Abstract
A genome wide analysis of the human fungal pathogen Cryptococcus neoformans var. grubii has revealed a number of duplications of highly conserved genes involved in morphogenesis. Previously, we reported that duplicate Cdc42 paralogs provide C. neoformans with niche-specific responses to environmental stresses: Cdc42 is required for thermotolerance, while Cdc420 supports the formation of titan cells. The related Rho-GTPase Rac1 has been shown in C. neoformans var. neoformans to play a major role in filamentation and to share Cdc42/Cdc420 binding partners. Here we report the characterization of a second Rac paralog in C. neoformans, Rac2, and describe its overlapping function with the previously described CnRac, Rac1. Further, we demonstrate that the Rac paralogs play a primary role in polarized growth via the organization of reactive oxygen species and play only a minor role in the organization of actin. Finally, we provide preliminary evidence that pharmacological inhibitors of Rac activity and actin stability have synergistic activity.
Keywords: Cryptococcus neoformans, Rac GTPase, paralogs, polarization, hyphal growth, ROS
Introduction
Gene and genome duplications offer potential benefits to microorganisms by facilitating adaptation to a variety of environments. In addition to providing protection against deleterious mutations, duplicated genes provide genetic plasticity through neofunctionalization, subfunctionalization, and gene conservation (Force et al., 1999; Hahn, 2009; Ohno, 1970). In the case of Saccharomyces cerevisiae, an ancient whole genome duplication may have allowed this fungal lineage to acquire the process of anaerobic fermentation (Wolfe and Shields, 1997). Experimental models of gene evolution in Salmonella enterica demonstrate similar examples of gene amplification and divergence in the development of metabolic pathways (Näsvall et al., 2012).
Beyond simple gene duplication events, genetic redundancy can also be provided by related gene families. For example, the related Rho-like GTPases Cdc42 and Rac serve distinct primary functions in divergent fungal species such as P. marneffei, U. maydis, and A. nidulans (Bassilana and Arkowitz, 2006; Boyce et al., 2001; Boyce et al., 2003; Harris, 2011; Mahlert et al., 2006; Virag et al., 2007). However, in each of these species, deletion of both of these genes is synthetically lethal, demonstrating their potential for functional overlap (Boyce et al., 2005; Mahlert et al., 2006; Virag et al., 2007).
In C. neoformans, several duplicated gene pairs control cellular processes required for pathogenesis and development. These gene duplication events appear to have arisen in the absence of a whole genome duplication event (Loftus et al., 2005). However, these duplications span multiple pathways and cellular processes (Ballou et al., 2010; Missal et al., 2005; Pukkila-Worley et al., 2005; Salas et al., 1996; Wang et al., 2002). For example, the cyclophillin-encoding genes CPA1 and CPA2, major and minor paralogs respectively, play shared and distinct roles in cell growth, mating, and virulence, in addition to their conserved roles in the detoxification of cyclosporine A (Wang et al., 2001). Similarly, duplicated Ras proteins are involved in mating, polarity, and thermotolerance (Alspaugh et al., 2000; Waugh et al., 2002). RAS1 appears to provide the majority of the inducible Ras activity required for survival within the infected host. However, the simultaneous deletion of both the RAS1 and the RAS2 genes results in a dramatic cell growth defect under otherwise permissive incubation conditions.
In addition to functional redundancy, duplicated genes in C. neoformans have undergone subfunctionalization. The highly conserved Rho-GTPase encoded by CnCDC42 is required for growth at 37°C and for virulence (Ballou et al., 2010). A paralogous gene, CnCDC420, is dispensable for growth at 37°C but is required for the induction of a novel virulence factor, the formation of titan cells, a morphology thought to have important consequences for the passage of C. neoformans into the brain (Okagaki et al., 2011).
A number of studies have suggested a model in which the duplicated C. neoformans Cdc42 paralogs function downstream of the duplicated Ras proteins to control various morphological events required for cell polarization, differentiation, and virulence (Nichols et al., 2007; Wang et al., 2002). Prior studies also suggest that a Rac protein functions in this signaling pathway (Nichols et al., 2007; Vallim et al., 2005). Both C. neoformans CDC42 and RAC1 over-expression suppress the ras1Δ mutant defect in high temperature growth, and rac1Δ mutants are defective in hyphal development, a process also regulated by the Ras paralogs (Nichols et al., 2007; Vallim et al., 2005; Waugh et al., 2002).
The maintenance of Rac in the genomes of C. neoformans and other fungi provides a potential for redundancy not observed in the simple yeasts Saccharomyces cerevisiae and Schizosaccharomyces pombe, which lack Rac homologs. However, previous efforts to explore the overlap between Rac and Cdc42 paralog function in C. neoformans raised a number of unanswered questions. For example, synthetic lethality between Cdc42 and the related Rho-type GTPase Rac in P. marneffei, U. maydis, and A. nidulans, and the lethality of cdc42Δ mutants in S. cerevisiae and S. pombe, has demonstrated the essential requirement for one or the other of these central regulators of morphogenesis (Adams et al., 1990; Boyce et al., 2005; Mahlert et al., 2006; Miller and Johnson, 1994; Virag et al., 2007). However, the lack of morphological defects observed in a C. neoformans rac1Δ cdc42Δ double mutant suggested that redundancy between these proteins was more complex in C. neoformans (Shen et al., 2011).
In order to explore the roles of gene duplications and related gene families in this morphogenesis signaling pathway, we performed a whole genome analysis of paralogous gene pairs. In addition to the duplicated RAS1 and CDC42 alleles, we identified a second cryptic paralog of RAC1, termed RAC2, in the genome of C. neoformans. Functional analysis of the two Rac paralogs in C. neoformans reveals conserved roles in hyphal development and ROS localization. Additionally, these Rac proteins appear to play significant roles in polarization during yeast phase growth.
Methods
Strains, media, and growth conditions
C. neoformans strains used in this study are listed in Table 2. Strains were incubated on YPD medium (Sherman, 1991), or MS mating medium(Murashige and Skoog, 1962). ERB033 was generated by mating between ERB032 and KN99a. ERB107 was generated by mating between ERB056 and KN99a. ERB025 was generated by mating between ERB011 and ERB033. ERB124 and ERB067 were generated via transformation of LCC1 or CNB45 with the plasmids pJN03 or pERB04, respectively. All other strains were generated via biolistic transformation according to the protocol developed by Toffaletti et al. (Toffaletti et al., 1993). Strains were confirmed by PCR and Southern analysis. Mating assays were performed by co-culturing strains of opposite mating type on MS mating medium in the dark at 25°C for 4 days. For morphogenesis experiments, cells were inoculated into liquid YPD (2% glucose) and grown to mid-log phase at 30°C, shaking at 150 RPM. Cultures were split and refreshed with media pre-warmed to 30 or 37°C, then incubated at the indicated temperature at 150 RPM for 4 hours or over night, as indicated. Mating assays were performed by co-culturing strains of opposite mating type on MS mating medium in the dark at 25°C.
Table 2.
| Strain | Genotype | Source |
|---|---|---|
| H99 | MATα | Perfect et al., 1980 |
| KN99a | MATa | Neilsen et al., 2003 |
| LCC1 | MATα ras1::ADE2 | Alspaugh et al., 2000 |
| CBN45 | MATα ras1::neo | Nichols et al., 2009 |
| ERB011 | MATα cdc42::nat cdc420::neo | Ballou et al., 2010 |
| ERB025 | MATα cdc42::nat cdc420::neo rac2::neo | This study |
| ERB027 | MATα rac2::nat | This study |
| ERB032 | MATα rac2::neo | This study |
| ERB033 | MATa rac2::neo | This study |
| ERB035 | MATα rac2::neo + pHRAC2-nat | This study |
| ERB056 | MATα rac1::hyg | This study |
| ERB067 | MATα ras1::neo + pHRAC2-nat | This study |
| ERB107 | MATa rac1::hyg | This study |
| ERB109 | MATα pHGRAC1-nat | This study |
| ERB124 | MATα ras1::ADE2 + pHRAC1-nat | This study |
| ERB130 | MATα pHGRAC1-neo | This study |
| ERB135 | MATα rac1::hyg + pHRAC1-nat | This study |
Disruption of RAC paralogs and generation of over-expression and GFP constructs
To generate the rac1Δ strain, the following primers with regions homologous to flanks surrounding the hygromycin resistance marker in pHyg-KB2 were used: AA1354:CGCCAACACTTGCTGCCGCTC; AA1814: GTCATAGCTGTTTCCTGTCTGGGCTGTTGCGCTATGCC; AA1815: GGCATAGCGCAACAGCCCAGACAGGAAACAGCTATGAC; AA1816: CCTCATTTGGCATTGCTCAGCGTAAAACGACGGCCAG; AA1817: CTGGCCGTCGTTTTACGCTGAGCAATGCCAAATGAGG; AA1359: GCAGGCGAAGAGCGGATGG. To generate the rac2Δ strains, the following primers with regions homologous to the m13 flanks surrounding the neomycin and nourseothricin resistance markers were used: AA3122: GTTGGGCCACCATCATTACT; AA3194:TACCATCATCCTCTCCTCCGTTGCTCTCGTTGAGGATTGA; AA319: TCAATCCTCAACGAGAGCAACGGAGGAGAGGATGATGGTA; AA3196: AGCTGTACCCTTGTCCGCTACGACAGCATCGCCAGTCACTA; AA3197: TAGTGACTGGCGATGCTGTCGTAGCGGACAAGGGTACAGCT; AA3127:CTCCTTCCACCCACCACTTA. To generate over-expression constructs, the open reading frames of C. neoformans Serotype A RAC1 and RAC2 were amplified by PCR using primers that incorporated the appropriate restriction sites in frame at the 5′ and 3′ ends. These primers were: For RAC1: AA1959:GCCGGTACCCGCCAACACTTGCTGCCGCTC; AA1960: GGCGAGCTCCTGGGCTGTTGCGCTATGCCG; For RAC2: AA1928: GGGCCCAGATCTATGGCCATGCAGAGTATC; AA3090: CCCGGGAGATCTGATCCGTGCTTGGTTTTTGT. Over-expression constructs were then generated by TA cloning and/or BamHI ligation into the plasmid pCN19, containing the HIS3 promoter sequence, the actin terminator sequence, and the nourseothricin selection marker, to generate the plasmids pJN03 and pERB04, respectively (Price et al., 2008).
Alignments
The identification of paralogs within the C. neoformans var. grubii genome was performed using BlastP (Altschul et al., 1990). The network was constructed using the freely accessible Cytoscape software package (Cline et al., 2007). CNAG numbers correspond to the Broad Institute H99 genome database (www.broadinstitute.org/annotation/genome/cryptococcus_neoformans). A high-resolution network image, as well as the entire network file and corresponding gene identities are included in Supplemental Figure 1. Sequence alignments were performed using the ClustalW algorithm available from the EMBL-EBI consortium (Larkin et al., 2007). The Rac phylogeny was constructed using the following DNA sequences, which were hand curated and aligned using the Muscle algorithm (Edgar, 2004) (C. n. grubii RAC1: BK008522, RAC2: BK008523; C. n. neoformans RAC1: AY780547, RAC2: XP_568391.1; C. gattii RAC1: XP_003193004.1; RAC2: XP_003197177.1; T. mesenterica RAC1: EIW72625.1, RAC2: EIW65787.1; C. cinerea XP_001828804.2; S. commune XP_003037339.1; U. maydis RAC1, AF495535; E. festucae RACA: AB360318; C. trifoli RAC1: AY325889; P. marneffei CLFB: AF515698; A. nidulans RACA: AN4743.2; Y. lipolytica RAC1: AF176831) (Boyce et al., 2003; Chen and Dickman, 2004; Hurtado et al., 2000; Tanaka et al., 2008; Vallim et al., 2005; Virag et al., 2007; Weinzierl et al., 2002). A neighbor-joining tree was generated using MEGA5 (Tamura et al., 2011). Bootstrap values represent 10000 iterations.
RT-PCR
Expression levels for each of the over-expression constructs was determined by RT-PCR. Mid-log phase cells incubated at 30°C in YPD were collected by centrifugation and flash frozen on dry ice. Total RNA was extracted from lyophilized cells using the Qiagen RNA extraction kit and the ‘Purification of total RNA from plant cells, tissue, and filamentous fungi’ protocol (2006). cDNA was prepared using the Clonetech Advantage RT-for-PCR kit (2006). Primer specificity was verified by qPCR in wild-type and deletion strains. RT-PCR was performed as described, with annealing at 50°C (Cramer et al., 2006). Primers sequences were as follows: RAC1 Forward: AA1853: CCGAACCAAATGGTATCCTG; Reverse: AA1854: TTAGGGTTGAGGACTGTCCG. RAC2 Forward: AA1851: TGTCAAAACTTGGATCCCCG; Reverse: AA3021: CAAGCCTTTTTGCGTCCGACTAGAAG. GPD Forward: AA301: AGTATGACTCCAACAATGGTCG; Reverse: AA302: AGACAAACATCGGAGCATCAGC. Expression levels were calculated using the ΔΔCt method, as described (Schmittgen and Livak, 2008). In order for replicates to be included in the final analysis, duplicate wells were required to have an SD <1.0
For the quantification of MFα1 expression, equal numbers of RAC2a and RAC2α or rac2a and rac2α cells were mixed and plated onto MS mating medium. The mating reaction was incubated for 48 hours in the dark at room temperature, and then cells were collected for RNA extraction. MFα1 was measured by RT-PCR relative to an internal standard (GPD1) in duplicate and in two independent experiments.
