Abstract
The unwinding of the parental DNA duplex during replication causes a positive linking number difference, or superhelical strain, to build up around the elongating replication fork. The branching at the fork and this strain bring about different conformations from that of (−) supercoiled DNA that is not being replicated. The replicating DNA can form (+) precatenanes, in which the daughter DNAs are intertwined, and (+) supercoils. Topoisomerases have the essential role of relieving the superhelical strain by removing these structures. Stalled replication forks of molecules with a (+) superhelical strain have the additional option of regressing, forming a four-way junction at the replication fork. This four-way junction can be acted on by recombination enzymes to restart replication. Replication and chromosome folding are made easier by topological domain barriers, which sequester the substrates for topoisomerases into defined and concentrated regions. Domain barriers also allow replicated DNA to be (−) supercoiled. We discuss the importance of replicating DNA conformations and the roles of topoisomerases, focusing on recent work from our laboratory.
A thorough understanding of DNA replication and recombination requires knowledge of the conformations and topology of replicating DNA. These are different from those of nonreplicating DNA. The action of DNA helicases, interruptions in replicated strands, and, most importantly, the uniquely branched structure of the replication fork itself, all contribute to these differences. In this review, we illustrate the major conformational differences between replicating and nonreplicating DNA and their physiological importance. We highlight the evidence for each structure in vitro and in vivo. In addition, we address how the links originally residing in the double helix of the parental duplex are fully resolved in bacteria by two type-2 topoisomerases, DNA gyrase and topoisomerase (topo) IV, to form two separate daughter molecules. Although we emphasize the situation in bacteria, we will also make generalizations applicable to the eukarya and archaea.
We begin by defining a few basic terms that form the language of DNA topology (1). The topology we will focus on are the links between the complementary Watson and Crick strands of an intact, topologically constrained piece of DNA. The simplest example is a closed circular DNA, as is found in plasmids and viruses, but the results can be generalized to linear chromosomes because of their organization into closed domains or loops. The intertwining of the complementary strands is described by the linking number (Lk), which is one-half of the signed number of times one strand crosses the other in any projection. According to the sign convention, the crossings in ordinary B-type DNA are (+). The crossings, or nodes, of the complementary strands can result from the local intertwining of the double helix itself, in which case they are measured by a parameter called twist (Tw). Alternatively, nodes result from one segment of the double helix crossing another, as measured by writhe (Wr). Lk is the sum of Tw and Wr. Notably, Lk is unaltered by any deformation short of DNA breakage and reunion. More important is the quantity ΔLk, the difference between Lk and Lk0, where Lk0 is the Lk of a relaxed DNA molecule. The strain on the DNA from a non-zero ΔLk often causes the DNA to supercoil, a form of writhe. Supercoiling can be either plectonemic (interwound) or solenoidal, as when DNA wraps around a protein. The most useful measure of the topological deviation of DNA from the relaxed state is its supercoiling density, or σ. Sigma is equal to ΔLk/Lk0 and is therefore independent of DNA length. Replication causes an increase in ΔLk, because separation of the parental strands lowers the value of Lk0. Therefore, the ΔLk of replication increases by about one for every ten base pairs of replicated DNA.
This review is divided into three parts. We begin by discussing the conformations of (−) supercoiled DNA that is not replicating, the form that DNA adopts away from the fork. Next, we discuss the three ways in which replicating DNA may differ from nonreplicating DNA: precatenanes, (+) supercoiled DNA, and the four-way junction at stalled forks. Finally, we will discuss topological domain barriers, which can sequester replicating DNA structures into limited regions of the chromosome where they can be processed more readily, allowing replication and chromosome segregation to proceed.
Conformations of Nonreplicating DNA
Several studies have shown that nonreplicating DNA with a (−) ΔLk has a characteristic branched, plectonemically supercoiled conformation both in vitro and in vivo. The initial results obtained with purified plasmid DNA have since been shown to apply well to plasmids in vivo and, ultimately, to the entire bacterial chromosome.
Conformations of Purified Plasmid DNA.
The (−) ΔLk of DNA in bacteria causes it to supercoil. This free (−) supercoiling is due to DNA gyrase, a type-2 topoisomerase unique in its ability to introduce (−) supercoils into relaxed or (+) supercoiled DNA. Although the enormous size of the Escherichia coli chromosome has limited studies on it, we now have a detailed picture of the conformational properties of the smaller, circular plasmid DNAs that are coresident with it in the cell. Electron microscopy (EM) allowed the direct visualization of the tightly intertwined, branched structure that is characteristic of (−) supercoiled molecules (1, 2). Beyond a visual image, EM also provided initial quantitative measurements of (−) supercoiled DNA structure (3). Furthermore, initial EM observations of the E. coli chromosome show very similar structures to those seen with plasmid DNA (ref. 4; C.D.H. and N.R.C., unpublished data).
A limitation of EM, however, is the distortion introduced by fixation on the grid and the poor control of the ionic conditions at the time of fixation. To understand better the conformation of (−) supercoiled DNA in solution, a study of the effect of ionic conditions on DNA conformation was undertaken (5, 6). By using sedimentation analysis and measures of the equilibrium formation of catenanes (interlinked circles) between (−) supercoiled circles and cyclizing linear DNAs, the effect of ionic conditions on the global and local conformation of supercoiled DNA, respectively, was measured. Importantly, the experimental results were also compared with those predicted from computer simulations and were found to be in excellent agreement.
All methods indicate that (−) supercoiled DNA has a compact, branched, plectonemic conformation over a range of σ, ionic conditions, and DNA length (7). As |σ| increases, a number of parameters remain constant: the extent of branching of the superhelix, the ratio of ΔWr to ΔTw of about 3, and the ratio of the length of the superhelix axis to DNA length of about 0.4. In contrast, the superhelix diameter decreases rapidly as σ increases. At physiological σ, around −0.06, the superhelix is tightly wound with a diameter of only about 100 Å, a feature key to the properties of (−) supercoiled DNA in the cell (reviewed in ref. 7). Ionic conditions strongly affect both the conformational and thermodynamic properties of (−) supercoiled DNA. Comparison of experimental and theoretical work has allowed the refinement of computer simulations such that we can now predict confidently the effects of mono-, di-, and trivalent ions, σ, and DNA length on supercoiled DNA conformations.
Plasmid DNA Conformations in Vivo.
Plasmids have proven invaluable not only for defining the structure of (−) supercoiled DNA in vitro, but also for comparing the structure of the same DNA outside and inside the cell. Site-specific recombination enzymes provided an assay that could be used in both environments. A number of these recombinases, including phage λ integrase (Int), the Gin invertase of phage Mu, and the resolvase of the Tn3 and γδ transposons, were shown to require a (−) supercoiled substrate in vitro, indicating that their in vivo substrates are similarly (−) supercoiled (8–11).
