Abstract
BACKGROUND
Low folate status increases colorectal cancer risk whereas abundant supplementation may paradoxically increase risk. The mechanisms are unknown.
OBJECTIVES
To define molecular pathways in the human colon altered by either dietary folate depletion (followed by repletion), or by supplementation.
METHODS
10 healthy volunteers consumed a low folate diet for 12 weeks. During the last 4 weeks, folic acid (1 mg/day) was administered. In a second study, 10 other subjects were provided supplemental folic acid for 8 weeks. Rectosigmoid biopsies were obtained at measured intervals in both studies for assessment of primary endpoints: genome-wide gene expression, genomic DNA methylation, promoter methylation (depletion study only) and p53 DNA strand breaks.
RESULTS
Serum and rectosigmoid folate concentrations accurately tracked all changes in folate delivery (p<0.05). Gene array analysis revealed that folate depletion downregulated genes involved in immunity, inflammation, cell cycle and mitochondrial energy pathways; repletion produced reversal in most instances. Similarly, supplementation upregulated multiple inflammatory- and immune-related pathways, and in addition altered several 1-carbon related enzymes (p<0.001). Neither genomic or promoter-specific DNA methylation changed over the course of the depletion/repletion protocol; nor did genomic methylation change due to supplementation. p53 strand breaks increased with depletion after 12 weeks.
CONCLUSIONS
Depletion downregulates, whereas repletion or supplementation upregulates, pathways related to inflammation and immune response. Supplementation also altered expression of several pivotal genes involved in 1-carbon metabolism. These changes occurred in the absence of changes in gene methylation. Modest changes in folate delivery create substantial changes in the molecular milieu of the human colon.
Keywords: Folic acid, folate depletion, folate supplementation, colonic carcinogenesis, one carbon metabolism, inflammation, gene expression profiling, DNA methylation
INTRODUCTION
Epidemiological observations and preclinical studies have demonstrated that habitually low intake of folate, or low levels of systemic folate, are associated with an increased risk of colorectal cancer, and possibly other organs (1–5). However, folate intake at supraphysiologic levels among individuals at an elevated risk of malignancy appears to promote cancer as, for example, among those who already possess an existing focus of dysplastic cells (reviewed in (1)).
Little is known about the specific cellular pathways through which either inadequate, or overly abundant, folate intake modulates the risk of colorectal cancer in the human. Folate is an essential co-factor that is central to DNA methylation and DNA synthesis & repair, cellular functions that are commonly implicated in colorectal carcinogenesis when they go awry. Rodent studies have demonstrated that a mild depletion of folate and other 1-carbon nutrients alone is sufficient to enhance throughput through certain pro-transformational signaling pathways in the colonic mucosa (6;7), presumably leading to neoplastic transformation when complemented by the presence of other pro-carcinogenic factors. The mechanistic basis for the pro-carcinogenic effect of excessive folate intake is also ill-defined, although it has been posited that an existing focus of neoplastic cells responds to increasing folate concentrations by accelerating DNA synthesis and cellular proliferation (8;9). An alternative explanation for the promotional effect of excess folate is based on a recent secondary analysis of the aspirin-folate polyp trial, and hypothesizes that folate supplementation attenuates the anti-inflammatory effect of aspirin (10).
Elucidating pathways through which folate modulates colorectal carcinogenesis will assist in the development of public health strategies that are both safe and effective in reducing colorectal cancer. We therefore sought to define molecular events that occur as a result of folate depletion/repletion and folate supplementation in two cohorts of human subjects at risk of colorectal cancer. For a depletion study we enrolled 10 folate-replete individuals and induced mild dietary folate depletion in the inpatient Metabolic Unit of The Rockefeller University Hospital, followed by repletion with folic acid supplements. The supplementation study enrolled a second group of 10 subjects and was conducted on an outpatient basis. The latter individuals were supplemented with daily folic acid pills over an 8 week period. Rectosigmoid biopsies and serum were obtained at measured intervals from both groups, and were used to monitor expression signatures, DNA methylation patterns and a representative example of gene-specific DNA strand breaks.
