Abstract
Modification of damaged replication forks is emerging as a crucial factor for efficient chromosomal duplication and the avoidance of genetic instability. The RecG helicase of Escherichia coli, which is involved in recombination and DNA repair, has been postulated to act on stalled replication forks to promote replication restart via the formation of a four-stranded (Holliday) junction. Here we show that RecG can actively unwind the leading and lagging strand arms of model replication fork structures in vitro. Unwinding is achieved in each case by simultaneous interaction with and translocation along both the leading and lagging strand templates at a fork. Disruption of either of these interactions dramatically inhibits unwinding of the opposing duplex arm. Thus, RecG translocates simultaneously along two DNA strands, one with 5′-3′ and the other with 3′-5′ polarity. The unwinding of both nascent strands at a damaged fork, and their subsequent annealing to form a Holliday junction, may explain the ability of RecG to promote replication restart. Moreover, the preferential binding of partial forks lacking a leading strand suggests that RecG may have the ability to target stalled replication intermediates in vivo in which lagging strand synthesis has continued beyond the leading strand.
Keywords: DNA repair, recombination, helicases
Efficient replication of the genome is essential for growth and survival of all organisms. Yet polymerase complexes assembled at origins of replication often fail to complete the task (1). Despite the intrinsic processivity of these complexes, the advance of replication forks is hindered by lesions or troublesome sequences in the template DNA and by protein complexes associated with gene expression and DNA packaging (2–4). Indeed, they often stall and may collapse to generate new DNA ends that provoke recombination and induce genomic rearrangements (5). Complete replication of the genome depends therefore on repair activities to remove or bypass lesions in the DNA, modulation of RNA polymerase to reduce conflicts with transcription, and recombination systems to rescue forks that have stalled or collapsed (6–8).
Recent studies in Escherichia coli have revealed that a major pathway for dealing with blocked replication complexes involves formation of a Holliday junction via regression of the fork (5, 8). This regression (Fig. 1A) may facilitate removal of the lesion and allow the damaged fork to be targeted by the Holliday junction specific helicase-endonuclease activity of RuvABC. Cleavage of the junction by RuvABC collapses the fork to generate a duplex DNA end [Fig. 1A (i)]. Processing of this DNA end by the helicase-exonuclease activity of RecBCD enzyme followed by RecA-mediated strand exchange with an intact homologous duplex (9) creates a recombination intermediate (D-loop structure) that can be bound by the primosome assembly factor PriA to catalyze assembly of the replicative apparatus (10, 11). Replication would therefore be reestablished. The potentially dangerous act of introducing a break into the chromosome may therefore be used to promote fork progression.
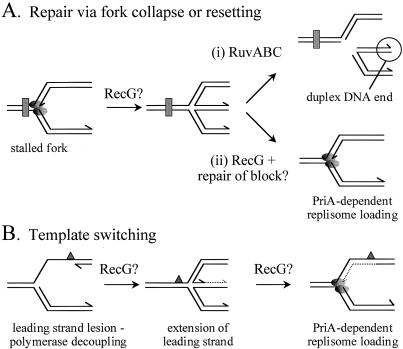
Figure 1.
Models of replication restart promoted by RecG and RuvABC. The shaded ovals represent the leading and lagging strand polymerases of a replication fork. The rectangle represents a lesion affecting both leading and lagging strand synthesis, whereas a lesion specifically in the leading strand template is shown as a triangle. New DNA synthesis utilizing the lagging strand as a template is shown as a dotted line. Arrowheads depict the 3′-OH groups of the leading and lagging strands. All other details are described in the text.
Formation of a Holliday junction by regression of a stalled fork requires unwinding of the newly replicated arms of the fork, annealing of the nascent strands, and reannealing of the parental strands. Studies of plasmid DNA replication in vitro have shown that such regression may occur spontaneously when positive torsional strain is chemically induced within the partially replicated plasmid (12). It may also be driven enzymatically. In vivo evidence suggests that regression of damaged forks to form Holliday junctions might involve the strand exchange activity of RecA (13). However, it is not known whether RecA-mediated strand exchange can catalyze the regression of stalled replication forks directly. A second enzymatic mechanism for fork regression utilizes the monomeric helicase RecG. We have shown that RecG can stimulate Holliday junction-specific endonucleases to cleave model fork structures in vitro, and that RecG is required to promote formation of Holliday junctions from damaged replication forks in vivo (8, 14). Moreover, we have also shown that RecG can unwind true replication forks reconstituted in vitro and that it can overcome the energetic barrier to fork regression when the fork is in a region of negative superhelicity (34). However, the role of RecG is not simply to process stalled forks into substrates for RuvABC [Fig. 1A (i)]. In conjunction with PriA, it appears to facilitate fork progression without the need for RuvABC-catalyzed cleavage of the chromosome and subsequent formation of a D-loop by recombination (8). This might involve RecG-catalyzed regression of a stalled fork to promote access of repair enzymes to the blocking lesion, followed by reversal of the regression to reestablish a fork structure that can be targeted directly by PriA [Fig. 1A (ii)]. RecG may also be important when a lesion affecting only a single strand of the parental duplex is encountered by a fork. If such a lesion is present on the lagging strand template then the replisome may prematurely terminate an Okazaki fragment at the lesion and continue synthesis downstream (15), leaving a gap that can then be repaired by RecA-dependent mechanisms (16). A lesion in the leading strand template presents a different problem because the two polymerases might decouple, allowing lagging strand synthesis to continue some way beyond the block (15, 17, 18). If such a process does occur, then the result would be a gap in the leading strand (Fig. 1B). In such a case, we have proposed that RecG might promote template switching via a mechanism in which regression of the stalled fork to form a Holliday junction allows the lagging strand to act as a template for leading strand synthesis (8). Assuming regression could be reversed, a normal fork might be reestablished via PriA. Such mechanisms would allow replication to proceed without the need for RuvABC-dependent cleavage of the fork.
Collapse of the fork appears to be essential when a fork encounters a lesion that blocks progression of the replicative helicase, DnaB, and which therefore might necessitate recombination with a sister duplex to bypass the lesion (8). However, chromosomal breakage is a dangerous process because there is a risk that the free DNA end will undergo illegitimate recombination leading to potentially fatal genome rearrangements. RecG provides the cell with a second pathway for replication restart that may allow replication to continue in the face of lesions affecting a single strand of the template, but which does not necessitate breakage of the fork (8).
