Abstract
To develop new therapies for inflammatory breast cancer (IBC) we have compared the effects of two hydroxamic acid-based histone deacetylase (HDAC) inhibitors, CG-1521 and Trichostatin A (TSA) on the biology of two IBC cell lines: SUM149PT and SUM190PT. CG-1521 and TSA induce dose (0−10 µM) and time-dependent (0−96 h) increases in the proportion of cells undergoing cell cycle arrest and apoptosis in the presence or absence of 17β-estradiol. In SUM 149PT cells, both CG-1521 and TSA increase the levels of acetylated α-tubulin; however the morphological effects are different: CG-1521 blocks mitotic spindle formation and prevents abscission during cytokinesis while TSA results in an increase in cell size. In SUM190PT cells CG-1521 does not cause an increase in acetylated-α-tubulin and even though TSA significantly increases the levels of acetylated tubulin, neither inhibitor alters the morphology of the cells. Microarray analysis demonstrates that CG-1521 modulates the expression of 876 mRNAs and 63 miRNAs in SUM149PT cells, and 1227 mRNAs and 35 miRNAs in SUM190PT cells. Only 9% of the genes are commonly modulated in both cell lines, suggesting that CG-1521 and TSA target different biological processes in the two cell lines most likely though the inhibition of different HDACs in these cell lines. Gene ontology (GO) analysis reveals that CG-1521 affects the expression of mRNAs that encode proteins associated with the spindle assembly checkpoint, chromosome segregation, and microtubule-based processes in both cell lines and has cell-type specific effects on lipid biosynthesis, response to DNA damage, and cell death.
Keywords: acetylation, midzone, abscission, chromosome misalignment, cell death, cell cycle
Introduction
Inflammatory breast cancer (IBC) accounts for less than 2% of all breast cancer casess. However it is a very aggressive and lethal disease, responsible for nearly 8% of breast cancer deaths.1,2 The median age at diagnosis is 55 y for IBC compared with 62 y of age for all other breast cancer cases, and as a result a significant proportion of women are pre-menopausal at the time of diagnosis.2 The median overall survival among women with IBC is approximately 2.9 y even with aggressive treatment including hormone therapy, Herceptin-based or anthracycline-based chemotherapy, surgery, radiation or a combination of these modalities. This disease is clearly much more aggressive than locally advanced breast cancer (LABC), which has a median overall survival of 6.4 y.3 IBC disproportionately afflicts Afro-American women who are diagnosed at an earlier age (48 y), have a higher proportion of advanced and aggressive tumors at diagnosis and have a shorter median survival of 2 y compared with 3 y for Caucasian women.2-4 In contrast to the incidence of ductal carcinoma in situ (DCIS) and LABC, which has decreased over the past 20 years, the overall incidence of IBC continues to increase.1,3
The molecular characteristics of IBC are heterogeneous: 45% of IBC tumors are Her2/neu-positive, one-third of tumors are estrogen receptor-positive (ER+) and one-third of tumors are classified as triple negative.2 The lack of obvious molecular targets for the treatment of IBC partially explains the poor responsiveness of IBC tumors to therapy and highlights the need to identify new therapeutic options for the treatment of the disease. Understanding of the molecular pathology of IBC has been hampered by the paucity of IBC-derived cell lines. Unlike other types of breast cancer, there are only two established cell lines, the SUM149PT and SUM190PT cell lines, that can be used to evaluate the response of IBC to experimental therapeutics. These two cell lines demonstrate some aspects of the molecular heterogeneity typical of IBC tumors. SUM149PT cells are triple negative, harbor a point mutation in the p53 gene, and are BRCA-1-negative. The SUM190PT cells overexpress Her2/neu, do not express functional p53 protein but express wild type BRCA-15,6 (Table S1). Neither cell line expresses ERα (annotated simply as ER− in Table S1), although they have both been reported to express ERβ.
Global gene expression is regulated mainly by changes in chromatin conformation, which is mediated by the coordinated efforts of histone acetyltransferases (HATs) and histone deacetylases (HDACs). HATs catalyze the acetylation of lysine residues in the histone tails resulting in an “open” chromatin conformation which facilitates and sustains gene transcription. The removal of acetyl groups by HDACs restores the “closed” chromatin conformation, effectively preventing transcription.7,8 The ability to modulate gene expression has made HATs and HDACs desirable targets for cancer drug development. There has been a concerted effort to develop novel HDAC inhibitors (HDACi) and a number of small molecules including SAHA (suberoylanilide hydroxamic acid, Vorinostat), TSA (Trichostatin A), and CG-1521 have been shown to be effective inhibitors of histone deacetylation and to induce changes in the behavior of cancer cells in vitro.9-14 While quantitatively the major function of HATs and HDACs is to regulate chromatin conformation, new data demonstrate that many transcription factors including p53, the estrogen receptor (ERα) and NFκB15-20 are acetylated and deacetylated by the same enzymes, suggesting that protein acetylation is important for the activity of transcription complexes.
We have previously compared the effects of several HDACi, including TSA and CG-1521, in LNCaP and PC-3 prostate cancer and MCF-7 breast cancer cell lines and have shown that they have distinctly different biological effects, despite the fact that they are regarded as pan-inhibitors of class 1 HDACs.13,21 The data presented here show that both TSA and CG-1521 induce cell cycle arrest and apoptosis in IBC cells. However, microarray analysis of mRNA and miRNA changes shows that CG-1521 modulates significantly different gene ontologies in the two cell lines, resulting in different biological effects that are most likely mediated by both transcriptional and post-transcriptional mechanisms. These differences undoubtedly reflect the differences in the molecular pathology but also provide a useful baseline for the development of personalized therapies incorporating HDACi and the identification of new druggable targets for the treatment of IBC.
Results
Growth response of IBC cell lines to CG-1521 and TSA
Since IBC is diagnosed in both pre- and post-menopausal women, the response of SUM149PT and SUM190PT cells to TSA and CG-1521 was assessed in the absence and presence of 10 nM E2. Both cell lines are sensitive to CG-1521, which causes a significant dose-dependent decrease in adherent cells after 48 h of treatment (Fig. 1A and C). SUM149PT and SUM190PT display similar sensitivities to CG-1521 (between 1–5 μM), however, the presence of E2 does not influence the sensitivity of either cell line to CG-1521. The SUM149PT and SUM190PT cells show marked differences in sensitivity to the well-known HDACi, TSA. SUM149PT cells are very sensitive to TSA and cell growth is arrested at doses as low as 50 nM (Fig. 1B). In contrast, SUM190PT cells are relatively insensitive to TSA, even at doses as high as 2 μM (Fig. 1D), regardless of whether E2 is present in the medium or not.
Figure 1. Inhibition of IBC cell growth by CG-1521 and TSA. SUM149PT cells (A and B) and SUM190PT cells (C and D) were treated with indicated doses of CG-1521 (A and C) or TSA (B and D) in the absence or presence of 10 nM E2 for 48 h. Adherent cells were measured using crystal violet as described in Methods. Results represent the mean (± SD) from three independent experiments assayed in quadruplicate. Comparisons between different treatments and vehicle control were analyzed using one-way ANOVA; differences were considered significant if P < 0.05 (*). NS, not significant.
