Abstract
The Epstein-Barr virus (EBV) is a ubiquitous B-lymphotropic herpesvirus associated with several malignant tumors, e.g., Burkitt's lymphoma and Hodgkin's disease, and is able to efficiently immortalize primary B lymphocytes in vitro. The growth program of infected B cells is initiated and maintained by the viral transcription factor EBV nuclear antigen 2 (EBNA2), which regulates viral and cellular genes, including the proto-oncogene c-myc. In our study, patterns of protein expression in B cells with and without EBNA2 were analyzed by two-dimensional polyacrylamide gel electrophoresis and mass spectrometry. For this purpose, we used a conditional immortalization system for EBV, a B cell line (EREB2-5) that expresses an estrogen receptor-EBNA2 fusion protein. In order to discriminate downstream targets of c-Myc from c-Myc-independent EBNA2 targets, we used an EREB2-5-derived cell line, P493-6, in which c-Myc is expressed under the control of a tetracycline-regulated promoter. Of 20 identified EBNA2 target proteins, 11 were c-Myc dependent and therefore most probably associated with proliferation, and one of these proteins was a posttranslationally modified protein, i.e., hypusinylated eIF5a. Finally, to estimate the relevance of EBNA2 targets during early EBV infection, we analyzed the proteomes of primary B cells before and after infection with EBV. The protein expression pattern induced upon EBV infection was similar to that following EBNA2 activation. These findings underscore the value of EREB2-5 cells as an appropriate model system for the analysis of early events in the process of EBV-mediated B-cell immortalization.
Epstein-Barr virus (EBV) is a ubiquitous B-lymphotropic herpesvirus that after a usually asymptomatic infection is carried by more than 90% of adults for the rest of their lives. When the primary infection is postponed to late childhood or adolescence, EBV can cause infectious mononucleosis. The virus has been linked to the development of several malignant tumors, including Burkitt's lymphoma, Hodgkin's disease, lymphoproliferative disease in immunosuppressed individuals, certain forms of T-cell lymphoma, and some lymphoepithelial tumors, such as nasopharyngeal carcinoma and a proportion of gastric cancers (reviewed in reference 4). Infection of primary B cells with EBV in vitro causes the activation of the cells and drives them into continuous proliferation, a process termed immortalization. Cell lines established by infection of primary human B cells with EBV are called lymphoblastoid cell lines (LCLs).
The first viral genes expressed in EBV-infected B cells are EBNA2 and EBNA-LP, two nuclear antigens that act as transcription activators in concert with each other and regulate the expression of viral as well as cellular genes involved in the initiation and maintenance of cell proliferation. EBNA2 in conjunction with EBNA-LP upregulates the expression of the viral membrane latent genes LMP1 and LMP2 and regulates the BamHI-C promoter driving the expression of all nuclear antigens (reviewed in references 10 and 27). EBNA2 also targets cellular genes such as c-myc (21), CD21, CD23 (14), and EBI1/BRL2 (11).
This EBV-induced growth program is also initiated during primary infection with EBV in vivo (3). In individuals with an intact immune system, proliferation of these highly immunogenic cells is controlled by cytotoxic T cells specific for a panel of EBV antigens (reviewed in reference 26). It represents, however, a life-threatening danger in immunosuppressed individuals, e.g., AIDS patients (reviewed in reference 40) or transplant recipients (posttransplantation lymphoproliferative disorders [reviewed in reference 41]).
In healthy individuals, EBV escapes from the immune system by persisting in resting memory B cells. In these cells, only a small subset of viral genes is expressed, and this subset includes LMP2A and maybe also EBNA1. EBNA1 evades immune recognition by expressing a glycine-alanine repeat which was shown to inhibit the presentation of major histocompatibility complex class I restricted cytotoxic T-cell epitopes in cis by abolishing antigen processing via the ubiquitin-proteasome pathway (31). Additionally, downregulation of the EBNA genes is achieved by methylation of the Cp promoter (47).
EBNA2-driven cell proliferation is believed to be required for the multiplication of virus-infected cells in the host in vivo and the efficient establishment of viral latency in a high number of B cells (3). It is so far unknown how the EBNA2-induced proliferation program is switched off in vivo. This step may involve either a physiological B-cell differentiation signal that shuts EBNA2 off or a selection mechanism for cells that have lost EBNA2 expression in a stochastic fashion.
While the EBNA2-dependent expression pattern of EBV genes is well understood, the pattern of cellular target genes remains poorly defined (reviewed in references 10 and 27). The most prominent cellular target gene is the proto-oncogene c-myc, which is induced to moderate levels by EBNA2 (21) as well as by LMP1 (U. Dirmeier, unpublished data). Overexpression of c-Myc to a much higher level than is observed in EBV-immortalized cells is sufficient to support B-cell proliferation independently of EBNA2 and LMP1 (45). The proliferation program imposed by enforced overexpression of c-Myc, however, differs substantially from that activated in EBV-immortalized cells. EBV-immortalized cells grow in large clumps, express a variety of activation markers and adhesion molecules, are highly resistant to apoptosis, and are characterized by pronounced immunogenicity of the cells, whereas c-Myc-driven cells grow as single cells in suspension, do not express the cell surface molecules expressed on EBV-immortalized cells, are highly sensitive to apoptosis, and are nonimmunogenic. The differences in the growth programs of cells driven into proliferation by EBV and c-Myc imply that there must be substantial differences in the target genes of EBNA2 and c-Myc. EBNA2 targets most likely include additional genes that are independent of c-Myc and are not involved in the regulation of proliferation but, rather, regulate EBV-induced B-cell activation, protection against apoptosis, immunogenicity, and cell-cell communication.
Most work aiming to identify differentially expressed genes in EBV-immortalized cells and Burkitt's lymphoma cells including EBNA2 target genes has been done by subtractive cDNA cloning (7, 11). Kaiser et al. (21, 22) again used subtractive cDNA cloning to identify genes induced by EBNA2 by making use of a cell line conditionally immortalized by EBV (EREB2-5) (23, 24). RNA-based methods, although superior from the point of view of feasibility, suffer from a number of disadvantages. The effector function of a gene depends not only on the level of mRNA present in a cell but also on posttranscriptional events like splicing, nuclear export, translation efficiency, and protein stability. Most importantly, RNA-based assays fail to detect posttranslational modifications which often account for changes in the activity and localization of mature proteins, such as phosphorylation, glycosylation, acetylation, methylation, and other more rare but functionally important modifications, such as, e.g., protein amidation, hypusinylation, and farnesylation. Indeed, proteome analysis of Saccharomyces cerevisiae (20) and mammalian cells (32) showed for many proteins only a poor correlation between the level of mRNA and the amount of the corresponding protein or even revealed the opposite relationship. Additionally, Gavioli and coworkers (17) showed that Burkitt's lymphoma cells proliferating under the control of c-Myc display impaired ubiquitin-proteasome-dependent proteolysis compared to that of LCLs proliferating under the control of EBNA2 and LMP1. As expression of some genes appears to be regulated mainly at the level of proteasomal degradation (e.g., HIF-1α, p27, and IκB-α), the comparison of mRNA expression patterns has obvious limitations.
We have used two-dimensional polyacrylamide gel electrophoresis (2D PAGE) and mass spectrometry to compare the protein expression pattern of cells newly infected with EBV with that of a conditionally immortalized cell line (EREB2-5) in which an estrogen receptor-EBNA2 fusion protein (23) whose activity can be switched on and off by the addition and withdrawal of estrogen is expressed. Making use of a derivative cell line of EREB2-5 (P493-6) in which EBNA2 is regulated by estrogen and c-myc expression is regulated by tetracycline (44, 50), we have been able to classify these proteins in c-Myc-dependent and -independent targets.
We show here that the protein expression pattern induced by EBNA2 activation is indeed very similar to that observed upon infection of primary B cells with EBV. Our findings reinforce the notion that the EREB2-5 cell line is a useful tool with which to recapitulate the early events of an EBV infection.
MATERIALS AND METHODS
Cell culture.
EREB2-5 and P493-6 cells were cultured as described previously (23, 44). EREB2-5 cells were depleted of estrogen 60 h before induction, and P493-6 cells were kept in medium containing 0.1 μg of tetracycline/ml 60 h before induction. A total of 107 cells were resuspended in 4 ml of cysteine- and methionine-free RPMI medium (Gibco) containing 10% dialyzed fetal calf serum (Gibco) and were stimulated either by 2 μM β-estradiol (Sigma) or by depletion of tetracycline for the time indicated. Two hours before harvest, 0.125 mCi of Trans35S (ICN Biomedicals)/ml (>1,000Ci/mmol) was added.
