Abstract
Burkholderia ubonensis is an environmental bacterium belonging to the Burkholderia cepacia complex (Bcc), a group of genetically related organisms that are associated with opportunistic but generally nonfatal infections in healthy individuals. In contrast, the near-neighbour species Burkholderia pseudomallei causes melioidosis, a disease that can be fatal in up to 95% of cases if left untreated. B. ubonensis is frequently misidentified as B. pseudomallei from soil samples using selective culturing on Ashdown’s medium, reflecting both the shared environmental niche and morphological similarities of these species. Additionally, B. ubonensis shows potential as an important biocontrol agent in B. pseudomallei-endemic regions as certain strains possess antagonistic properties towards B. pseudomallei. Current methods for characterising B. ubonensis are laborious, time-consuming and costly, and as such this bacterium remains poorly studied. The aim of our study was to develop a rapid and inexpensive real-time PCR-based assay specific for B. ubonensis. We demonstrate that a novel B. ubonensis-specific assay, Bu550, accurately differentiates B. ubonensis from B. pseudomallei and other species that grow on selective Ashdown’s agar. We anticipate that Bu550 will catalyse research on B. ubonensis by enabling rapid identification of this organism from Ashdown’s-positive colonies that are not B. pseudomallei.
Introduction
The Gram-negative Burkholderia spp. comprise an ecologically diverse group containing over 70 species (http://www.bacterio.cict.fr/b/burkholderia.html), some of which are pathogenic to humans, animals or plants [1], [2]. Burkholderia pseudomallei is the best-known member of the genus due to its ability to cause the potentially fatal disease melioidosis [3] and its biothreat potential [4]. B. pseudomallei was recently added as a Tier 1 Select Agent in the United States, a category that includes those organisms of greatest threat to human and animal health. B. pseudomallei is commonly recovered in the environment in northern Australia (particularly the “Top End“ of the Northern Territory) and north-eastern Thailand, but has also been described from a much wider endemic region including most other countries in Southeast Asia, the Indian subcontinent, Taiwan, southern China and Hong Kong [5]. The presence of B. pseudomallei in Africa and the Americas has also been described but the extent of its distribution remains unclear [6]. Several other soil-dwelling Burkholderia spp. reside in ecological niches where B. pseudomallei is present, and some of these species can also cause opportunistic, albeit less serious, infections in humans. Many of these species fall into the Burkholderia cepacia complex (Bcc), which contains at least 17 Burkholderia species, including Burkholderia ubonensis [7].
Misidentification of Burkholderia spp. has implications for environmental studies, clinical diagnosis and biosecurity responses [8], especially for B. pseudomallei, where false-negative and false-positive results may have serious consequences. Species misidentification can have an economic impact, as demonstrated by false-positive calls of near-neighbour species under the BioWatch program, which was introduced in the United States in 2003 to monitor aerosol samples for the presence of Select Agent organisms in the environment [9]. Detecting B. pseudomallei from clinical samples is also a nontrivial endeavour. Most hospital laboratories use standard culture media (e.g. MacConkey, horse blood and chocolate agars) for culturing of clinical specimens. Morphological identification of B. pseudomallei in non-endemic areas is therefore difficult due to unfamiliarity, a lack of selective media available for identification [10], and the frequent misidentification of B. pseudomallei using automated systems such as VITEK 2 [11]. In endemic regions, B. pseudomallei is typically enriched from environmental specimens using broth selection [12] followed by plating on Ashdown’s agar (ASA) [13]. However, no selective method is B. pseudomallei-specific. Indeed, many Burkholderia spp. residing in the same niches as B. pseudomallei, including B. ubonensis, are morphologically similar on ASA [10], [14], [15].
Since the “Burkholderia uboniae” species was first proposed in 2000 [16], little research has been conducted on B. ubonensis, despite being a potentially important biocontrol agent for B. pseudomallei [15]. Dideoxy sequencing-based genotyping approaches such as multilocus sequence typing (MLST), recA and 16S sequencing have been developed for Burkholderia spp. characterisation [8], [17], [18]. However, there are currently no cost-effective, rapid, and simple methods for detecting and differentiating B. ubonensis from other Burkholderia spp. including B. pseudomallei. For example, the type III secretion system 1 (TTS1) assay [19] only detects B. pseudomallei, and thus cannot further identify other species that grow on ASA. Therefore, the major aim of our study was to differentiate B. ubonensis from B. pseudomallei, with a secondary aim of differentiating B. ubonensis from other members of the Bcc and non-Burkholderiaecae organisms that also grow on ASA.
