Abstract
Although the B clade of human immunodeficiency virus type 1 (HIV-1) envelopes (Env) includes five highly variable regions, each of these domains contains a subset of sequences that remain conserved. The V3 loop has been much studied for its ability to elicit neutralizing antibodies, which are often restricted to a limited number of closely related strains, likely because a large number of antigenic structures are generated from the diverse amino acid sequences in this region. Despite these strain-specific determinants, subregions of V3 are highly conserved, and the effects of different portions of the V3 loop on Env tropism and immunogenicity have not been well delineated. For this report, selective deletions in V3 were introduced by shortening of the stem of the V3 loop. These mutations were explored in combination with deletions of selected V regions. Progressive shortening of the stem of V3 abolished the immunogenicity as well as the functional activity of HIV Env; however, two small deletions on both arms of the V3 stem altered the tropism of the dualtropic 89.6P viral strain so that it infected only CXCR4+ cells. When this smaller deletion was combined with removal of the V1 and V2 loops and used as an immunogen in guinea pigs, the antisera were able to neutralize multiple independent clade B isolates with a higher potency. These findings suggest that highly conserved subregions within V3 may be relevant targets for eliciting neutralizing antibody responses, affecting HIV tropism, and increasing the immunogenicity of AIDS vaccines.
Among the mechanisms used by human immunodeficiency virus (HIV) to avoid immune system recognition and antibody neutralization, the variable regions of the envelope play an important role in evasion. The envelope protein utilizes a variety of mechanisms to evade detection, including carbohydrate modification, conformational flexibility, and genetic variability between isolates (4, 5, 7, 9, 14, 23). Genetic diversity in specific segments of the viral Env protein gives rise to the variable regions. These regions serve to block access to the CD4 binding domain as well as the chemokine receptor binding site, in addition to influencing virus neutralization sensitivity and being responsible for the strain specificity of neutralizing antibodies (10, 19, 24). Although the variable regions have been defined by their genetic differences among alternative isolates, it is clear that there are subregions within the V loops that show some degree of conservation. This sequence homology is particularly evident in such regions as the tip of the V3 loop (12). Other motifs can also be identified in various virus strains. For example, specific N-linked glycosylation sites and sequences near the base of the V3 loop are well conserved (12). For this report, the fine specificity of the variable regions was explored in further detail. Specifically, the V3 loop was examined with regard to the contribution of the putative stem structures to viral tropism and immunogenicity. We found that a specific mutation that shortens the stem of the V3 loop can alter the tropism of the HIV envelope. This mutation, in combination with deletion of the V1 and V2 loops, further enhances the ability of the envelope to elicit a neutralizing antibody response.
MATERIALS AND METHODS
Antibody.
Anti-HIV type 1 (HIV-1) human monoclonal antibody 2F5 (20) and human HIV immunoglobulin G (IgG) were obtained from the AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, National Institutes of Health. Anti-HIV p24 antibody KC57-RD1 was obtained from Beckman Coulter, Inc.
Cell and virus stocks.
Human embryonic kidney 293 cells were purchased from the American Type Culture Collection and maintained in Dulbecco's modified Eagle's medium (Invitrogen, Carlsbad, Calif.) containing 10% fetal bovine serum (FBS) and 100 μg of penicillin-streptomycin/ml. The human T-cell leukemia cell line MT-2 and the HeLa-derived cell line MAGI-CCR5 were obtained from the AIDS Research and Reference Reagent Program.
HIV-1 isolates (ADA, JRCSF, JRFL, Bal, SF162, and 89.6) were obtained from the AIDS Research and Reference Reagent Program. Primary isolates 6101 (previously called P15) and 1168 were obtained from David Montefiori of Duke University (3). The viruses were expanded by two or three cycles of growth on phytohemagglutinin- and interleukin-2 (IL-2)-stimulated peripheral blood mononuclear cells (PBMC). For the production of the working stock virus, PBMC were exposed to undiluted virus for 2 h at a concentration of 107 cells/ml. IL-2 culture medium was added to bring the concentration to 106 cells/ml. The IL-2 culture medium was changed every 2 days, and supernatants were collected during the peak of p24 expression, usually 5 to 10 days after infection. Virus stocks were made cell free by centrifugation at 1,000 × g and filtration through a 0.45-μm-pore-size filter. In some cases, viral stocks were concentrated as much as 10-fold by using a 100-kDa cutoff polyethersulfone filter (Centricon Plus Biomax filter; Millipore, Bedford, Mass.) according to the manufacturer's instructions. Virus aliquots were stored in the vapor phase of liquid nitrogen. Viruses BL01 and BR07 were provided by Dana Gabuzda of the Dana-Farber Cancer Institute. Both are chimeric infectious molecular clones of HIV strain NL4-3 that contain the full-length env genes from primary HIV-1 isolates (18). After the initial plasmid transfection of 293 cells, these viruses were expanded in PBMC as described above.
Buoyant density gradient analysis of lentiviral vectors.