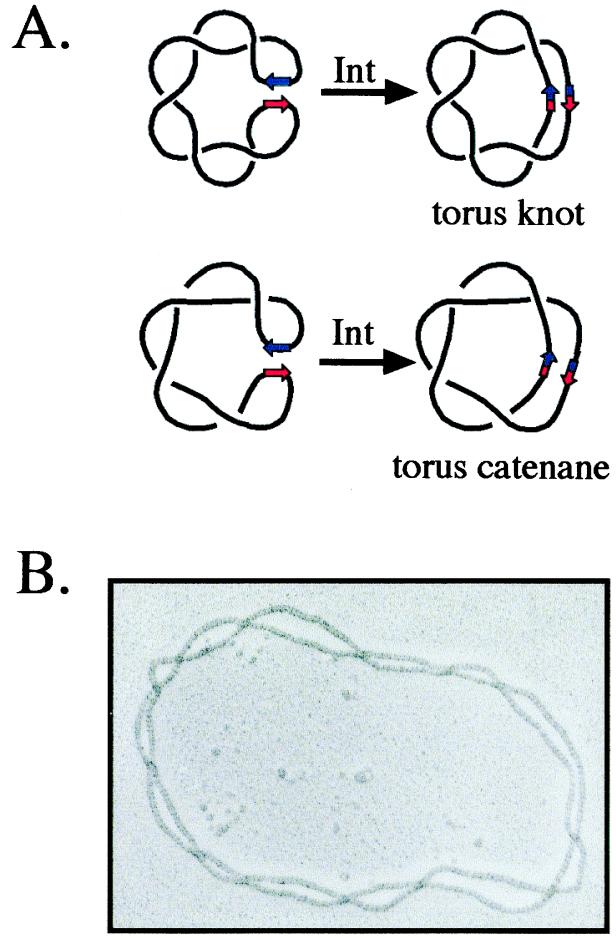
More direct evidence that plasmid DNA in the cell has the same plectonemic structure observed in vitro was first obtained with Int. Recombination between two Int binding sites (att) converts a labile geometric property of the DNA, supercoils, into a topological one, catenane or knot crossings (Fig. 1A; refs. 12 and 13). The in vitro products of Int recombination had been characterized by EM as right-handed knots and catenanes belonging to the torus family (ref. 14; Fig. 1B), reproducing the right-handed, plectonemic structure of (−) supercoiled DNA (15). When Int recombination was carried out in vivo, the products had the electrophoretic mobility characteristic of these torus catenanes and knots (16). Conversion of substrate plectonemes into knot and catenane nodes in vitro was similarly shown for Gin and resolvase by using a combination of EM and electrophoretic analyses (17–19).
Figure 1.
Conversion of plectonemic supercoils into knot and catenane nodes by Int site-specific recombination. (A) A (−) supercoiled Int substrate (black line) is shown with the att recombination sites represented by red and blue arrows. When the att sites are in inverse (head-to-head) orientation in the primary sequence, the recombination products are right-handed torus knots; these knots can be drawn without crossings on the surface of a torus- or doughnut-shaped object (Upper). When the sites are directly (head-to-tail) repeated, the product is a right-handed torus catenane (Lower). (B) An electronmicrograph of a 13-noded Int knot produced in vitro. The DNA was coated with RecA protein to help visualize the crossings. (Reprinted from Cell 276, Spergler, S. J., Stasiak, A. & Cozzarelli, N. R., “The stereostructure of knots and catenanes produced by phage lambda integrative recombination: 1985, Implications for mechanism and DNA structure,” 325–334, 1985, with permission from Elsevier Science; ref. 15.)
Int recombination also provided an estimate of the σ of plasmids in vivo. The number of crossings in the catenane products made by Int in vitro was shown to be linearly proportional to the σ of the substrate (16). This relationship was used to calibrate the effective σ, defined as the σ in vitro that has the same level of plectonemic supercoiling as that observed in vivo. The number of crossings in the catenanes made in cells in which the decatenating topisomerases were inhibited indicated that plasmids in vivo had about half of the number of plectonemic crossings obtained in vitro. The remaining ΔLk may be taken up by protein binding or by local changes in Tw. A similar fraction for the effective superhelical stress in vivo was obtained by comparing supercoiling-dependent transitions in vivo and in vitro, including cruciform extrusion, psoralen binding, and the transition from B to Z form DNA (20–22). Because the number of crossings and the superhelical stress both give a similar value for effective σ in vivo, a simple, but important conclusion can be drawn. Despite the enormous differences between the cellular milieu and the test tube, the basic properties of supercoiled DNA are the same in both. Thus, extrapolation from the detailed in vitro and in silico studies of supercoiling is justified.
Supercoiling of Chromosomal DNA.
Despite the 4.6-Mb size of the E. coli chromosome, several studies have indicated that it too has properties like those shown for plectonemically supercoiled DNA in vitro. Worcel and Burgi (23) found that the level of (−) supercoiling in isolated, intact chromosomes was similar to that observed for other closed circular DNAs as judged by the binding of the intercalating agent ethidium bromide (EtBr). As increasing amounts of EtBr bound to the chromosome, the sedimentation coefficient decreased because of relaxation of (−) supercoils and then increased as (+) supercoils formed. Supercoiling was also detected in the chromosome in vivo by measuring the number of trimethylpsoralen adducts bound to the chromosome upon photoactivation of the psoralen, a process with a linear dependence on (−) superhelicity (21).
As a complement to these physical methods, supercoiling-dependent gene expression showed functionally that the chromosome was globally (−) supercoiled in vivo (24). The lacZ gene expressed from a supercoiling-sensitive promoter was inserted at different chromosomal locations. Its expression varied only a few fold at different insertion sites, indicating that the degree of supercoiling was relatively constant throughout the chromosome. A study using site-specific recombination further demonstrated the plectonemic structure of the bacterial chromosome. Higgins et al. (25) inserted pairs of res sites, the sequence acted on by the resolvase recombinase, in a nonessential quadrant of the Salmonella chromosome. In vitro studies demonstrated that for recombination to occur, the two res sites must be (−) plectonemically interwound. Resolution was observed over the entire 6- to 100-kb range of res site separation tested, although the efficiency of recombination decreased with distance.
The Physiological Consequences of Conformations of (−) Supercoiled DNA in Chromosome Partitioning.
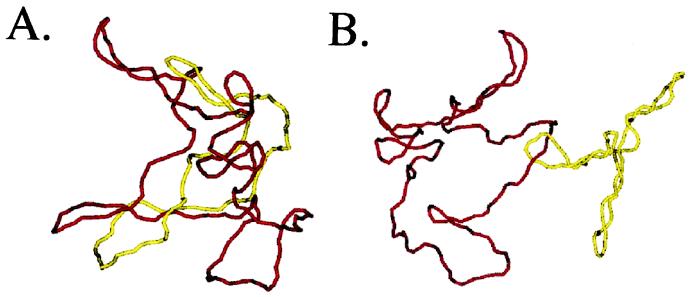
A critical role of (−) supercoiling is the lowering of the free energy necessary for denaturation of DNA. Equally essential for bacterial cell survival is the compaction of the chromosome by supercoiling in promoting partitioning to daughter cells. The first indication of this role for plectonemic supercoiling came from a combination of computer simulations and in vitro experiments (26). These studies demonstrated that the probability that two supercoiled plasmids will be catenated at thermodynamic equilibrium decreases exponentially with increasing |σ|. For 3.5-kb plasmids, physiological levels of supercoiling promoted decatenation by three orders of magnitude. Supercoiling promotes disentanglement because it is much harder to thread DNAs through each other when they are tightly plectonemically supercoiled than when they are relaxed (Fig. 2).
Figure 2.
Monte Carlo simulations of catenanes between relaxed and supercoiled DNA molecules. Simulation of a singly linked catenane between two relaxed plasmids (A) or between a relaxed and supercoiled plasmid (B). The yellow chain represents a 3.5-kb DNA, and the red chain a 7-kb DNA. Simulations courtesy of Alexander Vologodskii.