Materials and Methods
Ethics Statement
The Australian isolates used in our study were obtained from either private land or from Aboriginal communities. Prior to private land soil and water sampling, we obtained signed or verbal permission from land owners. Sampling permits were obtained from Northern Land Council (Northern Territory, Australia) prior to sample collection from Aboriginal communities. As per permit conditions, we obtained further permission from the community representatives prior to sampling. No specific permits were required for collection of the Thai isolates as they were obtained from unregulated public lands. Our field collection did not involve endangered or protected species.
Bacterial Isolates
Our laboratories have ongoing collections of isolates from soil and water samples obtained from both the Northern Territory and Thailand, comprising isolates that grow on ASA [13] yet are not B. pseudomallei according to the TTS1 assay [19]. These isolates were subjected to 16S sequencing, MLST, recA sequencing or whole-genome sequencing (WGS) as part of this and other studies to confirm genus and, where possible, for species assignment. All isolates were subcultured for purity on chocolate agar or ASA (Oxoid, Thebarton, SA, Australia) prior to DNA extraction. The Qiagen DNeasy kit (Qiagen, Doncaster, VIC, Australia) was used for DNA extraction as previously described [20]. DNA was diluted 1∶100 in molecular-grade H2O prior to PCR.
Bioinformatic Analysis to Identify B. ubonensis-specific Loci
Nineteen B. pseudomallei near-neighbour isolates were subjected to WGS: Burkholderia spp. MSMB175, Burkholderia spp. MSMB49, B. cenocepacia MSMB101, B. cenocepacia MSMB139, Burkholderia multivorans MSMB104, B. multivorans MSMB105, Burkholderia oklahomensis C6786, B. pseudomallei MSHR684, B. pseudomallei MSHR1079, Burkholderia thailandensis-like strain MSMB121, B. thailandensis MSMB59, B. ubonensis MSMB56, B. ubonensis MSMB106, B. ubonensis MSMB108, B. ubonensis MSMB145, B. ubonensis MSMB157, B. ubonensis MSMB166, B. ubonensis MSMB169 and B. ubonensis MSMB170. The Illumina GAIIx platform (Illumina, San Diego, CA, USA) was used to generate WGS data. An assembly of B. ubonensis MSMB170 was performed on paired-end Illumina v1.9 reads with Velvet v1.2.07 [21], using a kmer of 55. This assembly resulted in 836 contigs with an n50 of 101,278 bp. MSMB170 was subsequently used as a reference genome for read mapping with the Burrows-Wheeler Aligner (BWA) v0.5.9 [22]. The coverageBed module of BEDTools v2.15.0 [23] was used for presence/absence analysis based on a 1 kb window size. Candidate B. ubonensis-specific loci were identified by locating regions with 100% read coverage in all eight B. ubonensis strains but with <50% coverage in other Burkholderia species. Eleven candidate loci ≥5 kb were identified. One locus, Bu550, was chosen for real-time PCR assay design following confirmation of in silico specificity for B. ubonensis using Microbial Nucleotide BLAST.
B. ubonensis-specific Real-time PCR Assay Bu550
A fluorogenic probe-based real-time PCR assay (Bu550) was developed to target a candidate B. ubonensis-specific 7 kb locus. Four putative protein products are encoded within this locus; a major facilitator superfamily transporter (GenBank ID: WP_010089641), a hypothetical protein (WP_010089640), a carbamoyltransferase (WP_010089639) and a tannase (WP_010089638). Unlabelled primers Bu550-F (5′-ATGCCGTGATCGACAACGAT) and Bu550-R (5′-ACTCCAGAAACAGTTCAGGCGT) (Invitrogen, Mulgrave, VIC, Australia) were used to amplify a conserved 91-bp fragment within this locus. A Black Hole Quencher (BHQ) probe (5′-CAL Fluor Gold 540-CGGGTGATGTGGCGTGACATTTACAGA-BHQ1; Biosearch Technologies, Novato, CA, USA) was included to increase specificity. BLAST analysis was conducted on the primers and probes to ensure assay specificity and accuracy. Real-time PCR was performed in 384-well optical plates (Applied Biosystems, Foster City, CA, USA). Each 5 µL reaction contained 0.3 µM of each primer, 0.2 µM of probe, 1X TaqMan Environmental Master Mix (Applied Biosystems) and 1 µL genomic DNA, to a total reaction volume of 5 µL. We also tested 1X TaqMan Universal Master Mix (Applied Biosystems) to determine assay robustness across different mastermixes. The 306 isolates used in this study (Table 1) were tested in duplicate, and all runs contained appropriate positive control and no-template control reactions. Thermocycling was carried out under default conditions using an ABI PRISM 7900HT instrument (Applied Biosystems), using the TET channel for fluorescence detection.