293T cells (3 × 106) were transfected with 3 μg each of the relevant Gag and Env expression vectors in a 100-mm-diameter tissue culture dish with Dulbecco's modified Eagle's medium. Three days later, the cell supernatants were collected and mixed with 60% OptiPrep (iodixanol) medium (Invitrogen); the final concentration of OptiPre was adjusted to a 30% density gradient, formed by centrifugation at 45,000 × g for 6 h in a VTI50 rotor (used according to the manufacturer's instructions; Invitrogen); and each fraction was collected according to the indicated density. Lentiviral vector proteins were separated in a sodium dodecyl sulfate-4 to 15% polyacrylamide gel electrophoresis (SDS-4 to 15% PAGE) gel, transferred onto an Immobilon-P membrane, and blotted for the expression of Gag (human HIV IgG, used at 1:5,000) and Env (human HIV IgG, used at 1:5,000).
Construction of recombinant adenoviruses.
Adenovirus type 5 (Ad5)-based first-generation (ΔE1 and ΔE3) recombinant adenoviruses expressing different V loop deletions of gp140(ΔCFI) were constructed as described previously (1). In brief, PacI-linearized shuttle vectors containing V loop deletions of gp140(ΔCFI) were recombined with the right side of Ad5 genomic DNA carried in a cosmid by use of Cre recombinase (Novagen, Madison, Wis.). The resulting recombinants were ethanol precipitated, dissolved in Tris-EDTA, and transfected into 293 cells. Recombinant adenoviruses were observed based on plaque formation 10 to 14 days after transfection. Viruses were amplified, purified two times through a CsCl gradient, and stored in phosphate-buffered saline (PBS) plus 15% glycerol at −20°C.
Production of pseudotyped lentivirus.
HIV-Luc pseudotyped with HIV gp160(89.6P) and its V3 deletion mutants were prepared according to published methods (17). Briefly, the packaging vector pMD 8.2, pHR-Luciferase, and the envelope-expressing vector were transiently cotransfected into 293T cells by use of calcium phosphate. Supernatants were harvested 48 and 72 h after transfection, filtered, and stored at −80°C. Virus concentrations were determined by an enzyme-linked immunosorbent assay (ELISA) for the p24 antigen (Coulter). The same amount of virus was added onto MT-2 (X4 tropic) and MAGI-CCR5 (R5 tropic) cells, and cells were incubated for 2 h at 37°C. The cells were harvested 48 h after infection and lysed in cell culture lysis buffer (Promega). The luciferase assay was performed according to the manufacturer's recommendations (Promega).
Plasmid construction.
Plasmids pVRC1012-gp140(ΔCFI) (HXB2/BaL chimera) and pVRC1012-gp145(ΔCFI) (HXB2/BaL chimera) have been described previously (7). To make gp140(ΔCFI)(ΔV1V2) and gp145(ΔCFI)(ΔV1V2), we performed PCR to amplify an XbaI/NheI fragment covering ATG and the boundary of the V1 loop using primers 5′CCTCTAGACACCATGCGCGTGAAGGAGAAG3′ and 5′CCGCTAGCGTCGGTGCACTTCAGGCTCACGCACAGGGG3′ and an NheI/ApaI fragment covering the 3′ boundary of the V2 loop and the C3 region using primers 5′CCGCTAGCACCAGCTGCAACACCAGCGTGATCACCCAG3′ and 5′GGTGCAGGGGCCCTTGCCGTTGAACTTCTT3′. The XbaI/NheI- and NheI/ApaI-digested PCR fragments were cloned into XbaI/ApaI-digested pVRC1012-gp140(ΔCFI) and pVRC1012-gp145(ΔCFI). The resulting plasmids, pVRC1012-gp140(ΔV1V2) and pVRC1012-gp145(ΔCFI)(ΔV1V2), have deletions of the V1 and V2 loops as follows: CTDASTSC. Two extra amino acids (AS) were introduced due to the introduction of an NheI site. A similar approach was used to make other V loop deletion mutants of gp145ΔCFI (HXB2/BaL chimera) and gp140ΔCFI (HXB2/BaL chimera). The amino acid sequences of deleted V loops are as follows: ΔV1, CTDASKNC; ΔV2, CSFASTSC; ΔV3, CTRASAHC; and ΔV4, CNSASLPC.