B-cell preparation and virus infection.
Primary human B lymphocytes were isolated from routine adenoidectomies, and single-cell suspensions were generated. T cells were depleted by rosetting with whole sheep blood (Oxoid, Wesel, Germany). After centrifugation on a Ficoll cushion at 560 × g, the remaining erythrocytes were lysed in 10 mM KHCO3-155 mM NH4Cl-0.1 mM EDTA (pH 7.4). For virus infection, B lymphocytes (2 × 107 cells) were incubated with cell-free supernatant of B95-8 cells (38). Virus supernatant was generated by pelleting 1 × 107 B95-8 cells at 350 × g. Concentrated virus supernatant was obtained by ultracentrifugation of cell-free B95-8 supernatant for 2 h at 30,000 × g.
2D PAGE.
The 2D PAGE procedure followed the protocol of Görg et al. (19) with the modifications introduced by Bjellqvist and coworkers (8). Cells were rinsed in ice-cold phosphate-buffered saline (PBS) and once in half-concentrated PBS and were then lysed at room temperature in 400 μl of lysis buffer (10 M urea [Amersham], 1% 1,4-dithioerythritol [DTE; Merck], 4% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate [CHAPS; Sigma], 2.5 mM EDTA, 2.5 mM EGTA). The lysate was homogenized by using QIAshredder spin columns (QIAGEN) and centrifuged at 50,000 rpm (Ti75-rotor, 20°C) for 50 min. Isoelectric focusing (IEF) was performed with 370 μl of lysate containing 0.025% bromophenol blue and 2 μl of immobilized pH gradient buffer (pH 3 to 10; Amersham) in an IPGphor (Amersham) by using IEF strips (pH 3 to 10, nonlinear; Amersham) with the following voltage profile: 11 h at 30 V, a 10-min increase to 200 V, 30 min at 200 V, a 30-min increase to 500 V, 1 h at 500V, a 2.2-h increase to 2,000 V, 1 h at 2,000 V, a 2.5-h increase to 8,000 V, 8,000 V until 90 kVh are reached in total.
For protein identification, a lysate of 8 × 107 cells was applied to a laboratory-made IEF strip (pH 4.5 to 6.5) in a Multiphor as described previously (36).
Prior to the second dimension, strips were incubated for 15 min at room temperature in equilibration buffer (6 M urea, 2% sodium dodecyl sulfate, 50 mM Tris [pH 6.8], 29% glycerol), first with 2% (=130 mM) DTE (reductive) and second with 2.5% (=135 mM) iodoacetamide (alkylating).
Equilibrated strips were inserted onto sodium dodecyl sulfate-12% PAGE gels (20 by 22 by 0.75 cm) and sealed with 0.5% agarose, and 2D gels were run at 15°C (Bio-Rad Protean IIxi cell) for 7 h at approximately 20 mA per gel. Analytical gels were silver stained as described (9) and preserved by air drying between sheets of moistened cellophane, while preparative gels were Coomassie blue stained and not dried. Labeled gels were exposed to X-ray films, and films were analyzed with PDQuest software (Bio-Rad).
Two-dimensional gel quantification and analysis.
Two-dimensional gels with 35S-labeled proteins were exposed to X-ray films (Kodak X-OMAT AR). The films or silver-stained gels were scanned, and digital images were analyzed for changes in protein patterns with the image analysis software PDQuest (Bio-Rad). After background subtraction, spot intensities were measured as parts per million of volume, corresponding to pixel intensities integrated over the area of each spot and divided by the sum over all valid spots in the gel. Average relative values, standard deviations, and P values (Mann-Whitney U test and unpaired t test) of the normalized intensities for all spots at each time point were then calculated.
MALDI mass spectrometry.
Matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry (Reflex III; Bruker Daltonics GmbH, Bremen, Germany) was used to obtain peptide mass information for each unknown sample. Protein spots were cut from fixed Coomassie blue-stained (12% acetic acid, 0.4% Coomassie blue, 50% methanol) 2D gels; washed with water, 50% acetonitrile, and 50 mM NH4HCO3 (the last two steps were repeated once); and digested with 50 ng of porcine trypsin (Promega) in 50 mM NH4HCO3 for 5 h. Supernatants from tryptic digests were lyophilized and desalted by evaporation of the NH4HCO3 in the vacuum several times. Dry peptides were resolved in 20% acetonitrile-0.1% formic acid and mixed 1:1 with freshly prepared DHB matrix solution (18 mg of 2,5-dihydroxybenzoic acid [Sigma]/ml, 2 mg of 2-hydroxy-5-methoxybenzoic acid [Sigma]/ml, 20% acetonitrile, 0.1% formic acid) and spotted on anchor targets (Bruker). Spectra were analyzed, and peak lists were generated by using Xmass/XTOF software (Bruker). Proteins were identified by peptide mass fingerprinting with MSBiotools (MASCOT) by searching against all mammalian entries in the MSDB (the nonidentical protein sequence database of the Proteomics Department at the Hammersmith Campus of Imperial College London, July 2002). Theoretical pI and molecular mass information for each unknown protein was obtained from MSDB database entries.
PCR analysis.
The EBV status of each B-cell sample was assessed by PCR. Total DNA from freshly isolated B cells (6 × 106 cells) was isolated with lysis buffer (0.3 M sodium acetate in 8 M guanidine solution). DNA was amplified with primers specific for glycoprotein 85 of EBV (forward, 5′-TGTGGATGGGTTTCTTGGGC-3′; backward, 5′-TGGTCAGCAGCAATAGTGAACG-3′). PCR conditions consisted of 30 cycles of 94°C for 3 min, 60°C for 1 min, and 72°C for 1 min. Amplification products were separated in 1% agarose gel and visualized with ethidium bromide.
Immunofluorescence.
Cells were fixed with pure acetone (20 s) and incubated with EBNA2-specific antibodies (R3 and 1E6 hybridoma supernatants mixed 1:1; a kind gift of Elisabeth Kremmer, GSF) for 30 min at 37°C. After several washes with PBS, B cells were incubated with secondary goat anti-mouse immunoglobulin G (IgG) antibody conjugated with the Cy-3 fluorochrome (1:100 dilution; Dianova).
Immunoblot analysis.
Cells were lysed in PBS containing 1% Triton X-100 for 10 min, and the supernatant was used for immunoblotting analysis. Equal amounts of protein were resolved by PAGE, transferred on a polyvinylidene difluoride membrane (Immobilon-P, Millipore Corporation, Bedford), and probed with actin-specific (sc-1616; Santa Cruz), Nm23-H1-specific (sc-465; Santa Cruz), and EBNA2-specific antibodies (a kind gift of Elisabeth Kremmer, GSF) in combination with peroxidase-conjugated secondary antibodies and the ECL reagent (Amersham).
[3H]thymidine incorporation.
For the determination of DNA synthesis, 5 × 104 primary B lymphocytes were seeded in a volume of 100 μl/well of cell culture medium or supernatant of B95-8. Cells were kept at 37°C for up to 5 days. [3H]thymidine (0.5 μCi) was added for 4 h, and its incorporation was measured in harvested cells (57).
Real-time PCR analysis.
Total RNA was extracted from 6 × 106 B cells by using the Tripure isolation reagent (Roche) according to the manufacturer's protocol. cDNA synthesis was performed with 1 μg of isolated RNA and 200 U of Moloney murine leukemia virus reverse transcriptase (GIBCO/BRL)/μl. PCR was performed with a Lightcycler by rapid cycling in a reaction volume of 10 μl with 1 μmol of each primer per liter, 2 mM MgCl2, 10 ng of cDNA, and SYBR Green I (LC-FastStart DNA Master, catalog no. 2239264; Roche) according to the manufacturer's protocol. The following primer pairs were used: CD19, 5′ CTCCTTCTCCAACGCTGAGT 3′ and 5′ TGGAAGTGTCACTGGCATGT 3′; EBNA2, 5′ GGTTGCTGGAGAGGGCAAGG 3′ and 5′ GCCCAGAGGCTCCCATTCTC 3′; c-myc, 5′ TCCGTCCTCGGATTCTCTGC 3′ and 5′ CCAGTGGGCTGTGAGGAGGT 3′; Nm23, 5′ CCCATGGGAAGGAGGGGAAA 3′ and 5′ GCGGGGTCTTGTGGGAGAGA 3′. Cycling conditions were as follows: after an initial denaturation step at 95°C for 10 min, amplification was performed by using 45 cycles of denaturation (95°C, 1 s), annealing (65°C, 10 s), and extension (72°C, 20 s).
