Abstract
The structural proteins (SP) of the Togaviridae can be deleted in defective interfering RNAs. The dispensability of viral SP has allowed construction of noninfectious viral expression vectors and replicons from viruses of the Alphavirus and Rubivirus genera. Nevertheless, in this study, we found that the SP of rubella virus (RUB) could enhance expression of reporter genes from RUB replicons in trans. SP enhancement required capsid protein (CP) expression and was not due to RNA-RNA recombination. Accumulation of minus- and plus-strand RNAs from replicons was observed in the presence of SP, suggesting that SP specifically affects RNA synthesis. By using replicons containing an antibiotic resistance gene, we found 2- to 50-fold increases in the number of cells surviving selection in the presence of SP. The increases depended significantly on the amount of transfected RNA. Small amounts of RNA or templates that replicated inefficiently showed more enhancement. The infectivity of infectious RNA was increased by at least 10-fold in cells expressing CP. Moreover, virus infectivity was greatly enhanced in such cells. In other cells that expressed higher levels of CP, RNA replication of replicons was inhibited. Thus, depending on conditions, CP can markedly enhance or inhibit RUB RNA replication.
Rubella virus (RUB) is the sole member of Rubivirus genus in the Togaviridae family. RUB has a single-stranded, positive-sense RNA genome that is 9,762 nucleotides (nt) in length. The genome encodes two open reading frames (ORFs): the 5′-proximal ORF encodes viral nonstructural proteins (NSP) that are responsible for viral genome replication, while the 3′-proximal ORF encodes three virion structural proteins (SP), the capsid protein (CP), and the two envelope glycoproteins, E2 and E1.
A polyprotein, P200, is translated from the 5′-proximal ORF and is a major component of the replication complex for the synthesis of genomic minus-strand RNA, which serves as a template for the synthesis of plus-strand genomic and subgenomic RNA, the mRNA for the virion proteins. The switch of synthesis from minus-strand genomic RNAs to plus-strand RNA synthesis is dependent on the processing of P200 by the virally encoded protease (17), a mechanism similar to that observed in the NSP of Sindbis virus (16). Studies of RUB defective interfering (DI) RNAs have shown that DI RNAs generated during undiluted passaging of virus or persistently infected cells have most of the SP coding region removed, showing that most of the SP coding region is not required for replication of the viral genome (5, 6). RUB replicons have been constructed which have part of the SP coding region replaced by genes encoding the puromycin-resistance protein (PAC) (4) or green fluorescent protein (GFP) (28), respectively. Using the RUBrep/PAC, Chen et al. (4) identified the minimal region at the 3′ end of the RUB genome that is required for viral genome replication and measured the replication efficiency of various 3′ constructs. Using RUBrep/GFP, Tzeng et al. (28) showed that most of the NSP coding region was necessary for amplification of the replicon with the exception of a 500-nt region between two NotI sites within the P150 coding region: a replicon with this deletion only expressed GFP in the presence of helper virus (28). Tzeng and Frey have recently shown that CP is the major moiety responsible for the complementation of the defect in RUBrep/GFP_ΔNotI (27).
RUB CP interacts with viral genomic RNA, forming the nucleocapsid. Analysis of the amino acid sequence suggested that the N-terminal half of the CP interacts with RNA because it is hydrophilic and rich in prolines and arginines (reviewed in reference 9). The major RNA binding domain of RUB CP has been located within amino acid residues 28 to 56, but other regions, including the C terminus, might also be involved in enhancing the interaction (19). The C terminus of the CP is very hydrophobic and contains the putative signal peptide of the E2 protein (reviewed in reference 9). Therefore, the CPs are anchored to the cell membrane by their C termini, with the N-terminal region inside the viral envelope. This membrane-anchoring signal peptide is associated with apoptosis induction in RK-13 cells (7). The N-terminal region of the CP may interact with the cytoplasmic tail of E1 and may be involved in budding (12). Interaction of RUB CP through its N terminus with a mitochondrial protein, p32, has also been found (1).
In this study, we show that RUB SP and CP alone can modulate replication of RUB replicons with genetically crippling mutations and viruses, suggesting that CP exerts a general effect on virus replication. Cells expressing RUB CP developed in this study give enhanced replication of low levels of RUB and could prove very useful in isolating viruses from clinical specimens.
MATERIALS AND METHODS
Cell culture, viruses infection, infectious RNA plaque assay, and reverse plaque assay.
BHK-21 and Vero cell lines were maintained at 35°C under 5% CO2 in Dulbecco's minimal essential medium (DMEM) (Invitrogen, Carlsbad, Calif.) supplemented with 5% fetal bovine serum (FBS) and gentamicin (10 mg/ml; Invitrogen). RUB F-therien, BRD II, and RA27/3 strains were used in this study. The plaque assay was done as previously described (11). The protocol for reverse plaque assay, which is basically determining the number of puromycin resistant cells following transfection with a replicon which expresses puromycin acetyltransferase (PAC), has been described previously (4).
The infectious RNA plaque assay was done as follows. Robo402 RNA was serially diluted into water and transfected (see below) into Vero cells in the presence 20 μg of RUB C RNA or Sindbis virus C RNA. At the end of transfection, the plates were overlaid with agar and a plaque assay was done (11). A dilution of Robo402 and C RNA with about 100 plaques was compared with the equivalent dilution of Robo402 and Sindbis virus C RNA to control for the total amount of RNA.
Transfection and establishment of clonal cell lines.
Transfection was mediated by use of Lipofectamine 2000 (Invitrogen) transfection reagent, which was used according to the protocol provided by the manufacturer. Unless otherwise specified, 20 μg of RNA was used for transfection into cells in a 60-mm-diameter plate. At the end of the transfection, the transfection reagent-nucleic acid mixture was removed and cells were overlaid with 2% FBS-DMEM and incubated at 35°C under 5% CO2.