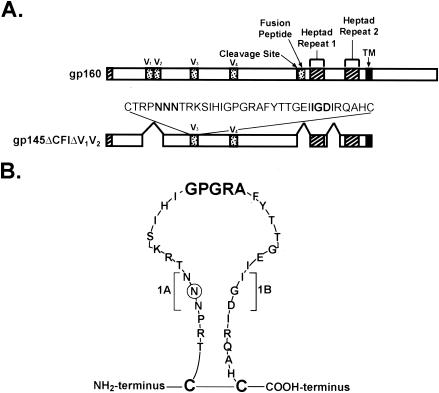
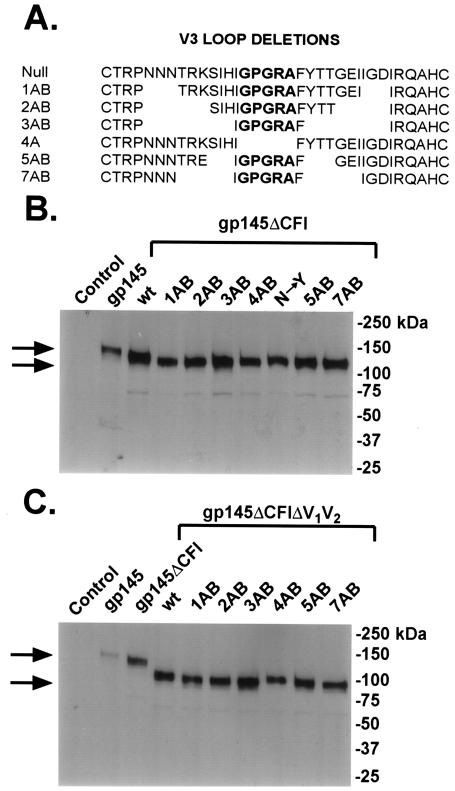
V3 deletion mutants were made by using the PCR-based Quickchange (Stratagene, La Jolla, Calif.) method according to the manufacturer's instructions. Each mutant was confirmed by double strain sequencing. An ApaI/SexAI fragment containing each confirmed V3 deletion was swapped with a corresponding fragment in pVRC1012-gp140(ΔCFI)(ΔV1V2) and pVRC1012-gp145(ΔCFI)(ΔV1V2). The cDNA encoding gp160(89.6P)(KB9) (11) was synthesized by using human preferred codons. Plasmids expressing different V3 deletion mutants of gp160(89.6P) were made similarly and are shown in Fig. 1. The details for each V3 mutant are listed in Fig. 2A.
FIG. 1.
Schematic representation of Env mutations. (A) The major structural motifs in HIV Env are shown, together with the selected expression vectors used in these studies. V1, V2, V3, and V4 indicate the respective variable regions, and the sequence of the relevant V3 loop (HXB2/BaL chimera) is indicated. (B) Schematic structure of the V3 loop (HXB2/BaL chimera) with V3 (1AB) stem-shortening mutations.
FIG. 2.
Mutations in the stem of the V3 loop and protein expression of various gp145ΔCFI (HXB2/BaL chimera) V3 deletion mutants. (A) Sequences of progressive V3 stem deletion mutations in Env from the HXB2/BaL chimera. (B) Protein expression of gp145ΔCFI (HXB2/BaL chimera) V3 deletion mutant expression vectors. The indicated mutations in the gp145ΔCFI constructs, described previously (7), were prepared and analyzed by SDS-PAGE followed by Western blot analysis with human monoclonal antibody 2F5. Plasmid expression vectors encoding the indicated mutants were transfected into 293 cells by use of calcium phosphate. Cell lysates were collected 48 h later. (C) Expression of gp145 (HXB2/BaL chimera) V3 deletion mutant vectors with the V1 and V2 regions deleted.
ELISA.
Guinea pig anti-HIV gp140(ΔCFI) ELISA titers were measured by using a modified lectin capture method. Briefly, Immunon 2HB ELISA plates (Thermo Labsystems, Franklin, Mass.) were coated with 100 μl of Galanthus nivalis lectin (Sigma, St. Louis, Mo.) (10 μg/ml in PBS)/well overnight at 4°C. The plates were blocked with 200 μl of PBS containing 10% FBS for 2 h at room temperature and washed twice with PBS containing 0.2% Tween 20 (PBS-T). One hundred microliters of tissue culture supernatant from pVRC 1012-gp140(ΔCFI)-transfected 293 cells was added to each well and incubated at room temperature for 1 h. The plates were washed five times with PBS-T. One-hundred-microliter serial dilutions of guinea pig immune serum in PBS containing 1% FBS were then added in triplicate and incubated for 1 h at room temperature. After five washes with PBS-T, 100 μl of horseradish peroxidase-conjugated F(ab)′2 donkey anti-guinea pig IgG (1:5,000) (Jackson ImmunoResearch Laboratories, West Grove, Pa.) in PBS plus 1% FBS was added to each well and incubated for 1 h at room temperature. The plates were washed five times with PBS-T, developed by the addition of 100 μl of o-phenylenediamine dihydrochloride (Sigma) (one gold and one silver tablet in 20 ml of water), and incubated at room temperature for 30 min. The reaction was stopped by the addition of 100 μl of 1 N H2SO4 to each well. The readout was measured at 450 nm by a SPECTRAmax plate reader (Molecular Devices, Sunnyvale, Calif.). The end-point dilution was calculated by picking the dilution for which the readout was above that of the 1:100 dilution of preimmune serum.
Neutralization assay.
The single-round intracellular p24 antigen flow cytometric HIV-1 neutralization assay has been described previously (16). Briefly, 40 μl of virus stock was incubated with 10 μl of heat-inactivated guinea pig immune serum (multiplicity of infection, approximately 0.1). After incubation for 30 min at 37°C, 20 μl of PBMC (1.5 × 105 cells) was added to each well. PBMC were maintained in IL-2 culture medium containing 1 μM indinavir, and the cells were fed on day 1 with 150 μl of IL-2 culture medium containing indinavir. One day after infection, cells were stained for intracellular p24 antigen with the KC57 anti-p24 antibody, followed by the quantitation of HIV-1-infected cells by flow cytometry. The percent neutralization was defined as the reduction in the number of p24-positive cells compared with the number for wells incubated with the corresponding preimmune serum.