The promotion of decatenation and disentanglement by supercoiling is not an in vitro curiosity. It plays an essential role in partitioning in vivo of both plasmids and the chromosome. Topo IV, and not gyrase, carries out decatenation in vivo (27, 28). Yet mutants in DNA gyrase have a classic chromosome partition defect (29). The interpretation is that (−) supercoiling by gyrase is necessary for the compaction needed for efficient decatenation. Direct evidence for this is that the rate of decatenation by topo IV in vivo is sharply reduced when supercoiling is reduced (30). Indeed, compaction by gyrase is more important than its promotion of fork movement by removing a (+) ΔLk because only gyrase can supercoil DNA, whereas topo IV can also support replication elongation (31). Finally, there is recent evidence regarding MukB, a protein involved in chromosome condensation and partitioning, concerning the relationship between supercoiling and partitioning. Sawitzke and Austin (32) showed that the severe partitioning defects of a mukB mutant are suppressed by a topA mutation, which decreases the activity of topo I and results thereby in increased (−) supercoiling. One model for what might be happening is that MukB condenses and organizes the DNA, making it clearer to decatenating enzymes which crossings should be removed for chromosome segregation to occur. This role of MukB apparently can be replaced by the role of (−) supercoiling, which is known to shift the equilibrium position in favor of decatenation. It appears that a major role of MukB and supercoiling is in condensing the chromosome and promoting decatenation (33).
Conformations of Replicating DNA
Whereas the DNA in the bulk of the bacterial chromosome is (−) supercoiled, the conformation around the replication fork is much more complicated. The parental strands of DNA can be topologically constrained, whereas the daughter strands cannot (Fig. 3A), because the free ends of the latter allow swiveling of the DNA at the branch point. Even if these ends were somehow constrained by the replication apparatus to form a topological domain barrier at the junction, the single-stranded regions between Okazaki fragments would prevent the lagging strand of the DNA behind the replication fork from being stably supercoiled.
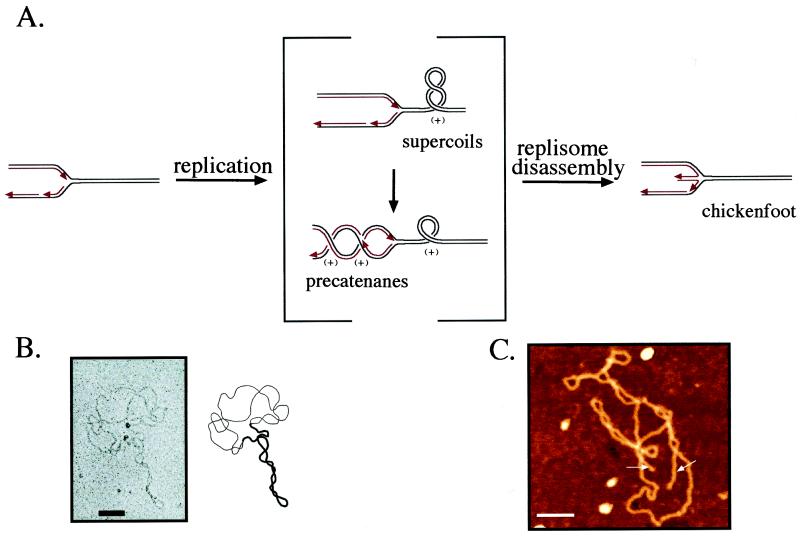
Figure 3.
Conformations of replicating DNA. (A) A replication fork is depicted at Left with the parental strands in black and the daughter strands in red. Red arrows denote 3′ ends. During replication, the denaturation of the parental duplex causes a (+) ΔLk in the replicating molecule (Center). This (+) ΔLk can be expressed either as (+) supercoiling of the parental duplex in front of the replication fork or (+) precatenanes between the replicated duplexes behind the fork. We have shown the (+) ΔLk in the usual fashion, which assumes that it is initially ahead of the fork and must diffuse past the replisome to generate precatenanes. It is, however, possible that the converse is true and that (+) precatenanes are the primary consequence of the (+) ΔLk from replication. Upon replisome dissociation (Right), the ends of the nascent strands will be free to base pair with each other, forming a four-way junction at the replication fork, the chickenfoot, that allows the replication fork to regress until the molecule is relaxed. (B) Electron micrograph of an in vitro replication intermediate with replication stalled by the Tus/ter complex. The molecule displays both supercoils in the unreplicated region (thick line) and precatenanes in the replicated region (thin line). (Reprinted from ref. 47, with permission from Elsevier Science.) (C) Scanning force microscopy of an in vivo replication intermediate incubated in ethidium bromide. This molecule displays the linear duplexes of the middle toe of the chickenfoot (white arrows) emerging from both the unidirectional origin of replication and the terminus. (Reprinted from ref. 60, with permission from the American Society for Biochemistry and Molecular Biology.) (For B and C, the scale bar is 100 nm.)
Replication generates a large (+) ΔLk from rapid fork movement coupled with denaturation of the parental strands. Therefore, the DNA in the vicinity of the fork is expected to have a (+) ΔLk rather than a (−) ΔLk. The mechanical strain imposed by this ΔLk can be relieved not only by (+) supercoiling of the unreplicated DNA, but also by intertwinings of the daughter duplexes behind the fork called precatenanes (Fig. 3 A and B). There is even a third possible conformation uniquely available to replicating DNA with a (+) ΔLk. In this structure, a regression of the fork and intertwining of the newly synthesized strands form a four-way junction that we call the chickenfoot (Fig. 3 A and C). We will next describe the conformations resulting from a ΔLk in replicating DNA and discuss how they may be processed inside the cell.
Often the terms ΔLk and supercoiling are used interchangeably, because a ΔLk in unreplicated DNA results in supercoiling. Because a ΔLk can result in two additional conformations of replicating DNA besides supercoiling, we will restrict the term “supercoiling” to the standard plectonemic conformations that we have just described in regions of DNA not containing a junction. Supercoiling will be present in the unreplicated arm of the junction and when DNA with a non-zero ΔLk is enclosed by topological constraints.
Precatenanes.
The relief of superhelical strain by topoisomerases becomes problematic as the replication fork nears the end of a domain. As the length of the unreplicated DNA becomes shorter, there is less room to contain the (+) supercoils resulting from fork movement, and less room for the topoisomerases to act to remove them. Furthermore, as the replicated region of DNA becomes larger, it titrates away the pool of active topoisomerases. The cell seems to have painted itself into a corner.
Champoux and Been (34) suggested a way out. They argued that mechanical stress caused by a (+) ΔLk need not be confined to the region in front of the fork. If the replication fork is free to rotate, the (+) superhelical stress in front of the fork can diffuse to create an interwinding of the daughter duplexes behind the fork (Fig. 3 A and B). The interwindings of the daughter duplexes were further defined and later termed “precatenanes” (35), because they will become catenane links if they are not removed by topoisomerases before the completion of replication. Precatenanes allow a solution to the topological problems of replicating DNA to the end of a domain by allowing links created by the movement of the replication fork to be removed behind it. Even though precatenanes and supercoils are alternative consequences of a ΔLk, there are striking differences between them. First, a free precatenated region will not be strained and thus precatenation, unlike supercoiling, will not affect helix unwinding. Second, although (+) supercoils can be removed by DNA gyrase in bacteria and by type-1B topoisomerases in eukaryotes, as well as by the typical type-2 topoisomerases throughout nature, only the latter can remove (+) precatenanes efficiently. Third, whereas supercoiling folds a DNA back on itself and provides a compaction that promotes partitioning (29), precatenation winds the daughter chromosomes around each other and thereby opposes partitioning. After discussing the convincing evidence that precatenanes are formed in purified replication intermediates, we will discuss the less direct, but largely persuasive evidence for their existence in vivo.