The modification of replication fork structures is emerging as a crucial factor in the maintenance of fork progression in E. coli. These processes may also be essential in eukaryotes. Holliday junctions have been directly observed in yeast within rDNA (19) and are coincident with the position of a preprogrammed replication fork block. Moreover, such blocks may coincide with hot spots of recombination and therefore of genome instability (20). Indeed, accumulating evidence suggests that genetic instabilities seen in certain human diseases may be attributable to aberrant processing of stalled replication forks (21).
The precise roles of RecG and RuvABC at stalled forks remain to be determined. Here we show that RecG actively unwinds the leading and lagging strands of partial fork structures in vitro by simultaneous translocation along the leading and lagging strand templates at the fork. Disruption of one of these translocation activities leads to a dramatic inhibition of the other, as revealed by the decrease in unwinding of the relevant duplex arm at the fork. Thus, RecG employs a mechanism that involves simultaneous tracking along two DNA strands with opposing polarities. In vivo, such a reaction may promote formation of a Holliday junction from a damaged fork. These data support a model in which RecG is a replication fork-specific helicase that modulates the structure of a stalled fork to facilitate replication restart.
Materials and Methods
DNA Substrates.
χSma DNA was prepared as described (8). Small forked DNA junctions were constructed by using oligonucleotides, one of which in each structure was labeled with [γ32P]ATP at the 5′ end, and purified by gel electrophoresis (22). Sequences of the oligonucleotides, written 5′-3′, are: (a) GTCGGATCCTCTAGACAGCTCCATGATCACTGGCACTGGTAGAATTCGGC; (b) CAACGTCATAGACGATTACATTGCTACATGGAGCTGTCTAGAGGATCCGA; (c) TGCCGAATTCTACCAGTGCCAGTGAT; (d) TAGCAATGTAATCGTCTATGACGTT; (e) CAACGTCATAGACGATTACATTGCTACATGGAGCTGTCTAGAGGATCCGA; (f) CAACGTCATAGACGATTACATTGCTACATGGAGCTGTCTAGAGGATCCGA; (g) GTCGGATCCTCTAGACAGCTCCATGATCACTGGCACTGGTAGAATTCGGC; (h) GTCGGATCCTCTAGACAGCTCCATGATCACTGGCACTGGTAGAATTCGGC; (i) ACGATTACATTGCTACATGGAGCTGTCTAGAGGATCCGA; (j) TACATTGCTACATGGAGCTGTCTAGAGGATCCGA; (k) GTCGGATCCTCTAGACAGCTCCATGATCACTGGCACTGGT; (l) GTCGGATCCTCTAGACAGCTCCATGATCACTGGCA; (m) TAGCAATGTAATCGTCTATG; (n) ATTCTACCAGTGCCAGTGAT. Italic nucleotides indicate those in which the polarity of the phosphate backbone was reversed with respect to the flanking sequences. These oligonucleotides were synthesized by using 5′ cyanoethyl phosphoramidites (Glen Research, Sterling, VA). Oligonucleotide d contained a 5′ terminal phosphate group made by using Phosphalink (Perkin–Elmer).
The small forks in Figs. 3 and 7 were made by annealing combinations of a, b, c, and d. Junctions having a region of reverse polarity in the lagging strand template (Fig. 4A) were made by using a, b, and c (no reversal of polarity); a, c, and e (six-base reversal); a, c, and f (12-base reversal). Junctions having a region of reverse polarity in the leading strand template (Fig. 4B) were made by using a, b, and d (no reversal); b, d, and g (six-base reversal); b, d, and h (12-base reversal). Junctions with truncations in the lagging strand template (Fig. 5A) were constructed by annealing a, b, and c (no truncation, 26-base arm); a, c, and i (15-base arm); a, c, and j (ten-base arm). Junctions with truncations in the leading strand template (Fig. 5B) were made by using a, b, and d (no truncation, 25-base arm); b, d, and k (15-base arm); b, d, and l (ten-base arm). Junctions used in Fig. 6A were constructed with (i) a, b, and d; (ii) a, b, and m; (iii) b, d, and l; and (iv) b, l, and m. Junctions in Fig. 6B were constructed with (i) a, b, and c; (ii) a, b, and n; (iii) a, c, and j; and (iv) a, j, and n.
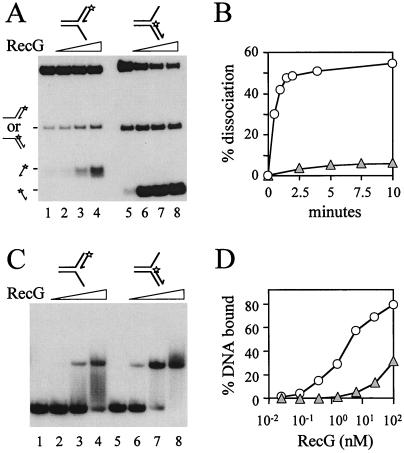
Figure 3.
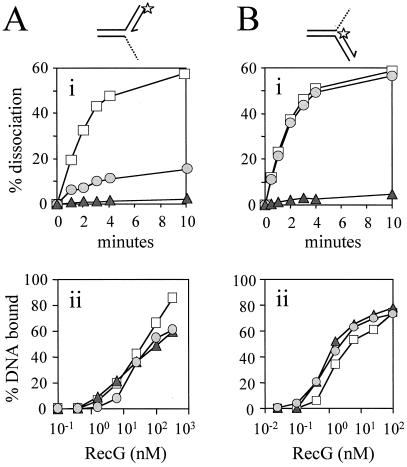
Targeting of forked DNA structures by RecG. (A) Unwinding of fork structures by 0, 0.5, 5, and 50 nM RecG. Junction DNA was present at an initial concentration of 20 nM. 3′ ends of oligonucleotides are indicated by arrows and the positions of 5′ 32P labels are marked with asterisks. The products of RecG catalysis are represented on the left. (B) Rates of RecG-catalyzed accumulation of DNA products from fork structures. Shaded triangles represent unwinding of the leading strand fork (A, lanes 1–4). Open circles represent unwinding of the lagging strand fork (A, lanes 5–7). The concentrations of DNA and of RecG were 20 nM and 5 nM, respectively. (C) Binding of partial forks by RecG. Band-shift assays were performed with 0.2 nM DNA and 0, 0.5, 5, and 50 nM RecG. (D) Quantification of binding of forks by RecG. Shaded triangles represent binding of the leading strand fork (C, lanes 1–4). Open circles represent binding of the lagging strand fork (C, lanes 5–8). DNA was present at 0.2 nM.