Table 1. Bacterial strain panel used in this study.
| Species | No. strainsa |
| Achromobacter spp. | 1 |
| Acidovorax caeni | 1 |
| Alcaligenes spp. | 1 |
| Burkholderia cenocepacia | 2 (1) |
| Burkholderia cepacia | 2 (16) |
| Burkholderia diffusa | 2 (1) |
| Burkholderia multivorans | 3 |
| Burkholderia pseudomallei | 75 (11) |
| Burkholderia pyrrocinia | 1 |
| Burkholderia thailandensis | 3 |
| Burkholderia thailandensis-likeb | 2 |
| Burkholderia ubonensis | 125 (15) |
| Burkholderia vietnamiensis | 1 |
| Burkholderia spp. BCCU6 | 1 |
| Burkholderia spp. M279 | 2 |
| Chromobacterium violaceum | 1 |
| Chryseobacterium spp. | 1 |
| Comamonas spp. | 1 |
| Cupriavidus spp. | 5 |
| Delftia spp. | 2 |
| Herbaspirillum seropedicae | 1 |
| Pandoraea spp. | 1 |
| Pigmentiphaga spp. | 1 |
| Pseudomonas aeruginosa | 1 |
| Ralstonia spp. | 8 |
| Staphylococcus epidermidis | 1 |
| Stenotrophomonas spp. | 1 |
| Unknown | 16 |
| TOTAL | 306 |
Numbers in parentheses indicate Thai strains; all other strains were isolated in the Northern Territory, Australia.
Species assignment based on [28].
Results
Phenotypic Diversity of B. Ubonensis Isolates on ASA
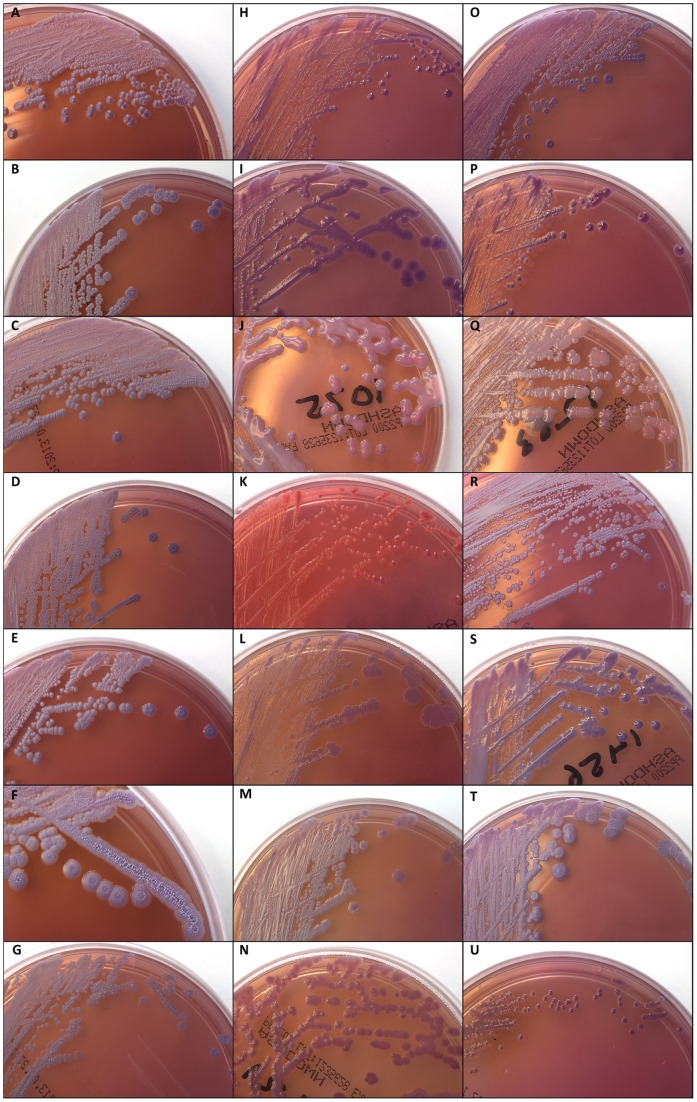
ASA is commonly used for isolation of B. pseudomallei from clinical and environmental isolates in endemic regions, and is commercially available in Australia. The isolates examined in our study contain a cross-section of isolates that grow on ASA but are TTS1-negative [19], and thus are not B. pseudomallei. Several species (e.g. Burkholderia diffusa, Chryseobacterium spp., Delftia spp., Ralstonia spp., Cupriavidus spp. and Acidovorax spp.) possessed morphologies (Figure 1) that were clearly distinguishable from the expected B. pseudomallei morphotypes [24]; in other cases, isolates were indistinguishable from B. pseudomallei. The latter category contained mostly Burkholderia spp., particularly B. ubonensis, which demonstrated multiple morphotypes, many of which resembled B. pseudomallei morphotypes (Figure 1). This inability to differentiate most B. ubonensis strains from B. pseudomallei on ASA provided the impetus for the rest of this study.