Effect of CG-1521 and TSA on cell cycle kinetics and apoptosis in IBC cells
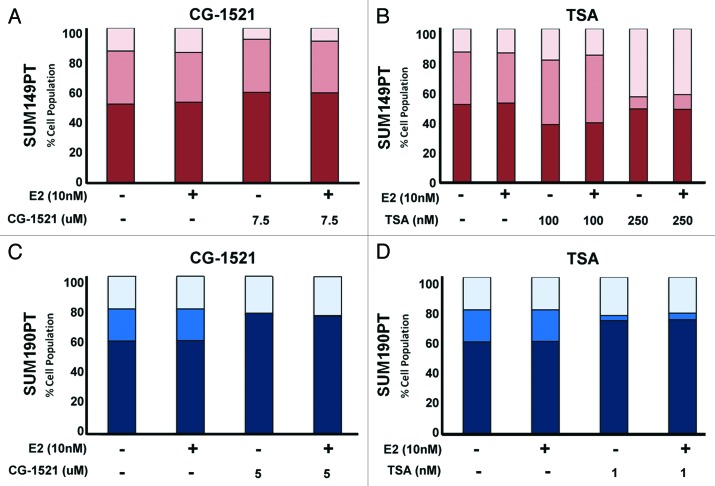
To investigate the underlying mechanism of cell growth repression by CG-1521 and TSA, the effects of the two HDACi on cell cycle progression and apoptosis were assessed by flow cytometry. Treatment of SUM149PT cells with CG-1521 for 48 h, results in the accumulation of cells in the G1 phase of the cell cycle with a concomitant reduction in the G2/M phase cell population (Fig. 2A). In contrast, CG-1521 causes accumulation of SUM190PT cells in the G0/G1 phase accompanied by the almost complete loss of cells in S phase (Fig. 2C). The effect of TSA on SUM149PT cells appears to be concentration dependent since 100 nM TSA induces a decrease in G1 with a corresponding increased accumulation of cells in S phase (Fig. 2B). In contrast, TSA at doses ≥250 nM causes a substantial increase in G2/M accumulation and concomitant decrease in S phase accumulation. In SUM190PT cells, TSA causes a marked increase in G1 accumulation with a significant decrease in the proportion of cells in S phase (Fig. 2D). The effects of CG-1521 and TSA on cell cycle progression are not affected by the absence or presence of E2 in either cell line.
Figure 2. Inhibition of cell cycle progression by CG-1521 and TSA in IBC cells. SUM149PT cells (A and B) and SUM190PT cells (C and D) were treated with indicated doses of CG-1521 (A and C) or TSA (B and D) in the absence or presence of 10 nM E2 for 48 h. Cell cycle kinetics were measured by flow cytometry using propidium iodide staining as described in Methods. SUM149PT cells were treated with 7.5 µM CG-1521 (A) or 100 nM or 250 nM TSA (B). SUM190PT cells were treated with 5 µM CG-1521 (C) or 1 µM TSA (D) for 48 h. For SUM149PT cells, red, G1; dark pink, S phase; light pink, G2/M phase. For SUM190PT cells, dark blue, G1; medium blue, S phase; light blue, G2/M phase. Results represent the mean of three experiments. The error bars are omitted for clarity.
The increased levels of DNA fragmentation in both SUM149PT and SUM190PT cells in the absence or presence of E2 (Fig. 3A and C) indicates CG-1521 induces apoptosis, although the SUM190PT cells are more sensitive to CG-1521 compared with SUM149PT cells. In contrast, the SUM149PT cells are highly sensitive while the SUM190PT cells are relatively resistant to TSA treatment (Fig. 3B and D). However, at doses greater than 250 nM, TSA appears to rapidly obliterate SUM149PT cells, leaving too few cells to determine whether there is evidence of DNA fragmentation (data not shown).
Figure 3. Induction of DNA fragmentation by CG-1521 and TSA in IBC cells. SUM149PT cells were treated with 7.5 µM CG-1521 (A) or 100 nM TSA (B); SUM190PT cells were treated with 5 µM CG-1521 (C) or 1 µM TSA (D) in the absence or presence of 10 nM E2 for 48 h. The percentage of cells displaying fragmented DNA was measured using Apo-BrdU staining as described in Methods. Results represent the mean (± SD) from three independent experiments. Comparisons between different treatment groups were analyzed using one-way ANOVA; differences were considered significant if P < 0.05 (*), NS, not significant.
Effect of CG-1521 and TSA on morphology of IBC cells
To examine the effects of CG-1521 and TSA on the cell biology of IBC cells, SUM149PT and SUM190PT cells were treated with the HDACi and examined by confocal microscopy to visualize the actin cytoskeleton and acetylated α-tubulin. CG-1521 treatment of SUM149PT cells does not induce a dramatic change in the actin cytoskeleton, which is diffusely distributed throughout the cell with a distinct localization on the plasma membrane. However in dividing SUM149PT cells, CG-1521 leads to the formation of extended midbody structures with increased acetylated α-tubulin staining which is indicative of a failure of abscission (Fig. 4A). In contrast, TSA does not significantly alter the actin cytoskeleton or cause a failure of abscission in SUM149PT cells. However, the level of acetylated α-tubulin increases and is also accompanied by a significant enlargement of the cells (Fig. 4A). In SUM190PT cells, treatment with CG-1521 does not increase the amount of acetylated α-tubulin staining, and there is no evidence of increased midbody length, abrogated abscission or increased cell size (Fig. 4B), As shown in Figure 4C, there is a significantly increase in the number of elongated midbodies in SUM149PT after 48 h of treatment with CG-1521, which is not evident in cells treated with TSA, suggesting that there may be a correlation between tubulin acetylation and the failure of abscission in this cell line.
Figure 4. Effect of CG-1521 and TSA on morphology of IBC cell lines. SUM149PT cells were treated with 7.5 µM or 100 nM TSA (A); SUM190PT cells were treated with 3 µM CG-1521 and 500 nM TSA (B) for 48 h then fixed and immunostained to visualize the actin cytoskeleton (green, left column in [A and B]), acetylated-α-tubulin (green, right column in [A and B]) and DNA (blue). White arrows indicate elongated midzone structures in SUM149PT cells (A). All images were captured under the same exposure settings. Scale bars: 25 microns. The number of elongated midzone structures was accessed manually as described in Methods (C).
Effect of CG-1521 and TSA on mRNA expression in IBC cells
Expression profiling analysis of SUM149PT and SUM190PT cells treated for 48 h with 7.5 µM and 5 µM CG-1521 respectively, in the presence and absence of 10 nM E2 was performed using Affymetrix Gene 1.0 ST whole genome arrays. In SUM149PT cells, 876 mRNAs are differentially regulated by CG-1521 (≥ 2.0 fold; p ≤ 0.05): 316 mRNAs are upregulated and 560 are downregulated (Fig. S1; Table S3). In SUM190PT cells, 1227 mRNAs are differentially regulated (≥2.0 fold; P ≤ 0.05) by CG-1521: 651 are upregulated and 576 are downregulated (Fig. S1; Table S4). Lists of CG-1521 modulated genes were analyzed using IPA and DAVID22,23 to identify GO terms or pathways which are significantly enriched by CG-1521 in SUM149PT cells (Table 1 and Fig. 5A) and SUM190PT cells (Table 2 and Fig. 5B). In SUM149PT cells, CG-1521 downregulates a significant number of genes associated with M phase (GO:0000279) and more specifically those associated with nucleosome organization (GO:0034728), chromosome segregation (GO:0007059), microtubule cytoskeleton organization (GO:0000226), mitotic cell cycle checkpoint (GO:0007093), and spindle checkpoint (GO:0031577). The effect of CG-521 on these ontologies is highly significant (Table 1), suggesting that the phenotypic effects of CG-1521 are not simply due to a pleitropic downregulation of transcription. CG-1521 also induced the expression of genes associated with lipid metabolic processes (GO:0006299), oxidoreductase activity (GO:0016491), electron carrier activity (GO:0009055), and steroid biosynthetic processes (GO:0006694) (Fig. 5A). In SUM190PT cells, CG-1521 also downregulates the expression of genes associated with M phase (GO:0000279), as well as lipid metabolic processes (GO:0006299), carboxylic acid metabolic processes (GO:0019752), and oxidoreductase activity (GO:0016616). CG-1521 also induced the expression of genes associated with the negative regulation of apoptosis (GO:0006629), response to unfolded protein (GO:0016491) and response to stress (GO:0009055) (Table 2 and Fig. 5B). The presence of E2 does not influence the steady-state level of the transcripts in untreated SUM149PT or SUM190PT cells, nor does it affect the response of either cell line treated with CG-1521as evidenced by the remarkable similarity in the heat map signatures from both cell lines in the absence and presence of estradiol (Fig. S1).