To obtain 50% inhibitory concentration (IC50) and IC80 data, we incubated serial dilutions of antiserum with the virus, as described above. Antiserum dose-response curves were fit with a nonlinear function, and the IC50 and IC80 of the virus were calculated by a least-squares regression analysis. Statistical analysis of IC50 titers was performed with the nonparametric Mann-Whitney rank-order test (GraphPad Prism software package 3.0; GraphPad Software Inc., San Diego, Calif.).
Vaccination.
Guinea pigs were immunized intramuscularly with 500 μg (in 400 μl of PBS) of the gp145 version of plasmid DNA at weeks 0, 2, and 6. At week 14, the guinea pigs were boosted with 1011 (in 400 μl of PBS) particles of recombinant Ad expressing the corresponding gp140 version of the protein. Sera were collected at weeks −2 and 16, divided into aliquots, and frozen at −20°C.
Western blotting.
293 cells were transfected with plasmid DNA expressing each immunogen by the calcium phosphate method performed according to the manufacturer's instructions (Invitrogen). Forty-eight hours after transfection, the cells were harvested, washed once with PBS, resuspended in lysis buffer (50 mM HEPES [pH 7.0], 150 mM NaCl, 1% NP-40, 1× proteinase inhibitor cocktail), and incubated on ice for 45 min. The cell lysate was centrifuged at 16,000 × g for 10 min at 4°C. The supernatant was collected, and the protein concentration was measured. Twenty micrograms of protein was mixed with 2× sample loading buffer (100 mM Tris, 4% SDS, 20% glycerol, 5% 2-mercaptoethanol, 0.2% bromophenol blue) and boiled for 5 min. The sample was then resolved by 4 to 15% gradient SDS-PAGE and transferred onto a nitrocellulose membrane (Bio-Rad). The membrane was blocked twice with Tris-buffered saline containing 0.3% Tween 20, 5% skim milk, and 1% bovine serum albumin at room temperature for 10 min, followed by incubation with the 2F5 antibody (1:2,500) in blocking buffer for 1 h at room temperature. The membrane was washed twice with 100 ml of Tris-buffered saline containing 0.3% Tween 20, followed by incubation with horseradish peroxidase-conjugated goat anti-human IgG (Chemicon, Temecula, Calif.) (1:5,000) for 30 min at room temperature. After two washes with 100 ml of washing buffer, the membrane was developed by use of ECL Western blotting detection reagents (Amersham, Piscataway, N.J.) and was exposed on Hyperfilm ECL (Amersham).
RESULTS
Expression of V3 region mutants.
Modifications of three regions of the HIV envelope, the cleavage site, fusion peptide, and interhelical coiled-coil domain (ΔCFI), were shown previously to enhance the ability of Env to elicit an antibody response (6). We evaluated additional mutations either in different V regions or through selective modifications of V3 (Fig. 1). The internal V3 loop deletions were made both in gp145ΔCFI (HXB2/BaL chimera), which was inserted into DNA expression vectors for primary immunization, and in gp140ΔCFI (HXB2/BaL chimera), which was placed into an adenoviral vector for boosting. These series of mutations were also introduced into the strain 89.6P Env (Fig. 2A), a dualtropic virus that was analyzed initially in functional pseudotyping assays for effects on tropism of different chemokine receptors. The expression of these progressive deletions of the V3 region was assessed by Western blot analysis. Immunoreactive proteins of the expected molecular weights were detected in cell lysates from 293T cells transfected with these expression vectors (Fig. 2B). These same mutations were also introduced into gp145ΔCFI (HXB2/BaL chimera) with the V1 and V2 regions deleted, and their protein expression was also confirmed (Fig. 2C).
Effects of V3 region mutations on Env function.
To evaluate the effects of progressive deletions in the V3 region, we analyzed the 89.6P Env mutants for the ability to mediate viral entry, using an HIV vector encoding a luciferase reporter gene. The abilities of these V3 variants to incorporate into the pseudotyped lentivirus were confirmed by buoyant density centrifugation (Fig. 3A). Because this Env is dualtropic, both a CXCR4+ cell line, MT-2, and a CCR5+ indicator cell line, MAGI-CCR5, were tested. Although longer deletions of the V3 region abolished the function of the 89.6 Env, the smallest deletion, which removed three amino acids from each side of the V3 loop, termed 1AB, preserved the ability of the Env to infect the CXCR4 target MT-2 cells (Fig. 3B, left panel). In contrast, even the smallest deletion of the V3 loop abolished its ability to infect MAGI-CCR5 cells (Fig. 3B, right panel). These data indicate that the length of the V3 loop or the deleted amino acids plays a critical role in determining its tropism for alternative chemokine receptors, and CCR5 tropism of this Env was more sensitive than its CXCR4 tropism.
FIG. 3.