The initial tests of the Champoux and Been proposal were very discouraging. Visualization of replicating DNA by EM showed supercoils in front of the replication forks, but no precatenanes (e.g., refs. 36 and 37). As a result, textbook descriptions of replication intermediates (38) left out any role for precatenanes in DNA replication.
The first indication that precatenanes might be important substrates for topoisomerase-mediated unlinking came from the work of Hiasa and Marians (39, 40). Using an in vitro system consisting entirely of purified components, they reconstituted the complete replication of plasmids from oriC, the chromosomal origin of replication in E. coli. Two experiments suggested that unlinking took place both in front of and behind the replication fork. First, they found that topo III was sufficient to support complete replication and segregation of oriC plasmids (39). Topo III cannot relax (+) supercoils because it is a type-1A topoisomerase (41). This result suggested that topo III acts behind the fork by removing (+) precatenanes at nicks or gaps caused by incomplete synthesis or ligation of Okazaki fragments.
The second experiment examined the activities of gyrase and topo IV. Gyrase efficiently removes (+) supercoils by converting them to (−) supercoils (42), but is a very poor decatenase. Topo IV was known then only to excel at decatenation, whereas it relaxed (−) supercoils poorly (43, 44). From the different salt optima of decatenation and relaxation, the authors sought to identify which activity of the enzymes was required to relieve the (+) ΔLk at different stages of plasmid replication. They concluded that gyrase is sufficient to support the early stages of replication, whereas topo IV becomes increasingly important in the terminal stages of replication, presumably by removing precatenanes (40).
Later work showed additional activities of topo III and topo IV (45, 46), which complicated the interpretation of these experiments, and the physiological relevance of these purified enzyme reactions was unclear. A direct proof of precatenanes was needed. It was provided by an investigation of the conformations of replication intermediates stalled at a unique position by the presence of a Tus protein on a ter site (47). These intermediates provided a uniform population of molecules that were readily analyzed. EM of plasmids partially replicated in vivo and in vitro revealed both precatenanes behind the fork and supercoils in front of it (Fig. 3B). Every molecule showed precatenane and supercoil crossings in a ratio that was in good agreement with energetic predictions. Moreover, the electrophoretic mobility of late replication intermediates had the steps-of-two spacing expected for precatenanes. The failure of previous EM studies to find precatenanes was demonstrated to be due to an artifact of the earlier spreading method used.
Sogo et al. were the first to demonstrate precatenanes in vivo (48). They used plasmids containing two opposed ColE1 origins that caused replication to stall. Random strand passages by topoisomerases in the replicated region of these partially replicated molecules created a series of knots whose topology indicated that the stalled plasmids had a precatenane region in vivo.
A caveat concerning these studies is that the precatenanes identified were (−) and not (+), the true substrates for unlinking during replication. This is because the systems used to produce the stalled replication intermediates contained sufficient DNA gyrase to (−) supercoil the intermediates before or after stalling. A related limitation is that the stalling of these forks may have disassembled the replication complex in preparation for restart, and this disassembly could be a prerequisite for the formation of the precatenanes.
Although they have never been directly visualized, the existence of (+) precatenanes in actively replicating DNAs is supported by the fact that (+) catenanes are produced as an intermediate in plasmid DNA replication in vivo when topo IV is inhibited (27), and these replication catenanes can have more than 30 crossings (49, 50). The most likely model is that replication catenanes arise from precatenanes in replication intermediates, and thus precatenanes can be a substrate for unlinking during replication. The alternative possibility is that catenane links arise from the denaturation of the terminal region before its replication. However, the denaturation of such a large region has not been observed, and it seems very unlikely that it would be a prerequisite for replication termination.
Positive Supercoils.
Early work on supercoiled DNA found that it was exclusively (−) in all organisms examined, even when the supercoiling was caused by winding around proteins, as in nucleosomes (51). There is, however, a growing appreciation for the importance of (+) supercoiling. Key sources of (+) supercoiling are tracking processes in which DNA is not free to rotate about its axis. This will generate “twin domains” of (−) and (+) supercoils (52). As a result, an important function of topoisomerases is to relax (+) as well as (−) supercoils.
Positive supercoils were known a priori to be important in DNA replication, and, in retrospect, it is odd that they have been underappreciated. Replication generates a (+) ΔLk. In eukarya and archaea the major role of topoisomerases must be to remove (+) supercoils produced by replication as these organisms have no DNA gyrase to reduce a (+) ΔLk by (−) supercoiling. Moreover, supercoils must contribute to the expression of a (+) ΔLk in eukaryotes, because type-1B topoisomerases, which readily relax (+) supercoils but not precatenanes, play an important role in replication fork elongation in these organisms (53).
In bacteria, the presence of DNA gyrase introduces another possibility. Gyrase could act so fast that even the DNA around the fork remains (−) supercoiled. Indeed, until recently, it was believed that DNA gyrase was the only enzyme that removed the (+) ΔLk generated during replication. We favor the view, however, that DNA gyrase is not able to act so quickly. Were this to be the case, topo IV could not promote replication elongation, and its sole contribution would be in decatenating daughter chromosomes before partitioning. However, if no (+) ΔLk built up because gyrase worked so quickly, then the catenanes that accumulate in the absence of topo IV would be (−). Instead, these catenanes are exclusively (+) (27). In addition, recent results from our laboratory provided direct evidence that topo IV can promote replication fork progression in addition to its role in decatenation (31). Topo IV can only contribute to fork progression if the ΔLk around the fork is (+). Removal of a (−) ΔLk by topo IV would, in fact, oppose replication fork progression by increasing Lk.
The first evidence for a role of topo IV during replication elongation in vivo came from the finding that mutational inactivation of DNA gyrase caused DNA replication to stop only slowly (31, 54–57). If DNA gyrase was solely responsible for chain growth, a fast stop is expected. This was just the result when topo IV was inhibited in addition, indicating that topo IV can support replication elongation in the absence of gyrase (50). A direct measure of fork movement by using DNA microarrays showed that topo IV supports replication elongation in vivo at one-third the normal rate in the absence of gyrase (31). These results are consistent with the fact that topo IV can support complete plasmid replication in vitro in the absence of gyrase (58).
Because topo IV can replace gyrase, it too must be able to remove the (+) ΔLk generated by replication in vivo. The (+) ΔLk can take the form of (+) supercoils or (+) precatenanes. The role of topo IV in decatenation in vivo and in vitro has been well documented (27, 28, 40, 43, 44). Therefore, it would be able to remove (+) precatenanes. It has recently been shown that topo IV can also efficiently remove (+) supercoils and thereby act in front of the replication fork.