Figure 1. Colony morphologies of various B. pseudomallei near-neighbour species on Ashdown’s agar.
Panels: A, Burkholderia ubonensis MSMB700; B, B. ubonensis MSMB704; C, B. ubonensis MSMB1138; D, B. ubonensis MSMB718; E, B. ubonensis MSMB1191; F, B. ubonensis MSMB1165; G, B. ubonensis MSMB1202; H, Pandoraea sp. MSMB824; I, Herbaspirillum seropedicae MSMB1000; J, Burkholderia diffusa MSMB1075; K, Chryseobacterium sp. MSMB1448; L, Cupriavidus metallidurans MSMB1495; M, Burkholderia vietnamiensis MSMB1224; N, Burkholderia multivorans MSMB1271; O, Burkholderia pyrrocinia MSMB1147; P, Delftia sp. MSMB943; Q, Ralstonia mannitolilytica MSMB1253; R, Burkholderia thailandensis MSMB1415; S, Burkholderia cepacia MSMB1456; T, B. cepacia MSMB1011; U, Acidovorax caeni MSMB1260. On this medium, Burkholderia ubonensis demonstrates similar morphological characteristics to its potentially deadly near-neighbour, Burkholderia pseudomallei, including uptake of crystal violet and neutral red, and wrinkling of colonies after ∼72 h growth [13], [24]. Molecular genotyping is therefore necessary for differentiation of B. ubonensis from other bacterial species that grow on Ashdown’s medium. Note the morphological differences among B. ubonensis strains; several morphotypes have also been observed in B. pseudomallei [24].
Identification of a B. Ubonensis-specific Target from Whole-genome Sequence Data
Comparative whole-genome analysis of 19 Burkholderia spp., which included eight B. ubonensis strains, was performed using B. ubonensis MSMB170 as the reference genome. With this approach, we identified a candidate 7 kb locus specific for B. ubonensis, corresponding to a region within B. ubonensis Bu contig PMP6xxBUBxxBu-101 (GenBank: ABBE01000101.1). Nucleotide BLAST analysis (http://blast.ncbi.nlm.nih.gov/) of 9,404 complete or draft microbial genomes confirmed specificity of this locus, with only a single significant hit occurring in B. ubonensis Bu (analysis performed 26-Mar-13).
