containing T7 promoter sequences flanking a cDNA fragment 200–1000 bp in length were generated by PCR. After in vitro transcription using the Megascript kit (Ambion), dsRNA was purified by ethanol precipitation and then dissolved in H2O. For RNAi, S2 cells were incubated with serum-free medium containing 25 μg/mL dsRNA for 3 h. Full-length mouse interleukin-1β dsRNA, which is not homologous to any Drosophila gene, was used for the mock RNAi treatment. Serum was then added back to the culture, and cells were grown for an additional 72 h prior to protein and RNA analyses.
Protein analysis
Whole-cell extracts for immunoblot analysis were prepared with lysis buffer (20 mM HEPES-KOH at pH 7.6, 150 mM NaCl, 10% glycerol, 1% Triton X-100, 25 mM β-glycerophosphate, 2 mM EDTA, and protease inhibitors) and then subjected to SDS-PAGE. Proteins transferred to nitrocellulose membrane were probed with antibodies directed against the RHD and the C-terminal domain of Relish (from Dan Hultmark, Umeå University, Umeå, Sweden), anti-Cactus (from Ruth Steward, Rutgers University, Piscataway, NJ), anti-PolII (from John Lis, Cornell University, Ithaca, NY), and anti-phospho-JNK (Promega) antibodies, and the immunoreactive species were visualized with the ECL Western blot reagent (Pierce). The immune complex kinase assay was carried out as previously described (Sluss et al. 1996). For the pulse-chase experiment, 107 S2 cells were pulse-labeled for 1 h with 200 μCi of [35S]methionine in 0.5 mL of methionine-free M3 medium (Sigma) and then chased by adding 0.5 mL of M3 medium supplemented with 100 mM unlabeled methionine. HA-Tak1 protein was immunoprecipitated from the whole-cell lysates with 2 μL of anti-HA antibody (Roche) and analyzed by SDS-PAGE and autoradiography.
RNA analysis
Total RNA was isolated from S2 cells or whole flies using the RNAwiz reagent (Ambion). For RNase protection assays, 10 μg of total RNA and 100,000 cpm of a 32P-labeled antisense probe with a length of ∼100 nt were hybridized, digested with a mix of RNases T1 and A, and recovered for gel electrophoresis using the RPA kit (Ambion). For real-time PCR analysis, cDNAs were synthesized with the Superscript II reverse transcriptase system (Invitrogen). An amount of cDNA equivalent to 0.2 μg of total RNA was subjected to 40 cycles of PCR amplification consisting of a 15-sec incubation at 95°C and a 1-min incubation at 60°C. Output was monitored using SYBR Green core reagents and the ABI Prism 7700 System (PE Applied Biosystems). All the results were normalized to the rp49 mRNA level. Individual primer sequences are available upon request.
Acknowledgments
We thank Dan Hultmark, John Lis, and Ruth Steward for providing antisera; Deborah Kimbrell, Dan Hultmark, and Bruno Lemaitre for the mutant fly lines; Tony Ip, Neal Silverman, Tom Maniatis, and Jun R. Huh for providing plasmids; Andrea D'Auria for technical assistance; and Paul Spellman for assistance and advice on microarray analysis. This work was supported by a BIOSTAR grant (00-10120) and an NIH grant GM50545 to S.A.W. and by NIH grants CA76188 and AI43477 to M.K. J.M.P. is supported by a postdoctoral fellowship from the Human Frontier Science Program. M.K. is an American Cancer Society Research Professor.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1168104.