The stable BHK-derived cell lines expressing RUB SP genes from pCI-Neo (Promega, Madison, Wis.) were established by transfection with plasmid DNA followed by Geneticin (Sigma, St. Louis, Mo.) selection at a concentration of 1 mg/ml, and single-cell colonies isolated. The clonal cells were maintained in DMEM supplemented with 5% FBS and Geneticin (0.5 mg/ml).
Detection of RUB-specific RNA species.
Intracellular RNA was prepared by using Tri-Reagent (Molecular Research Center Inc., Cincinnati, Ohio) according to the manufacturer's instructions. To detect RUB-specific minus-strand RNA, the intracellular RNA from one 60-mm-diameter plate was denatured in 80% dimethyl sulfoxide following 6 M glyoxal denaturation prior to electrophoresis and then blotted and probed with digoxigenin (DIG)-labeled RNA probes prepared by incorporating DIG-UTP during in vitro transcription using pGEM-GFP400. This plasmid contains the coding region of the GFP gene and the 3′-terminal 400 nt of the RUB genome. Excess RNA probe was removed by use of RNase (2 μg/ml) in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate).
RUB-specific positive or negative strand RNAs were detected by using an RNase protection assay as described (4, 29) except that the 35S-labeled strand-specific probe was transcribed from pGEM5Z-GFP220#2. RNA from one or 1/8 of a 60 mm plate was used for minus-and plus-strand RNA, respectively. This construct is similar to pGEM3Z-GFP220 (4) except that non-RUB sequences from the pGEM5Z vector were present after restriction enzyme digestion so that we could differentiate the products protected by RUBrep/GFP from undigested probe. The protected RNAs were resolved on an 8% Tris-borate-EDTA-polyacrylamide gel followed by autoradiography. For the two-cycle RNase protection assay, unlabeled RNA transcripts from pGEM-BglII, which contains 3′-terminal ∼4,400 nt of the RUB genome, were used as a protecting RNA in the first cycle (4).
Constructs.
The RUB infectious clone, Robo402, and the replicons, RUBrep/GFP, RUBrep/GFP_ΔNotI, and RUBrep/GFP_6512*, have been described elsewhere (24, 28). Site-directed mutagenesis was carried out by PCR using pairs of primers containing the desired mutations as previously described (3). The RUB replicon RUBrep/PAC and mutants derived from it have been previously described (4).
For expression of RUB P200, CP, and SP, constructs encoding genes of interest were created by PCR amplification using a forward primer containing a proper restriction enzyme sequence (SpeI or NheI) for cloning and the Kozak sequence (GCCGCCACC) prior to the start codon AUG, followed by 15 to 18 nt of the coding sequences of the gene of interest. The reverse primers usually contain complementary sequences of a restriction enzyme EcoRI sequence plus two UAG stop codons and the 3′ end of the gene of interest. The digested PCR fragments were ligated to NheI-EcoRI-digested pCI-Neo vector (Promega). The clones containing correct sequences were confirmed by restriction enzyme digestion and/or DNA sequencing. For example, using this protocol, the pCI-C construct contained nt 6505 through 7404 of the RUB genome.
Measurement of enhanced GFP expression.
GFP expressing RNA was transfected alone (control) or cotransfected with another RNA and the number of GFP expressing cells in fluorescent, digital images of the transfected monolayers determined by image analysis using Corel Draw (13). Any cell image which showed GFP fluorescence which was at least twice the background level or more over 10% or more of its area was considered positive.
Western blot and immunofluorescence assay (IFA).
The protocols for immunoprecipitation (24) and Western blot have been described previously (7). Approximately a 1/100 volume of cell lysate was analyzed on discontinuous sodium dodecyl sulfate-10% polyacrylamide gels and blotted with diluted RUB monoclonal antibodies (Viral Antigen Inc., Memphis, Tenn.). The chemiluminescence signal was detected using Western Lightning Chemiluminescence Reagent Plus (PerkinElmer Life Sciences, Inc., Boston, Mass.) and autoradiography.
Cells grown in eight-well chamber slides were fixed with 2% paraformaldehyde and permeabilized with methanol before being used in an IFA with specific monoclonal antibody (same antibodies as used in Western blot) The nuclei were then stained with propidium iodide and slides mounted with fluorescence mounting medium (Dako Cytomation, Glostrup, Denmark).
RESULTS
RUB SP enhance GFP expression from RUBrep/GFP and RUBrep/GFP_ΔNotI.
The ability of RUB RNAs expressing the RUB 5′ ORF (NSP/P200) or SP genes to enhance GFP expression from RUBrep/GFP_ΔNotI (with deletion of nt 1685 to 2192) or RUBrep/GFP in cotransfection experiments is shown in Fig. 1 Enhanced GFP expression was determined by measuring any increase in the number of GFP expressing cells. As shown in Fig. 1, no enhancement of GFP expression was detected in either P200 transfection (Fig. 1C and G), indicating that P200 failed to trans-complement RUBrep/GFP_ΔNotI efficiently or to enhance GFP expression from RUBrep/GFP. The translation and cleavage of NSP from the P200 RNA were confirmed in vitro (data not shown). The RUB SP gene enhanced GFP expression (Fig. 1D and H). Such enhancement could be detected as early as 18 h after transfection, and the number of GFP expressing cells was about the same as that found with helper virus (Fig. 1B and F). To address the possibility of low transfection efficiency in cotransfection experiments (<10%), similar experiments were done using cells stably expressing another RUB replicon, RUBrep/PAC (4), and similar results were obtained (data not shown). Note that GFP enhancement could have resulted from the SP gene or from SP involvement in RUB genome replication in trans (see below).