PCR Assay Design and Screening of the B. Ubonensis-specific Assay
A Black Hole Quencher (BHQ) probe-based real-time PCR assay [25] was designed based on a conserved region within the 7 kb B. ubonensis-specific locus, encoding the hypothetical protein BuboB_03639. We chose BHQ probe technology due to its comparatively low cost compared with conventional TaqMan minor groove-binding probes (Applied Biosystems). In addition, the Bu550 assay can be multiplexed in a single PCR tube with the existing B. pseudomallei TTS1 BHQ assay [19] due to compatible CAL Fluor 540 and 6FAM fluorophore chemistries. The Bu550 assay was screened for specificity using a diverse panel of bacterial species that grow on ASA, with greatest representation of B. ubonensis and B. pseudomallei isolates (Table 1). In total, 306 isolates were tested, including 140 B. ubonensis and 86 B. pseudomallei isolates from Australia and Thailand. As predicted from in silico analyses, the Bu550 assay demonstrated 100% specificity for B. ubonensis, with all other examined species failing to amplify (Figure 2; Table 1). We obtained similar results with the TaqMan Environmental and Universal Master Mixes, although the Environmental Master Mix provided more robust amplification (data not shown).
Figure 2. Bu550 B. ubonensis-specific real-time PCR.
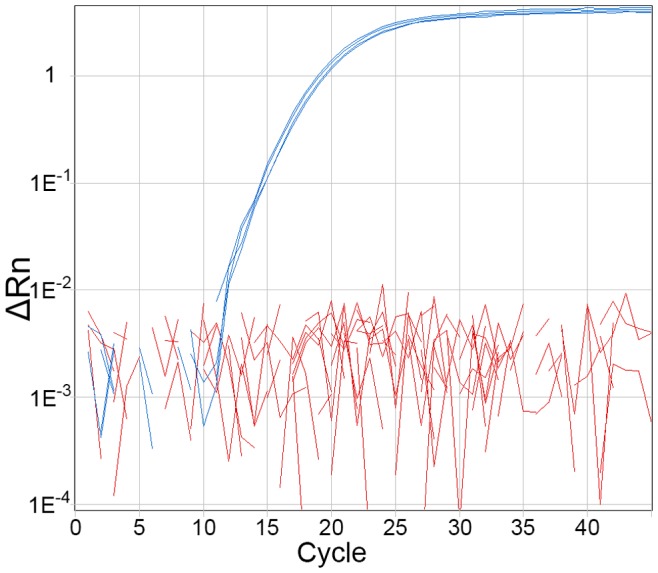
Bu550 differentiates Burkholderia ubonensis from other soil- and water-borne bacterial species that grow on Ashdown’s agar [13]. Only B. ubonensis (shown in blue) amplifies with this assay. Other species (shown in red) fail to amplify.
Discussion
Originally identified from soil collected in Ubon Ratchathani province, Thailand, in 1989 [16], Burkholderia ubonensis is now known to be an abundant environmental bacterium in northern Australia. Both regions are also endemic for B. pseudomallei, a pathogenic bacterium that causes significant morbidity and mortality, with up to 50% of infected individuals succumbing to disease [26]. Ashdown’s agar (ASA) is a common medium used in endemic regions to select B. pseudomallei from clinical and environmental specimens. However, ASA also supports the growth of other Burkholderia spp., several other Gram-negative bacteria and even some gentamicin-resistant strains of Gram-positive Staphylococcus spp. (Table 1; [27]). Of these non-B. pseudomallei species, we showed that B. ubonensis is the most frequently isolated in northern Australia due to its strong morphological resemblance to B. pseudomallei. Like B. pseudomallei, B. ubonensis possesses several morphotypes on ASA (Figure 1), many of which closely resemble B. pseudomallei morphotypes [24]. Therefore, it is impossible to differentiate B. ubonensis from B. pseudomallei based on morphological characteristics alone.
Due to the inherent difficulty in distinguishing B. ubonensis and B. pseudomallei based on morphology, we developed a cost-effective, rapid PCR-based method for differentiating B. ubonensis from other soil- and water-borne species that grow on ASA, particularly B. pseudomallei. Current methods of characterising Burkholderia spp. rely on dideoxy sequencing [8], [17], [18], which is an expensive and time-consuming endeavour for screening large isolate collections. We applied comparative whole-genome sequence analysis of 19 Burkholderia spp., including eight B. ubonensis isolates, to identify a locus specific for B. ubonensis. The specificity of one candidate locus, 550, was confirmed using in-depth in silico BLAST analysis. We subsequently developed a cost-effective real-time PCR assay, Bu550, for its interrogation. The performance of Bu550 was validated against a diverse collection of Burkholderiaceae and other bacterial species that grow on ASA, obtained from soil and water across the Northern Territory and Thailand. Our results indicate that Bu550 is highly specific towards B. ubonensis, with all other species we tested failing to amplify.