MATERIALS AND METHODS
Subjects
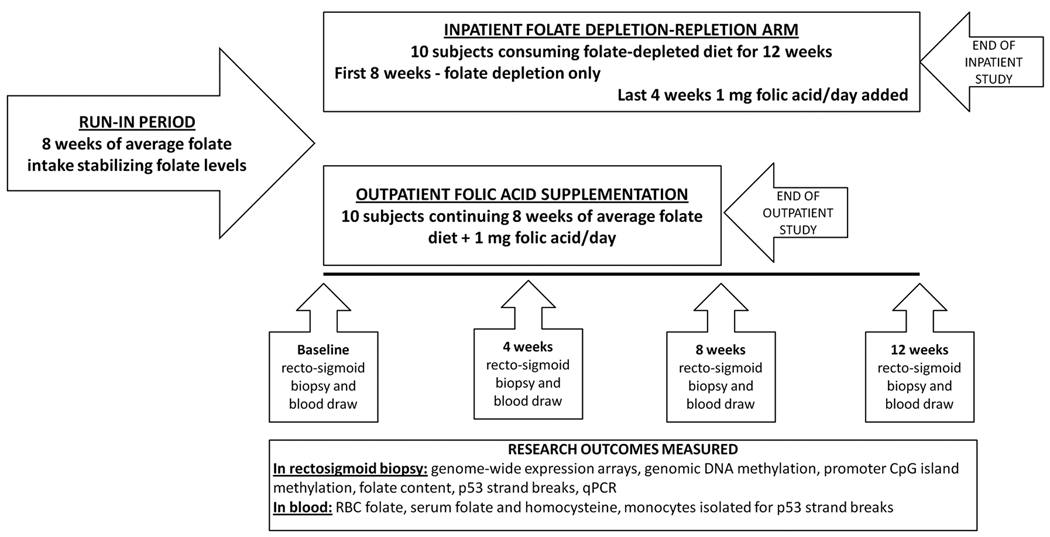
Twenty healthy subjects,12 men and 8 women, aged 44 to 72 years, 8 with a history of sporadic colorectal adenoma resection and 12 with a first degree relative with sporadic colorectal cancer or adenoma, were enrolled in 2 studies of 10 subjects each.(Figure 1 and Table 1). None of the subjects had either a history of multiple adenomas or multiple family members with colorectal cancer. No rectosigmoid adenomas or other macroscopic abnormalities were detected at study sigmoidoscopies. 12 subjects were Caucasian, 6 African-American and 2 of mixed racial background (Table 1). Subjects were excluded if they had a history of cancer other than non-melanoma skin cancer, a history of major intestinal surgery, inflammatory bowel disease, malabsorption, estrogen/progesterone replacement, supplemental vitamin D or regular use of non-steroidal anti-inflammatory drugs. Studies were approved by the IRB of Rockefeller University Hospital and the IRBs of the other collaborating institutions and informed consent was obtained from all subjects. The number of subjects was determined a priori, using sample size estimates derived from alterations in genomic DNA methylation of the colon observed in a prior pilot trials of folate supplementation (11). Subject enrollment and participation began in 2004 and ended in 2006.
Figure 1.
Figure shows a diagram of the 2 parallel folate intervention trials. Note that run-in period was identical for the two studies. Time-points of rectosigmoid biopsies collections are shown in the diagram; at these time-points blood endpoints were also measured.
TABLE 1.
Subjects characteristics and dietary intake
| SUBJECTS CHARACTERISTICS* | |||||||||||
| Age | Weight | Height | BMI | Gender | Ethnicity | ||||||
| years | kg. | cm | Kg/m2 | M | F | W | B | O | |||
| INPATIENT STUDY | Mean | 54.0 | 86.9 | 176.3 | 27.8 | 8 | 2 | 7 | 2 | 1 | |
| S.D. | 9.2 | 17.6 | 11.8 | 4.9 | |||||||
| OUTPATIENT STUDY | Mean | 57.6 | 86.5 | 171.0 | 29.6 | 4 | 6 | 5 | 4 | 1 | |
| S.D. | 7.3 | 16.7 | 6.2 | 5.7 | |||||||
| Mean ± SD for the 10 completed subjects; BMI: Body Mass Index; W- white; B-African American; O-other | |||||||||||
| DAILYAVERAGE OUTPATIENT DIETARY INTAKE | |||||||||||
| Energy | Fat | CHO | Protein | Fiber | Calcium | Folate | |||||
| Kcal | % | % | % | g | mg | mcg | |||||
| INPATIENT STUDY | Mean | 2635.0 | 35.1 | 49.9 | 15.0 | 21.0 | 1178.0 | 339.0 | |||
| S.D. | 522.0 | 3.6 | 1.4 | 3.0 | 7.6 | 706.0 | 126.0 | ||||
| OUTPATIENT STUDY | Mean | 2164.0 | 37.5 | 46.4 | 16.7 | 18.8 | 685.0 | 326.0 | |||
| S.D. | 342.0 | 8.2 | 11.0 | 5.0 | 2.7 | 309.0 | 69.8 | ||||
| Daily intake calculated for 3 day food records using ESHA food processing data base | |||||||||||
| RISK FACORS FOR COLON CANCER | |||||||||||
| INPATIENT STUDY | Family history of colorectal cancer or adenoma – first degree relative | 8 | |||||||||
| Personal history of adenoma | 2 | ||||||||||
| OUTPATIENT STUDY | Family history of colorectal cancer or adenoma – first degree relative | 4 | |||||||||
| Personal history of adenoma | 6 | ||||||||||
Subjects were excluded if they had a history of cancer other than non-melanoma skin cancer, previous major intestinal surgery, malabsorption or bleeding disorders, estrogen and/or progesterone replacement, supplemental vitamin D or non-steroidal anti-inflammatory drugs intake, systemic or bowel inflammatory disorder.