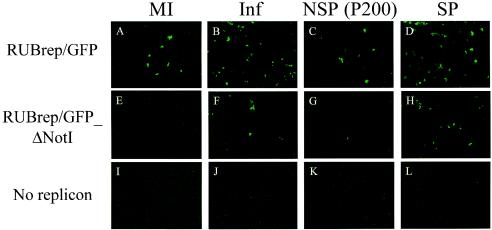
FIG. 1.
Enhanced GFP expression by RUB SP in trans; GFP expression in cotransfected cells. Vero cells were transfected with RUBrep/GFP (A to D) or RUBrep/GFP_ΔNotI (E to H) in the absence (MI) (A, E, and I) or presence (Inf) (B, F, and J) of helper virus or cotransfected with P200 RNA (C, G, and K) or SP RNA (D, H, and L). Infections were done with RUB F-therien at a multiplicity of infection of 1, and transfection was done at 24 h postinfection. GFP expression was examined 48 h after transfection.
The enhancement of GFP expression from RUBrep/GFP in three separate experiments as a result of cotransfection with SP RNA was 3.3-, 3.2-, and 4.3-fold (mean = 3.6-fold; standard deviation = 0.6-fold). When SP RNA was cotransfected with RUBrep/GFP_ΔNotI RNA, 91, 25, and 13 fluorescent cells were found, but no measure of the amount of enhancement could be made since no fluorescent cells were found with RUBrep/GFP_ΔNotI RNA alone in any of the three experiments.
Enhanced GFP expression by RUB SP was correlated with the accumulation of replicon genomes.
To investigate whether SP affected genome replication of RUB replicons, the accumulation of replicon genomes in the presence or absence of SP was examined by use of an RNase protection assay (Fig. 2A) or Northern hybridization (Fig. 2B). Intracellular RNA from transfected Vero cells was harvested after transfection and subjected to the RNase protection assay. The protected fragments of RNA species from the intracellular RNA pool corresponding to the replicon genomic (218 nt) and subgenomic (149 nt) RNAs and virus genomic (148 nt) and subgenomic (79 nt) RNAs are referred to as rG, rSG, vG, and vSG, respectively. A mutant, RUBrep/GFP_6512* (18, 28), which had a lethal mutation at the cleavage site between P150 and P90, was used as a control.
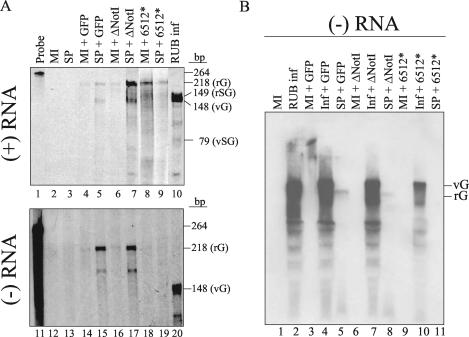
FIG. 2.
Accumulation of plus- and minus-strand genomic RNAs by RUB SP. The synthesis of replicon genome in the absence or presence of RUB SP was investigated by use of an RNase protection assay (A) and Northern blot analysis (B). (A) Intracellular RNA from transfected Vero cells with replicons with or without SP RNA cotransfection was harvested 18 h (for detecting minus-strand RNA; upper panel) or 72 h (for detecting plus-strand RNA; lower panel) after transfection and subjected to an RNase protection assay using 35S-labeled RNA probes. The RNase-protected fragments corresponding to the rG, rSG, vG, and vSG RNAs are indicated. (B) Intracellular RNA prepared from infected (RUB inf) or uninfected (MI) cells transfected with specific replicons with or without RUB SP gene was prepared at 18 h after transfection. RNA was denatured by dimethyl sulfoxide and glyoxal and electrophoresed on an 0.85% agarose gel followed by hybridization using a DIG-labeled pGEM-GFP400 probe of positive polarity. The RNA species corresponding to rG and vG RNA are indicated at the right.
As shown in the upper panel of Fig. 2A, in the presence of the RUB SP gene, more signal was observed from plus-strand genomic and subgenomic RNAs of RUBrep/GFP and RUBrep/GFP_ΔNotI. No increase in the amount of genomic or subgenomic RNA was detected with RUBrep/GFP_6512*. With the RUBrep/GFP_6512* construct, there is a decrease in signal from the replicon genomic RNA in the presence of SP for unknown reasons. However, the signal from subgenomic RNAs is a more reliable measure of de novo RNA synthesis because this signal is not affected by the transfected RNA, and since no subgenomic RNAs were detected with the RUBrep/GFP_6512* construct, we conclude that de novo RNA synthesis was not detected with this construct, with or without SP.
Accumulated minus-strand genomic RNA was found in the presence of RUB SP genes (Fig. 2A, lower panel). Comparison of the weak protected RNA signal from RUBrep/GFP or RUBrep/GFP_ΔNotI transfection in the absence of SP with RUBrep/GFP_6512* indicated that at least some RNA was synthesized de novo from these two replicons even in the absence of SP.
The presence of full-length minus-strand replicon RNA in the presence of SP genes was also confirmed by Northern hybridization. RUB minus-strand genomic RNA could be detected at a multiplicity of infection of 1 in cells infected with RUB F-therien strain (Fig. 2B). RNA species migrated faster than the genomic RNA were also detected, and this was likely the result of RNA degradation. In the presence of SP genes, the minus-strand genomic RNAs of RUBrep/GFP and RUBrep/GFP_ΔNotI) were detected but those of RUBrep/GFP_6512* were not (Fig. 2B). Note that the RNA of RUBrep/GFP_ΔNotI migrated faster than that of RUBrep/GFP because of the deletion in NSP coding region. In the absence of SP genes, no full-length RNA species was detected with RUBrep/GFP or RUBrep/GFP_ΔNotI, presumably because the amounts of minus-strand RNA were below the level detectable by Northern hybridization (Fig. 2B). The full-length replicon RNAs were not resolved when transfected into RUB-infected cells, because viral genome RNAs were present at much higher levels than replicon RNAs.