Microscopy
Differential interference microscopy (DIC) and fluorescent images were captured with a Zeiss Axio Imager A1 fluorescent microscope equipped with an AxioCam MRM digital camera. Cells were fixed with 9% microfiltered formaldehyde for 10 minutes, washed three times with 1XPBS, permeabilized with 1% Triton-PBS for 10 minutes, and washed three times with 1XPBS. Cells were stained with Rhodamine-conjugated Phalloidin or DAPI. For filaments, agar plugs from the edge of mating reactions were extracted, fixed as appropriate, washed as described above, and then stained with 9 μM DCHF-DA (Enzo Life Sciences) (unfixed) or DAPI. Agar slices were then embedded by slide squash on microscope slides and the hyphae were imaged.
The localization of GFP-Rac1 and GFP-Rac2 was visualized using a DeltaVision Elite deconvolution microscope equipped with a Coolsnap HQ2 high resolution CCD camera. Strains expressing GFP-Rac2 and GFP-Rac1 were cultured overnight at 30°C in YPD and used to seed cultures of YPD pre-warmed to 30°C or 37°C. Cultures were incubated for 4 hr and were washed 1 time with PBS prior to imaging. Samples grown at 37°C were maintained at 37°C during imaging.
Cell size counts
Because cell culture synchronization in C. neoformans requires exposure to low oxygen conditions, which may impact stress responses differentially in the tested strains, we compared unsynchronized paired mutant and wild type strains inoculated simultaneously and at the same density. Strains were allowed to grow overnight at 30°C to mid-log phase, diluted 1:10 in fresh YPD, and collected after 4 hours exposure to the indicated temperature. Cell area for more than 300 cells was measured using ImageJ software (Cramer et al., 2006; Schneider et al., 2012). Mann Whitney U Test was applied using Mathematica software (Wolfram Research, 2010).
Latrunculin B and EHT1864 sensitivity assays
To test for Latrunculin B sensitivity, the indicated strains were examined in the presence of the actin inhibitor Latrunculin B in a 96 well format. Cells (103 cells/well in YPD media) were incubated in the presence of decreasing concentrations of Latruculin B (100 μM to 0 μM) for 48 hours (duplicate wells, n=4). The minimum inhibitory concentration was noted for each strain. To test for synergistic effects between Latrunculin B and EHT1864, antifungal drug testing was performed according to CLSI standards with the modification of using YNB media for optimal C. neoformans growth conditions (Jessup, 1998). Growth inhibition of 104 cells/well C. neoformans was recorded after treatment with the actin inhibitor Latrunculin B, the Rac2 inhibitor EHT1864, and the combination of both drugs in a checkerboard MIC test (Odds, 2003). Growth inhibition by Latrunculin B was tested for the concentrations of 1.56–100 μM, and EHT1865 was tested for the concentrations of 0.97–500 μM. The fractional inhibitory concentration index (FICI) was used to calculate the synergistic relationship between the drugs, with synergy as defined by a FICI < 0.5 (Odds, 2003). MIC tests were performed in duplicate, and strains were incubated in the presence or absence of drug for 48 hours at 30°C.
Cell fusion assay
Cell fusion was performed as previously described (Alspaugh et al., 2000). Briefly, strains of opposite mating types with complementary markers (neomycin, hygromycin, or nourseothricin resistance) were co-incubated on MS medium. Equal numbers of cells (105/ml) were combined in sdH20, and 100 μl were spotted onto MS plates. The reactions were incubated at room temperature in the dark for 48 hours, after which the agar was excised and each patch was resuspended in 3 mL sdH20 and vortexed vigorously for 2 minutes. 100 μl from each suspension were plated onto double selection medium and incubated at 30°C for 48–72 hours to select for dikaryotic cells, at which point the plates were photographed.
Animal experiments
Using the murine inhalation model of systemic cryptococcosis, female A/Jcr mice were inoculated intra-nasally with 5 × 105 C. neoformans cells, as previously described (Cox et al., 2000). Briefly, groups of 10 mice were inoculated with one of five strains: H99 (RAC2 wild type), ERB027 (rac2Δ), or ERB035 (rac2Δ + RAC2 reconstituted). Mice were observed daily for signs of infection. Animals were sacrificed at predetermined clinical endpoints correlating with an imminent lethal infection. The statistical significance in the difference between the survival curves of the animals inoculated with each strain was evaluated using the log- rank test (JMP software; SAS Institute, Cary, NC). All studies were performed in compliance with institutional guidelines for animal experimentation.
Results
Whole genome analysis of paralogs
In searching for possible sources of redundancy among Rho-GTPases in C. neoformans var. grubii (H99), we first performed a whole genome analysis of paralogous proteins. Paralogs were broadly defined as those proteins with more than 70% similarity as assessed by blastP analysis, acknowledging that changing cutoff values will significantly effect the detection of paralogs. In S. cerevisiae, paralogous proteins resulting from the whole genome duplication ranged in similarity between 24% and 100%, although the average similarity was 63% (Wolfe and Shields, 1997). However, we opted to use the more stringent cut off of 70%, which was sufficient to separate paired proteins (e.g., Rac proteins, Cdc42 proteins, Rho proteins) from protein families (e.g., all Rho-like GTPases). Additionally, we included in our final analysis only those open reading frames expressed during yeast-phase growth, as documented by RNAseq experiments performed in our laboratory (O’Meara et al., 2010). Therefore, hyphal and spore specific proteins are also excluded from this analysis.
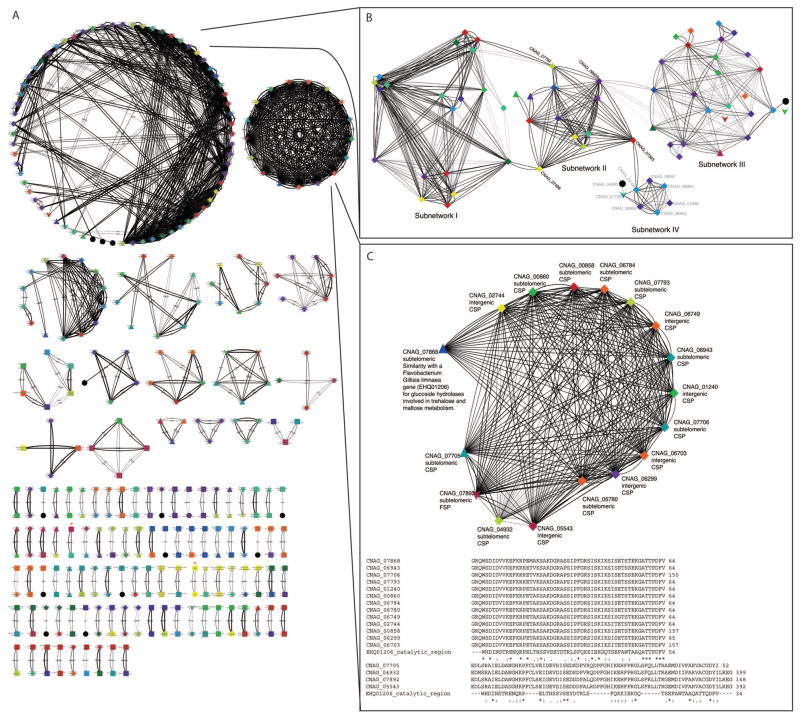
Paralogous proteins identified in this analysis can be seen in Figure 1 as a network model based on degree of similarity. A number of classes of proteins can be visualized using the network model, including two large paralog groups (Network 1 and Network 2) and several smaller networks composed of 3 to 14 proteins (Figure 1A). Duplicated protein pairs, including the Cdc42 and Ras pairs, are listed in Table 1. The paralogous gene sets represent several classes, without a particular pattern of involvement in any one cellular process. However, a large number of the identified paralogs were conserved, hypothetical, or predicted proteins with no available annotation within the Cryptococcus or other fungal lineages.
Figure 1. Analysis of paralogous proteins in the C. neoformans var. grubii genome.
A) Paralogous proteins with greater than 70% similarity whose corresponding gene expression was observed during yeast phase growth by RNAseq were visualized using the Cytoscape network building software. Paralogous proteins are represented by nodes, with the shape of the node indicating the classification of the paralog as an annotated, conserved, or predicted ORF, and the color indicating the chromosome position. The width of the edge indicates the percent similarity of the paralogs, ranging from 70% to 100%. Rac and Cdc42 paralogs are indicated by red stars (*). Two large paralog groups (B, C) identified in this manner were further examined for structure based on similarity to hub sequences that dictate relatedness within the group. Network 1 (B) showed little overall relatedness to conserved, annotated proteins, but could be further broken down into four sub-networks (I–IV) that related to each other through hub sequences, identified here in black by CNAG numbers corresponding to the Broad H99 genome database. A transposon-containing gene in subnetwork IV is indicated by a black star (*). (C) could be further broken down into proteins with 70% similarity to CNAG_07868. D) This similarity was correlated with the conservation of the catalytic domain of a protein from the flavobacterium Gillisea limnaea that is involved in trehalose and maltose metabolism (EHQ01206).
Table 1. Gene pairs identified by whole genome analysis.
Pairs correspond to those presented in Figure 1. Annotation was obtained from the H99 genome database hosted by the Broad Institute.
| Pair | CNAG | Gene 1 | CNAG | Gene 2 |
|---|---|---|---|---|
| 1 | 07406 | Alpha pheromone | 07407 | Fungal mating-type pheromone |
| 2 | 07042 | CHP | 07894 | CHO |
| 3 | 06978 | Cytoplasmic protein | 05657 | - |
| 4 | 06854 | PP | 07916 | PP |
| 5 | 06832 | Glucosidase | 06835 | glucosidase |
| 6 | 06805 | PP | 07828 | PP |
| 7 | 06802 | CHP | 07784 | CHP |
| 8 | 06801 | CHP | 07000 | CHP |
| 9 | 06748 | U3 sno-RNP-associated protein Utp7 | 07466 | WD repeat-containing protein 46 |
| 10 | 06536 | Monocarboxylic acid transporter | 06537 | Monocarboxylic acid transporter |
| 11 | 06338 | ABC transporter PMR5 | 06348 | ABC transporter PMR5 |
| 12 | 06134 | CHP | 00871 | - |
| 13 | 06101 | Eukaryotic ADP/ATP carrier | 06102 | Eukaryotic ADP/ATP carrier |
| 14 | 06026 | Aspartate transaminase | 06088 | Aspartate transaminase |
| 15 | 06010 | Fatty aldehyde dehydrogenase | 06018 | aldehyde dehydrogenase |
| 16 | 05992 | CHP | 07831 | PP |
| 17 | 05990 | PP | 07832 | PP |
| 18 | 05938 | CHP | 07501 | HP |
| 19 | 05914 | MFS maltose permease | 05929 | MFS maltose permease |
| 20 | 05832 | PP | 05833 | Allantoate transporter |
| 21 | 05817 | Nucleotide-sugar transporter | 03426 | - |
| 22 | 05668 | PP | 07396 | PP |
| 23 | 05450 | CHP | 05454 | CHP |
| 24 | 05448 | PP | 05455 | Euk translation initiation factor 1A |
| 25 | 05426 | PP | 07574 | PP |
| 26 | 05348 | CDC42 | 05968 | CDC420 |
| 27 | 05339 | CHP | 07785 | PP |
| 28 | 05327 | PP | 05989 | PP |
| 29 | 05325 | CHP | 05991 | Glycosyl hydrolase family 88 |
| 30 | 05263 | PP | 07425 | CHP |
| 31 | 05254 | UDP-galactose transporter | 07823 | PP |
| 32 | 04828 | Histone | 06745 | Histone H3 |
| 33 | 04794 | Spermine | 06119 | - |
| 34 | 04783 | Monosaccharide transporter | 04784 | Monosaccharide transporter |
| 35 | 04652 | Enoyl reductase | 06759 | Dehydrogenase |
| 36 | 04594 | Pro1 protein | 04618 | Pro1 protein |
| 37 | 04541 | Polyadenylation factor 64 kDa subunit | 04557 | PP |
| 38 | 04540 | ER receptor | 04556 | ER receptor |
| 39 | 04374 | CHP | 04394 | CHP |
| 40 | 03948 | Cytoplasmic protein | 03936 | - |
| 41 | 03904 | Endo-peptidase | 06333 | DNA replication factor |
| 42 | 03732 | Integral membrane protein | 03764 | Integral membrane protein |
| 43 | 03713 | Efflux protein EncT | 03719 | CHP |
| 44 | 03621 | Cyclophilin A | 03627 | Cyclophilin A |
| 45 | 03604 | No arches protein | 00520 | - |
| 46 | 03464 | LAC2 | 03465 | LAC1 |
| 47 | 03407 | PP | 03548 | PP |