Effects of mutations in the stem of the V3 loop on tropism of 89.6P Env. (A) Buoyant density sedimentation analysis of indicated V3 mutants in lentiviral vector particles, performed as described in Materials and Methods. (B) Effects of V3 mutations in strain 89.6P Env on infection of a CXCR4-tropic cell line, MT-2 (left), and a CCR-5-tropic indicator cell line, MAGI-CCR5 (right), using a luciferase reporter gene. The positions of the indicated V3 mutations in strain 89.6P Env are the same as those shown for the HXB2/BaL chimera (Fig. 2A). Both codon-modified and wild-type (wt) 89.6P Envs were used as positive controls.
Effect of V region mutations on immunogenicity.
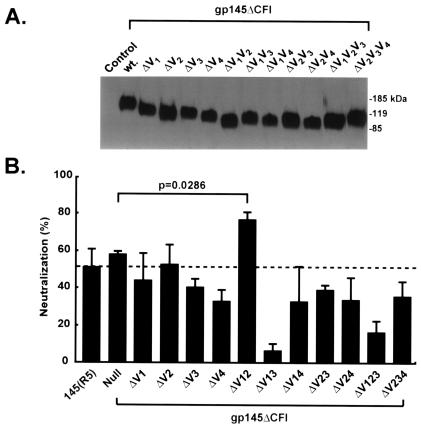
To evaluate the effects of these and other V region mutations on the elicitation of a neutralizing antibody response, we made deletions of different V regions in gp145ΔCFI (HXB2/BaL chimera) and gp140ΔCFI (HXB2/BaL chimera), individually or with combinations of V1 to V4. Expression of the mutants revealed similar levels of protein by Western blotting (Fig. 4A). These V region mutants were assessed for the ability to elicit neutralizing antibodies by using DNA/Ad immunization of guinea pigs. The elimination of specific V regions, particularly the combination of V1 and V3 or V1 and V4, markedly reduced their ability to induce a neutralizing antibody response. In contrast, vectors with specific combined deletions, including deletion of the V1 and V2 regions, increased the neutralizing antibody response to HIVBaL (Fig. 4B). The increased potency of the V1V2 deletion construct was confirmed in further experiments using nine additional primary HIV-1 isolates (data not shown); these data strongly suggested that this deletion construct provided better immunogenicity than the other constructs shown in Fig. 4B. Based on these analyses, additional V3 region mutations were made in the V1V2 deletion construct and the gp145ΔCFI envelope mutant. When tested against HIVBaL, the gp145ΔCFI immunogen elicited slightly increased neutralization compared to wild-type gp145, but it did not reach statistical significance (Fig. 4B and 5A). The less impressive response for V1V2 gp145ΔCFIΔ seen in Fig. 5A was due to a single nonresponder in a group of four animals. The actual values in Fig. 5A for gp145ΔCFI were 47, 60, 51, and 53, and those for V1V2 gp145ΔCFIΔ were 16, 71, 62, and 77%. However, we confirmed the improved immunogenicity of the ΔV1V2 mutant in numerous additional experiments. A further increment was suggested when the 1AB mutation was included in the V1V2 gp145ΔCFIΔ immunogen. When larger deletions of the V3 region were made, they became successively less able to elicit a neutralizing antibody response. In contrast, all mutants were able to elicit comparable antibody responses, as determined by ELISA end-point limiting dilution analysis (Fig. 5B).
FIG. 4.
Expression of different HIV gp145 (HXB2/BaL chimera) V region mutants and induction of neutralizing antibodies. (A) Expression of the indicated HXB2/BaL V region mutants was determined by SDS-PAGE followed by Western blotting with transfected 293 cells. (B) Neutralizing activity against BaL in sera from guinea pigs immunized with the indicated DNA/Ad expression vectors. Sera were tested at a 1:5 dilution. Results are the means ± standard errors of four guinea pig sera for each construct. The P value shown is the result of a Mann-Whitney test comparing the median neutralization value of the two groups indicated.
FIG. 5.
Characterization of antibody response induced by gp145 (HXB2/BaL chimera) ΔV1V2 and selected V3 deletion mutants. (A) Neutralization activity against BaL induced by immunization of guinea pigs with the indicated mutants, including the ΔV1V2 deletion mutants and the ΔV1V2V3(1AB) stem-shortening mutants. Sera were tested at a 1:5 dilution. Results are the means ± standard errors of four guinea pig sera for each construct. Results from one of two independent experiments are shown. The sera tested were independent from the sera tested for Fig. 4B. (B) Total ELISA titers in the same guinea pig sera are shown and are comparable between the different mutants.
Comparative neutralization profile of the V1V2 and V3 (1AB) deletion.