References
- Agaisse H., Petersen, U.-M., Boutros, M., Mathey-Prevot, B., and Perrimon, N. 2003. Signaling role of hemocytes in Drosophila JAK/STAT-dependent response to septic injury. Dev. Cell 5: 441–450. [DOI] [PubMed] [Google Scholar]
- Akira S., Takeda, K., and Kaisho, T. 2001. Toll-like receptors: Critical proteins linking innate and acquired immunity. Nat. Immunol. 2: 675–680. [DOI] [PubMed] [Google Scholar]
- Asai T., Tena, G., Plotnikova, J., Willmann, M.R., Chiu, W.L., Gomez-Gomez, L., Boller, T., Ausubel, F.M., and Sheen, J. 2002. MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415: 977–983. [DOI] [PubMed] [Google Scholar]
- Boutros M., Agaisse, H., and Perrimon, N. 2002. Sequential activation of signaling pathways during innate immune responses in Drosophila. Dev. Cell. 3: 711–722. [DOI] [PubMed] [Google Scholar]
- Chang L. and Karin, M. 2001. Mammalian MAP kinase signalling cascades. Nature 410: 37–40. [DOI] [PubMed] [Google Scholar]
- Chen W., White, M.A., and Cobb, M.H. 2002. Stimulus-specific requirements for MAP3Ks in activating the JNK pathway. J. Biol. Chem. 277: 49105–49110. [DOI] [PubMed] [Google Scholar]
- Choe K.M., Werner, T., Stöven, S., Hultmark, D., and Anderson, K.V. 2002. Requirement for a peptidoglycan recognition protein (PGRP) in Relish activation and antibacterial immune responses in Drosophila. Science 296: 359–362. [DOI] [PubMed] [Google Scholar]
- De Gregorio E., Spellman, P.T., Rubin, G.M., and Lemaitre, B. 2001. Genome-wide analysis of the Drosophila immune response by using oligonucleotide microarrays. Proc. Natl. Acad. Sci. 98: 12590–12595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Gregorio E., Spellman, P.T., Tzou, P., Rubin, G.M., and Lemaitre, B. 2002. The Toll and Imd pathways are the major regulators of the immune response in Drosophila. EMBO J. 21: 2568–2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y., Ren, X., Yang, L., Lin, Y., and Wu, X. 2003. A JNK-dependent pathway is required for TNFα-induced apoptosis. Cell 115: 61–70. [DOI] [PubMed] [Google Scholar]
- De Smaele E., Zazzeroni, F., Papa, S., Nguyen, D.U., Jin, R., Jones, J., Cong, R., and Franzoso, G. 2001. Induction of gadd45β by NF-κB downregulates pro-apoptotic JNK signalling. Nature 414: 308–313. [DOI] [PubMed] [Google Scholar]
- Dimarcq J.L., Imler, J.L., Lanot, R., Ezekowitz, R.A., Hoffmann, J.A., Janeway, C.A., and Lagueux, M. 1997. Treatment of l(2)mbn Drosophila tumorous blood cells with the steroid hormone ecdysone amplifies the inducibility of antimicrobial peptide gene expression. Insect Biochem. Mol. Biol. 27: 877–886. [DOI] [PubMed] [Google Scholar]
- Dushay M.S., Asling, B., and Hultmark, D. 1996. Origin of immunity: Relish, a compound Rel-like gene in the antibacterial defense of Drosophila. Proc. Natl. Acad. Sci. 93: 10343–10347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley E. and O'Farrell, P.H. 2003. Nitric oxide contributes to induction of innate immune responses to Gram-negative bacteria in Drosophila. Genes & Dev. 17: 115–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgel P., Naitza, S., Kappler, C., Ferrandon, D., Zachary, D., Swimmer, C., Kopczynski, C., Duyk, G., Reichhart, J.M., and Hoffmann, J.A. 2001. Drosophila immune deficiency (IMD) is a death domain protein that activates antibacterial defense and can promote apoptosis. Dev. Cell 1: 503–514. [DOI] [PubMed] [Google Scholar]
- Glise B., Bourbon, H., and Noselli, S. 1995. hemipterous encodes a novel Drosophila MAP kinase kinase, required for epithelial cell sheet movement. Cell 83: 451–461. [DOI] [PubMed] [Google Scholar]