RUB SP enhanced replication of 3′ mutants.
To simplify experiments evaluating the effect of SP on RUB replication, BHK clonal cell lines expressing the SP gene or P200 were established. We chose BHK cells because they are more susceptible than Vero cells to antibiotic selection (4). The cell line containing P200 construct was used as a control since it was created using the same expression vector and did not enhance replication (Fig. 1). Two SP clonal cell lines, SP#11 and SP#12, were chosen and the expression of RUB SP was confirmed by Western blot (Fig. 3A) and IFA (data not shown). Note that in SP#12 cells no E1 was detected, and an aberrant product that was ∼36 kDa in size was detected using the E2 monoclonal in Western blot (Fig. 3A). The expression of E2 and CP in both SP#11 and SP#12 was also examined by IFA and SP# 12 was stronger for both. By IFA, the amount of E1 expressed in SP#11 was about the same as E2 protein in SP#11 cells, and no E1 was detected in SP#12 cells.
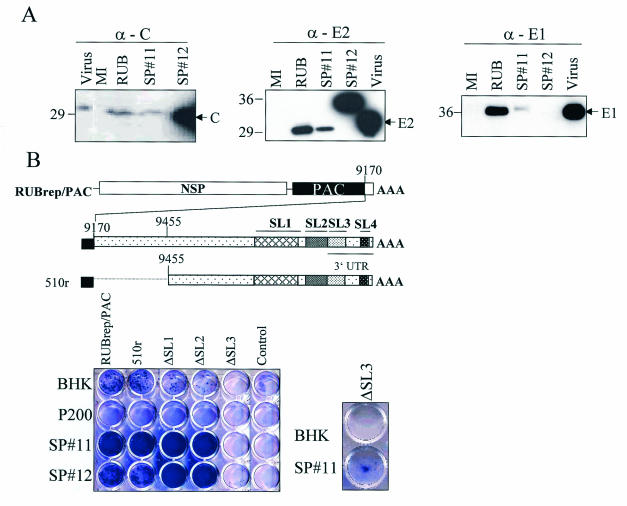
FIG. 3.
Replication of RUBrep/PAC and the 3′ mutants of RUBrep/PAC in BHK cells maintaining RUB NSP (P200) and SP. (A) Western blot showing the expression of RUB SP in SP#11 and SP#12. BHK clonal cell lines expressing RUB SP were obtained by antibiotic selection after transfecting a plasmid DNA containing RUB SP gene in pCI-Neo vector. Single colonies were isolated and maintained in media containing Geneticin. Cells grown in 10-cm2 plates were lysed in radioimmunoprecipitation buffer and 1/100 volume of the lysate were loaded on an sodium dodecyl sulfate-10% polyacrylamide gel. The proteins were transferred to a nitrocellulose membrane and blotted with a specific monoclonal antibody. Intracellular proteins from BHK cells infected with RUB F-therien (RUB) were harvested at 5 days postinfection. Virion proteins of RUB M33 (Virus) were purchased from Viral Antigens Inc. (B) Replication of 3′ mutants in cells expressing SP. BHK or clonal cell lines containing the indicated RUB genes were transfected with RUBrep/PAC or mutants with mutations at the 3′ end of RUBrep/PAC (unmodified RUBrepPAC is indicated). RNA transcripts (∼5 μg) were used for transfection to BHK or SP-expressing cells (∼106 cells). Cells were subjected to puromycin selection (5 μg/ml) 24 h after transfection and stained with crystal violet after 10 to 14 days of antibiotic selection. The control wells are cells without transfected RNA (Control). In order to visualize the small number of SP#11 cells containing the ΔSL3 construct surviving antibiotic selection, this culture and BHK control were passaged one more time.
Since minus-strand RNA accumulated when SP was present (Fig. 2), the effect of SP on previously constructed puromycin-resistant replicons with mutations in the 3′-terminal 600 nt was determined (4). These replicons allowed replication to be quantitated by counting puromycin-resistant cells. Replication of each RUBrep/PAC, mutated in the 3′-terminal 600 nt, in the BHK SP cell lines was examined by use of the reverse plaque assay (Fig. 3B). Cell lines expressing RUB SP greatly enhanced RUBrep/PAC replication as reflected by the fact that more puromycin-resistant colonies were observed. Those mutants include 510r, which maintains the 3′-terminal 305 nt as RUB DI (4), and ΔSL1, ΔSL2, and ΔSL3, which have the specific thermodynamically predicted 3′ SL structure deleted from RUBrep/PAC genome. The relative survival of 510r in BHK has been shown as twice as RUBrep/PAC while deletion of either SL1 or SL2 impaired, but not abolished replication (4). Deletion of SL3 was lethal in both infectious RNA and RUBrep/PAC (4, 5). Fewer colonies were recovered in P200-expressing cells than in control BHK cells suggesting that overexpression of RUB NSP may inhibit replication. In the presence of RUB SP, the replication was significantly improved with replicons in which the replication was impaired, such as ΔSL1, and ΔSL2. Moreover, replication of the lethal mutant, ΔSL3, could be detected in SP cells (Fig. 3B).
The synthesis of replicon genomes in both BHK and SP#11 cells was examined by Northern hybridization (data not shown). Both genomic and subgenomic RNAs of the replicons were synthesized in BHK or SP#11 cells. For mutants that replicated inefficiently in the absence of SP, such as ΔSL1 and ΔSL2, more replicon genomes were detected in SP#11 cells. The genome replication of ΔSL3, which was not viable in the absence of RUB SP, was also confirmed by Northern hybridization. The size of RUB replicon RNAs in BHK cells or SP#11 cells were very similar with each of the constructs, and thus, these results showed no evidence of RNA recombination.