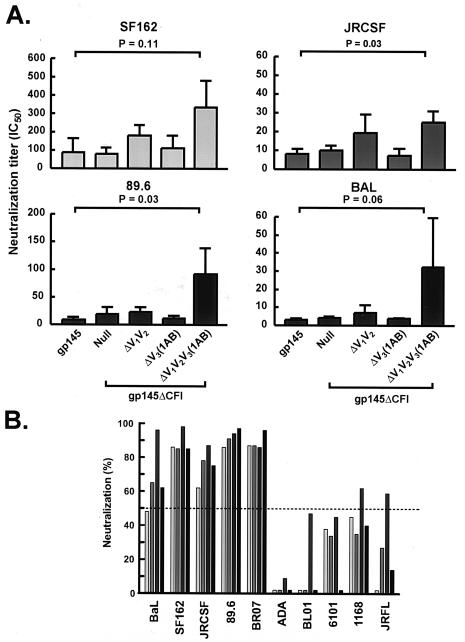
To more rigorously examine the effects of V3 deletion mutations on the breadth and potency of neutralization, we immunized guinea pigs with selected mutants of the gp145 DNA followed by a gp140 adenoviral vector boost, including the wild type or the ΔCFI or ΔV1V2V3(1AB)ΔCFI mutation. These modified Envs were compared for the ability to elicit a neutralizing antibody response. For a comparison of the potency of the neutralizing antibody response, serial dilutions of the guinea pig sera were tested against four primary viruses (BaL, JRCSF, 89.6, and SF162), and the respective IC50 values were calculated. Among the modified Env immunogens, the ΔV1V2V3(1AB)ΔCFI mutant was the most effective at inhibiting these four isolates. Compared to the wild type, the median IC50 of this construct was statistically significantly higher against two viruses (P = 0.03 for JRCSF and 89.6) and was close to significance for one virus (P = 0.06 for BaL) (Fig. 6A). Antisera elicited by this optimal immunogen, ΔV1V2V3(1AB)ΔCFI, were examined against a panel of 10 primary HIV-1 isolates. The antisera displayed reactivities against a number of unrelated HIV-1 strains. Five of the 10 viruses were moderately or strongly neutralized by a 1:5 dilution of guinea pig sera. Fifty percent neutralization was not achieved against three viruses (ADA, 6101, and BL01), and two viruses were neutralized at a low level (50 to 60%) by one of the four guinea pig sera; therefore, the immunogen remained limited in its breadth.
FIG. 6.
Comparison of breadth and potency of the antibody response induced by selected V region mutants. (A) IC50 titers against four clade B primary isolates are indicated (see Materials and Methods). Results are the means ± standard errors of four guinea pig sera for each construct. The statistically significant differences between ΔV1V2V3(1AB) and the wild-type gp140/145 are shown (Mann-Whitney test). (B) Four individual sera from ΔV1V2V3(1AB)-immunized guinea pigs were screened against a panel of 10 primary viruses. Sera were tested at a 1:5 dilution. Percentages of neutralization (compared with corresponding preimmune sera) are indicated. Data shown are averages of two experiments.
DISCUSSION
For this paper, we examined the ability of different V region mutations to alter the immunogenicity of the HIV envelope. Members of our laboratory have previously shown that mutations in the cleavage site, fusion domain, and interhelical coiled-coil region can enhance immunogenicity by improving the ability of Env vaccines to elicit an antibody response (7). Although improvements were observed with the ΔCFI mutations in their ability to elicit antibodies against Env, the enhancement of neutralizing antibodies was less striking. The goal of the present study was therefore to expand the potency and breadth of the neutralizing antibody response by including additional modifications and systematically evaluating the contributions of various V regions and V3 subregions. Truncations in the V3 region markedly altered the functional properties of the HIV89.6P Env. Mutations exceeding six amino acids, three each on opposite sides of the loop, abolished its functions of both CXCR4- and CCR5-mediated viral entry. In contrast, the smaller 1AB truncation eliminated the CCR5-tropic activity of this envelope but preserved its ability to target CXCR4+ cells. When the 1AB mutation made in gp145ΔCFI (HXB2/BaL chimera) was evaluated for its ability to elicit neutralizing antibodies after DNA priming and adenoviral vector boosting in guinea pigs, this mutation appeared to have the greatest efficacy in eliciting a neutralizing antibody response. This response was only seen with deletions of the V1 and V2 loops, as it was not observed in the gp140/145 ΔCFI background (Fig. 6A). After a comparison of the potencies of different immunogens, the breadth of the optimal candidate was determined against 10 representative clade B viral isolates. The ability of this antiserum to inhibit diverse strains increased, with higher IC50 titers and increased reactivities against different HIV isolates (Fig. 6). A previous study found differences in the immunogenicities of gp145 and gp145ΔCFI after DNA immunization (7), as detected by immunoprecipitation followed by Western blot analysis. Although the gp145 ΔCFI DNA immunogen induced an improved antibody response as a DNA vaccine, we show here that the ability of this cDNA insert to elicit neutralizing antibodies when used in a priming-boosting combination is not distinguishable. The results described here are consistent with those of the previous study in which neutralizing antibody responses for the mouse were not measured. It is likely that the differences observed with DNA vaccination are obscured by the potent DNA/Ad priming-boosting immunization, as is evident by the high ELISA titers. However, the additional V3 change on the ΔCFI backbone can further enhance the neutralizing antibody response.