Table 1. Gene set enrichment analysis of representative gene sets identified as significantly enriched after CG-1521 treatment in SUM149PT cells.
| Downregulated functional classes | |||
|---|---|---|---|
|
GO ID |
Term |
Genes |
P value |
| GO:0000279 |
M phase |
98 |
1.19E-68 |
| GO:0034728 |
Nucleosome organization |
36 |
5.07E-29 |
| GO:0007059 |
Chromosome segregation |
40 |
4.05E-37 |
| GO:0000226 |
Microtubule cytoskeleton organization |
29 |
1.51E-14 |
| GO:0007093 |
Mitotic cell cycle checkpoint |
11 |
6.21E-07 |
| GO:0031577 |
Spindle checkpoint |
6 |
1.94E-05 |
| Upregulated functional classes |
|
|
|
|
GO ID |
Term |
Genes |
P value |
| GO:0006629 |
Lipid metabolic process |
32 |
6.02E-06 |
| GO:0016491 |
Oxidoreductase activity |
28 |
1.85E-05 |
| GO:0009055 |
Electron carrier activity |
13 |
2.52 E-04 |
| GO:0006694 | Steroid biosynthetic process | 8 | 4.21E-04 |
The full list of genes associated with these ontologies is provided in Table S3.
Figure 5. Gene ontologies modulated by CG-1521 in IBC cells. Selected gene ontologies modulated by CG-1521 in both cell lines are represented. SUM149PT (A) and SUM190PT (B) cells were treated with 7.5 µM or 5 µM CG-1521, respectively. Functional annotation of each gene was assigned using DAVID. Red, genes upregulated by treatment (≥2.0-fold); Blue, genes downregulated by treatment (≤−2.0-fold).
Table 2. Gene set enrichment analysis of representative gene sets identified as significantly enriched after CG-1521 treatment in SUM190PT cells.
| Downregulated functional classes | |||
|---|---|---|---|
|
GO ID |
Term |
No. of genes |
P value |
| GO:0000279 |
M phase |
23 |
2.26E -11 |
| GO:0006629 |
Lipid metabolic process |
28 |
4.80E-07 |
| GO:0019752 |
Carboxylic acid metabolic process |
17 |
6.21E-04 |
| GO:0016616 |
Oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor |
8 |
1.52E-04 |
| Upregulated functional classes |
|
|
|
|
GO ID |
Term |
No. of genes |
P value |
| GO:0006629 |
Negative regulation of apoptosis |
18 |
1.95E-06 |
| GO:0016491 |
Response to unfold protein |
8 |
2.72E-05 |
| GO:0009055 | Response to stress | 40 | 7.58E-05 |
The full list of genes associated with these ontologies is provided in Table S3.
Quantitative PCR validation of microarray data
The effect of CG-1521 on the expression of selected transcripts associated with M phase, microtubule organization and chromosome segregation were validated in both cell lines and compared with the effects of TSA over a 48h time course. SUM149PT cells were treated for 12–48 h with 7.5 µM CG-1521 or 500 nM TSA; SUM190PT cells were treated for 12–48 h with 5 µM CG-1521 or 1 µM TSA. Treatment with either CG-1521 or TSA results in the time dependent repression of transcripts associated with M phase; specifically microtubule organization (NEK2, PRC1, KIF11, KIF23, KIF2C, KIF4A, KIF4B) and is most evident after 48 h, exemplified by KIF4A (Fig. 6); changes in the remaining mRNA transcripts are shown in Table S5. However, transcript levels are more dramatically modulated by TSA in the SUM149PT cells, whereas CG-1521 has a considerably larger effect on mRNA expression in SUM190PT cells. The presence of E2 does not significantly affect the regulation of genes by CG-1521 or TSA in either cell line (Fig. 6 and data not shown).

Figure 6. Validation of changes in KIF4A expression following treatment with CG-1521 and TSA in IBC cells. SUM149PT cells (A) were treated with 7.5 µM CG-1521 and 500 nM TSA alone and in the presence of 10 nM E2 for 12−48 h. SUM190PT cells (B) were treated with 5 µM CG-1521 and 1 µM TSA alone and in the presence of 10 nM E2 for 12−48 h. KIF4 expression was measured using SYBR Green assays with validated primers (Table S2) for both cells lines for 12−48 h. Four independent replicates were measured in duplicate for each treatment. Comparisons between different treatment groups were analyzed using one-way ANOVA; differences were considered significant if P < 0.05 (*), NS, not significant. For SUM149PT cells: red, CG-1521 (7.5 µM); red stripe, CG-1521 (7.5 µM) + E2 (10 nM); purple, TSA (500 nM); purple stripe, TSA (500 nM) + E2 (10 nM). For SUM190PT cells: blue, CG-1521 (5 µM); blue stripe, CG-1521 (5 µM) + E2 (10 nM); green, TSA (1 µM); green stripe, TSA (1 µM) + E2 (10 nM).
Effect of CG-1521 and TSA on miRNA expression in IBC cells
Interrogation of 866 miRNAs using Agilent Human miRNA v3 microarrays shows that CG-1521 differentially modulates 63 miRNAs in SUM149PT cells. Specifically, 35 miRNAs are upregulated and 28 are downregulated in SUM149PT cells (Fig. S2 and Table S6). In contrast 14 miRNAs are upregulated and 21 miRNAs downregulated in SUM190PT cells (Fig. S2 and Table S7). Validation of miRNA expression levels using TaqMan assays shows that changes in miRNA expression correlate to those measured by microarray, although the magnitude of the change is consistently less when measured by TaqMan. These changes are exemplified by miR-22 (Fig. 7) and changes in the remaining miRNAs are shown in Table S8. In SUM149PT cells, these changes are observed as early as 12 h, but occur much later in SUM190PT cells, reaching maximum levels after 72 h treatment.

Figure 7. Validation of changes in miR-22 expression following treatment with CG-1521 and TSA in IBC cells. SUM149PT cells (Panel A) were treated with 7.5 µM CG-1521 and 500 nM TSA alone and in the presence of 10 nM E2 for 12–72 h. SUM190PT cells (Panel B) were treated with 5 µM CG-1521 and 1 µM TSA alone and in the presence of 10 nM E2 for 12−72 h. Changes in expression of selected miRNAs were measured using TaqMan assays. Four independent replicates were measured in duplicate for each treatment. Comparisons between different treatment groups were analyzed using one-way ANOVA; differences were considered significant if p < 0.05 (*), NS: not significant. For SUM149PT cells: red, CG-1521 (7.5 µM); red stripe, CG-1521 (7.5 µM) + E2 (10 nM); purple, TSA (500 nM); purple stripe, TSA (500 nM) + E2 (10 nM). For SUM190PT cells: blue, CG-1521 (5 µM); blue stripe, CG-1521 (5 µM) + E2 (10 nM); green, TSA (1 µM); green stripe, TSA (1 µM) + E2 (10 nM).