The relative survival of each mutant was compared in the presence or absence of SP. The relative survival increased more significantly in the presence of SP in mutants with dramatic defects, such as ΔSL1 and ΔSL2. Replication of ΔSL1 was enhanced 58-fold, and replication of ΔSL2 by 15-fold. However, with mutants which could replicate efficiently in the absence of SP, such as RUBrep/PAC or 510r (4), the enhancement was not as dramatic as with those which could only replicate inefficiently, and only 1.2- and 0.8-fold increases were obtained.
Since the effect(s) of SP on genome replication seemed to be more significant on templates that did not replicate efficiently (Fig. 3B), we hypothesized that the enhancement depended on the amount of replicating templates present in the cytoplasmic pools rather than specific interaction with 3′ cis-acting elements. To test this possibility, the relative survival of RUBrep/PAC in BHK or SP#11 cells, using different amounts of this replicon RNA, was determined. Enhancement increased 28-fold in SP#11 cells in the presence of the lesser amount of template (2 μg RNA was used for transfection), while only 1.8-fold enhancement by SP was observed when 10-fold more template was transfected.
RUB CP is sufficient for enhancement.
Enhanced replication by SP#12 indicated that all three SP genes were not required to increase replication (i.e., E1 expression was not required.) (Fig. 3A). The minimal sequence/domain in RUB SP required for enhanced GFP expression was mapped by making various SP RNAs by runoff transcription from plasmid constructs made from the RUB SP in pCI-Neo vector (Promega), cotransfecting these RNAs with RUBrep/GFP_ΔNotI into Vero cells, and evaluating the expression of GFP (Fig. 4B). A similar strategy was applied to map the minimal domain required for enhancement within the C-coding region. The expected proteins from each truncated RNA shown in Fig. 4 were observed by in vitro translation (data not shown).
FIG. 4.
Mapping the minimal region of the SP coding region required for enhancing GFP expression from RUBrep/GFP_ΔNotI. (A) Schematic representation of the gene organization of the RUB SP gene. The numbers on the top left represent the distances from the start codon AUG while the numbers in parentheses indicate amino acid position. In the C gene, the solid box indicates the major RNA binding site (19), and the box with slashes indicates the E2 signal peptide. (B) GFP expression in Vero cells cotransfected with RUBrep/GFP_ΔNotI and various SP RNAs. The GFP expression was examined 48 h after transfection. The GFP expression results were evaluated as follows: +, enhanced number of cells with GFP expression (>10) was detected; −, no GFP expression was detected at 24 h after transfection; ±, only a few GFP-expressing cells (3 to 10 cells over the complete field) were detected. (C) Modifications in the C* construct are shown. The GFP expression following transfection with either uncapped C RNA or capped C* are shown.
Enhanced GFP expression was detected from cells with truncated SP. The RNAs that maintained the C- and E2-coding regions, such as NheI-, BsiWI-, and BamHI-digested templates (Fig. 4B), and those that had the C-coding region alone gave enhanced GFP expression. RNAs with E2, E1 or E2+E1 did not enhance expression of GFP.
Finer mapping of the region of the CP gene necessary for GFP enhancement was done, using RNAs transcribed from various constructs made with a pCI-C plasmid. GFP expression was significantly reduced with RNA from the 3′ terminus that was 90 nt shorter and resulted in deletion of the membrane domain of CP, (i.e., 30 amino acids were truncated from the C terminus of CP) (C_AscI) (Fig. 4B). Further truncations from the 3′ end did not restore activity (C_SalI and C_NotI) (Fig. 4B), indicating that maintenance of the 3′ end of the C gene was required. However, no GFP expression was detected in a mutant with the 5′ 87% of the C gene deleted [pCI-(CΔN262+E2)] (Fig. 4B), indicating that the 3′ end of the C gene alone, which is absent from C_AscI, was not sufficient. To determine if the sequences near the 5′ end of the C gene, which encodes the putative RNA binding domain of CP, was required, we tested two constructs, pCI-(CΔN147+E2), which had the 5′ half of the C gene deleted, and pCI-(CΔN27+E2), which had the 5′ 81 nt deleted but maintained the regions encoding the major RNA binding domain (19). Neither enhanced GFP expression (Fig. 4B). Taken together, these data suggest that the most of C gene is important for GFP enhancement. The 3′ end of the CP may be required because it encodes the putative E2 signal peptide which serves as the CP membrane anchor, or membrane domain.
trans-activation of transcription from one viral RNA species by other RNA species, such as with red necrotic mosaic virus (25) and flock house virus (8), has been recently observed. To investigate whether C RNA is directly involved in the GFP expression from RUBrep/GFP_ΔNotI, we constructed a mutant to disturb the reading frame. The mutant, C*, has two stop codons, UAG, inserted after the start codon AUG, followed by an extra G residue (Fig. 4C). Mutations in C* abolished translation from the first AUG in vitro (data not shown). RNA without the ability to be translated, including uncapped C RNA or capped C* RNA, failed to enhance GFP expression with either RUBrep/GFP or RUBrep/GFP_ΔNotI (Fig. 4C, subpanels C and D). The absence of enhancement of GFP expression by the C* gene presumably occurred with a stable RNA since this RNA was capped. These results indicate that CP, not the C gene, is required for enhanced GFP expression. In addition, these results provide further evidence that RNA-RNA recombination is not involved in enhanced GFP expression.
RUB CP increases infectivity of infectious RNA.
To test whether RUB CP also enhances replication of infectious RNA, serially diluted RUB infectious RNA transcripts (Robo402) were transfected to Vero cells, and the growth of the virus in the presence of cotransfected RUB C gene or Sindbis virus C gene was compared. In the presence of RUB C, but not Sindbis virus C, the number of plaques from transfected RNAs was increased by at least 10-fold on day 7 (data not shown). To compare the production of virus with and without CP, culture media from transfected cells were harvested at specific time points after transfection. When RUB C RNA was cotransfected, the titers of the virus were between 10- and 100-fold higher in the first 4 days after transfection; however, the viral titers 6 days after transfection were not affected by CP (data not shown).