| 48 | 03277 | Inositol oxidase | 05316 | Inositol oxidase |
| 49 | 03252 | CHP | 05305 | CHP |
| 50 | 03230 | CHP | 03233 | CHP |
| 51 | 03174 | Phosphopantetheinyl transferase | 04341 | - |
| 52 | 03092 | CHP | 07390 | CHP |
| 53 | 03084 | Endoribonuclease L-psp | 06589 | Endoribonuclease L-psp |
| 54 | 03060 | MDR protein | 03061 | MDR protein |
| 55 | 02896 | Hydroxymethylglutaryl-CoA synthase | 03311 | Hydroxymethylglutaryl-CoA synthase |
| 56 | 02883 | Rac1 | 05998 | Rac2 |
| 57 | 02863 | PP | 06672 | Formate dehydrogenase |
| 58 | 02814 | Glycerol-3-phosphate dehydrogenase | 02815 | Glycerol-3-phosphate dehydrogenase |
| 59 | 02798 | CHP | 05906 | CHP |
| 60 | 02752 | Short-chain dehydrogenase | 04926 | CHP |
| 61 | 02733 | Monosaccharide transporter | 07641 | Monosaccharide transporter |
| 62 | 02556 | Phytanoyl-CoA dioxygenase family protein | 04564 | Phytanoyl-CoA dioxygenase family protein |
| 63 | 02539 | amino acid transporter | 07902 | amino acid transporter |
| 64 | 02489 | mannitol-1-phosphate dehydrogenase | 07745 | mannitol-1-phosphate dehydrogenase |
| 65 | 02479 | monosaccharide transporter | 05340 | monosaccharide transporter |
| 66 | 02476 | CHP | 05328 | - |
| 67 | 02463 | CHP | 07386 | CHP |
| 68 | 02048 | Proline dehydrogenase | 02049 | Proline dehydrogenase |
| 69 | 01969 | Zn metalloprotease | 04524 | - |
| 70 | 01953 | CHP | 02299 | CHP |
| 71 | 01952 | Aryl-alcohol dehydrogenase | 02717 | Aryl-alcohol dehydrogenase |
| 72 | 01925 | CHP | 07869 | CHP |
| 73 | 01757 | CHP | 05185 | CHP |
| 74 | 01727 | HSC 70-4 | 01750 | Chaperone |
| 75 | 01648 | Histone H4 | 07807 | Histone H4 |
| 76 | 01635 | DUF1479 domain-containing protein | 04225 | DUF1479 domain-containing protein |
| 77 | 01446 | HP | 03143 | CHP |
| 78 | 01120 | Dihydrolipoyllysine-residue acetyltransferase | 03509 | Pyruvate dehydrogenase protein component |
| 79 | 00919 | Carboxypeptidase D | 01040 | Carboxypeptidase D |
| 80 | 00866 | transketolase | 03040 | transketolase |
| 81 | 00863 | Flavin-containing monooxygenase | 07389 | Flavin-containing monooxygenase |
| 82 | 00849 | PP | 07708 | CHP |
| 83 | 00598 | Nicotinamide mononucleotide permease | 01674 | MFS transporter |
| 84 | 00597 | Amino acid transporter | 07367 | Amino acid transporter |
| 85 | 00575 | Catalase 3 | 04981 | CAT1 |
| 86 | 00372 | Nuclear protein localization | 05757 | CHP |
| 87 | 00370 | Ubiquitin-carboxy extension protein fusion | 01920 | Poly-ubiquitin |
| 88 | 00260 | Transformer-2-beta isoform 3 | 00399 | Transformer-2-beta isoform 3 |
| 89 | 00128 | CHP | 06757 | CHP |
| 90 | 00047 | Kinase | 04108 | Kinase |
| 91 | 00039 | CHP | 02175 | Exo2 |
| 92 | 00036 | Sec14 cytosolic protein | 03153 | Sec14 cytosolic protein |
| 92 | 00005 | TPR domain-containing protein | 02012 | TPR domain-containing protein |
| 92 | 07876 | PP | 07877 | PP |
| 92 | 07532 | CHP | 03329 | - |
CHP = “conserved hypothetical protein”
HP = “hypothetical protein”
PP = “predicted protein”
- = no annotation
The two large networks were composed entirely of hypothetical proteins with no known function and only limited protein domain prediction, identifying them as “black box” networks. However, these networks could be further broken down into distinct gene families connected by “hub” sequences. For example, in Network 1, complex sequence similarities between 4 larger families can be traced to 4 central protein sequences (indicated by CNAG number) (Figure 1B, Figure S1). Those sub-networks can be further organized based on relatedness within the family. Overall, this network is composed of primarily sub-telomeric proteins with limited similarity to known, annotated sequences. All 14 chromosomes are represented, each at least twice, with the exception of chromosome 8 (CNAG_07729 in sub-network IV), which is related to the network only by an intergenic protein with no conserved domains and no homology to any other known sequences. However, limited structure is exhibited by each of the four sub-networks. For example, six of the eight proteins in Sub-network IV share an LTXXQ domain with similarity to bacterial proteins, and one element of this network, CNAG_06954, additionally maintains a DEAD-box domain, also with similarity to bacterial proteins. This entire sub-network, and the hub connecting it to the larger network, share homology with DNA helicase proteins such as RecQ, although the similarity is limited to regions outside of known functional domains. The other three sub-networks exhibit similar trends in protein sequence, with similarity to telomere-associated proteins, DNA binding proteins, and bacterial proteins, and one element of sub-network III, CNAG_00814, has been identified as a transposon-containing gene (UniProt: AFG24184.1). Overall, this network appears to describe the repetitive structure of C. neoformans sub-telomeric regions, rather than a particular functional protein family.
Closer examination of the tightly connected network of paralogs in Network 2 (Figure 1C) revealed several trends. First, the majority of these genes reside in subtelomeric regions (11 of 17) and show homology only to other hypothetical Cryptococcus specific proteins (CSPs) (16 of 17). Second, the network could be further divided into those genes with more than 70% similarity to CNAG_07868 (12 of 16). Blast analysis of CNAG_07868 identified low (52%) similarity with the catalytic region of a protein from the flavobacterium Gillisea limnaea that is involved in trehalose and maltose metabolism (UniProt: EHQ01206). An alignment of the corresponding protein sequences revealed that this catalytic domain is conserved in the CNAG_07868-like group but is absent in the remaining four paralogs, suggesting first that this domain helped drive the divergence of these two sub-networks, and second that this catalytic domain may be functionally significant in the larger of the two paralog subfamilies. We have therefore identified this sub-network as “Carbohydrate Metabolism Paralogs”. This analysis demonstrates the power of a network approach to paralog identification by facilitating the classification of related, conserved hypothetical proteins based on limited sequence similarity to other known protein sequences.
Identification of the Rac2 paralog
Using this network analysis, we identified a hypothetical protein (encoded by CNAG_005998) with 86% similarity and 74% identity to the Rho-GTPase Rac1. The C-terminus of this putative Rac paralog was non-cannonical, lacking a CAAX domain that is highly conserved among Rho-GTPases. Instead, the C-terminus had no homology to other known protein domains. Examination of the genomic sequence revealed an apparent alternative splice acceptor site for the terminal intron. Use of this alternate acceptor site for excision of the intron resulted in a frame shift that produced an alternate product with the canonical CAAX membrane localization sequence. In order to validate one acceptor site over the other, we mined RNAseq data from the H99 transcriptome (O’Meara et al., 2010). In this database, we detected only a single product, which corresponded to the production of the protein with the expected CAAX C-terminal sequence. We therefore predicted that CNAG_05998 encoded a paralog of Rac1 and termed this open reading frame RAC2.
CnRAC2 encodes a protein that is 87% similar and 71% identical to CnRac1. It is located on chromosome 12 and includes 6 exons and 5 introns. Both Rac1 and Rac2 sequences have been submitted to NCBI. The nucleotide sequence data reported are available in the Third Party Annotation Section of the DDBJ/EMBL/GenBank databases under the accession numbers TPA: BK008522 (RAC1) and TPA: BK008523 (RAC2).
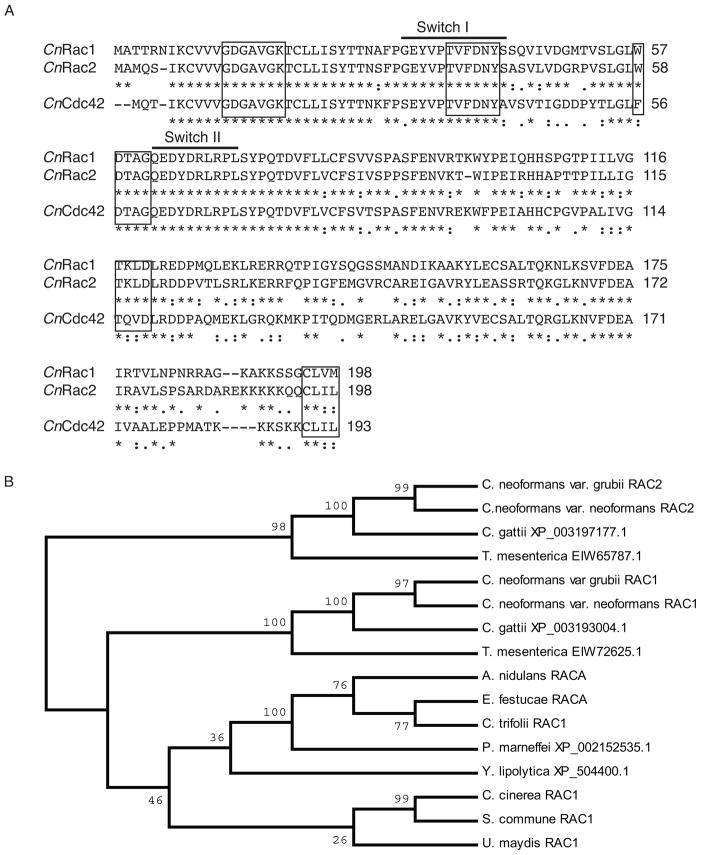
An alignment of CnRac1, CnRac2, and CnCdc42 can be seen in Figure 2a. Regions highly conserved among GTPases are identical in both Rac paralogs and Cdc42. All three maintain identical versions of the consensus elements GXXXXGK and DXXG, involved in interaction with GTP phosphates (GDGAVGK at CnRac2 residues 12–19, and DTAG at CnRac2 residues 59–62) (Valencia et al., 1991), and the sequence TVFDNY (CnRac2 37–42), involved in GAP interactions (Chen et al., 1993). Rac homologs are distinguished from Cdc42 homologs by tryptophan at position 59, present in both CnRac paralogs (Gao et al., 2001). Rac1 and Rac2, but not Cdc42, are identical in the switch I and II regions, which have been demonstrated to facilitate interaction with GEFs, GAPs, and GDIs. Among the GTPases, Rac proteins have a higher intrinsic rate of GDP release, and this has been linked to the sequence 116TKLD119, which is conserved in both C. neoformans Rac paralogs (Ménard et al., 1992). Finally, both Rac paralogs and Cdc42 maintain the consensus sequence CAAX (where A=aliphatic and X=any residue) at their C-termini. The terminal cysteine is a substrate for prenylation (Ménard et al., 1992), and this post-translational modification, coupled with the polybasic C-termini of these proteins, suggests that they are localized to the plasma membrane.
Figure 2. Alignment of Rac1, Rac2, and Cdc42 protein sequences.
A) The Rac2 protein sequence using the corrected splice site is provided in alignment with CnRac1 and CnCdc42. Key domains discussed in the text are indicated. B) A consensus NJ tree based on DNA sequences for the duplicate Rac paralogs in each of the Cryptococcus species complex members and Tremella mesenterica as well as single Rac paralogs identified in other fungal lineages. Bootstrap values represent 10,000 iterations.
Rac2 paralogs are present in both C. neoformans var. grubii and var. neoformans genomes
A search of closely related basidiomycete genomes revealed the presence of Rac1 and Rac2 paralogs in each of the members of the Cryptococcus species complex. Additionally, a search of the recently assembled draft genome of the closely related basidiomycete Tremella mesenterica identified two independent Rac-like proteins that likewise maintained all of the relevant Rac-defining residues, as well a single Cdc42 homolog, as previously reported (Ballou et al., 2010). A single Rac homolog was identified in the genomes of the basidiomycetes Ustilago maydis, Schizophyllum commune, and Coprinopsis cinerea, as well as in the genomes of the pathogenic ascomycetes Aspergillus nidulans, Epichlöe festucae, Colletotrichum trifolii, Yarrowia lipolytica, and Penicillium marneffei (Boyce et al., 2003; Chen and Dickman, 2004; Hurtado et al., 2000; Mahlert et al., 2006; Tanaka et al., 2008; Vallim et al., 2005; Virag et al., 2007; Weinzierl et al., 2002), suggesting that the Rac duplication may have occurred after the branching of the Tremella lineage (Figure 2B).