Discussion
Inflammatory breast cancer (IBC) is a very difficult disease to treat, largely due to the molecular heterogeneity of the tumors and lack of disease-specific targets. There has been a concerted effort to develop novel HDAC inhibitors as cancer therapies because of their ability to induce cell cycle arrest, differentiation and apoptosis as well as preventing metastasis.24 Currently, there are several HDAC inhibitors in phase I and II clinical trials as monotherapies or combined with other approved chemotherapeutics, for the treatment of hematological malignancies and advanced or metastatic breast and prostate cancer.25,26 A number of small molecules including SAHA (suberoylanilide hydroxamic acid, Vorinostat), TSA (Trichostatin A), and CG-1521 have been shown to be effective inhibitors of histone deacetylation and to induce changes in the behavior of breast cancer cells in vitro.21,27,28
The SUM149PT and SUM190PT cell lines reflect the heterogeneous molecular pathology of IBC (Table S1). Many IBC patients are younger and pre-menopausal when diagnosed, compared with patients with other forms of breast cancer, and a large proportion (30−55%) of IBC tumors do not express ERα and are classified as ER−, however both have been reported to express ERβ.2,29 For this reason we performed all experiments reported in this manuscript in the absence and presence of E2. Neither cell line responds to E2 in vitro, suggesting that loss of ERα-mediated signaling networks regulating proliferation and cell death is a common event. The data presented in this manuscript show that the cell lines respond to CG-1521 and TSA regardless of the presence of E2, establishing that ERβ-mediated signaling does not impinge on the responsiveness of the cell lines to either HDACi. Thus combining anti-estrogens or aromatase inhibitors with HDACi is unlikely to provide additional benefit over HDACi alone for the treatment of IBC tumors that are classified as ER-negative. On the other hand, both CG-1521 and TSA as monotherapies induce cell death in the SUM149PT (triple negative) and SUM190PT (HER2 amplified) cell lines, suggesting that they may be useful therapies alone or in combination with other current chemotherapies for the treatment of patients that are diagnosed with triple negative or HER2 amplified breast cancers, regardless of the menopausal status of the patients. The data presented in this manuscript suggest that that CG-1521 is effective in treating both triple negative and HER2 overexpressing IBCs, although TSA may prove to be more effective for triple negative breast cancer.
Both cell lines have similar sensitivities to CG-1521, demonstrating growth inhibition and induction of apoptosis when exposed to 1–5 μM CG-1521. SUM149PT cells are significantly more sensitive to TSA while SUM190PT cells are essentially resistant, suggesting that one or more of the HDACs targeted by TSA are not present in this cell line. Furthermore, the phenotypic differences exhibited between the two cell lines suggests that the SUM149PT and SUM190PT cell lines express different complements of HDACs and/or different downstream targets of the enzymes. While it is clear that CG-1521 and TSA induce cell cycle arrest and apoptosis, the underlying mechanisms for the effects appears to be different in the cell lines.
Flow cytometry indicates that CG-1521 induces G0/G1 arrest in the SUM149PT cells. In marked contrast, immunofluorescence microscopy shows clear evidence of elongated midbodies, abrogated abscission, and incomplete cytokinesis, suggesting G2/M arrest. The discordance between these data appears to be a methodological issue. It is likely that during the preparation of treated cells for flow cytometry, the delicate elongated midbody structures are ruptured producing two cells that appear to be arrested in G0/G1. Therefore in this context, flow cytometry does not provide a reliable measure of the effects of CG-1521 on cell cycle kinetics and we suggest that CG-1521 induces G2/M arrest by abrogation of abscission, resulting in the formation of elongated midbody structures. Failure to resolve the block in abscission subsequently induces mitotic catastrophe leading to death based on the recently delineated criteria.30
Originally, HDAC inhibitors were thought to stabilize histone acetylation and subsequently maintain the chromatin in an open conformation, resulting in sustained gene transcription. However, the microarray data presented here, and previously published data from our laboratory and others,10,31 demonstrate that the majority of transcripts are downregulated by HDACi treatment, suggesting an alternate mechanism. The downregulation of transcripts observed in this study is probably driven by two related yet independent mechanisms. First, CG-1521 may reduce transcription by altering the stability and/or activity of nuclear proteins including transcription factors. While the bulk of acetylation and deacetylation is directed toward histones; numerous studies demonstrate transcription factors such as p5315,17,32 and the estrogen receptor (ERα)19,33,34 are acetylated and deacetylated by HATs and HDACs, influencing their nuclear retention, protein binding, and DNA binding activity. Second, as the data in this manuscript demonstrate, HDACi including CG-1521 and TSA modulate the transcription of a small cohort of miRNAs, which alter the steady-state levels of a number of mRNAs, which encode proteins that are critical of M phase, mitotic spindle organization, chromosome segregation and abscission.
To determine the relevance of the inverse concordance between the changes in miRNA and mRNA expression documented above, the mRNA targets of the miRNAs affected by CG-1521 were identified using TargetScan (www.targetscan.org). Transcripts whose expression level changed significantly in the microarrays and showed an inverse concordance with miRNA expression were imported into DAVID to identify commonly targeted ontologies. The relationship between miRNAs and their presumptive targets are shown as Web diagrams for some of the most relevant ontologies in Figure 8. These in silico analyses reveal the different intricate relationships between the miRNAs and their target transcripts. For example, in SUM149PT cells there is an inverse relationship between 5 miRNAs and 31 predicted target transcripts associated with M phase (GO:0000279), roughly one-third of the genes in the ontology modulated by CG-1521. Most of the mRNA transcripts are targeted by 2 or 3 miRNAs, suggesting that the miRNAs coordinate subtle changes in the steady-state levels of the transcripts. Since the miRNAs target different sites in the 3′UTR in individual transcripts, this provides a versatile regulatory network that can additively or synergistically influence the steady-state mRNA levels. At the other end of the spectrum, 4 mRNA transcripts associated with Microtubule Cytoskeleton Organization (GO:0000226) (KIF2C, MAP2, MAP1B and UBE2B), are inversely modulated by 10 miRNAs and each transcript is targeted by 2−3 miRNAs, suggesting that miRNA-mediated degradation of these transcripts results in rapid changes in transcript levels. The data presented in this manuscript indicate that CG-1521 modulates the transcription of a small group of miRNAs that affect the stability of transcripts essential for progression through M phase.
Figure 8. Concordant changes in miRNA and mRNA levels induced by CG-1521 were interrogated using Target Scan and David Bioinformatics to generate miRNA interaction webs for selected Gene Ontologies influenced by CG-1521. MicroRNA-mRNA webs for enriched GO terms for SUM149PT cells treated with CG-1521 (A and B); miRNA-mRNA webs for enriched GO terms in SUM190PT cells treated with CG-1521 (C–E). Pentagons, miRNA molecules; rectangles, mRNA transcripts. Red, upregulated; blue, downregulated.