RESULTS
Identification of EBNA2 and c-Myc target proteins.
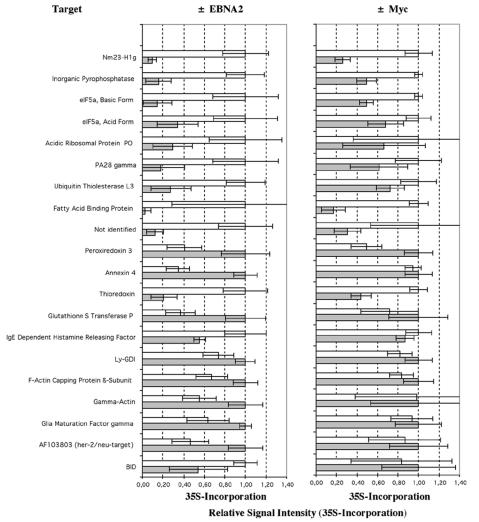
EBNA2 target proteins were analyzed by using the EREB2-5 cell line, in which the function of EBNA2 can be switched off and on by the withdrawal and readdition of estrogen. c-Myc target proteins were identified by using P493-6 cells, in which c-Myc expression is under the control of a tetracycline-responsive promoter. Overexpression of c-Myc by removal of tetracycline allows the cells to proliferate in the absence of estrogen, i.e., independently of EBNA2 and LMP1. We compared noninduced cells (estrogen-deprived EREB2-5 and tetracycline-supplemented P493-6 cells) with cells induced for 8 h by addition of estrogen (EREB2-5) or depletion of tetracycline (P493-6 cells), respectively. Thymidine incorporation assays performed before and 24 h after induction confirmed the appropriate cell cycle arrest and the reentry of the cells into the cell cycle (data not shown). A proteome analysis with [35S]methionine- and [35S]cysteine-labeled cells was performed before and 8 h after induction by using 2D PAGE as described in Materials and Methods. Gel spots from 35S autoradiographs or silver-stained gels were quantified by using PDQuest software and differentially regulated target proteins identified by MALDI-TOF mass spectrometry from tryptic digests in combination with peptide mass fingerprinting. Target proteins reproducibly regulated upon activation of EBNA2 are shown in autoradiographs of selected sections (pI 4 to 7.5) of 2D gels from 35S-labeled total cell lysates of EREB2-5 and P493-6 cells without or with estrogen (8 h) and with or without tetracycline (8 h), respectively (Fig. 1), and are described in Table 1.
FIG. 1.
Two-dimensional gel autoradiographs of 35S-labeled whole-cell lysates from EREB2-5 and P493-6 cells before (left panel) and 8 h after induction (right panel). Identified proteins are listed in Table 1. MW, molecular weight. Myc, c-Myc
TABLE 1.
EBNA2 target proteins obtained from analysis of 2D gel autoradiographs of 35S-labeled whole-cell lysatesa
| Spot | Function | Locus | Name (Mowse score) | Induction (fold)
|
|||
|---|---|---|---|---|---|---|---|
| 8 h, +EBNA2 (35S) | 8 h, +c-Myc (35S) | 3 days, EBV (35S) | 3 days, EBV (silver stain) | ||||
| 14 | DNA metabolism? | A33386 | Nm23-Hlg (150) | 10.53*** | 3.86** | 1.75** | 1.58* |
| 5 | Nucleotide metabolism | AAH01022 | PPase (210) | 6.40*** | 2.03** | 1.42 | 1.89* |
| 15.1 | mRNA transport | IF5A_HUMAN | elF5a, acid (nonhypusinylated) form (95) | 2.93*** | 1.47** | 1.06 | 0.81 |
| 15.2 | mRNA transport | IF5A_HUMAN | elF5a, basic (hypusinylated) form (95) | 6.83*** | 2.04** | 1.28 | 1.23 |
| 11 | Protein mabolism | R5HUPO | Acid ribosomal protein PO (175) | 3.39*** | 1.51 | 2.43 | 1.13 |
| 22 | Protein catabolism | A40085 | UCHL3 (137) | 3.64*** | 1.38* | 1.32* | 0.97 |
| 9 | Protein catabolism | AAH01423 | PA28γ (Ki autoantigen) (126) | 5.64*** | 1.63* | 1.67 | 1.95* |
| 27 | Lipid metabolism | 1B56 | Fatty acid binding protein (170) | 33.11** | 5.79** | 1.01 | 1.16 |
| 10 | Stress response | ANX4_HUMAN | Annexin IV (149) | 0.35*** | 0.94 | 0.54* | 0.76 |
| 20 | RedOx status | GTP_HUMAN | Glutathione S-transferase P (117) | 0.37*** | 0.72 | ND | ND |
| 21 | RedOx status | Q96HK4 | Peroxiredoxin 3 (122) | 0.41*** | 0.49** | 0.61* | 0.96 |
| 25 | RedOx status | THIO_HUMAN | Thioredoxin (132) | 4.83** | 2.27** | 0.56** | 0.75 |
| 1 | Immunology | S06590 | IgE-HRF (184) | 1.80*** | 1.15 | 0.81** | 1.05 |
| 4 | Cytoskeleton | AAA51580 | Gamma actin (105) | 0.55** | 0.98 | 1.34 | 0.79 |
| 6 | Cytoskeleton | CAPB_HUMAN | F-actin capping protein β-subunit (197) | 0.67*** | 0.84 | 0.83 | 0.84 |
| 19 | Cytoskeleton signaling | A47742 | Ly-GDI (142) | 0.74*** | 0.82* | 0.77** | 0.71 |
| 7 | Signaling? | Q9UKY7 | AF103803 (target mRNA of Her2/Neu) (92) | 0.47*** | 0.87 | 0.76 | 0.92 |
| Q96IP9 | Short AF103803 (98) | 0.47*** | 0.87 | 0.76 | 0.92 | ||
| 16 | Signaling? | GLMG_HUMAN | GMLFγ (96) | 0.63*** | 0.94 | 0.33** | 0.62** |
| 2 | Apoptosis | AAH09197 | Bid (103) | 1.85*** | 0.83 | ND | ND |
| 26 | Not identified | 8.27** | 3.24* | 1.43 | 0.71 | ||
Function and induction values were obtained from analysis of 2D gel autoradiographs of 35S-labeled whole-cell lysates (35S) or from silver-stained gels (silverstain). The induction level equals the value for the induced situation divided by that for the noninduced situation. Proteins were identified by peptide mass fingerprinting. The probability-based Mowse score is −10× log10(P), where P is the absolute probability that the observed match is a random event (defined by the Mowse algorithm). Scores greater than 69 are significant (P < 0.05). ND, not detectable. Spot 7 could be either long or short AF103803. ***, P ≤ 0.001; **, P ≤ 0.01; *, P ≤ 0.05 (Mann-Whitney U test and t test).
In EREB2-5 cells, 20 spots whose intensities increased or decreased reproducibly upon activation of EBNA2 were identified (Fig. 2). Of these spots, 12 were induced and 8 were repressed. Two spots (spots 15.1 and 15.2) represented the same protein (eIF5a) with different modifications (hypusinylated and nonhypusinylated forms [see reference 28]). Hypusinylation was confirmed by a shift of the protein from the position of the basic form (spot 15.2) to the position of the acidic form (spot 15.1) 3 days after the incubation of the cells with the hypusine synthase inhibitor diaminoheptane (Fig. 3). Of the proteins induced, two play roles in nucleotide metabolism (Nm23-H1 and inorganic pyrophosphatase [PPase]), four play roles in protein synthesis and degradation (eIF5a, acidic ribosomal protein P0, PA28γ, and ubiquitin thiolesterase L3 [UCHL3]), one plays a role in fatty acid metabolism (FABP5), one plays a role in redox metabolism (thioredoxin), one plays a role in regulation of apoptosis (Bid), and one plays a role in signal transduction (IgE-dependent histamine-releasing factor [IgE-HRF]). One protein could not be identified by mass spectrometry. Of those proteins that were downregulated, three are involved in the regulation of redox metabolism and apoptosis (peroxiredoxin 3, Annexin IV, and glutathione S-transferase P), three are involved in the organization of the cytoskeleton (F-actin capping protein beta subunit, gamma actin, and LY-GDI), and one is involved in signal transduction (glia maturation factor gamma). One has been reported to be a target gene of Her-2/Neu (AF103803).