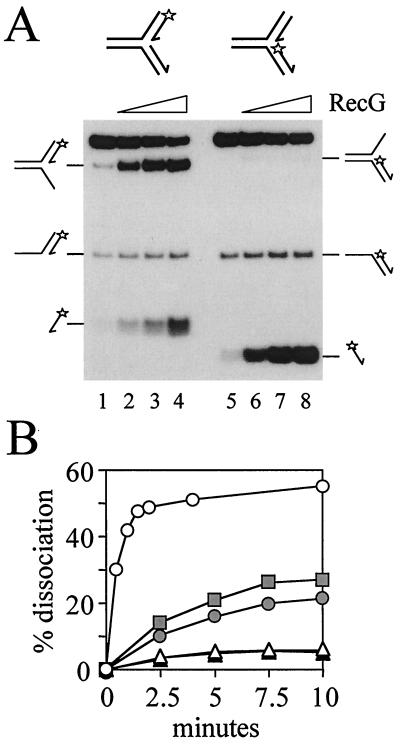
Figure 7.
Unwinding of fork structures containing both a leading and a lagging strand by RecG. (A) Forks containing both leading and lagging strand duplexes of 25 bp, having either a 5′ 32P-labeled leading (lanes 1–4) or lagging strand (lanes 5–8), were unwound by 0, 0.5, 5, or 50 nM RecG. Forks were at 20 nM. Products of RecG catalysis are indicated. (B) Rates of RecG-catalyzed accumulation of products of unwinding forks containing both a leading and a lagging strand (A, lanes 1–4). The rates of accumulation of the labeled three-strand product (shaded circle), produced by unwinding of the lagging strand, and the labeled single-stranded oligonucleotide (shaded triangle), produced by unwinding the leading strand, are indicated. The total accumulation of both products is also marked (shaded squares). For comparison, the rates of accumulation of labeled products by RecG catalysis at forks containing a leading strand only (open triangles; Fig. 3A, lanes 1–4) or a lagging strand only (open circles; Fig. 3A, lanes 5–8) are also shown. The concentrations of RecG and fork DNA were 5 and 20 nM, respectively.
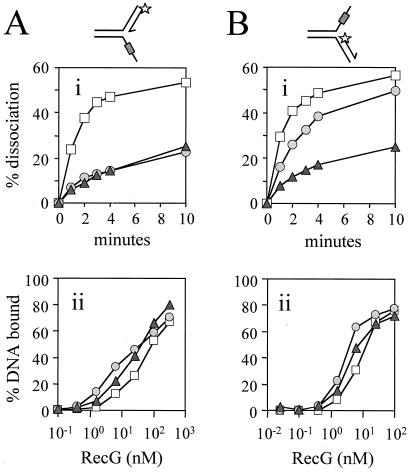
Figure 4.
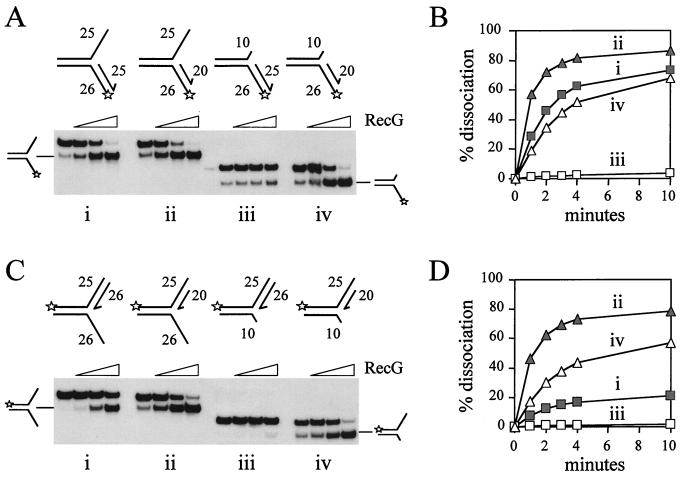
Effect of regions of reverse polarity in the template strands on unwinding of the opposing duplex arm. (A) Effect of regions of reverse backbone polarity within the lagging strand template. (i) Rate of RecG-catalyzed accumulation of unwound leading strand from junctions that had no bases (squares), six bases (circles), or 12 bases (triangles) of reverse polarity within the lagging strand template. Each region of reverse polarity was positioned ten bases away from the junction point. Junction DNA was at a concentration of 0.2 nM and RecG was at 100 nM. (ii) Binding of the forks used in (Ai) as measured by band-shift assays. Fork DNA was at a concentration of 0.2 nM. (B) Effect of regions of reverse backbone polarity within the leading strand template. (i) Rate of RecG-catalyzed accumulation of unwound lagging strand from junctions that had no bases (squares), six bases (circles), or 12 bases (triangles) of reverse polarity within the leading strand template. Each region of reverse polarity was positioned ten bases away from the junction point. Junction DNA was at a concentration of 0.2 nM and RecG was at 1 nM. (ii) Binding of the forks used in (Bi) as measured by band-shift assays. Fork DNA was at a concentration of 0.2 nM.
Figure 5.
Effect of truncation of template arms on unwinding of forks by RecG. (A) Truncation of the lagging strand template. (i) Rate of RecG-catalyzed accumulation of products of unwinding partial forks having a leading strand duplex of 25 bp with lagging strand template arms of 26, (squares), 15 (circles), and ten (triangles) bases. RecG and junction DNA were present at 50 nM and 0.2 nM respectively. (ii) Binding of the forks used in (Ai) as measured by band-shift assays using 0.2 nM fork DNA. (B) Truncation of the leading strand template. (i) Rate of RecG-catalyzed accumulation of products of unwinding partial forks having a lagging strand duplex of 25 bp with leading strand template arms of 25 (squares), 15 (circles), and ten (triangles) bases. RecG and junction DNA were both present at 0.2 nM. (ii) Binding of the forks used in (Bi) as measured by band-shift assays using 0.2 nM fork DNA.
Figure 6.