The proof of the ability of topo IV to efficiently remove (+) supercoils in vivo used the twin domain effect, in which transcription generates (+) and (−) supercoiled regions (52, 59). It had originally been thought that inhibition of gyrase was sufficient to cause a rapidly transcribed plasmid to become (+) supercoiled. Khodursky et al. showed that, instead, topo IV relaxed (+) supercoils so well in vivo that the transcribed plasmid became relaxed in the absence of gyrase (31). For the plasmid to become (+) supercoiled, it was necessary and sufficient also to inhibit topo IV.
These studies in vivo initially seemed at odds with early biochemical results that had shown that topo IV is very poor at relaxing (−) supercoils (43). The assumption was that (+) and (−) supercoil removal would be roughly equivalent. Instead, recent experiments showed that topo IV preferentially relaxes (+) supercoils (45), and, thus, the conflict is resolved. The decisive experiments analyzed the ability of topo IV to relax single DNA molecules. By rotating a magnetic bead attached to one end of a constrained piece of DNA, (−) and (+) supercoils could alternately be introduced into the same molecule. Positive supercoils were relaxed by topo IV at a 20-fold higher rate than (−) supercoils. In addition, topo IV was highly processive in relaxing (+) supercoils, but almost completely distributive with (−) supercoiled DNA.
The rate of (+) supercoil relaxation by topo IV measured in these experiments resolved a paradox concerning the action of topoisomerases in DNA replication. The rate of the replication fork in vivo requires the removal of 100 links/sec, or 3,000 strand passages/min, by topoisomerases at each fork. Because topo IV can support elongation at one-third the normal rate in the absence of gyrase, 1,000 strand passages/min would be needed at each fork (31). Yet, the much lower bulk rate of (−) supercoil relaxation by topo IV (43) implied that thousands of topoisomerase molecules would be needed to support replication elongation. Our single-molecule experiments indicated that a single topo IV could carry out 360 strand passages/min at 37°C to relax a (+) ΔLk. Therefore, only a few topo IV molecules would be sufficient at each fork. The single-molecule experiments may more accurately reflect the in vivo situation because they measure only the rate of active enzyme, whereas the rates from conventional methods are averages of active, inactive, unbound, and paused molecules. The preferential activity of topo IV on (+) supercoils has a very nice physiological consequence. Topo IV is efficient at removing (+) supercoils at replication forks, but will leave alone the important (−) supercoils elsewhere. Thus, the cell can have its cake and eat it too.
The Chickenfoot.
As noted, all of the stalled fork molecules studied had a (−) ΔLk and, therefore, (−) supercoils and precatenanes (47, 48). The true intermediate of replication has to have a (+) ΔLk. It was important to determine the structure of these (+) ΔLk molecules. We found that they have neither precatenanes nor supercoils, but a four-way junction, a finding with important physiological implications.
We introduced a (+) ΔLk into purified, protein-free replication intermediates by adding an intercalator. The (+) ΔLk results from a change in Tw imposed by the intercalator, which reduces Lk0. When a plasmid with a (−) ΔLk but no replication fork is exposed to the intercalator chloroquine, it becomes (+) supercoiled. Purified plasmids replicated in vivo in E. coli cells with replication forks stalled by the Tus/ter complex reacted quite differently to intercalators. They did not become (+) supercoiled or (+) precatenated. Instead, they comigrated upon electrophoresis with relaxed replication intermediates (60). The reason for the relaxation is that the replication fork regresses (Fig. 3A Right). The re-base pairing of the parental strands allows the complementary daughter strands to base pair. This process results in a four-way junction at the replication fork, referred to as a chickenfoot or a reversed fork (61, 62). The re-pairing of the parental strands replaces the (+) Wr of potential precatenanes or supercoils with increased Tw applied by the newly formed parental duplex. The resultant molecule has the same Lk, because no strands have been broken, but is completely relaxed.
The existence of this structure has been demonstrated by restriction enzyme amputation of the “middle toe” daughter–daughter duplex (60). The middle toe was cleaved off at unique restriction sites near both the replication terminus and the unidirectional origin of replication. Thus, chickenfeet can form at either junction, despite the differences in their properties. Whereas the replication fork is likely to have an incomplete Okazaki fragment, the junction at the origin will likely contain an RNA primer. The chickenfoot was directly imaged by scanning force microscopy, which showed chickenfeet at either one or both of the three-way junctions bounding the replicated region (Fig. 3C).
The chickenfoot forms readily in (+) ΔLk replication intermediates because it is energetically favorable as compared with both (+) supercoils and precatenanes. It has no torsional strain. All base pairs that are removed by fork reversal are reformed in the daughter–daughter duplex. The only thermodynamic penalty is from the formation of the fourth arm of the junction. However, this should not be energetically so different from a three-way junction and provides some enhanced entropic stabilization. The thermodynamic penalty for chickenfoot formation is so low that the structure prevails over supercoils and precatenanes in a replication intermediate with a ΔLk as low as (+) 1. Chickenfeet have even been observed with relaxed replication forks (60, 61), but a (−) ΔLk prevents chickenfoot formation (B.J.P. and N.R.C., unpublished data).
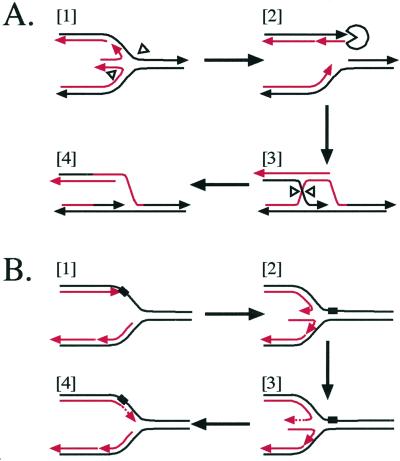
The chickenfoot has significance in living cells. There is a growing body of evidence suggesting that a four-way junction is a natural consequence of a stalled replication fork in vivo (63–65). The chickenfoot may be an intermediate in recombination-mediated replication restart (ref. 64; Fig. 4A), whose importance has been fully recognized only recently (66). In this model, Holliday junction resolving enzymes cleave the four-way junction, severing one of the replicated arms (Fig. 4A, [1]). This severed arm can then invade its sister duplex with the help of recombination enzymes (Fig. 4A, [2] and [3]), allowing a reassembly of the replication fork (Fig. 4A, [4]). The four-way junction may be further involved in allowing bypass of a lesion on the leading strand template (refs. 65 and 67; Fig. 4B). If the parental strand is an inappropriate template (Fig. 4B, [1]), the sister nascent strand can take its place, allowing replication on the leading strand to continue past the lesion (Fig. 4B, [2] and [3]). Reabsorption of the middle toe will allow the re-formation of the replication fork (Fig. 4B, [4]).
Figure 4.
Physiological implications of the chickenfoot. Replication forks are shown with parental strands in black and daughter strands in red, and with the 3′ end tipped with an arrowhead. (A) Recombination-mediated replication restart. The four-way junction of the chickenfoot can be cleaved by Holliday junction resolving enzymes such as RuvC (white arrowheads) [1]. Cleavage will sever one of the replicated arms, which can then be processed by the RecBCD complex (Pac-man) [2], allowing it to become a substrate for homologous recombination [3]. Recombination with the sister replicated arm re-forms the replication fork, allowing replication restart [4]. (B) Bypass of a lesion. An unpaired lesion (black rectangle) on the parental strand blocks leading strand replication, but lagging strand replication can continue for more than 500 nt [1]. Upon replisome dissociation, the two nascent strands can base pair, requiring the denaturation of more than 500 bp at the lagging strand [2]. Once a chickenfoot is formed, the lagging strand becomes a template for the leading strand, allowing leading strand replication to continue past the lesion [3]. Reabsorption of the chickenfoot re-forms the replication fork, allowing replication to continue and a second opportunity to repair the lesion [4].