Individual Rac paralogs are not essential and are dispensable for thermotolerance and virulence
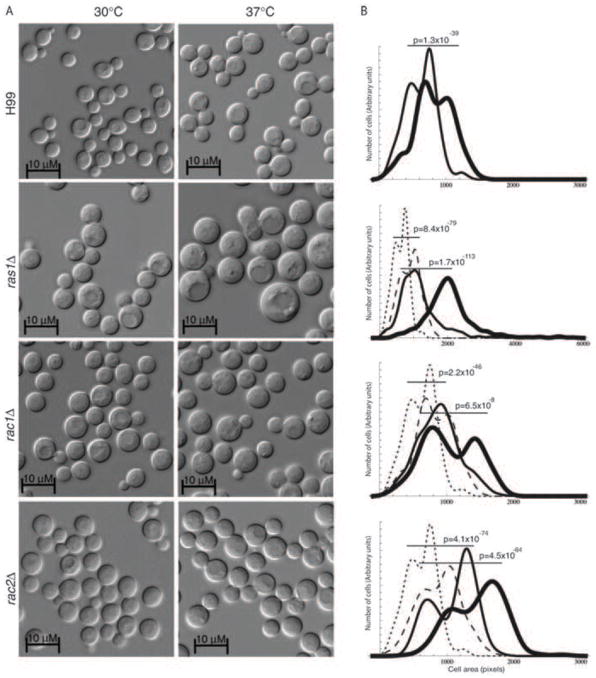
In order to examine the contribution of each of the Rac paralogs to morphogenesis, deletion constructs were generated in which the entire open reading frame of either RAC1 or RAC2 was replaced with a selectable marker. Single deletion of either the RAC1 or the RAC2 ORF using these constructs could be achieved with approximately 2% efficiency in either the H99 MATα or the KN99 MATa background. All strains were confirmed to be deletion mutants by PCR and Southern hybridization. These individual mutants exhibited no defects in growth at 30 or 37°C (Figure 3A), suggesting that the Rac paralog individually are not required for Ras-mediated thermotolerance. However, similar to the over-expression of Rac1, the over-expression of Rac2 was sufficient to restore high-temperature growth to the ras1Δ mutant.
Figure 3. Rac paralogs are dispensable for thermotolerance.
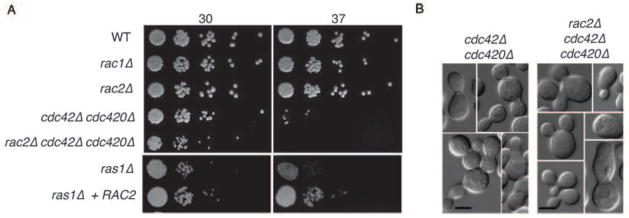
A) Indicated strains were spotted in 5-fold serial dilutions onto solid YPD medium and incubated at 30° or 37° as indicated for 48 hours. B) Double cdc42Δ cdc420Δ mutants and triple rac2Δ cdc42Δ cdc420Δ mutants incubated at 30°C exhibit similar morphological defects in polarized growth and cytokinesis.
We previously reported that the rac1Δ mutant is dispensable for virulence. Similarly, the loss of RAC2 had no impact on virulence in a murine model of infection. All mice infected with RAC2 or rac2Δ cells succumbed to the infection within 15 to 19 days of inoculation, with no statistical differences between infection groups.
Rac paralogs are synthetically essential
Rac transcripts are similarly expressed and are not induced in the opposite mutant (data not shown). In order to examine the interaction of the two Rac paralogs, we attempted to generate a rac1Δ rac2Δ double mutant. Although single deletion of either the RAC2 or the RAC1 ORF could be readily achieved in either the H99 MATα or the KN99 MATa background, no rac1Δ rac2Δ double mutants were identified after screening approximately 600 transformants. Therefore, we attempted to generate rac2Δ rac1Δ mutants via mating and spore dissection. Although rac1Δ mutant strains demonstrate delays in mating (Vallim et al., 2005), crosses between MATα rac1::NAT and MAT a rac2::NEO strains eventually produced viable spores. Among 36 viable spores, no rac1 rac2 double mutant strain were isolated, despite identifying other recombinant strains (MATa rac1Δ RAC2, MATα RAC1 rac2Δ, and RAC1 RAC2) in the expected proportions. Finally, we generated a rac2Δ cdc42Δ cdc420Δ triple mutant. Both the cdc42Δ cd420Δ mutant and the rac2Δ cdc42Δ cdc420Δ mutant fail to grow at 37°C, although the defect is slightly less severe in the cdc42Δ cdc420Δ mutant. The two mutants exhibit similar phenotypes at 30°C and have similar morphological defects, suggesting that Cdc42 paralogs do not compensate for the loss of Rac2 (Figure 3A,B) (Ballou et al., 2010). Together, these data suggest that the Rac paralogs together play an essential role in C. neoformans yeast-phase growth, and one for which the related Cdc42 proteins cannot compensate.
Rac paralogs contribute to cell size at 30°C and 37°C
The loss of RAS1 results in cells that have increased size at 30°C and 37°C, and this phenotype in both S. cerevisiae and C. neoformans has been connected to a defect in the polarization of actin following exposure to temperature stress (Ho and Bretscher; Nichols et al., 2007). Previously, Rac1 was identified as a suppressor of the ras1Δ mutant thermotolerance defect; however no role for Rac1 in yeast-phase morphogenesis was reported (Vallim et al., 2005). We therefore examined rac1Δ and rac2Δ cells for ras1Δ-like defects in cell size and the polarization of actin.
We determined the average cell size of wild type, ras1Δ, rac1Δ, and rac2Δ cells in log phase growth at 30°C and 37°C. The size of more than 500 cells was measured for each strain using ImageJ software, and pairwise comparisons were made using the Mann-Whitney U test (Figure 4). Because the synchronization of C. neoformans cultures requires exposure to O2 limiting conditions, and this condition has been reported to impact cell cycle, we chose instead to compare overall populations (Ohkusu et al., 2001; Ohkusu et al., 2004; Takeo et al., 1995). Instead, mutant and wild type cells were tightly paired for medium batch, growth time, temperature, and cell density.
Figure 4. Loss of Rac paralogs impacts cell size.
A) Cells lacking Ras1 or the Rac paralogs were grown in liquid YPD medium for 4 hours at 30 or 37°C. B) The area of more than 300 cells was measured for each mutant strain at 30°C (thin line) and 37°C (thick line). The resulting cell size distributions were compared to paired cultures of wild type cells incubated at 30°C (dotted line) and 37°C (dashed line) and analyzed using the Mann-Whitney U Test for non-parametric distributions (p<0.0001).
Upon exposure to 37°C, wild type cells increase in size, consistent with the temporary depolarization of actin (Figure 4A) (Nichols et al., 2007). As has been previously reported, the size of ras1Δ cells increases at both 30°C and 37°C relative to wild-type cells, and this difference was highly significant (Figure 4B) (Alspaugh et al., 2000). Similarly, both rac1Δ and rac2Δ cells increased in size compared to wild type cells (Figure 4 C, D). Despite this increase in overall cell size, no defects in bud development, morphology, or cytokinesis were observed.
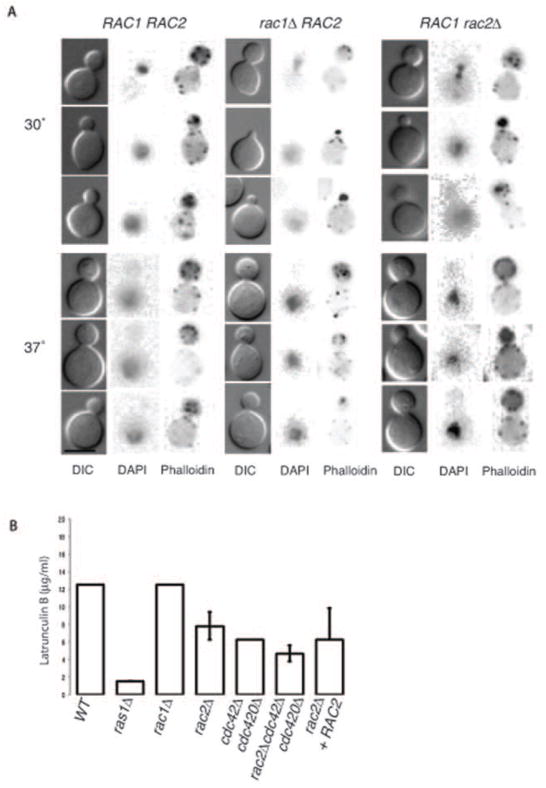
Consistent with the lack of morphological changes, we observed no defects in the localization of actin in the rac1Δ or rac2Δ mutants compared to wild type as assessed by rhodamine-conjugated phalloidin staining (Figure 5A). In both mutants and at both 30°C and 37°C, F-actin was appropriately polarized to the emerging bud in a manner that was not significantly different from wild type. This observation is in contrast to the marked defect in actin polarization in the ras1Δ and cdc42Δ mutant strains (Nichols et al., 2007). However, we did observe increased sensitivity in the rac2Δ mutant to the drug Latrunculin B, which disrupts actin filaments and sequesters actin monomers, thereby inhibiting the re-polymerization of actin filaments following exposure to stress (Ballou et al., 2010; Gibbon et al., 1999; Staiger et al., 2009) (Figure 5B). While the MIC for wild type and rac1Δ cells was 12.5 μM Latrunculin B, rac2Δ cells were inhibited at 6.25 μM, similar to the cdc42Δ cdc420Δ double mutant. The increased sensitivity of the rac2Δ mutant strain was restored by re-introduction of RAC2 under its native promoter. Additionally, the loss of RAC2 had an additive effect on the sensitivity of the cdc42Δ cdc420Δ mutant: cells lacking both RAC2 and the CDC42 paralogs were inhibited at 3.125 μM Latruculin B.
Figure 5. Rac paralogs play only a minor role in actin polarization.
A) Wild type, rac1Δ, and rac2Δ cells were grown in liquid YPD medium for 4 hours at 30 or 37°C and then stained for actin (Rhodamine-conjugated Phalloidin) and DNA (DAPI). Actin was observed polarized to bud tips in a majority of wild type and mutant cells (Scale bar = 5 μm). B) Wild type and the indicated mutant cells were grown in the presence of varying concentrations of the actin inhibitor Latrunculin B for 48 hours at 30°C. Bars indicate minimum inhibitory concentration (duplicate wells, n=4).
Together, these data suggest that Rac2, but not Rac1, plays a native role in actin polymerization. This observation is consistent with a role for Rac2 but not Rac1 in the re-polarization of actin in response to stress and may explain the larger size of the rac2Δ compared to the rac1Δ yeast phase cells. Additionally, the Rac paralogs by themselves are insufficient to compensate for the loss of Cdc42-paralog mediated actin polymerization/polarization.
A Rac inhibitor is synergistic with the inhibition of actin
These data suggest that Rac paralogs play a minor role in actin polarization, but may also play an actin-independent role in yeast-phase growth. Because we were unable to generate a rac1Δ rac2Δ double mutant, we took a chemical approach to characterizing the role of Rac paralogs in yeast-phase growth. EHT1864 is a specific inhibitor of Rac but not Cdc42 activity that is currently under investigation for the treatment of human cancers (Katz et al., 2012; Shutes et al., 2007; Takeo et al., 1995). When C. neoformans cells were exposed to high levels of EHT1864 (>500 μM), we observed only limited inhibition of growth (data not shown), suggesting that either C. neoformans Rac paralogs are insensitive to the drug or that Cdc42 activity might compensate for the loss of Rac activity in this context. We have previously demonstrated that Cdc42 paralogs are required for the re-polarization of actin in response to stress (Figure 5 and (Ballou et al., 2010)). However, the lack of actin localization defects and the relative insensitivity of the rac1Δ and rac2Δ mutants to Latrunculin B suggest that Rac paralogs play only a limited role, if any, in the re-polarization of actin in response to stress. We therefore hypothesized that Rac and Cdc42 paralogs might act in parallel pathways to modulate polarized growth and actin-dependent stress resistance.
In order to assess for synergistic effects between actin inhibition (Latrunculin B) and Rac inhibition (EHT1864), a checkerboard MIC assay was performed (Jessup, 1998; Odds, 2003). The minimal inhibitory concentration (MIC) for Latrunculin B alone against wild type C. neoformans was 100 μM, and the MIC for EHT1864 alone under the same conditions was 250μM. When the drugs were used in combination, we observed synergistic inhibition of wild type cell growth at otherwise sub-inhibitory concentrations. C. neoformans growth was inhibited with the combination of 6.25 μM Latrunculin B + 31.25μM EHT1864, giving a fractional inhibitory concentration (FIC) of <0.19, showing a strong synergistic effect. These data suggest that Rac paralogs are involved in both actin-dependent and actin-independent mechanisms in C. neoformans growth.
Rac paralogs are not required for cell cycle control
In C. neoformans, conditions that delay polarized growth, such as temperature stress, result in the accumulation of G2 stage cells that are larger than G1 cells (Nichols et al., 2007; Takeo et al., 2003). Similarly, in a number of yeast species, stable diploids are larger than haploid cells (Hickman et al., 2013; Mortimer, 1958). It was previously demonstrated that Rac1 plays a role in nuclear migration during hyphal development (Vallim et al., 2005), raising the possibility that Rac paralogs may be involved in nuclear dynamics during yeast phase growth. Given these observations, one possible explanation for the increased cell size of the rac1Δ and rac2Δ mutants is that they accumulate in G2, even in the absence of external stress. We therefore performed FACS analysis of the two mutant populations following growth at 30°C. However, we observed no difference in the proportion of G1 vs. G2 cells for either the rac1Δ or rac2Δ mutants compared to wild type haploid cells, nor did we observe the emergence of an apparent diploid population (data not shown).