RUB CP can enhance viral infectivity and the amount of CP affects replicon genome replication.
To examine whether exogenous CP affected virus infectivity, BHK clonal cell lines (C#26 and C#29) stably expressing the CP of the F-therien strain of RUB were established. The two cell lines had different CP expression levels: in C#26, CP was highly expressed as determined by Western blot analysis and IFA, while in C#29, CP was expressed at a moderate level by IFA and was barely detectable by immunoprecipitation with the CP monoclonal followed by Western blot with the same antibody (data not shown). Only ∼10% of C#29 cells gave detectable IFA signals. By reverse transcription-PCR, the amount of C gene in C#29 cells was shown to be 10- to 100-fold less than that in C#26 cells (data not shown).
Three RUB strains (F-therien [genotype I; laboratory strain], Brd II [genotype II; vaccine strain], and RA27/3 [genotype I; vaccine strain]) were used to infect BHK, C#26, and C#29 cells, and the growth of viruses was determined by the expression of E1 and E2 proteins by IFA (Fig. 5B). Four days after infection, enhanced IFA signals were detected in C#29 cells with all three RUB strains. The IFA signal was much weaker in infected C#26 cells, in which CP was expressed at high levels.
FIG. 5.
Viral infectivity regulation and replicon genome replication depends on the amount of CP. (A) RUB CP expression in C#26 and C#29 BHK clonal cell lines was determined by indirect immunofluorescent assay (IFA), using CP monoclonal antibody. A construct expressing RUB CP (F-therien strain) was created by cloning the RUB C gene, using pCI-Neo vector (Promega). C#26 and C#29 were isolated after transfection and selection. (B) Comparison of viral infectivity in cells expressing different amounts of CP. BHK, C#26, and C#29 were infected with three RUB strains (F-therien, Brd II, and RA27/3) at 10−1 PFU/ml. Four days after infection, virus growth was determined by IFA, using combined E1 and E2 monoclonal antibodies. MI, uninfected. (C) Comparison of RUBrep/PAC replication in cells expressing different amounts of CP. RUBrep/PAC RNA (5 μg) was transfected into BHK cells, C#26, or C#29 (∼106 cells) followed by selection with puromycin. The replication of RUBrep/PAC in all three cell lines was examined by reverse plaque assay. (D) Comparison of replication of RUBrep/GFP_ΔNotI in BHK, C#26, or C#29 cells. RUBrep/GFP_ΔNotI RNA was transfected into all three cell lines, and the GFP expression from the replicon was examined 48 h after transfection.
The results shown in Fig. 5B indicated that the amount of CP affected the replication of the virus genome. In other words, CP can either enhance or inhibit replication of the RUB genome. To further explore this, replicon replication was examined in C#26 and C#29 cells. Using RUBrep/PAC, the relative survival was significantly lower in C#26 cells compared to C#29 cells (Fig. 5C). Similarly, when using RUBrep/GFP_ΔNotI, fewer C#26 cells expressed GFP compared to C#29 cells (Fig. 5D). Each of the results with C#26 cells shown in Fig. 5B to D indicated that larger amounts of CP inhibited replication of RUB RNA. Furthermore, with all three systems, C#29 cells showed enhanced RUB RNA replication.
DISCUSSION
The present study showed that the RUB CP gene alone can modulate genome replication of replicons and viruses (Fig. 1, 3, and 5). Mapping experiments using various SP RNAs cotransfected with reporter replicons showed that the C gene was the major contributor to this effect. Expression of CP was required since constructs that lacked the initiation codon or uncapped C RNAs were unable to modulate replicon replication. The effect on replication was significantly larger when small amounts of RUB RNA were used. Modulation of RUB replication was found in the infectivity of three RUB strains (Fig. 5), in the expression of GFP from replicons with or without an in-frame deletion in the NSP region of the genome (Fig. 1), in the recovery of cells transfected with replicons that had deletions in the 3′-terminal 600 nt of the genome and that expressed an antibiotic-resistance gene (Fig. 3), and in the expression of RUB infectious RNAs in cotransfection experiments. In cell lines expressing CP, either enhancement or inhibition of replication was observed and the effect was inversely correlated with the amount of CP expressed (Fig. 5). Modulation of RUB replication was observed in both BHK cells and Vero cells. Together these results demonstrate that many RUB replication systems are modulated by the presence of RUB CP.
Mapping experiments presented here indicated that most of the CP was necessary for replication enhancement. In particular, both N-terminal residues and C-terminal residues were required. The requirement for the N terminus of the CP may not be related to the interaction with E1 proteins (12) because we found that E1 was dispensable (Fig. 4). Because the N termini of CP contain the major RNA-binding domain and are essential for the formation of nucleocapsid complexes during encapsidation (19) and because the C termini encode E2 signal peptide and function as a membrane anchor for CP (14), replication enhancement might be the result of formation of RNA-CP-membrane complexes. Although the major RNA-binding activity of CP was mapped within amino acid residues 28 to 56 in vitro, CP without the first 27 amino acids failed to enhance GFP expression from RUBrep/GFP_ΔNotI (Fig. 4). This result could be explained if the 27-amino-acid deletion resulted in a distorted conformation of the CP that had reduced RNA-binding activity.
The results presented here reinforce recent, similar results (27). In that report, RNAs, in which CP expression had been eliminated by site-direct mutagenesis of replicons expressing CP fused to a reporter protein, did not complement the NotI deletion in NSP in cotransfection experiments, indicating that CP rather than the C gene was responsible. In addition, by cotransfecting truncated C gene in plasmid cytomegalovirus constructs with RUBrep/GFP_ΔNotI transcripts, the region of the CP responsible for trans-complementation was mapped to the first 88 amino acids (27). The specific constructs used for C expression may explain the differences in the region of CP required observed here (Fig. 4B) (e.g., cytomegalovirus may express different levels of truncated C proteins leading to requirement of a smaller region of CP for complementation).