Although the chickenfoot structure is thermodynamically favorable in a (+) ΔLk replication intermediate, there are potential kinetic impediments to its formation. Lagging stand replication may continue for over 500 nucleotides past a lesion on the leading strand template that stops replication (68). Were a chickenfoot to form, a long stretch of double-stranded DNA involving the lagging strand would need to be denatured before the two nascent strands could base pair (see Fig. 4A, [1]). Although this should not affect the thermodynamics of chickenfoot formation, it would create a large kinetic obstacle. Under these conditions in vivo, therefore, additional factors may be needed to promote chickenfoot formation. Recent evidence has suggested that both RecG (65) and RecA (63) catalyze chickenfoot formation in vivo. The roles of these proteins may be to aid in denaturation or strand exchange during the formation of the chickenfoot. Once the chickenfoot is formed, branch migration promoting enzymes, such as RuvAB, may aid in further extrusion of the middle toe (64).
The chickenfoot, however, must not form at actively replicating forks. Such an event would place the nascent strands in the wrong place for replication to continue. The fact that chickenfeet do not occur during replication suggests that the replisome must provide a large kinetic barrier to their formation. Likely, it does this by holding the nascent strand ends against their parental templates to prevent them from base-pairing with each other.
An actively replicating domain should have a (+) ΔLk. When a replication fork stalls, the replisome may dissociate (69). If the nascent strands can be freed before topoisomerases can relieve the (+) superhelical strain, then the strong thermodynamic drive toward the chickenfoot would probably ensure its formation.
Domains
The bacterial chromosome is constantly replicating during rapid exponential growth. There will always be at least one pair of replication forks working its way through the chromosome. If a ΔLk could travel freely throughout the chromosome, these forks would have serious consequences for the cell. Because the daughter strands are not topologically constrained, they could not be supercoiled (see Fig. 3A). The bacterial σ, however, is controlled in a very narrow range, and deviations in either direction are lethal (70). In addition, bacterial origins of replication require (−) supercoiling to fire (38). Relaxation of the entire replicated region would prevent refiring before the first round of replication had been completed, which is not the case at maximum growth rates. Free migration of topology throughout the chromosome would create additional problems for replication itself. Precatenane links distributed throughout a large region of the chromosome would be very difficult to find by the decatenating enzymes. Sequestered, supercoiled precatenane links would be much easier to remove, as described above. In addition, any (−) links ahead of the replication fork could travel behind, creating (−) precatenane links. Removal of these links would increase the (+) ΔLk of replication intermediates, and thus act as an antiswivel, impeding progress of the replication fork. Finally, even in the absence of a replication fork, a single nick or double-strand break on the chromosome should wreak havoc on the carefully balanced topology of the DNA.
Fortunately, none of these dire consequences appears to be the case. There is evidence that the actively replicating part of the chromosome and any other insult to the DNA backbone are confined into topologically isolated domains, protecting the bulk of the chromosome from the hazards of relaxation and precatenation.
Topological domains are defined as regions of DNA that are topologically constrained at their ends, and therefore behave independently of the rest of the chromosome. They allow the unreplicated DNA to remain (−) supercoiled in the presence of (+) precatenanes and (+) supercoils in the replicating region and vice versa. Also, domains would prevent the relaxation resulting from DNA nicks from spreading through the entire bacterial chromosome. The evidence supporting the existence of domains and how they relate to the structure of the bacterial nucleoid has been discussed (71, 72). However, the exact nature of domain barriers, and the relationship between domains and higher order chromosome structure and replication forks, remain elusive. Domain barriers may result from protein, RNA, or attachment of the chromosome to the plasma membrane due to cotranscriptional translation of membrane proteins. Stable domain barriers would present a further problem during replication and must be displaced for the replication apparatus to pass through. Alternatively, barriers may be mobile and dissociate from a site before the fork reaches it. Whatever the relationship between domain barriers and the moving fork, domains prevent the relaxation and (+) supercoiling of replication from hindering most cell functions. Experiments suggest that the E. coli chromosome is composed of roughly 50–150 topologically closed loops (4, 25, 73, 74). Assuming that these are evenly spaced throughout the chromosome, each loop would contain about 50–100 kb of DNA. If a replication fork moves at about 1 kb/s in E. coli (75), the replicating domain should be relaxed or (+) supercoiled for about 1–2 min in vivo. That is probably not enough time to cause too many transcriptional or other problems. In addition, domains may provide structure for chromosomal DNA, allowing very long DNA molecules to be organized and replicated in the cell.
Eukaryotic cells have also been shown to possess topological domains (76–78), but the nature of the boundaries is as elusive as in bacteria. Nonetheless, it seems clear that the control of topology by domains evolved very early.
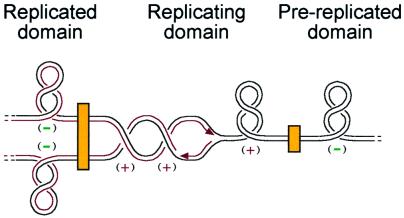
A model for how domain barriers organize the (+) ΔLk from replication is illustrated in Fig. 5. The boundaries may seal off manageable units of DNA and concentrate the type-2 topoisomerases at and behind the fork. This simplifies the enzymatic reactions that promote DNA replication and chromosome unlinking, and allows the cell to manage the topological problems posed by replicating DNA.
Figure 5.
Model for topology of the replicating chromosome. A segment of chromosomal DNA is depicted, with black lines as parental strands and red lines as nascent strands. In the bacterial chromosome, domain barriers (yellow boxes) isolate the topology around the fork from the rest of the chromosome, which is (−) supercoiled by DNA gyrase, as shown in the domains on either side of the replication domain. Replication creates a (+) ΔLk in the replicating domain (Center), which can cause (+) supercoils ahead of the fork and (+) precatenanes behind it. Thus, either gyrase or topo IV could support replication by removing (+) supercoils in front of the fork, and topo IV could also support replication by removing precatenanes behind the replication fork.
Acknowledgments
We thank Pat Higgins, Ken Kreuzer, Ken Marians, and Alex Vologodskii for critical comments on the manuscript. Work from this laboratory was supported by grants from the National Institutes of Health. C.D.H. is supported by a fellowship from the Howard Hughes Medical Institute.
Abbreviations
- Lk
linking number
- Tw
twist
- Wr
writhe
- EM
electron microscopy
Footnotes
This paper results from the National Academy of Sciences colloquium, “Links Between Recombination and Replication: Vital Roles of Recombination,” held November 10–12, 2000, in Irvine, CA.