Many of the genes that are downregulated by CG-1521 in SUM149PT cells are associated with the spindle checkpoint and organization of the microtubule cytoskeleton, which is concordant with the immunofluorescence data. CG-1521 downregulates numerous transcripts encoding proteins which are reported to accumulate at the midzone during late mitosis including: KIF23 (MKLP1), RACGAP1 (CYK4, MgcRacGAP), KIF20A (MKLP2), PLK1, Aurora B, PRC1, KIF4, ANLN (anillin), among others.35 Centralspindlin, composed of KIF23 and RACGAP1, is required for proper formation of the mitotic spindle and midbody. KIF20A binds the chromosomal passenger complex and plays an important role in the recruitment of PLK1 and Aurora B to the spindle.36 KIF4 was recently reported to mediate midzone elongation and localization of PRC1 at the midline. siRNA-mediated knockdown of KIF4 causes continued elongation of the midzone,37 similar to the phenotype observed in CG-1521 treated SUM149PT cells. Thus, the effects of CG-1521 appear to affect midzone length and subsequently prevent abscission in SUM149PT cells, most likely through the downregulation of several regulatory components necessary for these processes. These data also suggest that the effects of CG-1521 in these IBC cell lines is predominantly due to the impact on the steady-state levels of the transcripts involved in metaphase transition, rather than through the modulation of cyclins or cyclin dependent kinase inhibitors as is the case in many p53 wild-type cells.13,21 The fact that CG-1521 modulates the expression of fewer (and different genes) associated with M phase in SUM190PT is further indication that the drug targets different HDACs or influences different HDAC targets in the two cell lines.
In contrast to CG-1521, treatment with TSA does not induce the elongated midzone phenotype observed in SUM149PT cells. However, TSA induces significant cell death suggesting this effect is mediated by a different mechanism of action despite the fact that TSA modulates a similar complement of transcripts, albeit to a greater extent. In particular, Nek2 kinase is responsible for phosphorylation of Hec1, a component of the Ndc80 complex, which is required for normal chromosome alignment and signaling of the spindle assembly checkpoint.38 The absence of Nek2 kinase activity triggers defective chromosome alignment and cell death.38 The timing of these events may play a central role in selecting the central pathways leading to cell death; however the data are consistent with the notion that concerted transcriptional downregulation of genes responsible for spindle complex assembly and midbody formation by deacetylase inhibitors leads to abrogated abscission or defective chromosome alignment and cell death.
The increase in cell size in response to TSA treatment in SUM149PT cells has been observed in other cell lines, including NIH 3T3 cells. In these cells, TSA-induced inhibition of HDAC6 activity increased total adhesion area.18 HDAC6 is a member of the class II family of HDACs, whose major substrate is α-tubulin.39,40 This also highlights the fact that the phenotypic responses to HDAC inhibition are not necessarily mediated solely through transcriptional processes. In SUM149PT cells, treatment with either CG-1521 or TSA causes an increase in acetylated-α-tubulin suggesting that both compounds inhibit the activity of HDAC6, but elicit different biological responses. In SUM190PT cells, the failure to form elongated midbodies may be associated with minimal effects of the drug on HDAC6 in these cells. In addition, HDAC3-mediated deacetylation of Aurora B has been reported to cause a decline in mitotic kinase activity in PC-3 prostate cancer cells and knockdown of HDAC3 inhibits early mitotic progression and subsequently affects kinetochore-microtubule attachment resulting in chromosome misalignment.41 These data demonstrate that several HDACs are intimately involved in mitotic progression and highlight the need for comprehensive analysis of the changes in the acetyl proteome in response to HDAC inhibition in these cell lines.
In summary, CG-1521 induces cell death in both IBC cell lines, most likely through the transcriptional downregulation of a cohort of genes required for orderly spindle assembly, chromosome segregation and abscission. However, since histone deacetylases have been reported to target a significant number of non-histone proteins, including tubulin, it is likely that stabilization of acetylated components of the spindle assembly complex and midbody structures contributes to the failure to complete cytokinesis.42 Since CG-1521 demonstrates limited toxicity in vivo, it is a very promising HDAC inhibitor that has considerable potential for use in the treatment of IBC and other forms of breast cancer, including triple negative and HER2 amplified tumors.
Methods
Cell culture
SUM149PT and SUM190PT inflammatory breast cancer cell lines were purchased from Asterand Inc. SUM149PT cells were grown in Ham’s F-12 medium (Invitrogen) supplemented with 5% FBS (Sigma-Aldrich), 5 µg/mL insulin (Sigma-Aldrich), 1 µg/mL hydrocortisone (Sigma-Aldrich), 10 mM HEPES (Fisher Scientific, Pittsburgh, PA) and antimycotic/antibiotic (Sigma-Aldrich). SUM190PT cells were passaged and incubated in Ham’s F-12 medium (Invitrogen) supplemented with 2% FBS (Sigma-Aldrich), 5 µg/mL insulin (Sigma-Aldrich), 1 µg/mL hydrocortisone (Sigma-Aldrich), 10 mM HEPES (Fisher Scientific), 5 mM ethanolamine (Sigma-Aldrich), 5 µg/mL transferrin (Sigma-Aldrich), 10 nM triiodothyronine (Sigma-Aldrich), 50 nM sodium selenite (Sigma-Aldrich), 1 g/L bovine serum albumin (Sigma-Aldrich) and antimycotic/antibiotic (Sigma-Aldrich). After the cells attached to the culture dishes, cells were grown and maintained in the above media with no FBS. Cells were maintained at 37 °C in a humidified atmosphere of 95% air/5% CO2.
Crystal violet assay
SUM149PT and SUM190PT cells were seeded at 12 000 cells/well in 24-well plates for 48 h prior to treatment. Cells were treated with 0–2 µM Trichostatin A (TSA, Sigma-Aldrich) dissolved in DMSO and 0−10 µM CG-1521 (Errant Gene Therapeutics) dissolved in DMSO, alone or in combination with 10 nM 17β-estradiol (E2) (Sigma-Aldrich) dissolved in ethanol for 48 h. Cells were fixed with 2% glutaraldehyde (Fisher Scientific) in PBS for 20 min at room temperature, followed by staining with 0.1% crystal violet (Sigma-Aldrich) in distilled H2O for 30 min. After rinsing in distilled water, crystal violet stain was solubilized in 0.2% Triton X-100 (Sigma-Aldrich) in distilled water for 30 min with gentle shaking. The absorbance was measured with the Victor V 1420 Multilabel Counter (PerkinElmer Inc.) at 590 nm. Three independent biological replicates were analyzed in quadruplicate.
Flow cytometry
SUM149PT cells were treated with 7.5 µM CG-1521 or 100 nM TSA alone and in combination with 10 nM E2 for 24−48 h. SUM190PT cells were treated with 5 µM CG-1521 or 500 nM TSA alone and in combination with 10 nM E2 for 24−48 h. Cells treated with vehicle (EtOH or DMSO) served as the negative control. The apoptotic cell population was analyzed using Apo-BrdU staining of fragmented DNA. Treated cells were harvested by trypsinization and fixed with 4% formaldehyde in PBS for 20 min on ice, followed by permeabilization with 70% EtOH overnight at −20 °C. The 3′-OH ends of fragmented DNA were enzymatically labeled with bromodeoxyuridine triphosphate (Br-dUTP, Phoenix Flow Systems), using terminal transferase (Roche Applied Science) in TdT reaction buffer containing 2.5 mM cobalt chloride (Roche Applied Science) for 1 h at 37 °C. Br-dUTP labeled fragmented DNA was detected using a FITC-conjugated anti-BrdU monoclonal antibody (1:20, BD PharMingen). Cells were counterstained with 5 µg/mL propidium iodide (Sigma-Aldrich) in the presence of 0.015 U/mL RNase (Roche Applied Science) in PBS with 0.1% Triton X-100 for 30 min at room temperature. Samples were analyzed on a BD™ LSR II Flow Cytometer (BD Biosciences) within 4 h of labeling. Three independent biological replicates and a minimum of 10 000 events were analyzed for each experimental condition.