Effect of relative lengths of the template arm versus the duplex to be unwound. (A) Unwinding of lagging strand forks by RecG. Lengths of strands within each fork are indicated in nucleotides. The products of RecG catalysis are indicated. RecG was present at 0, 0.05, 0.5, and 5 nM and each junction was at a concentration of 1 nM. (B) Rates of RecG-catalyzed accumulation of products of unwinding lagging strands from the forks (i–iv) shown in A. The concentrations of RecG and fork DNA were 0.5 and 1 nM, respectively. (C) Unwinding of leading strand forks by RecG. Concentrations of RecG and junction DNA are as in A. Note that, because of the original design of these structures, although the leading strand oligonucleotide is 26 bases, the leading strand template is only 25 bases. Therefore, the leading strand duplex formed is 25 bp, which is identical to the full-length lagging strand duplex in A. (D) Rates of accumulation of products of unwinding leading strands from the forks (i–iv) shown in C. Concentrations of RecG and fork DNA are as in B.
Proteins.
Wild-type RecG and RecGK302A were purified as described (23). All concentrations are expressed in terms of protein monomer.
Enzyme Assays.
The stimulation of RuvC cleavage of χSma by RecG was measured as described (8). Reactions were incubated for 30 min at 37°C before deproteinization.
Dissociation of small oligonucleotide junction structures was performed as described (23), except that the buffer system was 50 mM Tris⋅acetate (pH 8), 20 mM potassium acetate, 1 mM DTT, and 0.1 mg/ml BSA. ATP and magnesium acetate were both used at a concentration of 5 mM. Reactions in which increasing concentrations of RecG were titrated against junction DNA were incubated for 30 min at 37°C before deproteinization. Band shift assays were used to measure binding of RecG to 0.2 nM fork structures in the presence of EDTA (23). All data shown are the means of at least two independent experiments.
Results
RecG Helicase Activity Is Required to Generate Holliday Junctions from Forked DNA.
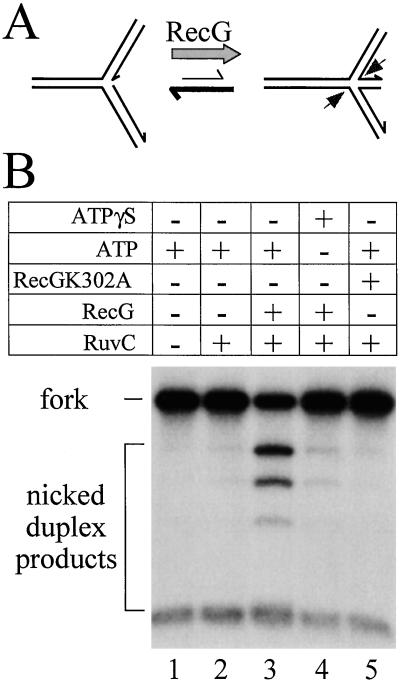
RecG has been shown to stimulate cleavage of forked DNA substrates by Holliday junction-specific endonucleases such as RuvC in vitro (8). This observation suggested that RecG might be able to unwind fork structures to generate Holliday junctions (Fig. 2A). However, it was also possible that RecG simply bound to and stabilized Holliday junctions formed spontaneously from forked DNA so as to facilitate cleavage by RuvC. To distinguish between these possibilities we tested whether ATP hydrolysis, and therefore helicase activity, was required to obtain RecG-specific stimulation of fork cleavage by RuvC. In the presence of ATP, RecG enhanced RuvC cleavage of χSma fork DNA ≈15-fold (Fig. 2B, lanes 2 and 3). However, this stimulation was completely abolished when ATP was replaced with the poorly hydrolyzable analogue ATPγS (Fig. 2B, lane 4). Moreover, a mutant RecG protein defective in ATP hydrolysis, but which retains the ability to bind to both fork and Holliday junction structures with the same affinity as wild-type enzyme (ref. 14 and data not shown) failed to stimulate cleavage by RuvC (Fig. 2B, lane 5). Thus, the observed stimulation of RuvC cleavage is not due to the binding and stabilization of preexisting Holliday junctions by RecG. It requires the active unwinding of the forked DNA substrate by RecG.
Figure 2.
Stimulation of RuvC cleavage of forked DNA requires RecG helicase activity. (A) Schematic representation of the junction structures that χSma DNA adopts in vitro. The junction is located within a 300-bp region of homology flanked by heterologous duplex arms of 0.8–1.6 kb. χSma preferentially adopts a fork structure, but the junction point has the ability to branch migrate within the homologous core to form a Holliday junction. RecG has been proposed to unwind the fork structure to generate a Holliday junction that can then be cleaved symmetrically (arrows) by junction-specific endonucleases such as RuvC. (B) Cleavage of χSma fork DNA by RuvC and the effects on cleavage of the presence or absence of RecG helicase activity. Reactions contained 5 mM ATP or ATPγS, 10 nM RuvC, 1 nM RecG, and 1 nM RecGK302A as indicated.
Unwinding of the Leading and Lagging Strands at Fork Structures by RecG.
The ability of RecG to generate a Holliday junction from fork DNA raised the possibility that it may be able to unwind both the leading and lagging strands of a replication fork. Simultaneous unwinding would facilitate reannealing of the template strands and annealing of the nascent strands. However, it is also possible that RecG unwinds just one of the nascent strands. If spontaneous branch migration then resulted in the second strand being displaced, the subsequent annealing of this strand with the first strand unwound would also generate a Holliday junction. To investigate these possibilities, small forks with only leading or lagging strands were constructed to determine whether RecG could target either strand. The junctions constructed did not contain any regions of homology, unlike fork structures in vivo. Thus, none of the unwound strands at these forks had the ability to anneal together. However, the use of heterologous forks was essential to prevent spontaneous branch migration and to ensure, therefore, that the junctions had a defined structure.
Both the leading and lagging strands could be unwound from the fork structures (Fig. 3A). This suggested that RecG could actively unwind both strands at a fork to generate a Holliday junction. However, with the particular substrates made, the lagging strand was unwound at a rate ≈40-fold higher than the leading strand (Fig. 3B). Indeed, RecG bound with higher affinity to the fork containing the lagging strand rather than the fork with the leading strand (Fig. 3 C and D), which correlated with the observed rates of unwinding the two forks. This raised the possibility that RecG specifically targets the lagging strand rather than the leading strand at a fork. Taken together, the data in Fig. 3 cannot distinguish between the two possible mechanisms, outlined above, by which RecG could generate Holliday junctions from forks. We therefore decided to pursue the strand specificity of RecG by analyzing the contribution of one arm of the fork to the unwinding of the opposing arm.