The design of the B. ubonensis-specific BHQ probe-based assay enables multiplex capability with the B. pseudomallei-specific assay, TTS1. B. ubonensis has demonstrated antagonistic activity towards B. pseudomallei and shows promise for biocontrol of naturally occurring B. pseudomallei, particularly in areas of high endemicity [15]. However, it is not yet known to what extent near-neighbour species such as B. ubonensis affect the natural prevalence of B. pseudomallei. Our future work will involve quantifying B. ubonensis and B. pseudomallei from soils obtained in B. pseudomallei-endemic regions to determine the relative abundance of these species, thereby improving our understanding of B. pseudomallei and B. ubonensis ecology. Bu550, in combination with TTS1, will be an important and valuable tool in such ecological studies.
Acknowledgments
We kindly thank Christopher Allender and Kevin Drees (Northern Arizona University) and Yuwana Podin and Glenda Harrington (Menzies School of Health Research) for helpful discussions on Burkholderia pseudomallei near-neighbours and for assistance with laboratory aspects of this work. We also wish to thank colleagues at the Wellcome Trust-Mahidol University-Oxford Tropical Medicine Research Programme for assistance with soil sampling in Thailand that yielded some of the isolates utilised in this study.
Funding Statement
The authors thank the Edgewood Chemical Biological Center (ECBC) for their contribution to genome sequencing efforts. This work was funded by a Menzies School of Health Research grant, by the US Department of Homeland Security Science and Technology Directorate through award HSHQDC-10-C-00139 and by the Defense Threat Reduction Agency award HDTRA1-12-C-0066. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Coenye T, Vandamme P (2003) Diversity and significance of Burkholderia species occupying diverse ecological niches. Environ Microbiol 5: 719–729. [DOI] [PubMed] [Google Scholar]
- 2. Gillis M, Tran Van V, Bardin R, Goor M, Hebbar P, et al. (1995) Polyphasic taxonomy in the genus Burkholderia leading to an emended description of the genus and proposition of Burkholderia vietnamiensis sp. nov. for N2-fixing isolates from rice in Vietnam. Int J Syst Evol Microbiol 2: 274–289. [Google Scholar]
- 3. Wuthiekanun V, Smith MD, Dance DA, White NJ (1995) Isolation of Pseudomonas pseudomallei from soil in north-eastern Thailand. Trans R Soc Trop Med Hyg 89: 41–43. [DOI] [PubMed] [Google Scholar]
- 4.Dance DAB (2005) Melioidosis and glanders as possible biological weapons. In: Bioterrorism and infectious agents A new dilemma for the 21st century. New York: Springer Science and Business Media. 99–145.