mRNA microarray analysis
SUM149PT and SUM190PT cells were plated at a density of 1.5 × 106 cells per 150 cm2 dish for 48 h prior to treatment with 7.5 µM and 5 µM CG-1521 respectively, either alone or in combination with 10 nM E2. After 48 h of treatment, cells were harvested by trypsinization and total RNA was extracted using mRNeasy mini kit (Qiagen). The quality and the concentrations of total RNA were assessed using the NanoDrop (Thermo Fisher Scientific) and Agilent Bioanalyzer (Agilent Technologies). Total RNA (100 ng) deemed to be of good quality (RIN greater than 8) was processed according to the standard Affymetrix Whole Transcript Sense Target labeling protocol (Affymetrix). The fragmented biotin-labeled cDNA from three independent biological replicates was hybridized over 16 h to Affymetrix Gene 1.0 ST arrays and scanned on an Affymetrix Scanner 3000 7G using AGCC software. The resulting CEL files were analyzed for quality using Affymetrix Expression Console software and were imported into GeneSpring GX11.5 (Agilent Technologies) where the data was quantile normalized using PLIER and baseline transformed to the median of the control samples. The probe sets were further filtered to exclude the bottom 20th percentile across all samples. The resulting entity list was then subjected to a unpaired t test with Benjamini-Hochberg false discovery rate correction and a 2.0-fold filter to identify differentially expressed transcripts between the conditions at a P value < 0.05. All data are MIAME compliant and the raw data has been deposited in Gene Expression Omnibus (GEO, GSE28542) as detailed on the Microarray Gene Expression Data Society (MGED) society website (http://www.mged.org/Workgroups/MIAME/miame.html). To identify pathways significantly modulated by CG-1521, data sets containing genes which passed the cut-off were uploaded into bioinformatics software [Ingenuity® Systems, www.ingenuity.com and the Database for Annotation, Visualization and Integrated Discovery (DAVID)]22,23 for analysis. A network was created for each cell line to illustrate significantly modulated genes within select GO terms regulated by CG-1521. The genes displayed within each GO term are based on relationships supported by curated knowledge bases from Ingenuity Pathway Analysis (IPA) and DAVID.
miRNA microarray analysis
SUM149PT and SUM190PT cells were plated at a density of 1.5 × 106 cells per 150 cm2 dish for 48 h prior to treatment with 7.5 µM and 5 µM CG-1521 respectively, either alone or in combination with 10 nM E2. After 48 h of treatment, cells were harvested by trypsinization and total RNA was extracted using miRNeasy mini kit (Qiagen). The quality and the concentrations of total RNA were assessed using the NanoDrop (Thermo Fisher Scientific) and Agilent Bioanalyzer (Agilent Technologies). Total RNA from four independent biological replicates was processed and hybridized to Agilent Human miRNA microarrays using standard protocols. Total RNA (100 ng) was dephosphorylated with calf intestinal phosphatase and end-labeled with Cy3-pCp by T4 RNA ligase prior to an overnight hybridization at 55 °C onto Agilent Human miRNA v3 (Sanger release 12.0) microarrays which contains probes sets for 866 human miRNAs. The arrays were washed and scanned on a high resolution GC2565CA Agilent Scanner using the manufacturer’s recommended settings. The raw data was extracted using Agilent Feature Extraction software v10.1.1 and imported into GeneSpring GX11.5 for further analysis. All data are MIAME compliant and the raw data has been deposited in Gene Expression Omnibus (GEO, GSE28543).
qPCR validation of mRNA and miRNA microarray data
To validate mRNA microarray data, candidate genes were selected and qPCR assays were conducted as described previously,43 using SYBR Green and validated primers (Table S2). Briefly, SUM149PT and SUM190PT cells were plated and treated for 12−72 h as previously described. Cells were harvested by trypsinization and total RNA was extracted using miRNeasy mini kit (Qiagen). Reverse transcription PCR reactions were performed with 1.5 μg total RNA using Taqman® Reverse Transcription Reagents (Applied Biosystems) to synthesize cDNA for mRNA expression analysis. The freaction mixture was incubated for 10 min at 25 °C, 1 h at 37 °C and 5 min at 95 °C and stored at -20 °C for further analysis. SYBR Green reactions with SYBR® Green PCR Master Mix (Applied Biosystems) were analyzed using the ABI 7900HT Fast Real-Time PCR System (Applied Biosystems): 50 °C for 2 min, 95 °C for 10 min, 95 °C for 15 sec and 60 °C for 1 min, repeated for 40 cycles. Relative expression levels of each gene compared with GAPDH in real time were analyzed using the 2-ΔΔCT method.44 Samples were assayed in duplicate from three or four independent sets of RNA preparations. Results are tabulated as mean (± SD) and are presented as fold change after transformation to show divergence from the vehicle control having no effect (zero fold change). To validate miRNA microarray data, candidate miRNAs were selected and qPCR assays were conducted using TaqMan® MicroRNA Assays according to the manufacturer’s protocol. Total RNA (10 ng) was used to make cDNA with TaqMan® MicroRNA Reverse Transcription Reagents (Applied Biosystems) for each selected miRNA. The reaction mixture was incubated for 30 min at 16 °C, 30 min at 42 °C and 5 min at 85 °C and kept at -20 °C for further analysis. TaqMan® reactions were analyzed using the ABI 7900HT Fast Real-Time PCR System: 95 °C for 10 min, 95 °C for 15 sec and 60 °C for 60 sec repeated for 40 cycles. Relative expression levels of each miRNA in real time were analyzed using the 2-ΔΔCT method with U6 snRNA as the endogenous control. Samples were assayed in duplicate from three or four independent sets of RNA preparations (RNA samples from mRNA qPCR validation). Results are tabulated as mean (± SD) and are presented as fold change after transformation to show divergence from the vehicle control having no effect (zero fold change).
Bioinformatics analysis of miRNA gene targets in CG-1521 treated IBC cells
After import into GeneSpring X11.5, the miRNA array data was log2 transformed and normalized to the 75th percentile. Entities with fold changes greater than 1.5 were considered significant (P < 0.05) using one way ANOVA with Benjamini-Hochberg FDR post-test correction. TargetScan® Human v6.6 from the Whitehead Institute for Biomedical Research was used to generate lists of theoretically conserved and non-conserved gene targets for each miRNA significantly regulated by CG-1521. The theoretical miRNA target lists were compared with the lists of mRNAs generated from the microarrays to identify transcripts inversely correlated with miRNA expression. The miRNA target list was then analyzed using DAVID Bioinformatics to identify pathways significantly affected by CG-1521 regulated miRNAs.45-49
Immunofluorescence microscopy
Cells were plated in chamber slides, treated as described above, fixed at room temperature for 10 min in 4% paraformaldehyde and permeabilized with 0.1% Triton X-100 in PBS for 5 min. Cells were stained with AlexaFluor 488 conjugated Phalloidin (Invitrogen) for 20 min at room temperature to visualize the actin cytoskeleton. After washing in PBS, slides were mounted with ProLong® Gold Reagent containing DAPI (Invitrogen).