RecG Translocates in a Polar Manner Along Both Template Arms of a Fork.
Helicases display a defined polarity with respect to the direction of movement along ssDNA to effect unwinding of partial duplex products. In such assays RecG translocates 3′-5′ along ssDNA (24). Translocation of RecG in a 3′-5′ direction along the lagging strand template may indeed explain its ability to unwind the lagging strand. However, the data in Fig. 3 imply that RecG may also move in a 5′-3′ direction with respect to the leading strand template to unwind the leading strand. Moreover, because RecG is a branched DNA-specific helicase (24), the implication is that RecG may interact with both arms of a fork simultaneously.
To test whether RecG can indeed move simultaneously along both arms of a fork with defined polarities, we embedded regions of reverse backbone polarity within the single-stranded arms of partial forks. A reversal of polarity should inhibit translocation if the polarity of the phosphate backbone of the DNA is important for translocation (25). Moreover, if RecG does move simultaneously along both arms of the fork, then blockage of translocation along the single-stranded template arm might be predicted to block translocation along and therefore unwinding of the opposing duplex arm. However, to test whether RecG did translocate along both arms of the fork it was important that the initial binding of RecG to these junctions was not inhibited. Because RecG binds specifically to branch points in DNA (26, 27), we located these reverse polarity blocks ten bases away from the junction point to try and avoid any inhibition of binding. Indeed, these blocks did not affect the initial binding affinity of RecG for each set of junctions (Fig. 4 Aii and Bii). However, as noted above (Fig. 3) the lagging strand forks in Fig. 4B were bound with higher affinity than the leading strand forks in 4A.
Reverse polarity regions on both lagging (Fig. 4Ai) and leading (Fig. 4Bi) strand templates reduced the rate of unwinding of the opposing duplex arm by RecG. Because the reverse polarity blocks did not affect the initial binding of RecG to the forks, this suggests that the reduction in unwinding activity must be due to inhibition of RecG translocation along the opposing template strand containing the block. Therefore, translocation along and unwinding of the leading or lagging strand duplexes from these forks depends on translocation of RecG along the opposing template strand. The corollary is that RecG has two essential but opposing polar interactions with the two fork DNA structures, 5′-3′ on the leading strand template and 3′-5′ on the lagging strand template.
It is also striking that reverse polarity blocks had a greater inhibitory effect when placed in the lagging strand template rather than the leading strand template (compare the effect of a six-base block on the lagging versus the leading strand template in Fig. 4 Ai and Bi). This suggests that the translocation of RecG along the two template arms of the fork may not be equivalent.
Coupling of Translocation Steps by RecG at a Fork.
The data above indicated that RecG couples unwinding of one duplex arm with translocation along the opposing template strand. To test this hypothesis, and to ensure that the previous observations were not artifacts of the use of reverse polarity regions, we utilized a second approach to selectively disrupt RecG translocation steps. If simultaneous translocation along both arms of a fork is essential for RecG catalysis, then truncation of one of these arms should inhibit translocation along the second arm.
Forks were constructed in which either the lagging (Fig. 5A) or leading (Fig. 5B) strand template arm was truncated by ten or 15 bases relative to the intact 25-bp duplex arm. Each set of junctions was bound with equal affinity by RecG (Fig. 5 Aii and Bii), although, as noted above, the lagging strand forks in Fig. 5B were bound with higher affinity than the leading strand forks in Fig. 5A. The initial binding of RecG to each set of junctions was therefore unaffected by the truncations. This correlated with the relative binding affinities displayed by the reverse polarity forks (Fig. 4 Aii and Bii) and therefore supports the conclusion that RecG initially binds to no more than ten bases of the single-stranded template arm at a fork. However, reducing the size of either template strand to ten bases, so that they were 15 bases shorter than the opposing duplex, drastically inhibited unwinding of that duplex (Fig. 5 Ai and Bi). Again these data reflect those seen with the reverse polarity forks in Fig. 4. Thus, inhibition of translocation along the lagging strand template of the fork inhibits unwinding of the leading strand duplex and vice versa. We conclude that RecG performs two translocation reactions at fork DNA and that these translocations are coupled.
It should also be noted that truncation of the lagging strand template had a more severe effect as compared with truncation of the leading strand template (compare the effect of a 15-base lagging strand template with that of the 15-base leading strand template in Fig. 5 Ai and Bi). This mirrors the data of Fig. 4 and confirms that translocation of RecG along the leading and lagging strand templates is not equivalent.
Relative Arm Length Is Crucial for RecG Catalysis.
The data in Figs. 4 and 5 support the conclusion that RecG translocates in a coupled manner along the two arms of a fork. They also demonstrate that this coupling is abolished if the length of the arms differ by more than ten to 15 bases (Fig. 5). This implies that reducing any differential between the length of the template strand and the opposing duplex strand to be unwound should increase the efficiency of unwinding by RecG. To test this hypothesis we utilized the observation that having a template strand of only ten bases inhibits unwinding of the opposing duplex if that duplex is 25 bp (Fig. 5). Or, put another way, if the opposing template strand is 15 bases shorter than the duplex to be unwound, unwinding by RecG is severely inhibited.
We shortened the lagging strand duplex arm to be unwound from 25 to 20 bp and asked whether unwinding was increased. With forks in which the leading strand template remained at 25 bases, shortening the duplex arm by 5 bp increased the rate of unwinding of this arm by RecG ≈2-fold (Fig. 6 A and B, compare i and ii), even though binding affinity was unaltered (data not shown). This increased rate can be ascribed, at least in part, to the low levels of processivity of RecG on in vitro DNA junctions (27). However, with a fork having a leading strand template of only ten bases, the 20-bp lagging strand duplex was unwound at an ≈50-fold higher rate than the 25-bp duplex (Fig. 6 A and B, compare iii and iv). This large rate increase cannot be ascribed solely to the direct effect of reducing the number of base pairs of duplex DNA that RecG must unwind given that there was only a 2-fold increase in the rate of unwinding of forks containing a 25-base leading strand template. This suggests that reduction of the difference in length between the lagging strand duplex and the leading strand template is a major contributor to the observed increase in the rate of unwinding.