FIG. 2.
EBNA2 target proteins. Induction values obtained from analysis of 2D gel autoradiographs of 35S-labeled whole-cell lysates (35S) from EREB2-5 and P493-6 cells. Gray bars represent relative levels of expression in uninduced cells (−EBNA2, −Myc), and white bars represent relative levels of expression in induced cells (+EBNA2, +Myc). The highest value is standardized to 1. Values and standard deviations were obtained from at least two independent experiments. Within one experiment, the lysates of three induced and three uninduced cell populations were analyzed.
FIG. 3.

eIF5a isoforms. Two-dimensional gel autoradiographs of 35S-labeled whole-cell lysates from EREB2-5 either with estrogen (+estr., top) or with estrogen and the hypusine synthase inhibitor diaminoheptane (+estr., +25 μM DAH, bottom). The arrows indicate eIF5a isoforms. The two basic forms disappear upon DAH incubation, while the acidic form is enriched, confirming that the acidic isoform is nonhypusinylated.
Most of the proteins strongly upregulated in EREB2-5 cells upon EBNA2 activation were also found to be significantly upregulated in P493-6 cells when c-Myc expression was switched on (Nm23-H1, PPase, eIF5a, UCHL3, PA28γ, fatty acid binding protein 5 [FABP5], and thioredoxin). Conversely, peroxiredoxin 3 and LY-GDI were downregulated under both conditions. This finding indicates that these proteins are indirect targets of EBNA2 and that their regulation is most likely associated with cell proliferation. The degree of regulation of all of these proteins was more pronounced in EREB2-5 cells than in P493-6 cells. This result is most likely due to the fact that EBNA2 and all of the EBNA2 target genes, including the c-myc gene, are more tightly regulated in the EREB2-5 cell system by estrogen than is c-Myc expression by tetracycline in P493-6 cells. This difference in the tightness of regulation is also evident from the thymidine incorporation assays, in which the difference between the uninduced and the induced states is consistently larger for EREB2-5 than for P493-6 cells (data not shown). All proteins identified are newly synthesized upon EBNA2 activation or c-Myc induction, as they were detected following 35S labeling.
Identification of EBNA2 target proteins that are not regulated by c-Myc in P493-6 cells.
Of the six proteins whose expression changed significantly in EREB2-5 but not in P493-6 cells, two were upregulated by EBNA2 (Bid and IgE-HRF) and four were downregulated (Annexin IV, gamma actin, GMFγ, and AF103803). Two of these proteins are involved in the regulation of apoptosis (Bid, Annexin IV), two are involved in signal transduction (IgE-HRF and GMFγ) and in cytoskeleton organization (gamma actin), and one has been identified as a target of the Her-2/Neu oncogene.
Infection of primary B cells with the EBV B95-8 strain.
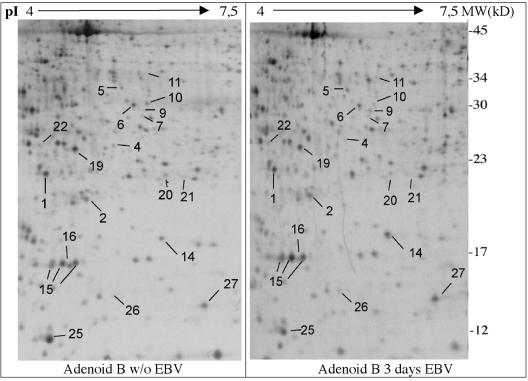
To evaluate whether estrogen-deprived EREB2-5 cells to which estrogen is readded reflect the early events following EBV infection of primary B cells, primary B cells were isolated from adenoids and infected with the EBV B95-8 strain in vitro. To exclude the possibility of a previous infection with EBV, DNA was isolated from a cell aliquot and a PCR analysis was performed with primers specific for the viral gp85 gene. All adenoids used in this study were EBV negative (data not shown). Infection of primary B lymphocytes was ascertained by monitoring EBNA2 expression by immunofluorescence staining (data not shown) and immunoblotting (Fig. 4) over a time period of 6 days. As expected, untreated B cells (control) yielded no EBNA2-specific fluorescence, whereas 3 days after infection, a substantial amount (55%) of the cells stained positive for EBNA2. At the same time point, EBNA2 was detected by immunoblotting (Fig. 4). To analyze the effect of EBV infection on DNA synthesis and proliferation, thymidine incorporation in primary B cells either kept untreated or infected with EBV over a time period of 5 days was assessed. Infection with EBV resulted in a continuous increase of thymidine incorporation (six- to eightfold at day 5 after infection) in comparison to noninfected primary B cells (data not shown). A substantial increase in thymidine incorporation was observed already at day 3 after infection. Based on these findings, the protein expression pattern of EBV-infected primary B cells was analyzed by using 2D PAGE 3 days postinfection (Fig. 5).
FIG. 4.
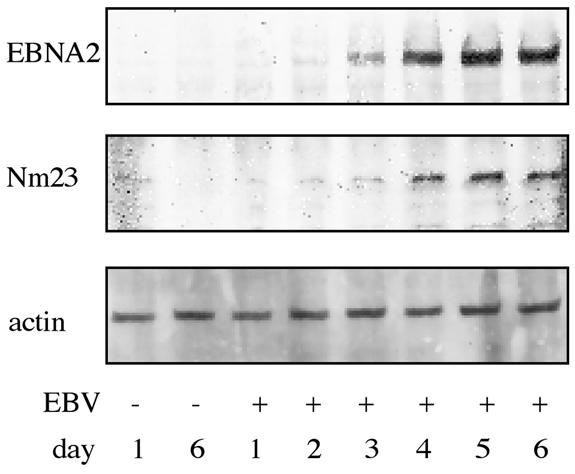
Increased Nm23-H1 levels after EBV infection. The kinetics of Nm23-H1 (Nm23) induction in primary B cells after EBV infection correlated with increased EBNA2 levels. B cells were exposed to B95-8 supernatant or left untreated (control days 1 and 6) for 6 days. The amounts of proteins were assessed every day after infection by immunoblotting with specific antibodies to Nm23-H1 (middle) and EBNA2 (top). Equal loading was confirmed by immunoblotting of the same membrane with an actin-specific antibody (bottom).
FIG. 5.
Two-dimensional gel autoradiographs of 35S-labeled whole-cell lysates from adenoid B cells before (left panel) and after infection (right panel) with EBV. Identified proteins are listed in Table 1. MW, molecular weight.
The pattern of EBV-induced proteins in primary B cells is similar to that in EREB2-5 cells upon activation of EBNA2.
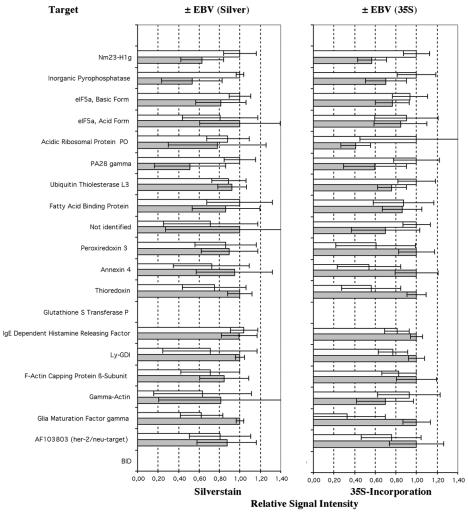
The proteomes of untreated and EBV-infected primary B cells (at day 3) were compared by 2D PAGE. Differentially expressed proteins were excised from the gels and analyzed by MALDI-TOF mass spectrometry following a trypsin in-gel digest. The majority of proteins identified in EBV-infected primary B cells correlated with targets identified in EREB2-5 cells. There were, however, also some differences. In primary B cells infected with EBV, proteins were induced to a lesser extent and the standard deviation was higher (Fig. 6 and Table 1). Nm23-H1, PPase, UCHL3, and PA28γ were also found to be upregulated in autoradiographs and/or silver stains, and peroxiredoxin 3, Annexin IV, Ly-GDI, and GMFγ were found to be downregulated under these conditions (Fig. 6, Table 1). However, thioredoxin and IgE-HRF displayed an inverse regulation, being downregulated in infected adenoid B cells and induced in EREB2-5 cells. FABP5 was strongly induced in EREB2-5 cells upon EBNA2 activation and did not exhibit any significant variation upon infection of B cells.