References
- 1.Vinograd J, Lebowitz J, Radloff R, Watson R, Laipis P. Proc Natl Acad Sci USA. 1965;53:1104–1111. doi: 10.1073/pnas.53.5.1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Upholt W B, Gray H B, Jr, Vinograd J. J Mol Biol. 1971;62:21–38. doi: 10.1016/0022-2836(71)90128-8. [DOI] [PubMed] [Google Scholar]
- 3.Boles T C, White J H, Cozzarelli N R. J Mol Biol. 1990;213:931–951. doi: 10.1016/S0022-2836(05)80272-4. [DOI] [PubMed] [Google Scholar]
- 4.Kavenoff R, Ryder O A. Chromosoma. 1976;55:13–25. doi: 10.1007/BF00288323. [DOI] [PubMed] [Google Scholar]
- 5.Rybenkov V V, Vologodskii A V, Cozzarelli N R. J Mol Biol. 1997;267:299–311. doi: 10.1006/jmbi.1996.0876. [DOI] [PubMed] [Google Scholar]
- 6.Rybenkov V V, Ullsperger C U, Vologodskii A V, Cozzarelli N R. Science. 1997;277:690–693. doi: 10.1126/science.277.5326.690. [DOI] [PubMed] [Google Scholar]
- 7.Vologodskii A V, Cozzarelli N R. Annu Rev Biophys Biomol Struct. 1994;23:609–643. doi: 10.1146/annurev.bb.23.060194.003141. [DOI] [PubMed] [Google Scholar]
- 8.Mizuuchi K, Gellert M, Nash H A. J Mol Biol. 1978;121:375–392. doi: 10.1016/0022-2836(78)90370-4. [DOI] [PubMed] [Google Scholar]
- 9.Mertens G, Hoffman A, Blocker H, Frank R, Kahmann R. EMBO J. 1984;3:2415–2421. doi: 10.1002/j.1460-2075.1984.tb02148.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Plasterk R H, Kanaar R, van de Putte P. Proc Natl Acad Sci, USA. 1984;81:2689–2692. doi: 10.1073/pnas.81.9.2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reed R R. Cell. 1981;25:713–719. doi: 10.1016/0092-8674(81)90178-1. [DOI] [PubMed] [Google Scholar]
- 12.Mizuuchi K, Gellert M, Weisberg R A, Nash H A. J Mol Biol. 1980;141:485–494. doi: 10.1016/0022-2836(80)90256-9. [DOI] [PubMed] [Google Scholar]
- 13.Pollock T J, Nash H A. J Mol Biol. 1983;170:1–18. doi: 10.1016/s0022-2836(83)80224-1. [DOI] [PubMed] [Google Scholar]
- 14.Vologodskii A V. Topology and Physics of Circular DNA. Boca Raton, FL: CRC Press; 1992. [Google Scholar]
- 15.Spengler S J, Stasiak A, Cozzarelli N R. Cell. 1985;42:325–334. doi: 10.1016/s0092-8674(85)80128-8. [DOI] [PubMed] [Google Scholar]
- 16.Bliska J B, Cozzarelli N R. J Mol Biol. 1987;194:205–218. doi: 10.1016/0022-2836(87)90369-x. [DOI] [PubMed] [Google Scholar]
- 17.Kanaar R, Klippel A, Shekhtman E, Dungan J M, Kahmann R, Cozzarelli N R. Cell. 1990;62:353–366. doi: 10.1016/0092-8674(90)90372-l. [DOI] [PubMed] [Google Scholar]
- 18.Krasnow M A, Stasiak A, Spengler S J, Dean F, Koller T, Cozzarelli N R. Nature (London) 1983;304:559–560. doi: 10.1038/304559a0. [DOI] [PubMed] [Google Scholar]
- 19.Wasserman S A, Dungan J M, Cozzarelli N R. Science. 1985;229:171–174. doi: 10.1126/science.2990045. [DOI] [PubMed] [Google Scholar]
- 20.Sinden R R, Broyles S S, Pettijohn D E. Proc Natl Acad Sci USA. 1983;80:1797–1801. doi: 10.1073/pnas.80.7.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sinden R R, Carlson J O, Pettijohn D E. Cell. 1980;21:773–783. doi: 10.1016/0092-8674(80)90440-7. [DOI] [PubMed] [Google Scholar]
- 22.Haniford D B, Pulleyblank D E. J Biomol Struct Dyn. 1983;1:593–609. doi: 10.1080/07391102.1983.10507467. [DOI] [PubMed] [Google Scholar]
- 23.Worcel A, Burgi E. J Mol Biol. 1972;71:127–147. doi: 10.1016/0022-2836(72)90342-7. [DOI] [PubMed] [Google Scholar]
- 24.Miller W G, Simons R W. Mol Microbiol. 1993;10:675–684. doi: 10.1111/j.1365-2958.1993.tb00939.x. [DOI] [PubMed] [Google Scholar]
- 25.Higgins N P, Yang X, Fu Q, Roth J R. J Bacteriol. 1996;178:2825–2835. doi: 10.1128/jb.178.10.2825-2835.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rybenkov V V, Vologodskii A V, Cozzarelli N R. J Mol Biol. 1997;267:312–323. doi: 10.1006/jmbi.1996.0877. [DOI] [PubMed] [Google Scholar]
- 27.Adams D E, Shekhtman E M, Zechiedrich E L, Schmid M B, Cozzarelli N R. Cell. 1992;71:277–288. doi: 10.1016/0092-8674(92)90356-h. [DOI] [PubMed] [Google Scholar]
- 28.Zechiedrich E L, Cozzarelli N R. Genes Dev. 1995;9:2859–2869. doi: 10.1101/gad.9.22.2859. [DOI] [PubMed] [Google Scholar]
- 29.Kato J-I, Nishimura Y, Suzuki H. Mol Gen Genet. 1989;217:178–181. doi: 10.1007/BF00330959. [DOI] [PubMed] [Google Scholar]
- 30.Zechiedrich E L, Khodursky A B, Cozzarelli N R. Genes Dev. 1997;11:2580–2592. doi: 10.1101/gad.11.19.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khodursky A B, Peter B J, Schmid M B, DeRisi J, Botstein D, Brown P O, Cozzarelli N R. Proc Natl Acad Sci USA. 2000;97:9419–9424. doi: 10.1073/pnas.97.17.9419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sawitzke J, Austin S. Proc Natl Acad Sci USA. 1999;97:1671–1676. doi: 10.1073/pnas.030528397. . (First Published January 31, 2000; 10.1073/pnas.030528397) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Holmes V F, Cozzarelli N R. Proc Natl Acad Sci USA. 2000;97:1322–1324. doi: 10.1073/pnas.040576797. . (First Published February 11, 2000; 10.1073/pnas.040576797) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Champoux J J, Been M D. In: Mechanistic Studies of DNA Replication and Genetic Recombination: ICN–UCLA Symposia on Molecular and Cellular Biology. Alberts B, editor. Vol. 19. New York: Academic; 1980. pp. 809–815. [Google Scholar]
- 35.Ullsperger C J, Vologodskii A V, Cozzarelli N R. In: Nucleic Acids and Molecular Biology. Lilley D M J, Eckstein F, editors. Vol. 9. Berlin: Springer-Verlag; 1995. pp. 115–142. [Google Scholar]
- 36.Sebring E D, Kelly T J, Thoren M M, Salzman N P. J Virol. 1971;8:478–490. doi: 10.1128/jvi.8.4.478-490.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fuke M, Inselburg J. Proc Natl Acad Sci USA. 1972;69:89–92. doi: 10.1073/pnas.69.1.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kornberg A, Baker T A. DNA Replication. New York: Freeman; 1992. [Google Scholar]