We also tested whether reducing the length differential between the lagging strand template and the leading strand duplex affected the rate of RecG-catalyzed unwinding of that duplex. Reduction of the length of the leading strand duplex from 25 to 20 bp while the lagging strand template remained at 26 bases did not alter the binding affinity of RecG (data not shown). However, this reduction did increase the rate of unwinding by RecG ≈6-fold (Fig. 6 C and D, compare i and ii). Again some or all of this increase may be attributed to the low processivity of RecG on in vitro forks. When a fork with a ten-base lagging strand template was used, the rate of RecG-catalyzed unwinding of the fork with the 20 as compared with the 25-bp leading strand duplex was also increased but by ≈80-fold (Fig. 6 C and D, compare iii and iv). The 80-fold increase, as compared with the 6-fold increase, again suggests that reduction of the number of base pairs that RecG must unwind is not the sole factor in the increased rate of unwinding. Reduction in the length differential between the two arms of the fork must be a major factor in the increased rate of unwinding.
The data in Fig. 6 indicate that the rate of unwinding of duplex DNA at a fork by RecG depends critically on the relative lengths of the duplex to be unwound and the opposing template strand. This length differential is equally important whether the leading or lagging strand duplex is being unwound. Taken together these findings support the conclusion that RecG translocates simultaneously and in a coupled manner along both arms of a fork.
Decreasing the length of the leading strand to be unwound also seems to have a greater stimulatory effect than an equivalent reduction in the length of the lagging strand to be unwound from the partial forks in Fig. 6. This can be seen in the 6-fold increase in the rate of unwinding the leading strand fork upon shortening the leading strand in Fig. 6D (compare i and ii), rather than the 2-fold increase seen upon shortening the lagging strand in Fig. 6B (again compare i and ii). This suggests that unwinding of the leading strand by RecG either faces a greater processivity problem and/or is more tightly coupled to translocation of RecG along the other arm of the fork as compared with unwinding of the lagging strand.
RecG Catalysis at Forks Containing Both Leading and Lagging Strand Duplexes.
The above data lead to the conclusion that to unwind the leading strand at a fork RecG must simultaneously translocate along the lagging strand template. Conversely, to unwind the lagging strand RecG must simultaneously translocate along the leading strand template. It is unlikely that the same helicase would be able to utilize two completely different mechanisms to unwind the leading and lagging strand forks used in Fig. 3. Therefore, the above findings suggest that RecG translocates simultaneously along the leading and lagging strand templates of both the fork structures used in Fig. 3 to effect unwinding of the leading and lagging strand duplexes. Can RecG perform such reactions when both leading and lagging strand duplexes are present at a fork? This was investigated by constructing two forks, both of which contained leading and lagging strand duplexes of 25 bp (Fig. 7). One contained the 32P label on the 5′ end of the leading strand (lanes 1–4), whereas the second was labeled at the 5′ end of the lagging strand (lanes 5–8). The most striking observation with both forks is that the presence of the leading strand inhibits unwinding of the lagging strand by RecG (Fig. 7B).
RecG catalysis at the fork containing the labeled leading strand produced both a labeled three-strand product and a free oligonucleotide (Fig. 7A, lanes 1–4). Production of the three-strand structure demonstrated that RecG could unwind the lagging strand from this fork, but that this was accomplished without simultaneous unwinding of the leading strand. The appearance of labeled free oligonucleotide indicated that the leading strand duplex could also be unwound. Moreover, the rate of accumulation of the leading strand was identical to the rate of accumulation of the leading strand from the partial fork (Fig. 7B). This would not be expected if removal of the leading strand from the complete fork occurred as a secondary reaction utilizing the three-strand product as substrate, because the reduced level of substrate in this case (three-strand product as compared with complete fork substrate) would be expected to lead to a reduced rate of accumulation of the free oligonucleotide product. This suggests that the leading strand was unwound from the complete fork structure in the absence of unwinding of the lagging strand. Furthermore, because the rates of unwinding the leading strand from the complete fork and from the partial fork are equal, this indicates that the presence of the lagging strand in the complete fork does not inhibit unwinding of the leading strand. This observation is in contrast to the inhibition of unwinding of the lagging strand when the leading strand is present (Fig. 7B). This finding emphasizes that the interactions between RecG and the leading and lagging strand arms of the fork are not equivalent. It is also striking that the labeled three-strand product accumulated to a much higher level than the labeled free oligonucleotide (Fig. 7A). This reflected the higher rates of unwinding of the lagging strand as compared with the leading strand in the partial forks (Fig. 7B) and indicated that RecG catalysis displayed a similar preference for unwinding of the lagging strand at all three forks.
When the label was placed on the lagging strand, the labeled product was free oligonucleotide (Fig. 7A, lanes 5–8). Very little labeled three-strand product could be detected. These data could be explained by the higher rate of unwinding the lagging strand duplex as compared with the leading strand, as seen in Figs. 3 and 7A (lanes 1–4). This explains the high level of labeled free oligonucleotide product. It also explains the low levels of labeled three-strand product, because the elevated rate of unwinding this structure (Fig. 3) would severely limit its accumulation.
RecG can therefore unwind the leading and lagging strands at forks containing both strands. Moreover, the patterns of unwinding are similar to those seen with the partial forks lacking either leading or lagging strands. This suggests that RecG utilizes the same mechanism to unwind complete and partial fork structures and that this mechanism involves simultaneous translocation of RecG along the leading and lagging strand templates. However, there is little evidence that RecG can couple unwinding of the leading and lagging strands at these static fork structures. The higher level of unwinding of the lagging strand as compared with the leading strand in Fig. 7 suggests that RecG unwinds the two strands of this fork primarily by two separate events. This was supported by our inability to detect significant coupling of unwinding of the two strands by RecG using excess competitor DNA to quench the reactions (data not shown). However, neither the template strands nor the leading and lagging strands could anneal together once unwound from these heterologous fork structures. Any effects this may have had on the possible coupling of unwinding of the leading and lagging strands remains unknown.
Discussion
The unwinding of stalled replication forks to form Holliday junctions, and the subsequent processing of these junctions, is essential for maintaining efficient replication fork progression (5, 8). We showed previously that RecG is critical in the formation of such Holliday junctions in vivo and that the enzyme can facilitate Holliday junction formation from fork structures in vitro (8). Here we show that formation of a Holliday junction from forked DNA requires the active unwinding of the fork by RecG (Fig. 2). The use of DNA forks with defined structures allowed us to demonstrate that RecG can unwind leading and lagging strands from partial fork substrates (Fig. 3). Moreover, these unwinding reactions depended on coupled translocation of RecG along the opposing template strand (Figs. 4 and 5). Because it is likely that RecG utilizes the same mechanism to unwind all branched DNA substrates, these data imply that RecG simultaneously translocates along both template strands to effect unwinding of both leading and lagging strand duplexes. Significantly, this model of RecG catalysis also explains the unwinding of Holliday junction DNA (28). Moreover, the dissociation of D-loops, R-loops, and three-strand and Y junctions (26, 29) can be explained by the same mechanism. Although it is unlikely that RecG targets such a wide range of substrates in vivo, the applicability of this mechanism to RecG catalysis at all of the junctions tested in vitro strongly supports the above model.