Rac proteins localize to the plasma membrane during yeast phase growth
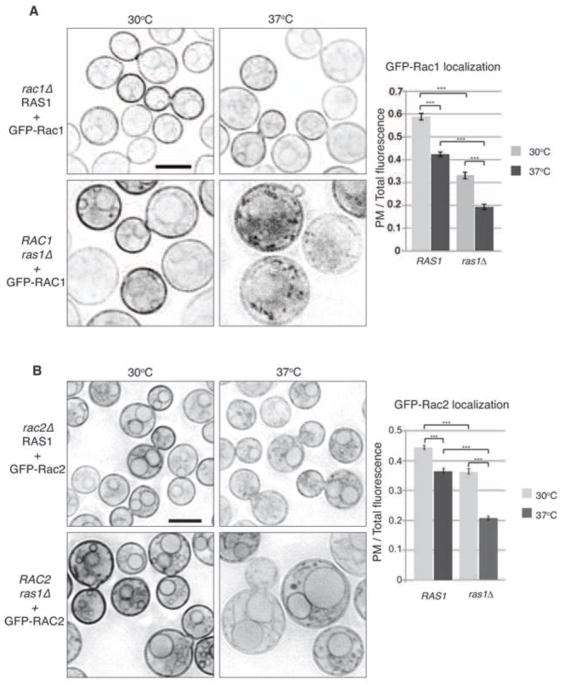
In order to observe the localization of the Rac paralogs in C. neoformans, we introduced GFP-Rac1 and GFP-Rac2 fusion constructs in wild type H99 background. Both GFP-Rac1 and GFP-Rac2 were localized to plasma membranes and endo-membranes at 30°C, consistent with their predicted localization based on the presence of C-terminal CAAX boxes (Figure 6A, B top left panels). After 4 hours incubation at 37°C, the localization of GFP-Rac1 and GFP-Rac2 to the plasma membrane decreased, with both proteins becoming more concentrated on endo-membranous structures and in the cytoplasm (Figure 6A, B top right panels). This decrease was quantified by calculating the total cell fluorescence and determining the percentage localized to the plasma membrane in each condition. In each case, the decrease was determined to be slight but statistically significant (p< 0.0001).
Figure 6. Rac paralog localization is dependent on temperature and Ras1.
A, B) The localization of GFP-Rac1 and GFP-Rac2 was observed by confocal microscope in wild type cells that had been incubated in liquid YPD medium for 4 hours at 30 or 37°C. The level of fluorescence in the plasma membrane was measured relative to overall fluorescence using ImageJ software. (p<0.001).
This temperature dependent localization of the GFP-Rac constructs coupled with the ability of Rac paralogs to suppress Ras1 defects in thermotolerance suggested that Ras1 might be involved in the temperature-dependent aspects of Rac paralog activity. We therefore assessed the localization of GFP-Rac1 and GFP-Rac2 in the ras1Δ background. Importantly, we specifically chose strains in which construct expression was documented by the appearance of GFP fluorescence and the restoration of hyphal growth to the ras1Δ mutant (data not shown), but in which the pHis-GFP-Rac1 and pHis-GFP-Rac2 constructs were not sufficiently overexpressed to suppress ras1Δ temperature sensitivity. In this way, the constructs could be used as markers of Rac1 and Rac2 localization independent of confounding gain-of-function effects resulting from the over-expression of these putative targets of Ras1.
At 30°C, the localization of GFP-Rac1 in the ras1Δ background was altered compared to GFP-Rac1 localization in RAS1 cells. Although the construct remained on cellular membranes, the percentage of fluorescence on the plasma membrane was decreased (36% compared to 43% in WT cells). Instead, a significant portion of the GFP-Rac1 appeared to be located on numerous intracellular vesicles (Figure 6A lower left panel). GFP-Rac1 in the ras1Δ mutant was further disrupted after 4 hr growth at 37°C. Under temperature stress, only 21% of the GFP-Rac1 fluorescence remained on the plasma membrane (p<0.0001). The remaining GFP-Rac1 showed punctate localization throughout the cytoplasm (Figure 6A lower right panel).
In the absence of Ras1, GFP-Rac2 remained primarily localized to the plasma membrane and endomembranes at 30°C (Figure 6B, lower left panel). This localization was visually similar to GFP-Rac2 in RAS1 cells. In contrast, the GFP-Rac2 localization in the ras1Δ mutant was drastically altered at 37°C. At this elevated temperature, GFP-Rac2 localization was more punctate, and the construct appeared to accumulate in the ER (Figure 6B, lower right panel). Quantification of this change in localization revealed that there was a 23% decrease in the percent of plasma membrane to total fluorescence, and this difference was highly significant (p<0.0001).
Together these data demonstrate that Rac1 and Rac2 are localized to cellular membranes, consistent with prenylation of their C-terminal CAAX box motifs, and suggest that their localization is dependent on both temperature and Ras1 activity. This raises the possibility that the large size of the ras1Δ mutants at 37°C may be the result of the aberrant localization of both Rac paralogs.
Rac paralogs play overlapping and separate roles in polarized growth during hyphal development
Together, our investigation of Rac paralog function in yeast phase cells suggests a role for Rac paralogs in polarized growth. We therefore examined the role of Rac paralogs in hyphal development during mating, fusion, MFα induction, and polarized growth.
Rac paralogs are required for hyphal development during mating
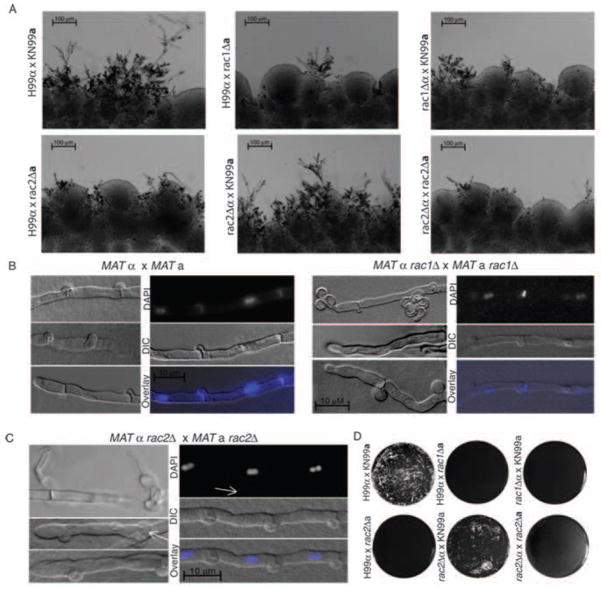
We have previously reported defects in hyphal development during bilateral rac1Δα × rac1Δa mating in C. neoformans var. neoformans (Vallim et al., 2005). Similarly, C. neoformans var. grubii Rac1 plays a significant role in hyphal development during mating. In unilateral rac1Δα × RAC1a and RAC1α × rac1Δa mating reactions, we observed a delay in hyphal development relative to wild type (Figure 7A). Additionally, close examination of rac1Δα × rac1Δa mating reactions often revealed hyphae that were altered in their appearance compared to wild type hyphae (Figure 7B). In some cases, we observed clamp cells that were truncated and had failed to fuse, similar to C. n. var. neoformans rac1Δ mating hyphae (Vallim et al., 2005). Additionally, MATα rac1Δ × MATa rac1Δ bilateral mating hyphae sometimes wandered, in contrast to the straight hyphae typical of wild type reactions (Figure 5B). This morphological abnormality was reminiscent of hyphae with defects in polarity maintenance, although we did not observe tip-splitting or increased branching phenotypes such as those exhibited by C. neoformans ste20Δ or pak1Δ mutants or racΔ mutants in other species (Araujo-Palomares et al., 2011; Nichols et al., 2004; Rolke and Tudzynski, 2008; Tanaka et al., 2008) Despite these defects, we were able to detect the presence of fused clamp cells and bi-nucleate hyphal compartments, indicative of a/α mating, and spores dissected from these crosses were distributed between a and α mating types (11a:22α).
Figure 7. Rac paralogs contribute to hyphal development during mating.
A) Equal numbers of cells for the indicated strains were mixed and incubated on MS mating medium for 5 days in the dark at room temperature. Filaments were imaged in situ. (Scale bar = 100 μm). H99α and KN99a are wild-type mating partner tester strains. B, C) Hyphae from mating reactions between the indicated strains were excised from MS mating plates and slide squashes were prepared as described. White arrows indicate B) abnormal clamp cells and C) haustoria arising from clamp cells (Scale bar = 10 μm for all images.) D) Fusion between the indicated strains was detected as described.
C. neoformans var. grubii Rac2 also plays a key role in mating: strains lacking RAC2 exhibit a delay in hyphal development (Figure 7A). In unilateral crosses with wild type cells, rac2Δa cells, but not rac2Δα cells, exhibit a slight delay in mating, and fewer, shorter hyphae are observed at day 5 than in a wild type control cross. Consistent with this unilateral delay, rac2Δα × rac2Δa cells in a bilateral cross exhibit a delay in the development of hyphae, with few foci forming after 5 days (Figure 7A). Despite the apparent delay in hyphal development, rac2Δa × rac2Δa mating reactions eventually (6 weeks) produce hyphae that extend into the medium to a similar degree to wild type reactions, and we observed fused clamp cells and bi-nucleate hyphae indicative of a/α mating (Figure 7C). However, rac2Δα × rac2Δa hyphae were broader than wild type hyphae, and produced a high proportion of haustoria, specialized hyphal mating structures of unknown significance. These defects did not appear to have an effect on the production of viable spores (25 of 30 dissected spores germinated).
Rac paralogs are required for fusion
We hypothesized that the delay in the development of hyphae observed in the unilateral and bilateral crosses using the rac1Δ and rac2Δ strains could be due to defects in initial fusion events. We therefore performed fusion assays to assess the rates of fusion in the unilateral and bilateral crosses (Figure 7D). This assay was performed by quantifying the formation of dikaryotic cells encoding two dominant selection markers that were individually present in the mutant mating partners (Nichols et al., 2004). In contrast to wild type mating reactions in which abundant double-resistant dikaryotic cells were evident after 24 hours, no fusion was detected after 24 or 48 hours in unilateral MATα rac1Δ × MATa RAC1, MATα RAC1 × MATa rac1Δ, MATα RAC2 × MATa rac2Δ or bilateral MATα rac2Δ × MATa rac2Δ crosses. Consistent with the more robust filament production observed in MATα rac2Δ × MATa RAC2 unilateral crosses after 5 days, this mutant cross demonstrated only a limited defect in fusion after 48 hours. This result was replicated using two independently generated MATα rac2Δ strains (data not shown). This observation suggests that the Rac paralogs may play differential roles in aspects of mating independent of polarization.
Rac paralogs are dispensable for the induction of mating pheromone
Several proteins in the Ras1 signal transduction cascade have been demonstrated to impact fusion during mating in different ways. For example, Ras1, but not the Rac/Cdc42 effector Pak1, has been linked to the induction of mating pheromone (MFα) in response to a mating partner. We therefore examined the role of the Rac paralogs in the induction of α pheromone in C. neoformans var. grubii by RT-PCR. The expression of α-pheromone by MATα cells persists at a basal level in the absence of a mating partner, and expression is induced by the presence of MATa cells. We measured the expression of MFα1 by wild type and Rac paralog mutant MATα cells after 12 hours of exposure to wild type and Rac paralog mutant MATa cells by RT-PCR relative to an internal control (GPD). We observed no significant reduction in the induction of MFα1 by either the rac1Δ or the rac2Δ cells in any of the unilateral crosses (< 2-fold). However, MFα1 induction was reduced in the rac1Δα × rac1Δa bilateral cross (5.9 fold decrease compared to wild type induction). In contrast, MFα1 induction was not significantly reduced in the rac2Δα × rac2Δa bilateral cross (1.1 fold decrease compared to wild type induction), suggesting that Rac1 may play a more significant role than Rac2 in the early events of mating.
Together, these data demonstrate that both Rac1 and Rac2 are required for hyphal development during mating, independent of the induction of MFα1, and suggest that Rac paralogs play a specific role in polarized growth and the cellular events related to fusion, rather than in the induction of a pheromone response cascade.
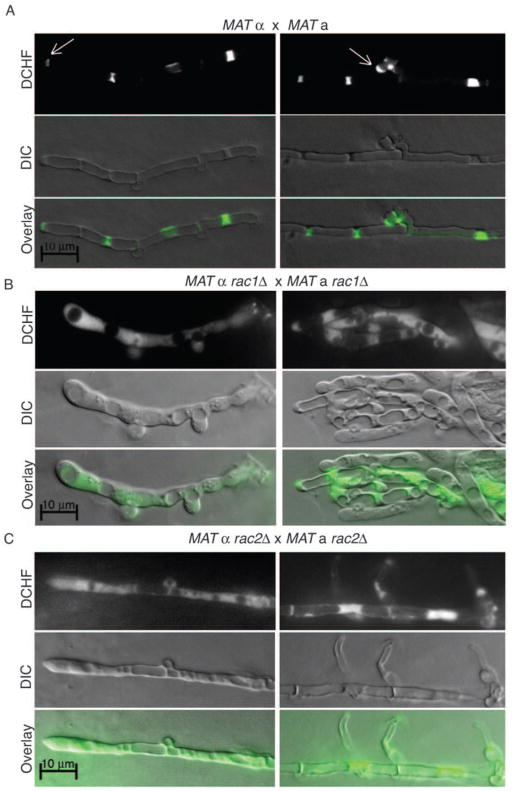
Rac paralogs are involved in the localization of ROS within hyphae
To explore the mechanisms by which Ras proteins direct polarized growth of C. neoformans cells, we examined localized ROS concentrations in mating hyphae (Figure 8). Rac GTPases have been shown to play a role in the regulation of reactive oxygen species (ROS), which has in turn been shown to be important for the regulation of hyphal development in a number of fungi. The presence of ROS was visualized by 2′,7′-dichlorodihydrofluorescein diacetate (DCHF-DA) fluorescence. DCFH-DA is a lipophilic, cell permeable molecule that is oxidized to DCF upon exposure to ROS. The degree and localization of the fluorescence is related to the concentration and distribution of the ROS (Bass et al., 1983). In wild type crosses, incubation of mating hyphae with DCFH-DA revealed a distinct staining pattern. The signal was localized to clamp cells and to the growing tip of the hypha (Figure 8A). This is consistent with observations in other fungi, in which ROS localizes to sites of polarized growth, including branch points and the hyphal tip (Chen and Dickman, 2004; Tanaka et al., 2008).