- 39.Hiasa H, Marians K J. J Biol Chem. 1994;269:32655–32659. [PubMed] [Google Scholar]
- 40.Hiasa H, Marians K J. J Biol Chem. 1996;271:21529–21535. doi: 10.1074/jbc.271.35.21529. [DOI] [PubMed] [Google Scholar]
- 41.Wang J C. Annu Rev Biochem. 1996;65:635–692. doi: 10.1146/annurev.bi.65.070196.003223. [DOI] [PubMed] [Google Scholar]
- 42.Brown P O, Cozzarelli N R. Science. 1979;206:1081–1083. doi: 10.1126/science.227059. [DOI] [PubMed] [Google Scholar]
- 43.Ullsperger C, Cozzarelli N R. J Biol Chem. 1996;271:31549–31555. doi: 10.1074/jbc.271.49.31549. [DOI] [PubMed] [Google Scholar]
- 44.Peng H, Marians K J. J Biol Chem. 1993;268:24481–24490. [PubMed] [Google Scholar]
- 45.Crisona N J, Strick T R, Bensimon D, Croquette V, Cozzarelli N R. Genes Dev. 2000;14:2881–2892. doi: 10.1101/gad.838900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Harmon F G, DiGate R J, Kowalczykowski S C. Mol Cell. 1999;3:611–620. doi: 10.1016/s1097-2765(00)80354-8. [DOI] [PubMed] [Google Scholar]
- 47.Peter B J, Ullsperger C, Hiasa H, Marians K J, Cozzarelli N R. Cell. 1998;94:819–827. doi: 10.1016/s0092-8674(00)81740-7. [DOI] [PubMed] [Google Scholar]
- 48.Sogo J M, Stasiak A, Martinez-Robles M L, Krimer D B, Hernandez P, Schvartzman J B. J Mol Biol. 1999;286:637–643. doi: 10.1006/jmbi.1998.2510. [DOI] [PubMed] [Google Scholar]
- 49.Sundin O, Varshavsky A. Cell. 1981;25:659–669. doi: 10.1016/0092-8674(81)90173-2. [DOI] [PubMed] [Google Scholar]
- 50.Khodursky A B, Zechiedrich E L, Cozzarelli N R. Proc Natl Acad Sci USA. 1995;92:11801–11805. doi: 10.1073/pnas.92.25.11801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bauer W R. Annu Rev Biophys Bioeng. 1978;7:287–313. doi: 10.1146/annurev.bb.07.060178.001443. [DOI] [PubMed] [Google Scholar]
- 52.Liu L F, Wang J C. Proc Natl Acad Sci USA. 1987;84:7024–7027. doi: 10.1073/pnas.84.20.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim R A, Wang J C. J Mol Biol. 1989;208:257–267. doi: 10.1016/0022-2836(89)90387-2. [DOI] [PubMed] [Google Scholar]
- 54.Drlica K. Microbiol Rev. 1984;48:273–289. doi: 10.1128/mr.48.4.273-289.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Orr E, Fairweather N F, Holland I B, Pritchard R H. Mol Gen Genet. 1979;177:103–112. doi: 10.1007/BF00267259. [DOI] [PubMed] [Google Scholar]
- 56.Filutowicz M. Mol Gen Genet. 1980;177:301–309. doi: 10.1007/BF00267443. [DOI] [PubMed] [Google Scholar]
- 57.Mirkin S, Shmerling Z. Mol Gen Genet. 1982;188:91–95. doi: 10.1007/BF00332999. [DOI] [PubMed] [Google Scholar]
- 58.Hiasa H, Marians K J. J Biol Chem. 1994;269:16371–16375. [PubMed] [Google Scholar]
- 59.Wu H-Y, Shyy S, Wang J C, Liu L F. Cell. 1988;53:433–440. doi: 10.1016/0092-8674(88)90163-8. [DOI] [PubMed] [Google Scholar]
- 60.Postow L, Ullsperger C, Keller R W, Bustamante C, Vologodskii A V, Cozzarelli N R. J Biol Chem. 2001;276:2790–2796. doi: 10.1074/jbc.M006736200. [DOI] [PubMed] [Google Scholar]
- 61.Viguera E, Hernandez P, Krimer D B, Lurz R, Schvartzman J B. Nucleic Acids Res. 2000;28:498–503. doi: 10.1093/nar/28.2.498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Michel B. Trends Biochem Sci. 2000;25:173–178. doi: 10.1016/s0968-0004(00)01560-7. [DOI] [PubMed] [Google Scholar]
- 63.Seigneur M, Ehrlich S D, Michel B. Mol Microbiol. 2000;38:565–574. doi: 10.1046/j.1365-2958.2000.02152.x. [DOI] [PubMed] [Google Scholar]
- 64.Seigneur M, Bidnenko V, Ehrlich S D, Michel B. Cell. 1998;95:419–430. doi: 10.1016/s0092-8674(00)81772-9. [DOI] [PubMed] [Google Scholar]
- 65.McGlynn P, Lloyd R G. Cell. 2000;101:35–45. doi: 10.1016/S0092-8674(00)80621-2. [DOI] [PubMed] [Google Scholar]
- 66.Cox M, Goodman M, Kreuzer K, Sherratt D, Sandler S, Marians K. Nature (London) 2000;404:37–41. doi: 10.1038/35003501. [DOI] [PubMed] [Google Scholar]
- 67.Higgins N P, Kato K, Strauss B. J Mol Biol. 1976;101:417–425. doi: 10.1016/0022-2836(76)90156-x. [DOI] [PubMed] [Google Scholar]
- 68.Cordeiro-Stone M, Makhov A M, Zaritskaya L S, Griffith J D. J Mol Biol. 1999;289:1207–1218. doi: 10.1006/jmbi.1999.2847. [DOI] [PubMed] [Google Scholar]
- 69.Kuzminov A. Recombinational Repair of DNA Damage. Georgetown, TX: R. G. Landes; 1996. [Google Scholar]
- 70.DiNardo S, Voelkel K A, Sternglanz R, Reynolds A E, Wright A. Cell. 1982;31:43–51. doi: 10.1016/0092-8674(82)90403-2. [DOI] [PubMed] [Google Scholar]
- 71.Drlica K, Rouviere-Yaniv J. Microbiol Rev. 1987;51:301–319. doi: 10.1128/mr.51.3.301-319.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pettijohn D E. In: Escherichia coli and Salmonella: Cellular and Molecular Biology. Neidhardt F C, editor. Vol. 1. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 158–166. [Google Scholar]
- 73.Lydersen B K, Pettijohn D E. Chromosoma. 1977;62:199–215. doi: 10.1007/BF00286044. [DOI] [PubMed] [Google Scholar]
- 74.Sinden R R, Pettijohn D E. Proc Natl Acad Sci USA. 1981;78:224–228. doi: 10.1073/pnas.78.1.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Helmstetter C. In: Escherichia coli and Salmonella: Cellular and Molecular Biology. Neidhardt F C, editor. Vol. 2. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 1627–1639. [Google Scholar]
- 76.Kramer P R, Sinden R R. Biochemistry. 1997;36:3151–3158. doi: 10.1021/bi962396q. [DOI] [PubMed] [Google Scholar]
- 77.Benyajati C, Worcel A. Cell. 1976;9:393–407. doi: 10.1016/0092-8674(76)90084-2. [DOI] [PubMed] [Google Scholar]
- 78.Cockerill P N, Garrard W T. Cell. 1986;44:273–282. doi: 10.1016/0092-8674(86)90761-0. [DOI] [PubMed] [Google Scholar]