Coupled translocation along the template strands of a fork suggests that RecG may also couple the unwinding of both leading and lagging strand duplexes. However, we failed to detect any significant levels of coupled unwinding with the forks used in this study (Fig. 7). This implies that RecG unwinds the two duplexes in two separate reactions and that Holliday junction formation occurs by the subsequent reannealing of the template strands and the annealing of the nascent strands at a fork in vivo. However, the arms of all of the forks used in this study were completely heterologous to generate substrates with defined branch points. Thus, upon unwinding the fork neither the template strands nor the leading and lagging strands had the ability to anneal. The importance of such annealing steps in the unwinding of fork DNA by RecG is unknown. However, regions of heterology are known to inhibit the unwinding of synthetic Holliday junctions by RecG (27). We cannot therefore exclude the possibility that coupling between the unwinding of the leading and lagging strand duplexes at the forks used in this study was inhibited by the inability of the unwound strands, or the template strands, to subsequently anneal. Whether RecG can couple the unwinding of homologous leading and lagging strands remains to be established.
Taken together, what do these findings tell us about the nature of RecG catalysis? Because RecG most probably acts as a monomer (14), a single RecG molecule must interact with at least three regions of a fork. The data above demonstrate that it binds to both template strands, while the known specificity of RecG for branched DNA molecules (24) indicates that it must also interact with the parental duplex DNA. Moreover, the interactions with the leading and lagging strand templates occur with opposing polarity with respect to the phosphodiester linkages of these strands. The mechanism of RecG catalysis must therefore be very different from those used by DNA helicases such as PcrA (30, 31) that track along single-stranded DNA and disrupt base pairing between the two strands of duplex DNA as they progress.
Although RecG can unwind the leading and lagging strands from fork structures in vitro (Figs. 3 and 7), the removal of the leading strand occurs at a much lower rate than unwinding of the lagging strand. However, the use of truncated leading and lagging strands (Fig. 6) suggests that this discrepancy may be due at least in part to a more severe processivity problem on the leading strand duplex. It is possible that such problems may be artifacts of the heterologous fork structures used here. Thus, the true relative rates of unwinding of the leading and lagging strands remains unknown. Despite these uncertainties, the initial binding of RecG to the lagging strand fork occurs with higher affinity than the leading strand fork regardless of the binding conditions used (Fig. 3 and data not shown). This implies that RecG does preferentially recognize forks in which there is a lagging strand end located at the fork. How could such structures arise at a stalled replication fork? Little is known about the local structure of the DNA at damaged forks. Moreover, it is likely to depend to a large extent on the nature of the damage. However, it has been proposed that a lesion on the leading strand template might completely block the leading strand polymerase, but allow lagging strand synthesis to continue some way beyond the block (15, 17). Thus, a block on the leading strand template may be one type of event that leads to a stalled replication fork having a local structure similar to the fork used in Fig. 3A, lanes 5–8. It may be no coincidence that this fork is bound preferentially by RecG. Recent studies suggest that stalled replication forks can be processed by formation of a Holliday junction to create a substrate that can be branch migrated and cleaved by the RuvABC complex (5). The free DNA end generated by this cleavage [Fig. 1A (i)] can then be acted on by RecBCD and RecA to generate a D-loop. This D-loop can then act as a target for PriA-mediated assembly of a new replication fork at the D-loop (1). Alternatively, the free DNA end spooled out by regression of a stalled fork may be acted on by RecBCD directly to allow RecA-catalyzed D-loop formation, followed by subsequent cleavage of the Holliday junction by RuvABC (5, 13). However, replication can also be reestablished in a manner that requires not only PriA but also RecG, and that can proceed without the need for RuvABC-directed cleavage of the regressed fork (8). How can replication be restarted from a stalled fork without formation of a D-loop? PriA preferentially binds to forks with the 3′ end of a leading strand present at the branch point (32). PriA can also assemble a competent replication complex that can utilize this 3′ end for priming of replication (11). However, in the absence of the 3′-OH group of a leading strand at a stalled fork there would be no means to prime leading strand synthesis. The conclusion that RecG preferentially binds forks that possess a lagging strand, whereas PriA has a higher affinity for forks with a leading strand, suggests that RecG may facilitate PriA-dependent replisome reloading when the stalled fork does not initially possess a 3′-OH group at the junction point to prime leading strand synthesis. How this might be achieved is not known. However, it has been suggested that RecG may promote a template switching reaction in which formation of a Holliday junction by RecG allows extension of the stalled leading strand by using the nascent lagging strand as a template (ref. 8; Fig. 1). The ability of RecG to unwind the leading and lagging strands at fork structures, together with the high initial binding affinity of RecG for forks possessing a lagging strand, support this model. Branch migration of the Holliday junction in the reverse direction would regenerate a fork that now had a leading strand 3′OH for binding by PriA and subsequent priming of leading strand synthesis by DNA polymerase III. Thus, the opposing binding affinities of RecG and PriA at fork structures might reflect the ability of RecG to bind and unwind stalled forks that cannot be directly targeted by PriA to reload an active replisome. We are currently investigating whether such a mechanism underlies the observed genetic interaction between RecG and PriA (33).
Acknowledgments
We thank Lynda Harris for outstanding technical support and Gary Sharples for critical reading of the manuscript. This work was supported by a program grant from the Medical Research Council to R.G.L. and Gary Sharples. P.M. is a Lister Institute–Jenner Research Fellow.
Footnotes
This paper results from the National Academy of Sciences colloquium, “Links Between Recombination and Replication: Vital Roles of Recombination,” held November 10–12, 2000, in Irvine, CA.