- Gottar M., Gobert, V., Michel, T., Belvin, M., Duyk, G., Hoffmann J.A., Ferrandon, D., and Royet, J. 2002. The Drosophila immune response against Gram-negative bacteria is mediated by a peptidoglycan recognition protein. Nature 416: 640–644. [DOI] [PubMed] [Google Scholar]
- Hedengren M., Asling, B., Dushay, M.S., Ando, I., Ekengren, S., Wihlborg, M., and Hultmark, D. 1999. Relish, a central factor in the control of humoral but not cellular immunity in Drosophila. Mol. Cell 4: 827–837. [DOI] [PubMed] [Google Scholar]
- Hoffmann J.A. and Reichhart, J.M. 2002. Drosophila innate immunity: An evolutionary perspective. Nat. Immunol. 3: 121–126. [DOI] [PubMed] [Google Scholar]
- Hultmark D. 2003. Drosophila immunity: Paths and patterns. Curr. Opin. Immunol. 15: 12–19. [DOI] [PubMed] [Google Scholar]
- Janeway C.A., and Medzhitov, R. 2002. Innate immune recognition. Annu. Rev. Immunol. 20: 197–216. [DOI] [PubMed] [Google Scholar]
- Karin M. and Lin, A. 2002. NF-κB at the crossroads of life and death. Nat. Immunol. 3: 221–227. [DOI] [PubMed] [Google Scholar]
- Khush R.S., Cornwell, W.D., Uram, J.N., and Lemaitre, B. 2002. A ubiquitin-proteasome pathway represses the Drosophila immune deficiency signaling cascade. Curr. Biol. 12: 1728–1737. [DOI] [PubMed] [Google Scholar]
- Kim D.H., Feinbaum, R., Alloing, G., Emerson, F.E., Garsin, D.A., Inoue, H., Tanaka-Hino, M., Hisamoto, N., Matsumoto, K., Tan, M.W., et al. 2002. A conserved p38 MAP kinase pathway in Caenorhabditis elegans innate immunity. Science 297: 623–626. [DOI] [PubMed] [Google Scholar]
- Kimbrell D.A. and Beutler, B. 2001. The evolution and genetics of innate immunity. Nat. Rev. Genet. 2: 256–267. [DOI] [PubMed] [Google Scholar]
- Kyriakis J.M. 2001. Life-or-death decisions. Nature 414: 265–266. [DOI] [PubMed] [Google Scholar]
- Leulier F., Rodriguez, A., Khush, R.S., Abrams, J.M., and Lemaitre, B. 2000. The Drosophila caspase Dredd is required to resist Gram-negative bacterial infection. EMBO Rep. 1: 353–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leulier F., Vidal, S., Saigo, K., Ueda, R., and Lemaitre, B. 2002. Inducible expression of double-stranded RNA reveals a role for dFADD in the regulation of the antibacterial response in Drosophila adults. Curr. Biol. 12: 996–1000. [DOI] [PubMed] [Google Scholar]
- Leulier F., Parquet, C., Pili-Floury, S., Ryu, J.H., Caroff, M., Lee, W.J., Mengin-Lecreulx, D., and Lemaitre, B. 2003. The Drosophila immune system detects bacteria through specific peptidoglycan recognition. Nat. Immunol. 4: 478–484. [DOI] [PubMed] [Google Scholar]
- Liu Z.G. 2003. Adding facets to TNF signaling: The JNK angle. Mol. Cell 12: 795–796. [DOI] [PubMed] [Google Scholar]
- Lu Y., Wu, L.P., and Anderson, K.V. 2001. The antibacterial arm of the Drosophila innate immune response requires an IκB kinase. Genes & Dev. 15: 104–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda S., Chang, L., Li, Z.-W., Luo, J.-L., Leffert, H., and Karin, M. 2003. IKKβ is required for prevention of apoptosis mediated by cell-bound but not by circulating TNFα. Immunity 19: 725–737. [DOI] [PubMed] [Google Scholar]
- Mallo G.V., Kurz, C.L., Couillault, C., Pujol, N., Granjeaud, S., Kohara, Y., and Ewbank, J.J. 2002. Inducible antibacterial defense system in C. elegans. Curr. Biol. 12: 1209–1214. [DOI] [PubMed] [Google Scholar]
- Martin-Blanco E., Gampel, A., Ring, J., Virdee, K., Kirov, N., Tolkovsky, A.M., and Martinez-Arias, A. 1998. puckered encodes a phosphatase that mediates a feedback loop regulating JNK activity during dorsal closure in Drosophila. Genes & Dev. 12: 557–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naitza S., Rosse, C., Kappler, C., Georgel, P., Belvin, M., Gubb, D., Camonis, J., Hoffmann, J.A., and Reichhart, J.-M. 2002. The Drosophila immune defense against Gram-negative infection requires the death protein dFADD. Immunity 17: 575–581. [DOI] [PubMed] [Google Scholar]