To observe acetylated-α-tubulin in cells treated with CG-1521 and TSA, cells were fixed at room temperature for 10 min with 4% paraformaldehyde in PBS, permeabilized with 0.1% Triton X-100 in PBS for 5 min and blocked in protein block (DAKO) for 1h at room temperature. Human acetylated tubulin was detected using a mouse monoclonal antibody (1:100, Abcam, Cambridge, MA) and secondary tagged with AlexaFluor 488 F(ab’)2 Goat anti-mouse IgG (1:200, Invitrogen). After washing in PBS, slides were mounted with ProLong® Gold Reagent containing DAPI (Invitrogen). All imaging was performed using the Leica TCS SP5 Confocal Microscopy System. Images were captured under the same conditions to compare intensities with a scanning speed of 8000 Hz and image resolution of 512 × 512 pixels, and images were line averaged 32 times. The excitation power of the UV diode and argon laser was maintained at 20% and the gain, offset and pinhole were kept constant throughout the imaging.
To quantify elongated midzone structures, SUM149PT and SUM190PT cells were treated, stained with phalloidin and imaged as described above. Identifiers for each confocal image were removed and midzone structures were manually counted for three regions on two slides for each treatment. Images were imported into ImageJ software (http://rsb.info.nih.gov/ij/) and converted to binary images. The threshold was set to highlight nuclei and the “Analyze Particle” plug-in was used to count nuclei based on size. The number of elongated midzones is represented by the number of structures per 100 nuclei.
Supplementary Material
Acknowledgments
NC would like to thank the University at Albany Cancer Research Center’s Fund for Memory and Hope for support. WLWW would like to acknowledge the Congressionally Directed Medical Research Program for Prostate Cancer for Pre-doctoral support (W81XWH-11-1-0587).The authors acknowledge the outstanding help from David Frank and Marcy Kuentzel in the Center Functional Genomics, University at Albany.
Glossary
Abbreviations:
- IBC
inflammatory breast cancer
- HDAC
histone deacetylase
- HDACi
histone deacetylase inhibitor
- TSA
Trichostatin A
- SAHA
suberoylanilide hydroxamic acid
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/cbt/article/25088
References
- 1.Levine PH, Veneroso C. The epidemiology of inflammatory breast cancer. Semin Oncol. 2008;35:11–6. doi: 10.1053/j.seminoncol.2007.11.018. [DOI] [PubMed] [Google Scholar]
- 2.Robertson FM, Bondy M, Yang W, Yamauchi H, Wiggins S, Kamrudin S, et al. Inflammatory breast cancer: the disease, the biology, the treatment. CA Cancer J Clin. 2010;60:351–75. doi: 10.3322/caac.20082. [DOI] [PubMed] [Google Scholar]
- 3.Hance KW, Anderson WF, Devesa SS, Young HA, Levine PH. Trends in inflammatory breast carcinoma incidence and survival: the surveillance, epidemiology, and end results program at the National Cancer Institute. J Natl Cancer Inst. 2005;97:966–75. doi: 10.1093/jnci/dji172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wingo PA, Jamison PM, Young JL, Gargiullo P. Population-based statistics for women diagnosed with inflammatory breast cancer (United States) Cancer Causes Control. 2004;15:321–8. doi: 10.1023/B:CACO.0000024222.61114.18. [DOI] [PubMed] [Google Scholar]
- 5.Forozan F, Veldman R, Ammerman CA, Parsa NZ, Kallioniemi A, Kallioniemi OP, et al. Molecular cytogenetic analysis of 11 new breast cancer cell lines. Br J Cancer. 1999;81:1328–34. doi: 10.1038/sj.bjc.6695007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hollestelle A, Nagel JHA, Smid M, Lam S, Elstrodt F, Wasielewski M, et al. Distinct gene mutation profiles among luminal-type and basal-type breast cancer cell lines. Breast Cancer Res Treat. 2010;121:53–64. doi: 10.1007/s10549-009-0460-8. [DOI] [PubMed] [Google Scholar]
- 7.Fukuda H, Sano N, Muto S, Horikoshi M. Simple histone acetylation plays a complex role in the regulation of gene expression. Brief Funct Genomic Proteomic. 2006;5:190–208. doi: 10.1093/bfgp/ell032. [DOI] [PubMed] [Google Scholar]
- 8.Horn PJ, Peterson CL. Molecular biology. Chromatin higher order folding--wrapping up transcription. Science. 2002;297:1824–7. doi: 10.1126/science.1074200. [DOI] [PubMed] [Google Scholar]
- 9.Arnold NB, Arkus N, Gunn J, Korc M. The histone deacetylase inhibitor suberoylanilide hydroxamic acid induces growth inhibition and enhances gemcitabine-induced cell death in pancreatic cancer. Clin Cancer Res. 2007;13:18–26. doi: 10.1158/1078-0432.CCR-06-0914. [DOI] [PubMed] [Google Scholar]
- 10.Chang J, Varghese DS, Gillam MC, Peyton M, Modi B, Schiltz RL, et al. Differential response of cancer cells to HDAC inhibitors trichostatin A and depsipeptide. Br J Cancer. 2012;106:116–25. doi: 10.1038/bjc.2011.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma X, Fang Y, Beklemisheva A, Dai W, Feng J, Ahmed T, et al. Phenylhexyl isothiocyanate inhibits histone deacetylases and remodels chromatins to induce growth arrest in human leukemia cells. Int J Oncol. 2006;28:1287–93. [PubMed] [Google Scholar]
- 12.Marrocco DL, Tilley WD, Bianco-Miotto T, Evdokiou A, Scher HI, Rifkind RA, et al. Suberoylanilide hydroxamic acid (vorinostat) represses androgen receptor expression and acts synergistically with an androgen receptor antagonist to inhibit prostate cancer cell proliferation. Mol Cancer Ther. 2007;6:51–60. doi: 10.1158/1535-7163.MCT-06-0144. [DOI] [PubMed] [Google Scholar]
- 13.Roy S, Packman K, Jeffrey R, Tenniswood M. Histone deacetylase inhibitors differentially stabilize acetylated p53 and induce cell cycle arrest or apoptosis in prostate cancer cells. Cell Death Differ. 2005;12:482–91. doi: 10.1038/sj.cdd.4401581. [DOI] [PubMed] [Google Scholar]
- 14.Vanhaecke T, Henkens T, Kass GEN, Rogiers V. Effect of the histone deacetylase inhibitor trichostatin A on spontaneous apoptosis in various types of adult rat hepatocyte cultures. Biochem Pharmacol. 2004;68:753–60. doi: 10.1016/j.bcp.2004.05.022. [DOI] [PubMed] [Google Scholar]
- 15.Gu W, Roeder RG. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/S0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 16.Roy S, Tenniswood M. Site-specific acetylation of p53 directs selective transcription complex assembly. J Biol Chem. 2007;282:4765–71. doi: 10.1074/jbc.M609588200. [DOI] [PubMed] [Google Scholar]
- 17.Sykes SM, Mellert HS, Holbert MA, Li K, Marmorstein R, Lane WS, et al. Acetylation of the p53 DNA-binding domain regulates apoptosis induction. Mol Cell. 2006;24:841–51. doi: 10.1016/j.molcel.2006.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tang Y, Zhao W, Chen Y, Zhao Y, Gu W. Acetylation is indispensable for p53 activation. Cell. 2008;133:612–26. doi: 10.1016/j.cell.2008.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang C, Fu M, Angeletti RH, Siconolfi-Baez L, Reutens AT, Albanese C, et al. Direct acetylation of the estrogen receptor alpha hinge region by p300 regulates transactivation and hormone sensitivity. J Biol Chem. 2001;276:18375–83. doi: 10.1074/jbc.M100800200. [DOI] [PubMed] [Google Scholar]