Previous studies have indicated that subregions of V3 may be conserved among various isolates and may affect Env functioning. This conservation is evident in specific sequences in this region, for example in the tip of the V3 loop (12). Although the V3 region has been shown to affect the tropism of HIV for the chemokine receptor (2), the effects of progressive deletions in the V3 loop and its selective effect on CXCR4 targeting have not been previously appreciated. Recently, it was suggested that conserved conformational determinants are present in the V3 loop of diverse isolates that show similar sensitivities to neutralizing antibodies (8, 13, 21). For example, the ability of the monoclonal antibody 447-52D and related V3 monoclonal antibodies to inhibit different strains with disparate V3 sequences suggests that common determinants may be shared by genetically disparate strains (8, 9). The enhanced immunogenicity of the V1V2 mutations in this study suggests that there may be a masking of the V3 loop by V1 and V2 in HIVBaL (6, 15, 25). Deletion of the V2 region has been suggested in previous studies to improve the antibody response (22), but it is not certain whether similar mechanisms are responsible for those effects and the observations noted here in a different strain in combination with V3 partial deletions, since the additional V1 deletion and the 1AB mutation in V3 further enhance the immunogenicity. The increased breadth of this response suggests that common antigenic determinants are shared by many, though not all, clade B viruses. Taken together, these conserved regions reflect underlying functional requirements and structural homologies between different viruses and raise the possibility that families of V3 determinants may be targeted to expand the breadth of the neutralizing antibody response.
Acknowledgments
We thank Tina Suhana and Ati Tislerics for manuscript preparation, Karen Stroud for figure preparation, Mark Louder and Anna Sambor for technical support, and members of the G. Nabel lab for helpful discussions.
REFERENCES
- 1.Aoki, K., C. Barker, X. Danthinne, M. J. Imperiale, and G. J. Nabel. 1999. Efficient generation of recombinant adenoviral vectors by Cre-lox recombination in vitro. Mol. Med. 5:224-231. [PMC free article] [PubMed] [Google Scholar]
- 2.Briggs, D. R., D. L. Tuttle, J. W. Sleasman, and M. M. Goodenow. 2000. Envelope V3 amino acid sequence predicts HIV-1 phenotype (co-receptor usage and tropism for macrophages). AIDS 14:2937-2939. [DOI] [PubMed] [Google Scholar]
- 3.Bures, R., A. Gaitan, T. Zhu, C. Graziosi, K. M. McGrath, J. Tartaglia, P. Caudrelier, R. El Habib, M. Klein, A. Lazzarin, D. M. Stablein, M. Deers, L. Corey, M. L. Greenberg, D. H. Schwartz, and D. C. Montefiori. 2000. Immunization with recombinant canarypox vectors expressing membrane-anchored glycoprotein 120 followed by glycoprotein 160 boosting fails to generate antibodies that neutralize R5 primary isolates of human immunodeficiency virus type 1. AIDS Res. Hum. Retrovir. 16:2019-2035. [DOI] [PubMed] [Google Scholar]
- 4.Burns, D. P., and R. C. Desrosiers. 1994. Envelope sequence variation, neutralizing antibodies, and primate lentivirus persistence. Curr. Top. Microbiol. Immunol. 188:185-219. [DOI] [PubMed] [Google Scholar]
- 5.Burton, D. R. 2002. Antibodies, viruses and vaccines. Nat. Rev. Immunol. 2:706-713. [DOI] [PubMed] [Google Scholar]
- 6.Cao, J., N. Sullivan, E. Desjardin, C. Parolin, J. Robinson, R. Wyatt, and J. Sodroski. 1997. Replication and neutralization of human immunodeficiency virus type 1 lacking the V1 and V2 variable loops of the gp120 envelope glycoprotein. J. Virol. 71:9808-9812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chakrabarti, B. K., W. P. Kong, B.-Y. Wu, Z.-Y. Yang, J. Friborg, Jr., X. Ling, S. R. King, D. C. Montefiori, and G. J. Nabel. 2002. Modifications of the human immunodeficiency virus envelope glycoprotein enhance immunogenicity for genetic immunization. J. Virol. 76:5357-5368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gorny, M. K., T. C. VanCott, C. Hioe, Z. R. Israel, N. L. Michael, A. J. Conley, C. Williams, J. A. Kessler, P. Chigurupati, S. Burda, and S. Zolla-Pazner. 1997. Human monoclonal antibodies to the V3 loop of HIV-1 with intra- and interclade cross-reactivity. J. Immunol. 159:5114-5122. [PubMed] [Google Scholar]