FIG. 6.
Induction values obtained from analysis of 2D gel analysis of 35S-labeled (35S; right panel) and silver-stained (silver stain; left panel) whole-cell lysates from adenoid B cells before and after infection with EBV. Gray bars represent induction values of noninfected cells (−EBV), and white bars represent induction values of infected cells (+EBV). The highest value is standardized to 1. Values and standard deviations were obtained from three different patient samples.
Sequential expression of EBNA2, c-Myc, and Nm23-H1 during early EBV infection.
One target, Nm23-H1, was analyzed in more detail with respect to its activation kinetics in response to EBNA2 following EBV infection of primary B cells. Immunoblot analysis revealed an increase in Nm23-H1 protein levels which correlated with the regulation of EBNA2 (Fig. 4). Additionally, the mRNA levels of EBNA2, c-Myc, and Nm23-H1 were assessed by real-time PCR. EBNA2 mRNA levels gradually increased during the first 3 days after infection. EBNA2 mRNA preceded the advent of c-myc mRNA, and c-myc mRNA again preceded Nm23-H1 mRNA (data not shown). The sequential appearance of c-myc and Nm23-H1 mRNA in estrogen-deprived EREB2-5 cells was also observed at early time points after the readdition of hormone. c-myc mRNA became detectable 1 h after the addition of estrogen and peaked 8 h after the addition of hormone, whereas the induction of Nm23-H1 RNA lagged slightly behind and reached its maximum at 12 h (Fig. 7). This finding is consistent with the notion that c-myc is a direct target gene of EBNA2 (21) and that Nm23-H1 is a target gene of c-Myc (49, 51).
FIG. 7.

Real-time reverse transcription-PCR kinetics for c-Myc (MYC) and Nm23-H1 (Nm23) mRNA in EREB2-5 cells after induction with estrogen. Levels of induction (fold) were normalized on CD19 mRNA expression. Maxima were adjusted to 100%. Nm23-H1 induction followed MYC induction with a 3-h delay, but its maximum was prolonged.
DISCUSSION
Only limited information about cellular targets of the viral transcription factor EBNA2 is available. To gain further insight into the cellular changes induced by EBNA2, we have compared the effect of EBNA2 activation in conditionally EBV-immortalized EREB2-5 cells with the early events following the infection of primary B cells with EBV. In contrast to the functional analysis of an oncogene in the background of an already-existing tumor cell line, the use of the EREB2-5 cell system ensures that EBNA2 is indeed studied in its physiological context, i.e., in resting B cells. Apart from identifying EBNA2 target proteins that support cell proliferation, we were also interested in the identification of cellular proteins or pathways that have been usurped by EBV to induce B-cell activation and intercellular adhesion. Since strong overexpression of one of EBNA2's direct target genes, c-myc, is sufficient to induce the proliferation of EREB2-5 cells in the absence of estrogen (45), we reasoned that c-Myc-independent EBNA2 targets are likely candidates to play a role in B-cell activation and intercellular adhesion.
We were particularly interested in posttranslational modifications that might play a role in the process of B-cell immortalization and therefore decided to use a proteome approach rather than RNA-based technologies such as gene arrays and standard agarose gel electrophoresis. In EREB2-5 cells, 20 proteins whose expression changed compared to that in hormone-deprived EREB2-5 cells upon addition of estrogen were identified. Twelve of these proteins were induced, and eight were repressed. One of the changes observed, i.e., the hypusinylation of eIF5a, was indeed a posttranslational modification. EIF5a is the only protein known to bear hypusine, an unusual amino acid synthesized from lysine by two successive enzymatic modifications involving the addition of spermidine to lysine. The biological role of eIF5a hypusinylation is only poorly understood. It is believed to be of primordial importance based on the findings that eIF5a and its specific modification are conserved throughout evolution from yeast to humans (reviewed in reference 12). EIF5a was proposed to be an essential export regulator of some specific mRNAs derived from the human immunodeficiency virus genome and the CD83 mRNA (6, 29). Hypusinylated eIF5a contributes essentially to the life cycle of human immunodeficiency virus by interacting with the REV protein and by mediating the export of unspliced viral mRNA to the cytoplasm (6). Since these findings cannot explain the highly conserved expression, the biological role of eIF5a hypusinylation in the physiology of the cell still remains elusive. In higher organisms, the enzymatic steps involved in the modification of lysine to hypusine appear to be at least in part under the control of c-Myc (5). ODC, the rate-limiting enzyme in spermidine synthesis, is a well known c-Myc target, and other enzymes in the polyamine pathway are also transcripitionally activated by c-Myc (49). This finding is consistent with the notion that the increase in hypusinylated eIF5a is not mediated by EBNA2 directly but is a consequence of c-Myc activation by EBNA2 (Fig. 2).
The most strongly induced proteins are involved in cell growth and metabolism. PPase) is expressed in a variety of tissues (15) and catalyses the hydrolysis of pyrophosphate (PPi) to orthophosphate. PPi is generated as a by-product of nucleoside triphosphate- and deoxynucleoside triphosphate-dependent biosynthetic reactions and is formed in large amounts during mRNA and DNA synthesis. Depletion of PPi by PPase allows these biosynthetic reactions to proceed. PPase was found to be upregulated in adenocarcinomas (13), most likely as a consequence of c-Myc overexpression (48) (Fig. 2).
Other strongly induced proteins are involved in protein synthesis (acidic ribosomal protein P0, which is part of the 60S subunit of eukaryotic ribosomes) or degradation (PA28γ and ubiquitin thiolesterase 3). PA28γ (also known as Ki-autoantigen) is a coregulator of the 20S proteasome. In contrast to the gamma interferon-inducible subunits of heteroheptamers forming PA28α/β, which are important for the formation of the “immunoproteasome,” PA28γ forms homoheptamers, which are predominantly localized in the nucleus and in microtubule-like extensions in the cytoplasm and support trypsin-like proteasome activity (25, 55). Experiments with K-ras temperature-sensitive mutant cell lines (42) and with PA28γ knockout mice (39) indicate that PA28γ expression might be correlated with the regulation of cell growth and cellular transformation.
Nm23-H1 is a nucleotide diphosphate kinase that has also been described to suppress tumor cell metastasis (30). It was recently shown to be a part of the SET complex and to mediate granzyme A-induced caspase-independent cell death characterized by single-stranded DNA nicks and other features of apoptosis (16). Since Nm23-H1 is highly upregulated at the protein level as well as at the mRNA level in cells driven into proliferation by c-Myc (49), Nm23-H1 expression seems to correlate with proliferation rather than with a suppressor function. Nm23-H1 was also reported to interact with EBNA3C, a viral downstream target of EBNA2. EBNA3C, by interacting with Nm23-H1, was reported to reverse Nm23-H1's ability to suppress the migration of Burkitt's lymphoma cells (52). In addition, EBNA3C overexpression leads to nuclear translocation of Nm23-H1. Interestingly, its homolog Nm23-H2 was reported to bind to the c-myc P1 promoter and to be necessary for efficient P1 and P2 transcription initiation in vitro (46).
Changes in the redox status of EREB2-5 cells were reflected by the regulation of thioredoxin, peroxiredoxin 3 and glutathione S-transferase P. Thioredoxin, a small molecule involved in a multitude of redox reactions, was strongly upregulated in EREB2-5 cells by estrogen. It is involved in the control of apoptosis and in the activation of factors involved in transcription and repair, like AP1, NF-κB, Ref-1, and p53 (reviewed in reference 1). In its secreted form, thioredoxin also potentiates the action of cytokines and acts as a growth factor for EBV- and human adult T-cell leukemia virus-immortalized cells (54).
Genes or gene products regulated in EREB2-5 cells by hormone may be either direct or indirect targets of EBNA2. Since c-Myc is a direct target gene of EBNA2 and an important mediator of proliferation, we were interested in classifying EBNA2 targets with respect to c-Myc. To this end, we made use of the P493-6 cell line, a cell line that is conditional for both EBNA2 (regulated by estrogen) and c-Myc (regulated by tetracycline) (44, 49, 50). Using 2D PAGE and MALDI-TOF analysis, we were able to show that all of the targets mentioned above are indeed c-Myc dependent. Notably, the induction rates upon switching c-Myc on were consistently lower than the induction rates in EREB2-5 cells after the activation of EBNA2. This finding is presumably due to the leakiness of the tetracycline regulation system. In fact, P493-6 cells maintain clearly detectable c-myc mRNA levels in the presence of tetracycline and may therefore be regarded as preactivated compared to EREB2-5 cells, in which EBNA2 activity (and thus also c-Myc) is tightly regulated by hormone.