Although modulation of viral genome replication by CP has not been reported in RUB or other animal alphaviruses, nucleocapsid proteins have been found to be involved in the genomic replication of other viruses. The mechanisms include protein-protein interaction either with virally encoded polymerase (reviewed in references 23 and 26) or cellular proteins (33), viral genome-protein interaction to facilitate viral genome replication (10, 22), regulating translation-transcriptional control (31), melting RNA secondary structure (32), and trafficking the viral genome to replication machinery (reviewed in references 23 and 30). In avian myeloblastosis virus, for example, it was found that the coat protein was required to initiate viral genome replication, a process called genome activation, through the interaction with the 3′ terminus of the viral genome, and led to plus-strand RNA accumulation (reviewed in reference 2). Based on our observation of accumulated minus- and plus-strand RNA as well as the increase yield of virus in the presence of CP, the CP may also exert the effect at the early stage of RUB life cycle. CP may enhance genome replication by enhancing NSP expression, stabilizing the viral genome, directing viral RNA to microsomal vesicles for replication (20), or by interacting with cellular proteins such as p32 which can enhance viral infectivity (1, 21).
A simple model to explain our results presented here is as follows. When RUB RNA levels are low (e.g., mutations in the NSP or 3′ untranslated region), low levels of extra CP enhance RNA replication, (e.g., by increasing RNA stability). When larger amounts of RNA are present, such as when large amounts of intracellular RNA are being replicated, processes such as RNA degradation are overwhelmed by the large amount of RUB RNA present, even without CP. When there is a large excess of CP, replication is hindered, possibly because another process, such as RUB RNAs being sequestered into the assembly pathway, comes into play.
The results in the present study should be considered in light of recent observations concerning CP-RNA interactions during assembly of RUB, in which hypophosphorylated CP results in stronger CP-RNA interactions, putatively promoting assembly of nucleocapsids (15). We have found that cell lines expressing CP in which S46 was replaced with A, which was shown by Law et al. (15) to produce a hypophosphorylated form of CP, were unable to enhance GFP expression from RUBrep/GFP_ΔNotI. A reasonable explanation for these results is that when the assembly of nucleocapsids is enhanced by preventing phosphorylation or by providing excess capsid protein (e.g., in C#26 cells), enhancement of RUB RNA replication by CP is reduced. We suggest that there are at least two CP-related processes which affect RUB RNA replication: one is CP enhancement of RNA replication, and the other is CP-RNA interactions leading to assembly, which reduce RNA replication.
Finally, the results of this study have practical applications. For example, the enhancement of the infectivity of infectious RNA and virus by CP should allow the development of improved assays to isolate RUB from clinical specimens, especially when low amounts of virus are present. Such assays may facilitate control of rubella in countries where it and congenital rubella syndrome are still endemic.
Acknowledgments
We thank the Core Facility at the CDC (Atlanta, Ga.) for help with oligosynthesis and Stephanie Liffick (Measles Virus Section, CDC) for automatic DNA sequencing. We thank Teryl Frey (Georgia State University) for providing RUBrep/GFP_6512*, RUBrep/PAC, and 3′ mutants of RUBrep/PAC and for facilitating M.-H.C.'s financial support.
M.-H.C. was supported by an Interagency Personnel Agreement between the CDC and Georgia State University. M.-H.C.'s contributions to the work presented here were made entirely at the CDC. Primary funding for this research was from the CDC.
REFERENCES
- 1.Beatch, M. D., and T. C. Hobman. 2000. Rubella virus capsid associates with host cell protein p32 and localizes to mitochondria. J. Virol. 74:5569-5576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bol, J. F. 1999. Alfalfa mosaic virus and ilarviruses: involvement of coat protein in multiple steps of the replication cycle. J. Gen. Virol. 80:1089-1102. [DOI] [PubMed] [Google Scholar]
- 3.Chen, M. H., and T. K. Frey. 1999. Mutagenic analysis of the 3′ cis-acting elements of the rubella virus genome. J. Virol. 73:3386-3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, M. H., I. Frolov, J. Icenogle, and T. K. Frey. 2004. Analysis of the 3′ cis-acting elements of rubella virus using replicons expressing a puromycin resistance gene. J. Virol. 78:2553-2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Derdeyn, C. A. 1994. The characterization of rubella virus defective-interfering RNAs generated during serial undiluted passage and persistent infection. Ph.D. thesis. Georgia State University, Atlanta.