- 9.Gorny, M. K., C. Williams, B. Volsky, K. Revesz, S. Cohen, V. R. Polonis, W. J. Honnen, S. C. Kayman, C. Krachmarov, A. Pinter, and S. Zolla-Pazner. 2002. Human monoclonal antibodies specific for conformation-sensitive epitopes of V3 neutralize human immunodeficiency virus type 1 primary isolates from various clades. J. Virol. 76:9035-9045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guillon, C., M. Schutten, P. H. Boers, R. A. Gruters, and A. D. Osterhaus. 2002. Antibody-mediated enhancement of human immunodeficiency virus type 1 infectivity is determined by the structure of gp120 and depends on modulation of the gp120-CCR5 interaction. J. Virol. 76:2827-2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karlsson, G. B., M. Halloran, J. Li, I. W. Park, R. Gomila, K. A. Reimann, M. K. Axthelm, S. A. Iliff, N. L. Letvin, and J. Sodroski. 1997. Characterization of molecularly cloned simian-human immunodeficiency viruses causing rapid CD4+ lymphocyte depletion in rhesus monkeys. J. Virol. 71:4218-4225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Korber, B. T., K. J. Kunstman, B. K. Patterson, M. Furtado, M. M. McEvilly, R. Levy, and S. M. Wolinsky. 1994. Genetic differences between blood- and brain-derived viral sequences from human immunodeficiency virus type 1-infected patients: evidence of conserved elements in the V3 region of the envelope protein of brain-derived sequences. J. Virol. 68:7467-7481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krachmarov, C. P., S. C. Kayman, W. J. Honnen, O. Trochev, and A. Pinter. 2001. V3-specific polyclonal antibodies affinity purified from sera of infected humans effectively neutralize primary isolates of human immunodeficiency virus type 1. AIDS Res. Hum. Retrovir. 17:1737-1748. [DOI] [PubMed] [Google Scholar]
- 14.Kwong, P. D., M. L. Doyle, D. J. Casper, C. Cicala, S. A. Leavitt, S. Majeed, T. D. Steenbeke, M. Venturi, I. Chaiken, M. Fung, H. Katinger, P. W. Parren, J. Robinson, D. Van Ryk, L. Wang, D. R. Burton, E. Freire, R. Wyatt, J. Sodroski, W. A. Hendrickson, and J. Arthos. 2002. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature 420:678-682. [DOI] [PubMed] [Google Scholar]
- 15.Ly, A., and L. Stamatatos. 2000. V2 loop glycosylation of the human immunodeficiency virus type 1 SF162 envelope facilitates interaction of this protein with CD4 and CCR5 receptors and protects the virus from neutralization by anti-V3 loop and anti-CD4 binding site antibodies. J. Virol. 74:6769-6776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mascola, J. R., M. K. Louder, C. Winter, R. Prabhakara, S. C. DeRosa, D. C. Douek, B. J. Hill, D. Gabuzda, and M. Roederer. 2002. Human immunodeficiency virus type 1 neutralization measured by flow cytometric quantitation of single-round infection of primary human T cells. J. Virol. 76:4810-4821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Naldini, L., U. Blomer, P. Gallay, D. Ory, R. Mulligan, F. H. Gage, I. M. Verma, and D. Trono. 1996. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science 272:263-267. [DOI] [PubMed] [Google Scholar]
- 18.Ohagen, A., A. Devitt, K. J. Kunstman, P. R. Gorry, P. P. Rose, B. Korber, J. Taylor, R. Levy, R. L. Murphy, S. M. Wolinsky, and D. Gabuzda. 2003. Genetic and functional analysis of full-length human immunodeficiency virus type 1 env genes derived from brain and blood of patients with AIDS. J. Virol. 77:12336-12345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parren, P. W. H. I., J. P. Moore, D. R. Burton, and Q. J. Sattentau. 1999. The neutralizing antibody response to HIV-1: viral evasion and escape from humoral immunity. AIDS 13:S137-S162. [PubMed] [Google Scholar]
- 20.Purtscher, M., A. Trkola, A. Grassauer, P. M. Schulz, A. Klima, S. Dopper, G. Gruber, A. Buchacher, T. Muster, and H. Katinger. 1996. Restricted antigenic variability of the epitope recognized by the neutralizing gp41 antibody 2F5. AIDS 10:587-593. [DOI] [PubMed] [Google Scholar]
- 21.Schreiber, M., C. Wachsmuth, H. Muller, S. Odemuyiwa, H. Schmitz, S. Meyer, B. Meyer, and J. Schneider-Mergener. 1997. The V3-directed immune response in natural human immunodeficiency virus type 1 infection is predominantly directed against a variable, discontinuous epitope presented by the gp120 V3 domain. J. Virol. 71:9198-9205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Srivastava, I. K., K. VanDorsten, L. Vojtech, S. W. Barnett, and L. Stamatatos. 2003. Changes in the immunogenic properties of soluble gp140 human immunodeficiency virus envelope constructs upon partial deletion of the second hypervariable region. J. Virol. 77:2310-2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wei, X., J. M. Decker, S. Wang, H. Hui, J. C. Kappes, X. Wu, J. F. Salazar-Gonzalez, M. G. Salazar, J. M. Kilby, M. S. Saag, N. L. Komarova, M. A. Nowak, B. H. Hahn, P. D. Kwong, and G. M. Shaw. 2003. Antibody neutralization and escape by HIV-1. Nature 422:307-312. [DOI] [PubMed] [Google Scholar]
- 24.Wyatt, R., and J. Sodroski. 1998. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science 280:1884-1888. [DOI] [PubMed] [Google Scholar]
- 25.Zwick, M. B., R. Kelleher, R. Jensen, A. F. Labrijn, M. Wang, G. V. Quinnan, Jr., P. W. Parren, and D. R. Burton. 2003. A novel human antibody against human immunodeficiency virus type 1 gp120 is V1, V2, and V3 loop dependent and helps delimit the epitope of the broadly neutralizing antibody immunoglobulin G1b12. J. Virol. 77:6965-6978. [DOI] [PMC free article] [PubMed] [Google Scholar]