Annexin IV, AF103803, and GMFγ (downregulated) and IgE-HRF and Bid (upregulated) were regulated by EBNA2 in a c-Myc-independent fashion. The membrane surface protein Annexin IV is known to be upregulated during the induction of apoptosis by either CD95/Fas (18) or other mechanisms. In the present study, Annexin IV was induced following estrogen starvation, reflecting the cellular stress imposed on these cells by hormone withdrawal and the spontaneous induction of apoptosis within 3 to 4 days. Bid is a protein that is present in the cell in an inactive native form. Upon proapoptotic stimulation, Bid is proteolytically cleaved and converted into its active form. Overexpression of Bid may render EREB2-5 cells susceptible to killing through CD95/Fas ligand and partially explain differences observed between LCLs and Burkitt's lymphoma cells with respect to sensitivity to apoptosis. AF103803 is a cDNA that has been described to be upregulated by the proto-oncogene Her2/Neu (43); however, its biological function is still unknown.
Glia maturation factor gamma (hGMFγ), a homolog of the glia maturation factor-β gene (2), was identified as a gene expressed in human CD34+ hematopoietic stem-progenitor cells (37). Northern blot analysis of rats indicated that the rat GMFγ mRNA is predominantly expressed in the thymus, the testis, and the spleen (53). The function of hGMFγ has not been analyzed so far. Its homolog GMFβ was found to be an intracellular regulator of signal transduction pathways involving p38 and NF-κB (33, 34). Recently, Zaheer and coworkers (56) observed that overexpression of GMFβ in astrocytes resulted in the secretion of granulocyte-macrophage colony-stimulating factor, which stimulated microglia to express major histocompatibility complex class II and interleukin-1β. Soluble recombinant GMFβ inhibits the proliferation of neoplastic cells in culture by arresting them in the G0/G1 phase (35). It may be speculated that hGMFγ might fulfill similar functions during B-cell immortalization by EBV. If so, downregulation of hGMFγ by EBNA2 might relieve a negative growth signal and may thus allow EREB2-5 cells to proliferate in response to EBNA2 activation.
It is an absolutely crucial question whether the changes observed during the reactivation of EBNA2 in hormone-deprived EREB2-5 cells do indeed reflect events observed early during infection of primary B cells by EBV. This issue has been addressed here for the first time. Primary human B cells were infected with the EBV B95-8 strain, and gene products differentially expressed after infection were assessed by 2D PAGE, excision from the gel, and mass spectrometry. The majority of proteins identified in EBV-infected primary B cells early after infection correlated with the EBNA2 targets identified in EREB2-5 cells. However, the induction rates were lower and the standard deviation was higher. This finding is not surprising when it is taken into account that only approximately 55% of the cells were infected by EBV. Nm23-H1, PPase, UCHL3, and PA28γ were upregulated upon infection of primary B cells with EBV, whereas peroxiredoxin 3, Annexin IV, Ly-GDI, and GMFγ were reproducibly downregulated (Table 1 and Fig. 6). The time at which differentially expressed gene products were identified in virus-infected cells coincided with an increase in DNA synthesis. Western blot analysis of EBNA2 and Nm23-H1, as well as real-time PCR analysis of EBNA2, c-myc, and Nm23-H1 mRNA expression in EBV-infected primary B cells, revealed a tight correlation between the expressions of these three genes. In looking at the time course of induction, it became evident that EBNA2 expression preceded the induction of c-myc RNA and the appearance of Nm23-H1 (data not shown). The time course of induction of EBNA2, c-myc, and Nm23-H1 was also studied for EREB2-5 cells. Real-time PCR confirmed the induction of c-myc mRNA within 1 h and confirmed that Nm23-H1 RNA lagged slightly behind (Fig. 7). These data support the notion that c-myc is a direct target of EBNA2 and that Nm23-H1 is regulated by c-Myc (49).
There are three exceptions that do not follow this pattern of regulation by EBNA2 and viral infection in primary B cells. First, the level of thioredoxin increased in EREB2-5 cells upon EBNA2 activation but decreased in virus-infected primary B cells. Second, IgE-HRF was induced in EREB2-5 cells upon EBNA2 activation and was unchanged or slightly downregulated in EBV-infected primary B cells. Third, FABP5 was strongly induced in EREB2-5 upon EBNA2 activation but remained virtually unaltered after virus infection of primary B cells. It is not clear whether these exceptions do indeed represent true biological differences. It is also conceivable that primary adenoid B cells isolated ex vivo express certain levels of these proteins and that these levels decreased more rapidly in the noninfected cell population than they increased in the infected cells. In the case of thioredoxin, the ratio of oxidized to reduced protein is presumably biologically more relevant than the total amount of thioredoxin protein, which has been determined here.
Keeping these differences in mind, we conclude that the EREB2-5 cell system is indeed a very useful cell system that mimics essential steps in the immortalization of primary B cells by EBV that are initiated by EBNA2. The system offers the great advantage that the cells are derived from a common origin and may thus be regarded as genetically homogenous. Most importantly, the cells can be expanded to virtually unlimited levels before estrogen is withdrawn and readded, so that biochemical analyses focusing on the kinetics of consecutive events become feasible.
A limitation of the present study is that only highly abundant proteins could be identified when 2D PAGE and mass spectrometry were applied to the analysis of whole-cell extracts. This is probably the reason for the fact that most of the proteins identified as differentially expressed in EREB2-5 cells upon EBNA2 activation were also found to be differentially expressed in P493-6 cells driven into proliferation by c-Myc. The proteome approach has thus selected for proteins differentially expressed in proliferating cells versus resting cells and has only poorly discriminated between the c-Myc- and EBV-driven proliferation programs. For a more detailed view on the proteomes of the respective proliferation programs, more refined techniques, e.g., analysis of subcellular compartments and protein complexes that can be purified by appropriate tags, now have to be applied. In addition, these studies have to be complemented by other strategies that shed light on the transcriptomes of the cells in different conditions, e.g., subtractive hybridization and cloning strategies (e.g., PCR-Select or standard gel electrophoresis) or differential hybridization by use of microarrays. Such experiments with P493-6 and EREB2-5 cells have been performed in our laboratory to identify c-Myc and EBNA2 target genes and have resulted in the accumulation of a wealth of information regarding c-Myc- and EBV-induced proliferation (21, 49; M. Schlee, A. Rosenwald, L. M. Staudt, and G. W. Bornkamm, unpublished data). The great challenge is now to develop high-throughput functional knockout or knock-down systems (e.g., small interfering RNA) for the functional analysis of this large number of potentially interesting target genes and to unravel the chain of events that follow c-Myc or EBNA2 activation.
Acknowledgments
This work was supported by grants to G.W.B, W.H. (grant SFB 455), and J.L. (Sachbeihilfe LO 746/1-1) from Deutsche Forschungsgemeinschaft, Fonds der Chemischen Industrie and Deutsche Krebshilfe.
We thank Michael Hölzel (GSF) and D. Hölzel (Institute for Medical Informatics, Biometry and Epidemiology, University Munich) for support with the statistical analysis.