Diet and Study Arms
Research dieticians instructed study subjects to maintain their pre-study self-selected diet for at least 8 weeks prior to each study (‘run-in’ period). These diets provided sufficient calories and nutrients to maintain energy balance and contained quantities of calcium and vitamin D that approximate current recommended intakes (Table 1) and no subject was taking a multivitamin preparation containing folic acid prior to the run-in period. In order to facilitate folate depletion in the depletion protocol, subjects were instructed to avoid folate-containing vitamin supplements and fortified ready-to-eat cereals during the run-in phase, since these are the two greatest sources of folic acid in the American diet (12). Average dietary intakes during the run-in phase were calculated based on validated 3-day questionnaires and 24-hour recalls which were randomly checked during telephone calls at which time the diet of each volunteer was reviewed. The run-in diets contained a calculated daily mean of 327 mcg of folate. All subjects maintained their pre-study weights within 1.5 percent of the basal value. At the end of run in period subjects entered either the 12-week inpatient folate depletion-repletion study arm or the 8-week outpatient folic acid supplementation study arm (Figure 1).
Inpatient folate depletion-repletion protocol
During the 12-week residence in the Metabolic Unit of The Rockefeller University Hospital; all meals were consumed in the Unit. The folate depletion diet consisted of 3 different daily menus that were rotated and prepared specifically to meet each volunteer’s caloric needs. All the diets were analyzed for macronutrient and micronutrient content at Covance Laboratories (Princeton, NJ). Folate content was independently verified by the Mason laboratory using the microtiter plate L. casei assay, as previously described (13). The mean folate contents of the three rotating diets were 92±6 (SEM) and 73±2 mcg/day, as measured by the Mason and Covance laboratories, respectively. The diets of all subjects were supplemented with vitamins and nutrients other than folic acid to meet their daily requirements. After subjects consumed the low folate diet for 8 weeks, they were administered 1 mg of folic acid orally daily for 4 weeks (Barr Laboratories, Pomona, NY) while remaining on the low folate diet.
Supplementation protocol
After the 8-week run-in, subjects were given 1 mg of folic acid orally daily for 8 weeks (Barr Laboratories, Pomona, NY) while maintaining their run-in diet. The dose of 1 mg was selected as an amount that is commonly used for pharmacologic purposes in clinical settings and clinical trials. Compliance was maximized by selection of motivated volunteers and by close monitoring of all meal and supplement consumption by the inpatient nursing staff and by outpatient nutritionist.
Experimental end-points
After the 8 week run-in period, subjects underwent a baseline blood sample collection and rectosigmoid biopsies and then were either admitted for an inpatient stay of 12 weeks (10 subjects, depletion/repletion study) or given 1 mg of folic acid daily for 8 weeks as outpatients (supplementation study). Samples for experimental endpoints were collected at baseline, and then again after 4 and 8 weeks in both protocols. In addition, samples were collected at 12 weeks (i.e.: after 4 weeks of folate repletion) in the depletion protocol). Blood draws were performed via venipucture. Blood mononuclear cells were isolated by Ficoll gradient centrifugation. Serum, plasma and mononuclear cell samples were immediately frozen and blood samples for clinical laboratory studies were processed immediately. Flexible proctosigmoidoscopy was performed after a 60ml tap water enema and approximately 12 biopsies of rectosigmoid mucosa were taken from 4 quadrants at 10–15 cm from the anal verge, placed in NUNC cryotubes and immediately frozen in liquid nitrogen. Biopsies were taken after close inspection to exclude any mucosal abnormalities. Biopsies were subsequently extracted for RNA (using 2 separate biological duplicates), as well as for DNA and the determination of folate concentrations.
DNA Extraction
DNA was extracted from human biopsies using Qiagen DNeasy Kit as described by the manufacturer (Quiagen, Valencia, CA). Frozen mucosal biopsies and mononuclear cells were lysed in DNA extraction buffer and subjected to digestion by proteinase K. DNA was purified and extracted using silica spin columns. DNA was extracted by a conventional phenol-chloroform method and purities were checked by a spectrophotometric 260:280 ratio, with all samples having a ratio >1.8.