Figure 8. Rac paralogs contribute to the localization of ROS during hyphal growth.
Hyphae from wild type (A), rac1Δ bilateral (B) and rac2Δ bilateral (C) crosses that had been incubated in the dark for 1 week were excised from MS mating plates and co-incubated with 9 μM DCHF-DA for 10 minutes to detect ROS localization and intensity. Slide squashes were prepared as described. (Scale bar = 10 μm for all images.)
The loss of either Rac paralog impacted ROS localization, with Rac1 playing a more significant role (Figure 8B, C). In rac1Δα × rac1Δa hyphae, DCF fluorescence was diffusely localized along the length of the hypha and only minimally concentrated at septa. In rac2Δα × rac2Δa hyphae, in contrast, DCF fluorescence remained concentrated around septa but was more diffuse than in wild type hyphae and was inappropriately punctate. These data suggest that Rac1 and Rac2 play roles in the regulation of ROS, either through the localization of Nox proteins, as has been reported for Cdc42 in A. nidulans, or through the regulation of Nox activity, as has been reported in E. festucae and C. purpurea (Rolke and Tudzynski, 2008; Semighini and Harris, 2008; Tanaka et al., 2008).
Discussion
Here we have taken a whole genome approach to the identification of duplicated genes in the genome of the human fungal pathogen C. neoformans var. grubii. Using this approach, we identified two large networks of paralogous gene families that likely speak to the structure of C. n. var. grubii sub-telomeric regions, as well as a number of smaller networks and pairs likely to have functional significance. We validated this approach to paralog identification by undertaking the characterization of the cryptic Rac paralog Rac2 and here describe its contributions to polarized growth during both yeast and hyphal phase C. neoformans morphogenesis.
For human pathogens, adaptation to the environment of the host can have important consequences for virulence and pathogenicity. Recently, a number of mechanisms by which this adaptation can be achieved have been described. Plasticity at the genome level has been observed for Candida albicans, in which the emergence of isochromosome 5 facilitates resistance to fluconazole (Selmecki et al., 2009). In Cryptococcus neoformans, multiple disomic chromosomes temporarily emerge in response to fluconazole exposure in patients, suggesting that the plasticity of the C. neoformans genome may provide similar flexibility in response to stress environments (Sionov et al., 2010). At the genetic level, duplicated genes have long been proposed to support niche adaptation, and recent work in Salmonella enterica demonstrated this experimentally: the duplication of single genes with multiple functions resulted in gene amplification, functional divergence, and selective advantage over time (Näsvall et al., 2012).
Sub-telomeric regions have been shown to be enriched in genes that facilitate adaptation to environmental niches (Adams et al., 1992; Christiaens et al., 2012; de Bruin et al., 1994; Linardopoulou et al., 2001), and although little is known about the structure of C. neoformans sub-telomeres specifically, these regions have been identified as sites of rapid gene evolution (Morrow et al., 2012). Using a whole genome approach to the analysis of paralogs in C. neoformans, we identified of a sub-telomeric family of carbohydrate metabolism genes with conserved catalytic domains related to trehalose and maltose metabolism, possibly indicating another example of this potential for niche adaptation. Future work will examine the role of metabolic paralogs in niche adaptation.
The identification of Rac paralogs
In addition to these larger networks, we here report the identification of the cryptic Rac paralog Rac2. Homologs of the Rac GTPase were first identified in human myeloid cells, and early work in mammalian systems suggested roles for Rac in the generation of reactive oxygen species (ROS), the organization of actin filaments, and secretion (Abo et al., 1991; Didsbury et al., 1989; Price et al., 1995; Ridley et al., 1992). Recently, it was demonstrated that Rac is the originating Rho-GTPase, with Cdc42 emerging much later in the eukaryotic lineage (Boureux et al., 2007). As such, Rac proteins play highly conserved roles in basic morphological processes throughout eukaryotes. Because the model yeasts S. cerevisiae and S. pombe lack Rac homologs, efforts to examine their role in fungal morphogenesis have been focused on the filamentous and dimorphic fungi. The mammalian roles for Rac proteins have been upheld in these fungi, suggesting that these emerging model systems may aid efforts to understand Rac function. CnRac1 was first identified as a suppressor of ras1Δ defects in thermotolerance and a modulator of hyphal development (Vallim et al., 2005). More recently, CnRac1 was demonstrated to play a role in secretion (Shen et al., 2011). However, CnRac1’s roles in the generation of ROS and the organization of actin remained unclear. Additionally, there had been no description of its role in yeast-phase morphogenesis.
The data presented here suggest that Rac paralogs regulate polarity during both yeast-like and hyphal growth in C. neoformans. Mating cell fusion defects, aberrant hyphal morphology, and increased yeast cell size in the rac1Δ and rac2Δ single mutants, coupled with the apparent synthetic lethality of the rac1Δ rac2Δ double mutant, are consistent with a role for the Rac paralogs in polarized growth and are consistent with the roles reported for Rac homologs in other fungi. For example, in N. crassa, Rac1 was identified in a screen for genes required for fusion, and both Cnrac1Δ and Cnrac2Δ mutants exhibit defects in fusion of mating partners (Fu et al., 2011). Similarly, the loss of Rac in a number of filamentous fungi negatively impacts hyphal morphology (Chen and Dickman, 2004; Rolke and Tudzynski, 2008; Tanaka et al., 2008).
Based on the overlap in phenotypes between the rac1Δ and rac2Δ mutants, we hypothesize that the Rac paralogs are largely redundant. However, subtle phenotypic differences between the single mutants may indicate that a more dominant role is played by Rac1 during mating and hyphal growth and by Rac2 during yeast-phase growth. These differences may provide insight into the signaling cascades utilized in the two morphological forms. For example, morphogenesis may be mediated by Rac-dependent MAP kinase cascades, as likely occurs in C. trifoli and C. albicans (Chen and Dickman, 2004; Hope et al., 2010), or through the activation and regulation of ROS, either directly or via Ste20, as occurs in Epichloe festucae and Claviceps purpurea (Rolke and Tudzynski; Tanaka et al., 2008). Rac proteins have additionally been shown to regulate the fungal polarisome, independent of any role in the organization of the cytoskeleton. In Ustilago maydis, the polarized localization of Spa2, a polarisome component, depends on Rac1 but is independent of actin and microtubule function (Carbó and Pérez-Martín, 2008). Elements of the C. neoformans polarisome have not yet been identified.
Possible downstream targets: PAKs and Wsp1
There are several candidates for the immediate downstream effectors of Rac function. The conserved PAK Pak1, the Cla4-like PAK Ste20, and the WASP homolog Wsp1 have been demonstrated by yeast two hybrid to interact with Rac1 from C. neoformans var. neoformans (Shen et al.; Vallim et al., 2005; Wang et al., 2002). However, the loss of either Ste20 or Wsp1 does not entirely phenocopy the loss of either Rac paralog, either because the two paralogs are redundant or as a result of overlap with Cdc42 paralog functions. During mating, both ste20Δ and wsp1Δ mutants exhibit reductions in fusion that are not as severe as those exhibited by rac1Δ or rac2Δ mutants, suggesting that the Rac paralogs may have multiple targets during fusion (Nichols et al., 2004). Likewise, ste20Δ hyphae exhibit a distinctive “tip-splitting” morphology that is more severe than the morphological defects exhibited by rac1Δ or rac2Δ bilateral mutant crosses and is indicative of defects in polarity maintenance (Nichols et al., 2004). This difference in the severity of hyphal defects may suggest that Ste20 is a shared target of both Rac paralogs during hyphal growth but not during fusion. Additionally, both wsp1Δ and ste20Δ cells are temperature sensitive, unlike the rac1Δ or rac2Δ mutants but more similar to the cdc42Δ mutant, suggesting an underlying specificity in Rho-GTPase signal transduction (Ballou et al., 2010; Shen et al., 2011; Wang et al., 2002).
A negative regulatory relationship between UmRac1 and the PAK UmCla4 was recently demonstrated in the basidiomycete Ustilago maydis. During mating in U. maydis, activated Rac1 is regulated via a PAK-dependent negative feedback loop that triggers the degradation of Cdc24, the Rac1 GEF (Frieser et al., 2011). A similar relationship may exist in C. neoformans: While ras1Δ and racΔ mutants are deficient in polarized growth, ste20Δ yeast-phase cells become hyper-polarized at 39°C, (Wang et al., 2002). The importance of this regulation is highlighted by the observation that the expression of inducible dominant active UmRac1 is lethal (Mahlert et al.). Similarly, dominant active Rac2 appears to be lethal in C. neoformans (Ballou, unpublished data).
In contrast to the limited evidence of functional overlap between C. neoformans Rac and Ste20 proteins, Rac and Pak1 exhibit significant functional overlap. Like Rac1 and Rac2, CnPak1 is not primarily involved in thermotolerance, since the pak1Δ mutant grows well at 37°C (Wang et al., 2002). Additionally, CnPak1 has been previously demonstrated to control the mating fusion event in a manner independent of the induction of MFα pheromone, similar to Rac1 and Rac2 (Nichols et al., 2004).
Despite these similarities in mutant phenotypes, there are subtle complexities in the interaction of the related Rac and Cdc42 GTPases and the downstream Pak1 and Ste20 kinases. Although pak1Δ mutants are most phenotypically similar to the rac1Δ and rac2Δ mutants, and ste20Δ mutants are most similar to cdc42Δ mutants, yeast two hybrid studies have demonstrated potential interactions between Pak1 and Cdc420, as well as between Rac1 and Ste20 (Vallim et al., 2005; Wang et al., 2002). Additionally, the hyphal defects of pak1Δα × pak1Δa bilateral crosses, including failed spore production and defects in clamp-cell fusion, are more typical of cdc42Δ mutant mating defects than those observed in racΔ mutants crosses (Ballou et al., 2010; Nichols et al., 2004). It is possible that there is some degree of co-activation of downstream effectors by the related Rac and Cdc42 proteins. Additionally, the presence of the second Rac/Cdc42 paralog may activate PAK kinases, in a native or non-native way, in the individual mutant strains. Future work will examine the specific roles of the Rac and Cdc42 paralogs in the activation of these conserved downstream effectors.
Possible Rac targets: Nox proteins
The data presented here suggest that Rac paralogs regulate polarity via the localization of Reactive Oxygen Species (ROS). Although ROS have been previously thought to be toxic by-products of mitochondrial function, a number of recent works have highlighted their role as secondary signaling molecules in fungal and mammalian systems (Chen and Dickman; Hamanaka et al., 2013; Li et al., 2011; Semighini and Harris, 2008). Importantly, the mammalian Rac GTPase was first identified an essential component of the NADPH oxidase complex, a multi-component enzyme responsible for the generation of the oxidative burst (Abo et al., 1991; Lambeth). In this complex, Rac interacts with p67phox to facilitate electron transfer via the catalytic subunit cytochrome b (p22phox plus gp91phox) from NADPH to FAD, eventually resulting in the production of ROS (Diebold and Bokoch). Similarly, in filamentous fungi Rac interacts with Nox (gp91phox) and NoxR (p67phox), and a growing body of work suggests that this interaction plays important roles in development, differentiation, and morphogenesis. Work in A. fumigatus and A. nidulans has demonstrated the specific importance of the generation of ROS to polarized growth and has revealed roles for both Rac and Cdc42 in Nox activation (Li et al., 2011; Semighini and Harris, 2008). For example, in A. fumigatus, ΔracA hyphal cells with the characteristic swelling of mutants with polarity defects exhibit decreased ROS production at hyphal tips and accumulate ROS at sites of branching (Li et al., 2011). Treatment of wild type A. fumigatus cells with an NADPH oxidase inhibitor phenocopies the polarity defects of the ΔracA mutant, suggesting that that ROS is required for, rather than a by-product of, RacA-mediated polarized growth.
Moreover, the interaction appears to be highly conserved. In E. festucae, the loss of RacA phenocopies the loss of the NADPH oxidase NoxA, and RacA physically interacts with NoxR to modulate polarized growth and mutualism with perennial ryegrass (Takemoto et al., 2006; Tanaka et al., 2008). In C. purpurea, Rac and its target Cla4 are required for the maintenance of a polarity axis and for pathogenesis. Additionally, CpRac and CpCla4 play a key role in the transcriptional regulation of the NADPH oxidase Nox1, which is likewise required for pathogenesis (Giesbert et al., 2008; Rolke and Tudzynski, 2008). Similar regulatory roles for Rac in polarity and the generation of ROS have also been described in Colletotrichum trifoli (Chen and Dickman; Li et al., 2011; Semighini and Harris, 2008). To date, no elements of the Nox complex have been identified in C. neoformans, and the mechanisms by which the Rac paralogs regulate ROS localization remain unclear. Future work will examine the interaction of the Rac paralogs with elements of the CnNox complex.