- 20.Patel J, Pathak RR, Mujtaba S. The biology of lysine acetylation integrates transcriptional programming and metabolism. Nutr Metab (Lond) 2011;8:12. doi: 10.1186/1743-7075-8-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Knutson AK, Welsh J, Taylor T, Roy S, Wang WL, Tenniswood M. Comparative effects of histone deacetylase inhibitors on p53 target gene expression, cell cycle and apoptosis in MCF-7 breast cancer cells. Oncol Rep. 2012;27:849–53. doi: 10.3892/or.2011.1590. [DOI] [PubMed] [Google Scholar]
- 22.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 23.Huang W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Close P, Creppe C, Gillard M, Ladang A, Chapelle JP, Nguyen L, et al. The emerging role of lysine acetylation of non-nuclear proteins. Cell Mol Life Sci. 2010;67:1255–64. doi: 10.1007/s00018-009-0252-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frew AJ, Johnstone RW, Bolden JE. Enhancing the apoptotic and therapeutic effects of HDAC inhibitors. Cancer Lett. 2009;280:125–33. doi: 10.1016/j.canlet.2009.02.042. [DOI] [PubMed] [Google Scholar]
- 26.Linares A, Dalenc F, Balaguer P, Boulle N, Cavailles V. Manipulating protein acetylation in breast cancer: a promising approach in combination with hormonal therapies? J Biomed Biotechnol. 2011;2011:856985. doi: 10.1155/2011/856985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Duong V, Bret C, Altucci L, Mai A, Duraffourd C, Loubersac J, et al. Specific activity of class II histone deacetylases in human breast cancer cells. Mol Cancer Res. 2008;6:1908–19. doi: 10.1158/1541-7786.MCR-08-0299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robertson FM, Woodward WA, Pickei R, Ye Z, Bornmann W, Pal A, et al. Suberoylanilide hydroxamic acid blocks self-renewal and homotypic aggregation of inflammatory breast cancer spheroids. Cancer. 2010;116(Suppl):2760–7. doi: 10.1002/cncr.25176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anderson WF, Schairer C, Chen BE, Hance KW, Levine PH. Epidemiology of inflammatory breast cancer (IBC) Breast Dis. 2005-2006;22:9–23. doi: 10.3233/bd-2006-22103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Galluzzi L, Vitale I, Abrams JM, Alnemri ES, Baehrecke EH, Blagosklonny MV, et al. Molecular definitions of cell death subroutines: recommendations of the Nomenclature Committee on Cell Death 2012. Cell Death Differ. 2012;19:107–20. doi: 10.1038/cdd.2011.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roy S, Jeffrey R, Tenniswood M. Array-based analysis of the effects of trichostatin A and CG-1521 on cell cycle and cell death in LNCaP prostate cancer cells. Mol Cancer Ther. 2008;7:1931–9. doi: 10.1158/1535-7163.MCT-07-2353. [DOI] [PubMed] [Google Scholar]
- 32.Dai C, Gu W. p53 post-translational modification: deregulated in tumorigenesis. Trends Mol Med. 2010;16:528–36. doi: 10.1016/j.molmed.2010.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Popov VM, Wang C, Shirley LA, Rosenberg A, Li S, Nevalainen M, et al. The functional significance of nuclear receptor acetylation. Steroids. 2007;72:221–30. doi: 10.1016/j.steroids.2006.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vigushin DM, Coombes RC. Targeted histone deacetylase inhibition for cancer therapy. Curr Cancer Drug Targets. 2004;4:205–18. doi: 10.2174/1568009043481560. [DOI] [PubMed] [Google Scholar]
- 35.Steigemann P, Gerlich DW. Cytokinetic abscission: cellular dynamics at the midbody. Trends Cell Biol. 2009;19:606–16. doi: 10.1016/j.tcb.2009.07.008. [DOI] [PubMed] [Google Scholar]
- 36.Douglas ME, Mishima M. Still entangled: assembly of the central spindle by multiple microtubule modulators. Semin Cell Dev Biol. 2010;21:899–908. doi: 10.1016/j.semcdb.2010.08.005. [DOI] [PubMed] [Google Scholar]
- 37.Hu CK, Coughlin M, Field CM, Mitchison TJ. KIF4 regulates midzone length during cytokinesis. Curr Biol. 2011;21:815–24. doi: 10.1016/j.cub.2011.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wei R, Ngo B, Wu G, Lee WH. Phosphorylation of the Ndc80 complex protein, HEC1, by Nek2 kinase modulates chromosome alignment and signaling of the spindle assembly checkpoint. Mol Biol Cell. 2011;22:3584–94. doi: 10.1091/mbc.E11-01-0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hubbert C, Guardiola A, Shao R, Kawaguchi Y, Ito A, Nixon A, et al. HDAC6 is a microtubule-associated deacetylase. Nature. 2002;417:455–8. doi: 10.1038/417455a. [DOI] [PubMed] [Google Scholar]
- 40.Matsuyama A, Shimazu T, Sumida Y, Saito A, Yoshimatsu Y, Seigneurin-Berny D, et al. In vivo destabilization of dynamic microtubules by HDAC6-mediated deacetylation. EMBO J. 2002;21:6820–31. doi: 10.1093/emboj/cdf682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fadri-Moskwik M, Weiderhold KN, Deeraksa A, Chuang C, Pan J, Lin SH, et al. Aurora B is regulated by acetylation/deacetylation during mitosis in prostate cancer cells. FASEB J. 2012;26:4057–67. doi: 10.1096/fj.12-206656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gabrielli B, Brown M. Histone deacetylase inhibitors disrupt the mitotic spindle assembly checkpoint by targeting histone and nonhistone proteins. Adv Cancer Res. 2012;116:1–37. doi: 10.1016/B978-0-12-394387-3.00001-X. [DOI] [PubMed] [Google Scholar]
- 43.Wang WL, Chatterjee N, Chittur SV, Welsh J, Tenniswood MP. Effects of 1α,25 dihydroxyvitamin D3 and testosterone on miRNA and mRNA expression in LNCaP cells. Mol Cancer. 2011;10:58. doi: 10.1186/1476-4598-10-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 45.Perreard L, Fan C, Quackenbush JF, Mullins M, Gauthier NP, Nelson E, et al. Classification and risk stratification of invasive breast carcinomas using a real-time quantitative RT-PCR assay. Breast Cancer Res. 2006;8:R23. doi: 10.1186/bcr1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cui W, Taub DD, Gardner K. qPrimerDepot: a primer database for quantitative real time PCR. Nucleic Acids Res. 2007;35(Database issue):D805–9. doi: 10.1093/nar/gkl767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhu C, Zhao J, Bibikova M, Leverson JD, Bossy-Wetzel E, Fan J-B, et al. Functional analysis of human microtubule-based motor proteins, the kinesins and dyneins, in mitosis/cytokinesis using RNA interference. Mol Biol Cell. 2005;16:3187–99. doi: 10.1091/mbc.E05-02-0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nabetani A, Koujin T, Tsutsumi C, Haraguchi T, Hiraoka Y. A conserved protein, Nuf2, is implicated in connecting the centromere to the spindle during chromosome segregation: a link between the kinetochore function and the spindle checkpoint. Chromosoma. 2001;110:322–34. doi: 10.1007/s004120100153. [DOI] [PubMed] [Google Scholar]
- 49.Horvath S, Zhang B, Carlson M, Lu KV, Zhu S, Felciano RM, et al. Analysis of oncogenic signaling networks in glioblastoma identifies ASPM as a molecular target. Proc Natl Acad Sci U S A. 2006;103:17402–7. doi: 10.1073/pnas.0608396103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.