References
- 1.Sandler S J, Marians K J. J Bacteriol. 2000;182:9–13. doi: 10.1128/jb.182.1.9-13.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lindahl T. Philos Trans R Soc London B. 1996;351:1529–1538. doi: 10.1098/rstb.1996.0139. [DOI] [PubMed] [Google Scholar]
- 3.Samadashwily G M, Raca G, Mirkin S M. Nat Genet. 1997;17:298–304. doi: 10.1038/ng1197-298. [DOI] [PubMed] [Google Scholar]
- 4.Krasilnikova M M, Samadashwily G M, Krasilnikov A S, Mirkin S M. EMBO J. 1998;17:5095–5102. doi: 10.1093/emboj/17.17.5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seigneur M, Bidnenko V, Ehrlich S D, Michel B. Cell. 1998;95:419–430. doi: 10.1016/s0092-8674(00)81772-9. [DOI] [PubMed] [Google Scholar]
- 6.Lindahl T, Wood R D. Science. 1999;286:1897–1905. doi: 10.1126/science.286.5446.1897. [DOI] [PubMed] [Google Scholar]
- 7.Tornaletti S, Hanawalt P C. Biochimie. 1999;81:139–146. doi: 10.1016/s0300-9084(99)80046-7. [DOI] [PubMed] [Google Scholar]
- 8.McGlynn P, Lloyd R G. Cell. 2000;101:35–45. doi: 10.1016/S0092-8674(00)80621-2. [DOI] [PubMed] [Google Scholar]
- 9.Kowalczykowski S C. Trends Biochem Sci. 2000;25:156–165. doi: 10.1016/s0968-0004(00)01569-3. [DOI] [PubMed] [Google Scholar]
- 10.Liu J, Xu L, Sandler S J, Marians K J. Proc Natl Acad Sci USA. 1999;96:3552–3555. doi: 10.1073/pnas.96.7.3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu J, Marians K J. J Biol Chem. 1999;274:25033–25041. doi: 10.1074/jbc.274.35.25033. [DOI] [PubMed] [Google Scholar]
- 12.Postow L, Ullsperger C, Keller R W, Bustamante C, Vologodskii A V, Cozzarelli N R. J Biol Chem. 2000;276:2790–2796. doi: 10.1074/jbc.M006736200. [DOI] [PubMed] [Google Scholar]
- 13.Seigneur M, Ehrlich S D, Michel B. Mol Microbiol. 2000;38:565–574. doi: 10.1046/j.1365-2958.2000.02152.x. [DOI] [PubMed] [Google Scholar]
- 14.McGlynn P, Mahdi A A, Lloyd R G. Nucleic Acids Res. 2000;28:2324–2332. doi: 10.1093/nar/28.12.2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Svoboda D L, Vos J M. Proc Natl Acad Sci USA. 1995;92:11975–11979. doi: 10.1073/pnas.92.26.11975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.West S C, Cassuto E, Howard-Flanders P. Nature (London) 1981;294:659–662. doi: 10.1038/294659a0. [DOI] [PubMed] [Google Scholar]
- 17.Cordeiro-Stone M, Zaritskaya L S, Price L K, Kaufmann W K. J Biol Chem. 1997;272:13945–13954. doi: 10.1074/jbc.272.21.13945. [DOI] [PubMed] [Google Scholar]
- 18.Cordeiro-Stone M, Makhov A M, Zaritskaya L S, Griffith J D. J Mol Biol. 1999;289:1207–1218. doi: 10.1006/jmbi.1999.2847. [DOI] [PubMed] [Google Scholar]
- 19.Zou H, Rothstein R. Cell. 1997;90:87–96. doi: 10.1016/s0092-8674(00)80316-5. [DOI] [PubMed] [Google Scholar]
- 20.Defossez P A, Prusty R, Kaeberlein M, Lin S J, Ferrigno P, Silver P A, Keil R L, Guarente L. Mol Cell. 1999;3:447–455. doi: 10.1016/s1097-2765(00)80472-4. [DOI] [PubMed] [Google Scholar]
- 21.Chakraverty R K, Hickson I D. BioEssays. 1999;21:286–294. doi: 10.1002/(SICI)1521-1878(199904)21:4<286::AID-BIES4>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 22.Parsons C A, Kemper B, West S C. J Biol Chem. 1990;265:9285–9289. [PubMed] [Google Scholar]
- 23.McGlynn P, Lloyd R G. Nucleic Acids Res. 1999;27:3049–3056. doi: 10.1093/nar/27.15.3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Whitby M C, Vincent S, Lloyd R G. EMBO J. 1994;13:5220–5228. doi: 10.1002/j.1460-2075.1994.tb06853.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Amaratunga M, Lohman T M. Biochemistry. 1993;32:6815–6820. doi: 10.1021/bi00078a003. [DOI] [PubMed] [Google Scholar]
- 26.McGlynn P, Al-Deib A A, Liu J, Marians K J, Lloyd R G. J Mol Biol. 1997;270:212–221. doi: 10.1006/jmbi.1997.1120. [DOI] [PubMed] [Google Scholar]
- 27.Whitby M C, Lloyd R G. J Biol Chem. 1998;273:19729–19739. doi: 10.1074/jbc.273.31.19729. [DOI] [PubMed] [Google Scholar]
- 28.Lloyd R G, Sharples G J. EMBO J. 1993;12:17–22. doi: 10.1002/j.1460-2075.1993.tb05627.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vincent S D, Mahdi A A, Lloyd R G. J Mol Biol. 1996;264:713–721. doi: 10.1006/jmbi.1996.0671. [DOI] [PubMed] [Google Scholar]
- 30.Soultanas P, Dillingham M S, Wiley P, Webb M R, Wigley D B. EMBO J. 2000;19:3799–3810. doi: 10.1093/emboj/19.14.3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Velankar S S, Soultanas P, Dillingham M S, Subramanya H S, Wigley D B. Cell. 1999;97:75–84. doi: 10.1016/s0092-8674(00)80716-3. [DOI] [PubMed] [Google Scholar]
- 32.Nurse P, Liu J, Marians K J. J Biol Chem. 1999;274:25026–25032. doi: 10.1074/jbc.274.35.25026. [DOI] [PubMed] [Google Scholar]
- 33.Al-Deib A A, Mahdi A A, Lloyd R G. J Bacteriol. 1996;178:6782–6789. doi: 10.1128/jb.178.23.6782-6789.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McGlynn P, Lloyd R G, Marians K J. Proc Natl Acad Sci USA. 2001;98:8235–8240. doi: 10.1073/pnas.121007798. [DOI] [PMC free article] [PubMed] [Google Scholar]