REFERENCES
- 1.Arrigo, A. P. 1999. Gene expression and the thiol redox state. Free Radic. Biol. Med. 27:936-944. [DOI] [PubMed] [Google Scholar]
- 2.Asai, K., K. Fujita, M. Yamamoto, T. Hotta, M. Morikawa, M. Kokubo, A. Moriyama, and T. Kato. 1998. Isolation of novel human cDNA (hGMF-gamma) homologous to glia maturation factor-beta gene. Biochim. Biophys. Acta 1396:242-244. [DOI] [PubMed] [Google Scholar]
- 3.Babcock, G. J., D. Hochberg, and A. D. Thorley-Lawson. 2000. The expression pattern of Epstein-Barr virus latent genes in vivo is dependent upon the differentiation stage of the infected B cell. Immunity 13:497-506. [DOI] [PubMed] [Google Scholar]
- 4.Baumforth, K. R., L. S. Young, K. J. Flavell, C. Constandinou, and P. G. Murray. 1999. The Epstein-Barr virus and its association with human cancers. Mol. Pathol. 52:307-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bello-Fernandez, C., G. Packham, and J. L. Cleveland. 1993. The ornithine decarboxylase gene is a transcriptional target of c-Myc. Proc. Natl. Acad. Sci. USA 90:7804-7808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bevec, D., H. Jaksche, M. Oft, T. Wohl, M. Himmelspach, A. Pacher, M. Schebesta, K. Koettnitz, M. Dobrovnik, R. Csonga, F. Lottspeich, and J. Hauber. 1996. Inhibition of HIV-1 replication in lymphocytes by mutants of the Rev cofactor eIF-5A. Science 271:1858-1860. [DOI] [PubMed] [Google Scholar]
- 7.Birkenbach, M., K. Josefsen, R. Yalamanchili, G. Lenoir, and E. Kieff. 1993. Epstein-Barr virus-induced genes: first lymphocyte-specific G protein-coupled peptide receptors. J. Virol. 67:2209-2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bjellqvist, B., C. Pasquali, F. Ravier, J. C. Sanchez, and D. Hochstrasser. 1993. A nonlinear wide-range immobilized pH gradient for two-dimensional electrophoresis and its definition in a relevant pH scale. Electrophoresis 14:1357-1365. [DOI] [PubMed] [Google Scholar]
- 9.Blum, H., H. Beier, and H. J. Gross. 1987. Improved silver staining of plant proteins, RNA and DNA in polyacrylamide gels. Electrophoresis 8:93-99. [Google Scholar]
- 10.Bornkamm, G. W., and W. Hammerschmidt. 2001. Molecular virology of Epstein-Barr virus. Philos. Trans. R. Soc. Lond. B 356:437-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burgstahler, R., B. Kempkes, K. Steube, and M. Lipp. 1995. Expression of the chemokine receptor BLR2/EBI1 is specifically transactivated by Epstein-Barr virus nuclear antigen 2. Biochem. Biophys. Res. Commun. 215:737-743. [DOI] [PubMed] [Google Scholar]
- 12.Caraglia, M., M. Marra, G. Giuberti, A. M. D'Alessandro, A. Budillon, S. del Prete, A. Lentini, S. Beninati, and A. Abbruzzese. 2001. The role of eukaryotic initiation factor 5A in the control of cell proliferation and apoptosis. Amino Acids 20:91-104. [DOI] [PubMed] [Google Scholar]
- 13.Chen, G., T. G. Gharib, C. C. Huang, D. G. Thomas, K. A. Shedden, J. M. Taylor, S. L. Kardia, D. E. Misek, T. J. Giordano, M. D. Iannettoni, M. B. Orringer, S. M. Hanash, and D. G. Beer. 2002. Proteomic analysis of lung adenocarcinoma: identification of a highly expressed set of proteins in tumors. Clin. Cancer Res. 8:2298-2305. [PubMed] [Google Scholar]
- 14.Cordier, M., A. Calender, M. Billaud, U. Zimber, G. Rousselet, O. Pavlish, J. Banchereau, T. Tursz, G. Bornkamm, and G. M. Lenoir. 1990. Stable transfection of Epstein-Barr virus (EBV) nuclear antigen 2 in lymphoma cells containing the EBV P3HR1 genome induces expression of B-cell activation molecules CD21 and CD23. J. Virol. 64:1002-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fairchild, T. A., and G. Patejunas. 1999. Cloning and expression profile of human inorganic pyrophosphatase. Biochim. Biophys. Acta 1447:133-136. [DOI] [PubMed] [Google Scholar]
- 16.Fan, Z., P. J. Beresford, D. Y. Oh, D. Zhang, and J. Lieberman. 2003. Tumor suppressor NM23-H1 is a granzyme A-activated DNase during CTL-mediated apoptosis, and the nucleosome assembly protein SET is its inhibitor. Cell 112:659-672. [DOI] [PubMed] [Google Scholar]
- 17.Gavioli, R., T. Frisan, S. Vertuani, G. W. Bornkamm, and M. G. Masucci. 2001. c-myc overexpression activates alternative pathways for intracellular proteolysis in lymphoma cells. Nat. Cell Biol. 3:283-288. [DOI] [PubMed] [Google Scholar]
- 18.Gerner, C., U. Frohwein, J. Gotzmann, E. Bayer, D. Gelbmann, W. Bursch, and R. Schulte-Hermann. 2000. The Fas-induced apoptosis analyzed by high throughput proteome analysis. J. Biol. Chem. 275:39018-39026. [DOI] [PubMed] [Google Scholar]
- 19.Görg, A., W. Postel, A. Domscheit, and S. Gunther. 1988. Two-dimensional electrophoresis with immobilized pH gradients of leaf proteins from barley (Hordeum vulgare): method, reproducibility and genetic aspects. Electrophoresis 9:681-692. [DOI] [PubMed] [Google Scholar]
- 20.Gygi, S. P., Y. Rochon, B. R. Franza, and R. Aebersold. 1999. Correlation between protein and mRNA abundance in yeast. Mol. Cell. Biol. 19:1720-1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaiser, C., G. Laux, D. Eick, N. Jochner, G. W. Bornkamm, and B. Kempkes. 1999. The proto-oncogene c-myc is a direct target gene of Epstein-Barr virus nuclear antigen 2. J. Virol. 73:4481-4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaiser, C., O. von Stein, G. Laux, and M. Hoffmann. 1999. Functional genomics in cancer research: identification of target genes of the Epstein-Barr virus nuclear antigen 2 by subtractive cDNA cloning and high-throughput differential screening using high-density agarose gels. Electrophoresis 20:261-268. [DOI] [PubMed] [Google Scholar]
- 23.Kempkes, B., M. Pawlita, U. Zimber-Strobl, G. Eissner, G. Laux, and G. W. Bornkamm. 1995. Epstein-Barr virus nuclear antigen 2-estrogen receptor fusion proteins transactivate viral and cellular genes and interact with RBP-J kappa in a conditional fashion. Virology 214:675-679. [DOI] [PubMed] [Google Scholar]
- 24.Kempkes, B., D. Spitkovsky, P. Jansen-Durr, J. W. Ellwart, E. Kremmer, H. J. Delecluse, C. Rottenberger, G. W. Bornkamm, and W. Hammerschmidt. 1995. B-cell proliferation and induction of early G1-regulating proteins by Epstein-Barr virus mutants conditional for EBNA2. EMBO J. 14:88-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khan, S., M. van den Broek, K. Schwarz, R. de Giuli, P. A. Diener, and M. Groettrup. 2001. Immunoproteasomes largely replace constitutive proteasomes during an antiviral and antibacterial immune response in the liver. J. Immunol. 167:6859-6868. [DOI] [PubMed] [Google Scholar]
- 26.Khanna, R., and S. R. Burrows. 2000. Role of cytotoxic T lymphocytes in Epstein-Barr virus-associated diseases. Annu. Rev. Microbiol. 54:19-48. [DOI] [PubMed] [Google Scholar]
- 27.Kieff, E. 1996. Epstein-Barr virus and its replication, p. 2343-2396. In B. N. Fields, D. M. Knipe, P. M. Howley, R. M. Chanock, T. P. Monath, J. L. Melnick, B. Roizman, and S. E. Strauss (ed.), Fields virology. Lippincott-Raven Publishers, Philadelphia, Pa.