- Ramet M., Manfruelli, P., Pearson, A., Mathey-Prevot, B., and Ezekowitz, R.A. 2002. Functional genomic analysis of phagocytosis and identification of a Drosophila receptor for E. coli. Nature 416: 644–648. [DOI] [PubMed] [Google Scholar]
- Riesgo-Escovar J.R., Jenni, M., Fritz, A., and Hafen, E. 1996. The Drosophila Jun-N-terminal kinase is required for cell morphogenesis but not for DJun-dependent cell fate specification in the eye. Genes & Dev. 10: 2759–2768. [DOI] [PubMed] [Google Scholar]
- Rutschmann S., Jung, A.C., Zhou, R., Silverman, N., Hoffmann, J.A., and Ferrandon, D. 2000. Role of Drosophila IKKγ in a toll-independent antibacterial immune response. Nat. Immunol. 1: 342–347. [DOI] [PubMed] [Google Scholar]
- Sakon S., Xue, X., Takekawa, M., Sasazuki, T., Okazaki, T., Kojima, Y., Piao, J.H., Yagita, H., Okumura, K., Doi, T., et al. 2003. NF-κB inhibits TNF-induced accumulation of ROS that mediate prolonged MAPK activation and necrotic cell death. EMBO J. 22: 3898–3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman N. and Maniatis, T. 2001. NF-κB signaling pathways in mammalian and insect innate immunity. Genes & Dev. 15: 2321–2342. [DOI] [PubMed] [Google Scholar]
- Silverman N., Zhou, R., Stöven, S., Pandey, N., Hultmark, D., and Maniatis, T. 2000. A Drosophila IκB kinase complex required for Relish cleavage and antibacterial immunity. Genes & Dev. 14: 2461–2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman N., Zhou, R., Erlich, R.L., Hunter, M., Bernstein, E., Schneider, D., and Maniatis, T. 2003. Immune activation of NF-κB and JNK requires Drosophila TAK1. J. Biol. Chem. 278: 48928–48934. [DOI] [PubMed] [Google Scholar]
- Sluss H.K., Han, Z., Barrett, T., Davis, R.J., and Ip, Y.T. 1996. A JNK signal transduction pathway that mediates morphogenesis and an immune response in Drosophila. Genes & Dev. 10: 2745–2758. [DOI] [PubMed] [Google Scholar]
- Stöven S., Ando, I., Kadalayil, L., Engstrom, Y., and Hultmark, D. 2000. Activation of the Drosophila NF-κB factor Relish by rapid endoproteolytic cleavage. EMBO Rep. 1: 347–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stöven S., Silverman, N., Junell, A., Hedengren-Olcott, M., Erturk, D., Engstrom, Y., Maniatis, T., and Hultmark, D. 2003. Caspase-mediated processing of the Drosophila NF-κB factor Relish. Proc. Natl. Acad. Sci. 100: 5991–5996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stronach B.E. and Perrimon, N. 1999. Stress signaling in Drosophila. Oncogene 18: 6172–6182. [DOI] [PubMed] [Google Scholar]
- Sun H., Bristow, B.N., Ou, G., and Wasserman, S.A. 2002. A heterodimeric death domain complex in Toll signaling. Proc. Natl. Acad. Sci. 99: 12871–12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang G., Minemoto, Y., Dibling, B., Purcell, N.H., Li, Z., Karin, M., and Lin, A. 2001. Inhibition of JNK activation through NF-κB target genes. Nature 414: 313–317. [DOI] [PubMed] [Google Scholar]
- Tzou P., De Gregorio, E., and Lemaitre, B. 2002. How Drosophila combats microbial infection: A model to study innate immunity and host–pathogen interactions. Curr. Opin. Microbiol. 5: 102–110. [DOI] [PubMed] [Google Scholar]
- Vidal S., Khush, R.S., Leulier, F., Tzou, P., Nakamura, M., and Lemaitre, B. 2001. Mutations in the Drosophila dTAK1 gene reveal a conserved function for MAPKKKs in the control of rel/NF-κB-dependent innate immune responses. Genes & Dev. 15: 1900–1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasserman S.A. 2000. Toll signaling: The enigma variations. Curr. Opin. Gen. Dev. 10: 497–502. [DOI] [PubMed] [Google Scholar]