Ras-dependent Rac localization
Finally, we report here that Rac paralog localization is dependent on Ras1. The localization of both Rac1 and Rac2 to the plasma membrane is consistent with Rac localization in other yeast-like fungi and with their predicted prenylation sequences (Hope et al., 2010; Wright and Philips, 2006). In addition, the interaction of Ras- and Rho- class GTPases has been previously demonstrated to facilitate function: In S. cerevisiae, the Ras-family GTPase Rsr1 and the Rho-GTPase Cdc42 physically interact and this interaction is required for proper bud site selection (Kang et al., 2010; Kozminski et al., 2003; Park et al., 2002; Zheng et al., 1995). The Ras-dependence of Rac localization may partially explain the ras1Δ mutant phenotypes, including the increase in cell size at 30°C and the loss of polarized growth at 37°C; in the absence of Ras1 activity, the function of both Rac paralogs may be simultaneously impaired. The functional consequences of aberrant Rac localization have been demonstrated in C. albicans. The dynamics of Rac1 localization to the plasma membrane of C. albicans are dependent on the Rac1 GEF Dck1 and the Rac1 target Lmo1, an ELMO-type adaptor protein required for activation of MAP kinase cascades regulating filamentation and cell wall integrity (Hope et al., 2010). In the absence of any element of this module, C. albicans is unable to make the transition to filamentous growth so crucial to its pathogenicity (Bassilana and Arkowitz, 2006; Hope et al., 2008; Hope et al., 2010). It remains unclear whether an analogous module exists in C. neoformans, as further effort will be required to determine the identities of any Rac paralog-specific GEFs or their specific effectors. However, Ras1 will likely play a key role in such regulation.
Significance and larger context
For human pathogens, adaptation to the environment of the host can have important consequences for virulence and pathogenicity, and the existence of duplicated genes has been shown to facilitate this adaptation in C. neoformans (Ballou et al., 2010; Okagaki et al., 2011). It was recently demonstrated that C. neoformans is able to modulate the behavior of human brain microvascular epithelial cells (HBMECs) via phospholipase B1- mediated re-arrangement of the host actin cytoskeleton, which in turn is regulated by human Rac1 (Chang et al., 2004; Maruvada et al., 2012). This modulation facilitates the uptake of C. neoformans cells and transport across the blood/brain barrier in mice. Treatment of infected mice with an inhibitor of Rac activity decreased passage of C. neoformans across the blood brain barrier. Although no role for CnRac1 was observed in this process, the data we present here, including the identification of a second Rac paralog, Rac2, and the synergistic effect of Rac and actin inhibitors on C. neoformans growth, further underscores both the importance of gene duplications in fungal pathogenesis and the potential of Rac inhibitors for anti-fungal therapy.
Supplementary Material
Metadata associated with Figure 1 can be accessed at the provided links. The data are provided as a .cys file, which can be opened using the freely available Cytoscape software (www.cytoscape.org) according to the tutorial on the Cytoscape website {Cline, 2007}. These data were generated using Cytoscape3 (2013). Information for each node and edge is listed in the Table Panel. Selecting a particular node will provide the gene number as identified by the Broad database for C. neoformans H99 (http://www.broadinstitute.org/annotation/genome/cryptococcus_neoformans/MultiHome.html), as well as chromosome number and the genome annotation. Under the VizMapper menu, ensure that Node Label (under the Node Visual Properties menu) is set at Gene_name. This will pass annotation information in the network table through to the nodes so that gene names are displayed on the nodes. Similarly, setting the Edge Label (under the Edge Visual Properties menu) at Percent_Identity will display the percent identity between two nodes on the edge connecting those nodes.
Highlights.
A whole genome analysis of C. neoformans paralogs revealed a second Rac, Rac2.
Rac paralogs contribute to yeast cell size and exhibit Ras-dependent localization.
C. neoformans is synthetically sensitive to Rac and actin inhibitory agents.
Racs contribute to ROS localization within hyphae and are required for fusion.
Acknowledgments
We would like to thank Sam Johnson of the Duke University Light Microscopy Core Facility for his assistance with confocal imaging and analysis. We would like to thank Michael Cooke at the Duke University Flow Cytometry Shared Resource for his assistance with FACS optimization. Additionally, we would like to thank Lukasz Kozubowski for thoughtful comments and advice. This manuscript was supported by PHS grant AI050128.
Footnotes
The authors declare that there are no conflicts of interest associated with this work.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abo A, et al. Activation of the NADPH oxidase involves the small GTP-binding protein p21rac1. Nature. 1991;353:668–670. doi: 10.1038/353668a0. [DOI] [PubMed] [Google Scholar]
- Adams AE, et al. Cdc42 and Cdc43, Two additional Genes Involved in Budding and Establishment of Cell Polarity in the Yeast Saccharomyces cerevisiae. J Cell Biology. 1990;111:131–142. doi: 10.1083/jcb.111.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams J, et al. Adaptation and major chromosomal changes in populations of Saccharomyces cerevisiae. Current Genetics. 1992;22:13–19. doi: 10.1007/BF00351736. [DOI] [PubMed] [Google Scholar]
- Alspaugh JA, et al. RAS1 regulates filamentation, mating and growth at high temperature of Cryptococcus neoformans. Molecular Microbiology. 2000;36:352–365. doi: 10.1046/j.1365-2958.2000.01852.x. [DOI] [PubMed] [Google Scholar]
- Altschul SF, et al. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Araujo-Palomares CL, et al. Functional characterization and cellular dynamics of the CDC-42 - RAC - CDC-24 module in Neurospora crassa. PLoS ONE. 2011;6:e27148. doi: 10.1371/journal.pone.0027148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballou ER, et al. Two CDC42 paralogues modulate Cryptococcus neoformans thermotolerance and morphogenesis under host physiological conditions. Molecular Microbiology. 2010;75:763–780. doi: 10.1111/j.1365-2958.2009.07019.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass DA, et al. Flow cytometric studies of oxidative product formation by neutrophils: a graded response to membrane stimulation. Journal of immunology (Baltimore, Md: 1950) 1983;130:1910–1917. [PubMed] [Google Scholar]
- Bassilana M, Arkowitz RA. Rac1 and Cdc42 have different roles in Candida albicans development. Eukaryotic Cell. 2006;5:321–329. doi: 10.1128/EC.5.2.321-329.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boureux A, et al. Evolution of the Rho family of ras-like GTPases in eukaryotes. Molecular biology and evolution. 2007;24:203–216. doi: 10.1093/molbev/msl145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyce KJ, et al. The CDC42 homolog of the dimorphic fungus Penicillium marneffei is required for correct cell polarization during growth but not development. Journal of bacteriology. 2001;183:3447–3457. doi: 10.1128/JB.183.11.3447-3457.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyce KJ, et al. Control of morphogenesis and actin localization by the Penicillium marneffei RAC homolog. Journal of Cell Science. 2003;116:1249–1260. doi: 10.1242/jcs.00319. [DOI] [PubMed] [Google Scholar]
- Boyce KJ, et al. The Ras and Rho GTPases genetically interact to co-ordinately regulate cell polarity during development in Penicillium marneffei. Molecular Microbiology. 2005;55:1487–1501. doi: 10.1111/j.1365-2958.2005.04485.x. [DOI] [PubMed] [Google Scholar]
- Carbó N, Pérez-Martín J. Spa2 is required for morphogenesis but it is dispensable for pathogenicity in the phytopathogenic fungus Ustilago maydis. Fungal genetics and biology: FG & B. 2008;45:1315–1327. doi: 10.1016/j.fgb.2008.06.010. [DOI] [PubMed] [Google Scholar]
- Chang YC, et al. Cryptococcal yeast cells invade the central nervous system via transcellular penetration of the blood-brain barrier. Infection and Immunity. 2004;72:4985–4995. doi: 10.1128/IAI.72.9.4985-4995.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Dickman MB. Dominant active Rac and dominant negative Rac revert the dominant active Ras phenotype in Colletotrichum trifolii by distinct signalling pathways. Molecular Microbiology. 2004;51:1493–1507. doi: 10.1111/j.1365-2958.2003.03932.x. [DOI] [PubMed] [Google Scholar]
- Chen W, et al. The CDC42 homologue from Caenorhabditis elegans. Complementation of yeast mutation. The Journal of biological chemistry. 1993;268:13280–13285. [PubMed] [Google Scholar]
- Christiaens JF, et al. Functional divergence of gene duplicates through ectopic recombination. EMBO Reports. 2012;13:1145–1151. doi: 10.1038/embor.2012.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline M, et al. Integration of biological networks and gene expression data using Cytoscape. Nature Protocols. 2007;2:2366–2382. doi: 10.1038/nprot.2007.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox GM, et al. Urease as a virulence factor in experimental cryptococcosis. Infection and Immunity. 2000;68:443–448. doi: 10.1128/iai.68.2.443-448.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer KL, et al. Transcription factor Nrg1 mediates capsule formation, stress response, and pathogenesis in Cryptococcus neoformans. Eukaryotic Cell. 2006;5:1147–1156. doi: 10.1128/EC.00145-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruin D, et al. The polymorphic subtelomeric regions of Plasmodium falciparum chromosomes contain arrays of repetitive sequence elements. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:619–623. doi: 10.1073/pnas.91.2.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didsbury J, et al. rac, a novel ras-related family of proteins that are botulinum toxin substrates. The Journal of biological chemistry. 1989;264:16378–16382. [PubMed] [Google Scholar]
- Diebold BA, Bokoch GM. Molecular basis for Rac2 regulation of phagocyte NADPH oxidase. Nature immunology. 2001;2:211–215. doi: 10.1038/85259. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004:5. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Force A, et al. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999;151:1531–1545. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frieser SH, et al. Cla4 kinase triggers destruction of the Rac1-GEF Cdc24 during polarized growth in Ustilago maydis. Molecular Biology of the Cell. 2011;22:3253–3262. doi: 10.1091/mbc.E11-04-0314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu C, et al. Identification and characterization of genes required for cell-to-cell fusion in Neurospora crassa. Eukaryotic Cell. 2011;10:1100–1109. doi: 10.1128/EC.05003-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, et al. Trp(56) of rac1 specifies interaction with a subset of guanine nucleotide exchange factors. The Journal of biological chemistry. 2001;276:47530–47541. doi: 10.1074/jbc.M108865200. [DOI] [PubMed] [Google Scholar]
- Gibbon BC, et al. Latrunculin B has different effects on pollen germination and tube growth. The Plant Cell. 1999;11:2349–2363. doi: 10.1105/tpc.11.12.2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giesbert S, et al. The NADPH oxidase Cpnox1 is required for full pathogenicity of the ergot fungus Claviceps purpurea. Molecular plant pathology. 2008;9:317–327. doi: 10.1111/j.1364-3703.2008.00466.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MW. Distinguishing among evolutionary models for the maintenance of gene duplicates. The Journal of Heredity. 2009;100:605–617. doi: 10.1093/jhered/esp047. [DOI] [PubMed] [Google Scholar]
- Hamanaka RB, et al. Mitochondrial Reactive Oxygen Species Promote Epidermal Differentiation and Hair Follicle Development. Sci Signal. 2013;6:ra8. doi: 10.1126/scisignal.2003638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris SD. Cdc42/Rho GTPases in fungi: variations on a common theme. Molecular Microbiology. 2011;79:1123–1127. doi: 10.1111/j.1365-2958.2010.07525.x. [DOI] [PubMed] [Google Scholar]
- Hickman MA, et al. The “obligate diploid” Candida albicans forms mating-competent haploids. Nature. 2013;494:55–59. doi: 10.1038/nature11865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho J, Bretscher A. Ras regulates the polarity of the yeast actin cytoskeleton through the stress response pathway. Molecular Biology of the Cell. 2001;12:1541–1555. doi: 10.1091/mbc.12.6.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hope H, et al. Activation of Rac1 by the guanine nucleotide exchange factor Dck1 is required for invasive filamentous growth in the pathogen Candida albicans. Molecular Biology of the Cell. 2008;19:3638–3651. doi: 10.1091/mbc.E07-12-1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hope H, et al. The Candida albicans ELMO homologue functions together with Rac1 and Dck1, upstream of the MAP Kinase Cek1, in invasive filamentous growth. Molecular Microbiology. 2010;76:1572–1590. doi: 10.1111/j.1365-2958.2010.07186.x. [DOI] [PubMed] [Google Scholar]