- 28.Klier, H., R. Csonga, H. C. Joao, C. Eckerskorn, M. Auer, F. Lottspeich, and J. Eder. 1995. Isolation and structural characterization of different isoforms of the hypusine-containing protein eIF-5A from HeLa cells. Biochemistry 34:14693-14702. [DOI] [PubMed] [Google Scholar]
- 29.Kruse, M., O. Rosorius, F. Kratzer, D. Bevec, C. Kuhnt, A. Steinkasserer, G. Schuler, and J. Hauber. 2000. Inhibition of CD83 cell surface expression during dendritic cell maturation by interference with nuclear export of CD83 mRNA. J. Exp. Med. 191:1581-1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leone, A., U. Flatow, C. R. King, M. A. Sandeen, I. M. Margulies, L. A. Liotta, and P. S. Steeg. 1991. Reduced tumor incidence, metastatic potential, and cytokine responsiveness of nm23-transfected melanoma cells. Cell 65:25-35. [DOI] [PubMed] [Google Scholar]
- 31.Levitskaya, J., A. Sharipo, A. Leonchiks, A. Ciechanover, and M. G. Masucci. 1997. Inhibition of ubiquitin/proteasome-dependent protein degradation by the Gly-Ala repeat domain of the Epstein-Barr virus nuclear antigen 1. Proc. Natl. Acad. Sci. USA 94:12616-12621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lewis, T. S., J. B. Hunt, L. D. Aveline, K. R. Jonscher, D. F. Louie, J. M. Yeh, T. S. Nahreini, K. A. Resing, and N. G. Ahn. 2000. Identification of novel MAP kinase pathway signaling targets by functional proteomics and mass spectrometry. Mol. Cell 6:1343-1354. [DOI] [PubMed] [Google Scholar]
- 33.Lim, R., and A. Zaheer. 1996. In vitro enhancement of p38 mitogen-activated protein kinase activity by phosphorylated glia maturation factor. J. Biol. Chem. 271:22953-22956. [DOI] [PubMed] [Google Scholar]
- 34.Lim, R., A. Zaheer, M. A. Yorek, C. J. Darby, and L. W. Oberley. 2000. Activation of nuclear factor-kappaB in C6 rat glioma cells after transfection with glia maturation factor. J. Neurochem. 74:596-602. [DOI] [PubMed] [Google Scholar]
- 35.Lim, R., W. X. Zhong, and A. Zaheer. 1990. Antiproliferative function of glia maturation factor beta. Cell Regul. 1:741-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lovric, J., S. Dammeier, A. Kieser, H. Mischak, and W. Kolch. 1998. Activated raf induces the hyperphosphorylation of stathmin and the reorganization of the microtubule network. J. Biol. Chem. 273:22848-22855. [PubMed] [Google Scholar]
- 37.Mao, M., G. Fu, J. S. Wu, Q. H. Zhang, J. Zhou, L. X. Kan, Q. H. Huang, K. L. He, B. W. Gu, Z. G. Han, Y. Shen, J. Gu, Y. P. Yu, S. H. Xu, Y. X. Wang, S. J. Chen, and Z. Chen. 1998. Identification of genes expressed in human CD34(+) hematopoietic stem/progenitor cells by expressed sequence tags and efficient full-length cDNA cloning. Proc. Natl. Acad. Sci. USA 95:8175-8180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miller, G., and M. Lipman. 1973. Release of infectious Epstein-Barr virus by transformed marmoset leukocytes. Proc. Natl. Acad. Sci. USA 70:190-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Murata, S., H. Kawahara, S. Tohma, K. Yamamoto, M. Kasahara, Y. Nabeshima, K. Tanaka, and T. Chiba. 1999. Growth retardation in mice lacking the proteasome activator PA28gamma. J. Biol. Chem. 274:38211-38215. [DOI] [PubMed] [Google Scholar]
- 40.Nador, R. G., A. Chadburn, G. Gundappa, E. Cesarman, J. W. Said, and D. M. Knowles. 2003. Human immunodeficiency virus (HIV)-associated polymorphic lymphoproliferative disorders. Am. J. Surg. Pathol. 27:293-302. [DOI] [PubMed] [Google Scholar]
- 41.Nalesnik, M. A. 2002. Clinicopathologic characteristics of post-transplant lymphoproliferative disorders. Recent Results Cancer Res. 159:9-18. [DOI] [PubMed] [Google Scholar]
- 42.Nikaido, T., K. Shimada, Y. Nishida, R. S. Lee, A. B. Pardee, and Y. Nishizuka. 1989. Loss in transformed cells of cell cycle regulation of expression of a nuclear protein recognized by SLE patient antisera. Exp. Cell Res. 182:284-289. [DOI] [PubMed] [Google Scholar]
- 43.Oh, J. J., D. R. Grosshans, S. G. Wong, and D. J. Slamon. 1999. Identification of differentially expressed genes associated with HER-2/neu overexpression in human breast cancer cells. Nucleic Acids Res. 27:4008-4017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pajic, A., D. Spitkovsky, B. Christoph, B. Kempkes, M. Schuhmacher, M. S. Staege, M. Brielmeier, J. Ellwart, F. Kohlhuber, G. W. Bornkamm, A. Polack, and D. Eick. 2000. Cell cycle activation by c-myc in a Burkitt lymphoma model cell line. Int. J. Cancer 87:787-793. [DOI] [PubMed] [Google Scholar]
- 45.Polack, A., K. Hortnagel, A. Pajic, B. Christoph, B. Baier, M. Falk, J. Mautner, C. Geltinger, G. W. Bornkamm, and B. Kempkes. 1996. c-myc activation renders proliferation of Epstein-Barr virus (EBV)-transformed cells independent of EBV nuclear antigen 2 and latent membrane protein 1. Proc. Natl. Acad. Sci. USA 93:10411-10416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Postel, E. H., S. J. Berberich, S. J. Flint, and C. A. Ferrone. 1993. Human c-myc transcription factor PuF identified as nm23-H2 nucleoside diphosphate kinase, a candidate suppressor of tumor metastasis. Science 261:478-480. [DOI] [PubMed] [Google Scholar]
- 47.Robertson, K. D., S. D. Hayward, P. D. Ling, D. Samid, and R. F. Ambinder. 1995. Transcriptional activation of the Epstein-Barr virus latency C promoter after 5-azacytidine treatment: evidence that demethylation at a single CpG site is crucial. Mol. Cell. Biol. 15:6150-6159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schleger, C., C. Verbeke, R. Hildenbrand, H. Zentgraf, and U. Bleyl. 2002. c-MYC activation in primary and metastatic ductal adenocarcinoma of the pancreas: incidence, mechanisms, and clinical significance. Mod. Pathol. 15:462-469. [DOI] [PubMed] [Google Scholar]
- 49.Schuhmacher, M., F. Kohlhuber, M. Holzel, C. Kaiser, H. Burtscher, M. Jarsch, G. W. Bornkamm, G. Laux, A. Polack, U. H. Weidle, and D. Eick. 2001. The transcriptional program of a human B cell line in response to Myc. Nucleic Acids Res. 29:397-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schuhmacher, M., M. S. Staege, A. Pajic, A. Polack, U. H. Weidle, G. W. Bornkamm, D. Eick, and F. Kohlhuber. 1999. Control of cell growth by c-Myc in the absence of cell division. Curr. Biol. 9:1255-1258. [DOI] [PubMed] [Google Scholar]
- 51.Schuldiner, O., S. Shor, and N. Benvenisty. 2002. A computerized database-scan to identify c-MYC targets. Gene 292:91-99. [DOI] [PubMed] [Google Scholar]
- 52.Subramanian, C., M. A. Cotter II, and E. S. Robertson. 2001. Epstein-Barr virus nuclear protein EBNA-3C interacts with the human metastatic suppressor Nm23-H1: a molecular link to cancer metastasis. Nat. Med. 7:350-355. [DOI] [PubMed] [Google Scholar]
- 53.Tsuiki, H., K. Asai, M. Yamamoto, K. Fujita, Y. Inoue, Y. Kawai, T. Tada, A. Moriyama, Y. Wada, and T. Kato. 2000. Cloning of a rat glia maturation factor-gamma (rGMFG) cDNA and expression of its mRNA and protein in rat organs. J. Biochem. (Tokyo) 127:517-523. [DOI] [PubMed] [Google Scholar]
- 54.Wakasugi, N., Y. Tagaya, H. Wakasugi, A. Mitsui, M. Maeda, J. Yodoi, and T. Tursz. 1990. Adult T-cell leukemia-derived factor/thioredoxin, produced by both human T-lymphotropic virus type I- and Epstein-Barr virus-transformed lymphocytes, acts as an autocrine growth factor and synergizes with interleukin 1 and interleukin 2. Proc. Natl. Acad. Sci. USA 87:8282-8286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wilk, S., W. E. Chen, and R. P. Magnusson. 2000. Properties of the nuclear proteasome activator PA28gamma (REGgamma). Arch. Biochem. Biophys. 383:265-271. [DOI] [PubMed] [Google Scholar]
- 56.Zaheer, A., S. N. Mathur, and R. Lim. 2002. Overexpression of glia maturation factor in astrocytes leads to immune activation of microglia through secretion of granulocyte-macrophage-colony stimulating factor. Biochem. Biophys. Res. Commun. 294:238-244. [DOI] [PubMed] [Google Scholar]
- 57.Zeidler, R., P. Meissner, G. Eissner, S. Lazis, and W. Hammerschmidt. 1996. Rapid proliferation of B cells from adenoids in response to Epstein-Barr virus infection. Cancer Res. 56:5610-5614. [PubMed] [Google Scholar]