- 5. Currie BJ, Dance DA, Cheng AC (2008) The global distribution of Burkholderia pseudomallei and melioidosis: an update. Trans R Soc Trop Med Hyg 102 Suppl 1S1–4. [DOI] [PubMed] [Google Scholar]
- 6. Limmathurotsakul D, Dance DA, Wuthiekanun V, Kaestli M, Mayo M, et al. (2013) Systematic Review and Consensus Guidelines for Environmental Sampling of Burkholderia pseudomallei . PLoS Negl Trop Dis 7: e2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Vanlaere E, Lipuma JJ, Baldwin A, Henry D, De Brandt E, et al. (2008) Burkholderia latens sp. nov., Burkholderia diffusa sp. nov., Burkholderia arboris sp. nov., Burkholderia seminalis sp. nov. and Burkholderia metallica sp. nov., novel species within the Burkholderia cepacia complex. Int J Syst Evol Microbiol 58: 1580–1590. [DOI] [PubMed] [Google Scholar]
- 8. Baldwin A, Mahenthiralingam E, Thickett KM, Honeybourne D, Maiden MC, et al. (2005) Multilocus sequence typing scheme that provides both species and strain differentiation for the Burkholderia cepacia complex. J Clin Microbiol 43: 4665–4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kman NE, Bachmann DJ (2012) Biosurveillance: a review and update. Adv Prev Med 2012: 301408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Glass MB, Beesley CA, Wilkins PP, Hoffmaster AR (2009) Comparison of four selective media for the isolation of Burkholderia mallei and Burkholderia pseudomallei . Am J Trop Med Hyg 80: 1023–1028. [PubMed] [Google Scholar]
- 11. Lowe P, Engler C, Norton R (2002) Comparison of automated and nonautomated systems for identification of Burkholderia pseudomallei . J Clin Microbiol 40: 4625–4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mayo M, Kaesti M, Harrington G, Cheng AC, Ward L, et al. (2011) Burkholderia pseudomallei in unchlorinated domestic bore water, Tropical Northern Australia. Emerg Infect Dis 17: 1283–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ashdown LR (1979) An improved screening technique for isolation of Pseudomonas pseudomallei from clinical specimens. Pathology 11: 293–297. [DOI] [PubMed] [Google Scholar]
- 14. Levy A, Merritt AJ, Aravena-Roman M, Hodge MM, Inglis TJ (2008) Expanded range of Burkholderia species in Australia. Am J Trop Med Hyg 78: 599–604. [PubMed] [Google Scholar]
- 15. Marshall K, Shakya S, Greenhill AR, Padill G, Baker A, et al. (2010) Antibiosis of Burkholderia ubonensis against Burkholderia pseudomallei, the causative agent for melioidosis. Southeast Asian J Trop Med Public Health 41: 904–912. [PubMed] [Google Scholar]
- 16. Yabuuchi E, Kawamura Y, Ezaki T, Ikedo M, Dejsirilert S, et al. (2000) Burkholderia uboniae sp. nov., L-arabinose-assimilating but different from Burkholderia thailandensis and Burkholderia vietnamiensis . Microbiol Immunol 44: 307–317. [DOI] [PubMed] [Google Scholar]
- 17. Godoy D, Randle G, Simpson AJ, Aanensen DM, Pitt TL, et al. (2003) Multilocus sequence typing and evolutionary relationships among the causative agents of melioidosis and glanders, Burkholderia pseudomallei and Burkholderia mallei . J Clin Microbiol 41: 2068–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Payne GW, Vandamme P, Morgan SH, Lipuma JJ, Coenye T, et al. (2005) Development of a recA gene-based identification approach for the entire Burkholderia genus. Appl Environ Microbiol 71: 3917–3927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Novak RT, Glass MB, Gee JE, Gal D, Mayo MJ, et al. (2006) Development and evaluation of a real-time PCR assay targeting the type III secretion system of Burkholderia pseudomallei . J Clin Microbiol 44: 85–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Currie BJ, Gal D, Mayo M, Ward L, Godoy D, et al. (2007) Using BOX-PCR to exclude a clonal outbreak of melioidosis. BMC Infect Dis 7: 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zerbino DR, Birney E (2008) Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18: 821–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Quinlan AR, Hall IM (2010) BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chantratita N, Wuthiekanun V, Boonbumrung K, Tiyawisutsri R, Vesaratchavest M, et al. (2007) Biological relevance of colony morphology and phenotypic switching by Burkholderia pseudomallei . J Bacteriol 189: 807–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sowers B, Reddington M, Lyttle M, Johansson MK, Cook R (2004) CAL Fluor™ dyes: Multiplexing reporters for real-time PCR instruments and probe designs. Quantitative PCR. San Diego, CA: Biosearch Technologies.
- 26. White NJ (2003) Melioidosis. Lancet 361: 1715–1722. [DOI] [PubMed] [Google Scholar]
- 27. Peacock SJ, Chieng G, Cheng AC, Dance DA, Amornchai P, et al. (2005) Comparison of Ashdown’s medium, Burkholderia cepacia medium, and Burkholderia pseudomallei selective agar for clinical isolation of Burkholderia pseudomallei . J Clin Microbiol 43: 5359–5361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gee JE, Glass MB, Novak RT, Gal D, Mayo MJ, et al. (2008) Recovery of a Burkholderia thailandensis-like isolate from an Australian water source. BMC Microbiol 8: 54. [DOI] [PMC free article] [PubMed] [Google Scholar]