- Hurtado CA, et al. A rac homolog is required for induction of hyphal growth in the dimorphic yeast Yarrowia lipolytica. Journal of bacteriology. 2000;182:2376–2386. doi: 10.1128/jb.182.9.2376-2386.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessup CJ. Fluconazole Susceptibility Testing of Cryptococcus neoformans: Comparison of Two Broth Microdilution Methods and Clinical Correlates among Isolates from Ugandan AIDS Patients. Journal of Clinical Microbiology. 1998;36:2874–2876. doi: 10.1128/jcm.36.10.2874-2876.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang PJ, et al. The Rsr1/Bud1 GTPase interacts with itself and the Cdc42 GTPase during bud-site selection and polarity establishment in budding yeast. Molecular Biology of the Cell. 2010;21:3007–3016. doi: 10.1091/mbc.E10-03-0232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz E, et al. Targeting of Rac GTPases blocks the spread of intact human breast cancer. Oncotarget. 2012;3:608–619. doi: 10.18632/oncotarget.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozminski KG, et al. Interaction between a Ras and a Rho GTPase couples selection of a growth site to the development of cell polarity in yeast. Molecular Biology of the Cell. 2003;12:4958–4970. doi: 10.1091/mbc.E03-06-0426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambeth JD. NOX enzymes and the biology of reactive oxygen. Nature Reviews Immunology. 2004;4:181–189. doi: 10.1038/nri1312. [DOI] [PubMed] [Google Scholar]
- Larkin M, et al. ClustalW and ClustalX version 2. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Li H, et al. The small GTPase RacA mediates intracellular reactive oxygen species production, polarized growth, and virulence in the human fungal pathogen Aspergillus fumigatus. Eukaryotic Cell. 2011;10:174–186. doi: 10.1128/EC.00288-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linardopoulou E, et al. Transcriptional activity of multiple copies of a subtelomerically located olfactory receptor gene that is polymorphic in number and location. Human Molecular Genetics. 2001;10:2373–2383. doi: 10.1093/hmg/10.21.2373. [DOI] [PubMed] [Google Scholar]
- Loftus BJ, et al. The genome of the basidiomycetous yeast and human pathogen Cryptococcus neoformans. Science (New York, NY) 2005;307:1321–1324. doi: 10.1126/science.1103773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahlert M, et al. Rac1 and Cdc42 regulate hyphal growth and cytokinesis in the dimorphic fungus Ustilago maydis. Molecular Microbiology. 2006;59:567–578. doi: 10.1111/j.1365-2958.2005.04952.x. [DOI] [PubMed] [Google Scholar]
- Maruvada R, et al. Cryptococcus neoformansphospholipase B1 activates host cell Rac1 for traversal across the blood-brain barrier. Cellular Microbiology. 2012;14:1544–1553. doi: 10.1111/j.1462-5822.2012.01819.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ménard L, et al. Rac1, a low-molecular-mass GTP-binding-protein with high intrinsic GTPase activity and distinct biochemical properties. European journal of biochemistry/FEBS. 1992;206:537–546. doi: 10.1111/j.1432-1033.1992.tb16957.x. [DOI] [PubMed] [Google Scholar]
- Miller PJ, Johnson DI. Cdc42p GTPase Is Involved in Controlling Polarized Cell Growth in Schizosaccharomyces pombe. Molecular and Cellular Biology. 1994;14:1075–1083. doi: 10.1128/mcb.14.2.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Missal TA, et al. Two glutathione peroxidases in the fungal pathogen Cryptococcus neoformans are expressed in the presence of specific substrates. Microbiology (Reading, England) 2005;151:2573–2581. doi: 10.1099/mic.0.28132-0. [DOI] [PubMed] [Google Scholar]
- Morrow CA, et al. A unique chromosomal rearrangement in the Cryptococcus neoformans var. grubii type strain enhances key phenotypes associated with virulence. mBio. 2012:3. doi: 10.1128/mBio.00310-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer RK. Radiobiological and genetic studies on a polyploid series (haploid to hexaploid) of Saccharomyces cerevisiae. Radiat Res. 1958;9:312–326. [PubMed] [Google Scholar]
- Murashige T, Skoog F. A Revised Medium for Rapid Growth and Bio Assays with Tobacco Tissue Cultures. Physiologia Plantarum. 1962;15:473–497. [Google Scholar]
- Näsvall J, et al. Real-time evolution of new genes by innovation, amplification, and divergence. Science. 2012;338:384–287. doi: 10.1126/science.1226521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols CB, et al. PAK kinases Ste20 and Pak1 govern cell polarity at different stages of mating in Cryptococcus neoformans. Molecular Biology of the Cell. 2004;15:4476–4489. doi: 10.1091/mbc.E04-05-0370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols CB, et al. A Ras1-Cdc24 signal transduction pathway mediates thermotolerance in the fungal pathogen Cryptococcus neoformans. Molecular Microbiology. 2007;63:1118–1130. doi: 10.1111/j.1365-2958.2006.05566.x. [DOI] [PubMed] [Google Scholar]
- O’Meara TR, et al. Interaction of Cryptococcus neoformans Rim101 and protein kinase A regulates capsule. PLoS Pathogens. 2010;6:e1000776. doi: 10.1371/journal.ppat.1000776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odds FC. Synergy, antagonism, and what the chequerboard puts between them. J Antimicrob Chemother. 2003:52. doi: 10.1093/jac/dkg301. [DOI] [PubMed] [Google Scholar]
- Ohkusu M, et al. Deficit in oxygen causes G2 budding and unbudded G2 arrest in Cryptococcus neoformans. FEMS Microbiology Letters. 2001;204:29–32. doi: 10.1111/j.1574-6968.2001.tb10857.x. [DOI] [PubMed] [Google Scholar]
- Ohkusu M, et al. Induced synchrony in Cryptococcus neoformans after release from G2-arrest. Antonie van Leeuwenhoek. 2004;85:37–44. doi: 10.1023/B:ANTO.0000020272.19569.15. [DOI] [PubMed] [Google Scholar]
- Ohno S. Evolution by gene duplication. Springer-Verlag; New York: 1970. [Google Scholar]
- Okagaki LH, et al. Cryptococcal titan cell formation is regulated by G-protein signaling in response to multiple stimuli. Eukaryotic Cell. 2011;10:1306–1316. doi: 10.1128/EC.05179-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H-o, et al. Localization of the Rsr1/Bud1 GTPase involved in selection of a proper growth site in yeast. The Journal of biological chemistry. 2002;277:26721–26724. doi: 10.1074/jbc.C200245200. [DOI] [PubMed] [Google Scholar]
- Price LS, et al. The small GTPases Rac and Rho as regulators of secretion in mast cells. Current biology: CB. 1995;5:68–73. doi: 10.1016/s0960-9822(95)00018-2. [DOI] [PubMed] [Google Scholar]
- Price MS, et al. The Cryptococcus neoformans Rho-GDP dissociation inhibitor mediates intracellular survival and virulence. Infection and Immunity. 2008;76:5729–5737. doi: 10.1128/IAI.00896-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pukkila-Worley R, et al. Transcriptional network of multiple capsule and melanin genes governed by the Cryptococcus neoformans cyclic AMP cascade. Eukaryotic Cell. 2005;4:190–201. doi: 10.1128/EC.4.1.190-201.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridley AJ, et al. The small GTP-binding protein rac regulates growth factor-induced membrane ruffling. Cell. 1992;70:401–410. doi: 10.1016/0092-8674(92)90164-8. [DOI] [PubMed] [Google Scholar]
- Rolke Y, Tudzynski P. The small GTPase Rac and the p21-activated kinase Cla4 in Claviceps purpurea: interaction and impact on polarity, development and pathogenicity. Molecular Microbiology. 2008;68:405–423. doi: 10.1111/j.1365-2958.2008.06159.x. [DOI] [PubMed] [Google Scholar]
- Salas SD, et al. Effect of the laccase gene CNLAC1, on virulence of Cryptococcus neoformans. The Journal of experimental medicine. 1996;184:377–386. doi: 10.1084/jem.184.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nature protocols. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- Schneider CA, et al. NIH Image to ImageJ: 25 years of image analysis. Nature Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selmecki AM, et al. Acquisition of aneuploidy provides increased fitness during the evolution of antifungal drug resistance. PLoS Genetics. 2009:5. doi: 10.1371/journal.pgen.1000705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semighini CP, Harris SD. Regulation of apical dominance in Aspergillus nidulans hyphae by reactive oxygen species. Genetics. 2008;179:1919–1932. doi: 10.1534/genetics.108.089318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen G, et al. Wsp1, a GBD/CRIB domain-containing WASP homolog, is required for growth, morphogenesis, and virulence of Cryptococcus neoformans. Eukaryotic Cell. 2011;10:521–529. doi: 10.1128/EC.00274-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F. Getting started with yeast. Methods in enzymology. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- Shutes A, et al. Specificity and mechanism of action of EHT 1864, a novel small molecule inhibitor of Rac family small GTPases. The Journal of Biological Chemistry. 2007;282:35666–35678. doi: 10.1074/jbc.M703571200. [DOI] [PubMed] [Google Scholar]
- Sionov E, et al. Cryptococcus neoformans overcomes stress of azole drugs by formation of disomy in specific multiple chromosomes. PLoS Pathogens. 2010;6:e1000848. doi: 10.1371/journal.ppat.1000848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staiger CJ, et al. Actin filament dynamics are dominated by rapid growth and severing activity in the Arabidopsis cortical array. The Journal of Cell Biology. 2009;184:269–280. doi: 10.1083/jcb.200806185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemoto D, et al. A p67Phox-like regulator is recruited to control hyphal branching in a fungal-grass mutualistic symbiosis. The Plant cell. 2006;18:2807–2821. doi: 10.1105/tpc.106.046169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeo K, et al. Effects of growth temperature upshift on cell cycle progression in Cryptococcus neoformans. Mycoscience. 2003;44:465–471. [Google Scholar]
- Takeo K, et al. Unbudded G2 as well as G1 arrest in the stationary phase of the basidiomycetous yeast Cryptococcus neoformans. FEMS Microbiology Letters. 1995;129:231–235. doi: 10.1111/j.1574-6968.1995.tb07585.x. [DOI] [PubMed] [Google Scholar]
- Tamura K, et al. MEGA5: Molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Molecular Biology and Evolution. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka A, et al. NoxA activation by the small GTPase RacA is required to maintain a mutualistic symbiotic association between Epichloë festucae and perennial ryegrass. Molecular Microbiology. 2008;68:1165–1178. doi: 10.1111/j.1365-2958.2008.06217.x. [DOI] [PubMed] [Google Scholar]
- Toffaletti DL, et al. Gene transfer in Cryptococcus neoformans by use of biolistic delivery of DNA. Journal of bacteriology. 1993;175:1405–1411. doi: 10.1128/jb.175.5.1405-1411.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valencia A, et al. The ras protein family: evolutionary tree and role of conserved amino acids. Biochemistry. 1991;30:4637–4648. doi: 10.1021/bi00233a001. [DOI] [PubMed] [Google Scholar]
- Vallim MA, et al. A Rac homolog functions downstream of Ras1 to control hyphal differentiation and high-temperature growth in the pathogenic fungus Cryptococcus neoformans. Eukaryotic Cell. 2005;4:1066–1078. doi: 10.1128/EC.4.6.1066-1078.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virag A, et al. Regulation of hyphal morphogenesis by cdc42 and rac1 homologues in Aspergillus nidulans. Molecular Microbiology. 2007;66:1579–1596. doi: 10.1111/j.1365-2958.2007.06021.x. [DOI] [PubMed] [Google Scholar]
- Wang P, et al. Two cyclophilin A homologs with shared and distinct functions important for growth and virulence of Cryptococcus neoformans. EMBO reports. 2001;2:511–518. doi: 10.1093/embo-reports/kve109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, et al. Mating-type-specific and nonspecific PAK kinases play shared and divergent roles in Cryptococcus neoformans. Eukaryotic Cell. 2002;1:257–272. doi: 10.1128/EC.1.2.257-272.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waugh MS, et al. Ras1 and Ras2 contribute shared and unique roles in physiology and virulence of Cryptococcus neoformans. Microbiology (Reading, England) 2002;148:191–201. doi: 10.1099/00221287-148-1-191. [DOI] [PubMed] [Google Scholar]
- Weinzierl G, et al. Regulation of cell separation in the dimorphic fungus Ustilago maydis. Molecular Microbiology. 2002;45:219–231. doi: 10.1046/j.1365-2958.2002.03010.x. [DOI] [PubMed] [Google Scholar]
- Wolfe KH, Shields DC. Molecular evidence for an ancient duplication of the entire yeast genome. Nature. 1997;387:708–713. doi: 10.1038/42711. [DOI] [PubMed] [Google Scholar]
- Wolfram Research, I. Mathematica Edition: Version 8.0. Wolfram Research, Inc; Champaign, Illinois: 2010. [Google Scholar]
- Wright LP, Philips MR. Thematic review series: lipid posttranslational modifications. CAAX modification and membrane targeting of Ras. Journal of lipid research. 2006;47:883–891. doi: 10.1194/jlr.R600004-JLR200. [DOI] [PubMed] [Google Scholar]
- Zheng Y, et al. Interactions among proteins involved in bud-site selection and bud-site assembly in Saccharomyces cerevisiae. The Journal of biological chemistry. 1995;270:626–630. doi: 10.1074/jbc.270.2.626. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Metadata associated with Figure 1 can be accessed at the provided links. The data are provided as a .cys file, which can be opened using the freely available Cytoscape software (www.cytoscape.org) according to the tutorial on the Cytoscape website {Cline, 2007}. These data were generated using Cytoscape3 (2013). Information for each node and edge is listed in the Table Panel. Selecting a particular node will provide the gene number as identified by the Broad database for C. neoformans H99 (http://www.broadinstitute.org/annotation/genome/cryptococcus_neoformans/MultiHome.html), as well as chromosome number and the genome annotation. Under the VizMapper menu, ensure that Node Label (under the Node Visual Properties menu) is set at Gene_name. This will pass annotation information in the network table through to the nodes so that gene names are displayed on the nodes. Similarly, setting the Edge Label (under the Edge Visual Properties menu) at Percent_Identity will display the percent identity between two nodes on the edge connecting those nodes.