- 6.Derdeyn, C. A., and T. K. Frey. 1995. Characterization of defective-interfering RNAs of rubella virus generated during serial undiluted passage. Virology 206:216-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duncan, R., A. Esmaili, L. M. Law, S. Bertholet, C. Hough, T. C. Hobman, and H. L. Nakhasi. 2000. Rubella virus capsid protein induces apoptosis in transfected RK13 cells. Virology 275:20-29. [DOI] [PubMed] [Google Scholar]
- 8.Eckerle, L. D., and L. A. Ball. 2002. Replication of the RNA segments of a bipartite viral genome is coordinated by a transactivating subgenomic RNA. Virology 296:165-176. [DOI] [PubMed] [Google Scholar]
- 9.Frey, T. K. 1994. Molecular biology of rubella virus. Adv. Virus Res. 44:69-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hatton, T., S. Zhou, and D. N. Standring. 1992. RNA- and DNA-binding activities in hepatitis B virus capsid protein: a model for their roles in viral replication. J. Virol. 66:5232-5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hemphill, M. L., R. Y. Forng, E. S. Abernathy, and T. K. Frey. 1988. Time course of virus-specific macromolecular synthesis during rubella virus infection in Vero cells. Virology 162:65-75. [DOI] [PubMed] [Google Scholar]
- 12.Hobman, T. C., M. L. Lundstrom, C. A. Mauracher, L. Woodward, S. Gillam, and M. G. Farquhar. 1994. Assembly of rubella virus structural proteins into virus-like particles in transfected cells. Virology 202:574-585. [DOI] [PubMed] [Google Scholar]
- 13.Knight, A. W., and N. Billinton. 2001. Distinguishing GFP from cellular autofluorescence. Biophotonics Int. 8:42-50. [Google Scholar]
- 14.Law, L. M., R. Duncan, A. Esmaili, H. L. Nakhasi, and T. C. Hobman. 2001. Rubella virus E2 signal peptide is required for perinuclear localization of capsid protein and virus assembly. J. Virol. 75:1978-1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Law, L. M., J. C. Everitt, M. D. Beatch, C. F. Holmes, and T. C. Hobman. 2003. Phosphorylation of rubella virus capsid regulates its RNA binding activity and virus replication. J. Virol. 77:1764-1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lemm, J. A., T. Rumenapf, E. G. Strauss, J. H. Strauss, and C. M. Rice. 1994. Polypeptide requirements for assembly of functional Sindbis virus replication complexes: a model for the temporal regulation of minus- and plus-strand RNA synthesis. EMBO J. 13:2925-2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liang, Y., and S. Gillam. 2000. Mutational analysis of the rubella virus nonstructural polyprotein and its cleavage products in virus replication and RNA synthesis. J. Virol. 74:5133-5141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liang, Y., J. Yao, and S. Gillam. 2000. Rubella virus nonstructural protein protease domains involved in trans- and cis-cleavage activities. J. Virol. 74:5412-5423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu, Z., D. Yang, Z. Qiu, K. T. Lim, P. Chong, and S. Gillam. 1996. Identification of domains in rubella virus genomic RNA and capsid protein necessary for specific interaction. J. Virol. 70:2184-2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Magliano, D., J. A. Marshall, D. S. Bowden, N. Vardaxis, J. Meanger, and J. Y. Lee. 1998. Rubella virus replication complexes are virus-modified lysosomes. Virology 240:57-63. [DOI] [PubMed] [Google Scholar]
- 21.Mohan, K. V., B. Ghebrehiwet, and C. D. Atreya. 2002. The N-terminal conserved domain of rubella virus capsid interacts with the C-terminal region of cellular p32 and overexpression of p32 enhances the viral infectivity. Virus Res. 85:151-161. [DOI] [PubMed] [Google Scholar]
- 22.Patton, J. T., M. T. Jones, A. N. Kalbach, Y. W. He, and J. Xiaobo. 1997. Rotavirus RNA polymerase requires the core shell protein to synthesize the double-stranded RNA genome. J. Virol. 71:9618-9626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Portela, A., and P. Digard. 2002. The influenza virus nucleoprotein: a multifunctional RNA-binding protein pivotal to virus replication. J. Gen. Virol. 83:723-734. [DOI] [PubMed] [Google Scholar]
- 24.Pugachev, K. V., E. S. Abernathy, and T. K. Frey. 1997. Improvement of the specific infectivity of the rubella virus (RUB) infectious clone: determinants of cytopathogenicity induced by RUB map to the nonstructural proteins. J. Virol. 71:562-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sit, T. L., A. A. Vaewhongs, and S. A. Lommel. 1998. RNA-mediated trans-activation of transcription from a viral RNA. Science 281:829-832. [DOI] [PubMed] [Google Scholar]
- 26.Tacken, M. G., P. J. Rottier, A. L. Gielkens, and B. P. Peeters. 2000. Interactions in vivo between the proteins of infectious bursal disease virus: capsid protein VP3 interacts with the RNA-dependent RNA polymerase, VP1. J. Gen. Virol. 81:209-218. [DOI] [PubMed] [Google Scholar]
- 27.Tzeng, W. P., and T. K. Frey. Complementation of a deletion in the rubella virus P150 nonstructural protein by the viral capsid protein. J. Virol. 77:9502-9510. [DOI] [PMC free article] [PubMed]
- 28.Tzeng, W. P., M. H. Chen, C. A. Derdeyn, and T. K. Frey. 2001. Rubella virus DI RNAs and replicons: requirement for nonstructural proteins acting in cis for amplification by helper virus. Virology 289:63-73. [DOI] [PubMed] [Google Scholar]
- 29.Wang, X., and S. Gillam. 2001. Mutations in the GDD motif of rubella virus putative RNA-dependent RNA polymerase affect virus replication. Virology 285:322-331. [DOI] [PubMed] [Google Scholar]
- 30.Zhang, J., and C. S. Crumpacker. 2002. Human immunodeficiency virus type 1 nucleocapsid protein nuclear localization mediates early viral mRNA expression. J. Virol. 76:10444-10454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang, J., O. Yamada, H. Yoshida, T. Iwai, and H. Araki. 2002. Autogenous translational inhibition of core protein: implication for switch from translation to RNA replication in hepatitis C virus. Virology 293:141-150. [DOI] [PubMed] [Google Scholar]
- 32.Zhang, W. H., C. K. Hwang, W. S. Hu, R. J. Gorelick, and V. K. Pathak. 2002. Zinc finger domain of murine leukemia virus nucleocapsid protein enhances the rate of viral DNA synthesis in vivo. J. Virol. 76:7473-7484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang, X., C. Glendening, H. Linke, C. L. Parks, C. Brooks, S. A. Udem, and M. Oglesbee. 2002. Identification and characterization of a regulatory domain on the carboxyl terminus of the measles virus nucleocapsid protein. J. Virol. 76:8737-8746. [DOI] [PMC free article] [PubMed] [Google Scholar]