Rectal biopsy folate content
Rectosigmoid mucosal folate concentrations were measured as described previously (14;15). Three rectosigmoid biopsies from each subject at each time point were combined, rapidly weighed and tissue folates were immediately extracted in 20 volumes of freshly prepared folate extraction buffer [5 mM β-mercaptoethanol and 0.1 M sodium ascorbate in 0.1 M bis(2-hydroxyethyl)imino-tris(hydroxymethyl) methane (pH 7.85)] at 95°C for 20 mins, which extracts more than 95% of tissue folates. Extracts were then treated with chicken pancreas conjugase to convert folylpolyglutamates to diglutamate derivatives, which was then subjected to the microbiological assay.
Genomic DNA methylation
Genomic DNA methylation was assessed by a validated quantitative liquid chromatography/mass spectrophotometric (LC/MS) method as previously described (16). The isotopomers 15N3 2’-deoxycytidine and methyl-D3, ring-6-D1 5-methyl-2’-deoxycytidine (Cambridge Isotope Laboratories) were used as internal standards. DNA methylation status was defined as a percentage: 5-methylcytosine divided by the total of cytosine plus 5-methylcytosine (16).
Promoter Methylation
For the depletion/repletion protocol, promoter methylation of 432 genes known to be abnormally methylated in human cancers were also assessed in DNA extracted from colorectal mucosa biopsies using a promoter methylation bead array. Briefly, a universal bead array system, developed by Illumina, was utilized to examine the methylation of 1505 CpG sites contained within promoter (and related 5’ UTR) regions, as described previously (17). These analyses were conducted at the USC Epigenome Center, University of Southern California. The assay procedure is similar to that described for standard SNP genotyping and gene expression profiling using universal bead arrays (18), except that four oligonucleotides, two allele-specific oligonucleotides (ASOs), and two locus-specific oligonucleotides (LSOs) are required for each assay site rather than three. Briefly, bisulfite-treated, biotinylated genomic DNA (gDNA) was immobilized on paramagnetic beads. Pooled query oligonucleotides were annealed to the gDNA under a controlled hybridization program, and then washed to remove excess or mishybridized oligonucleotides. Hybridized oligonucleotides were then extended and ligated to generate amplifiable templates. Requiring the joining of two fragments to create a PCR template in this scheme provided an additional level of locus specificity. It is unlikely that any incorrectly hybridized ASOs and LSOs will be adjacent, and therefore would be able to ligate after ASO extension. A PCR reaction was performed with fluorescently labeled universal PCR primers. The methylation status of an interrogated CpG site was determined by calculating a β-value, which is defined as the ratio of the fluorescent signal from the methylated allele to the sum of the fluorescent signals of both methylated and unmethylated alleles. The β-value provides a continuous measure of levels of DNA methylation in samples, ranging from 0 in the case of completely unmethylated sites to 1 in completely methylated sites.
p53 strand breaks
Exons 6 and 8 of the p53 hypermutable region (exons 5–8) were chosen because previous animal studies have shown that these regions are susceptible to strand breakage due to dietary folate depletion (7;19). The detection of p53-specific DNA strand breaks were determined by a previously described quantitative PCR method (20). Assay validation was achieved by testing the effect of increasing amounts of template and of restriction digest on product formation. Strand breaks at p53 exon 6 and 8 are reported as a 4ΔCt value (Ct p53 exon 6 or 8 – Ct β-actin), with a higher ΔCt indicating a lower template integrity. This assay has been validated as a sensitive means of detecting DNA breaks in the human p53 gene (20) but is not entirely specific since abasic sites, bulky adducts or DNA cross-links may inhibit amplification as well and be detected by this method.
RNA extraction for expression analyses
Frozen human biopsies were maintained in liquid nitrogen until total RNA extraction using the Trizol method (Invitrogen). Trizol extracted RNA was further purified using Qiagen RNEasy kits (Qiagen Inc. Valencia, CA), yielding high quality RNA suitable for microarray analyses. RNA quality was verified by analysis on an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA) and RNA was quantified by NanoDrop (NanoDrop Technologies, Wilmington, DE). Tissue or biological duplicates (i.e.: biopsies taken from 2 different mucosal areas from a subject at the same time) were used for gene expression end-points. Additionally, one technical replicate (RNA from one of the 2 tissue or biological replicates) was used for baseline and 8 weeks of outpatient study to generate array data in order to increase the power of the analyses at these time-points. 500 ng of total RNA was used for in vitro transcription and cRNA amplification and labeling using Ambion’s Illumina kit according to the manufacturers’ instruction. Biotin labeled cRNA was labeled with fluorescent dye, hybridized onto Sentrix Human Ref-8 24K Expression Array Bead Chip (Illumina, San Diego, CA) and scanned. More advanced Version 2 of this array was used for depletion-repletion study.
RT-PCR analysis
For RT-PCR, duplicate 1 µg samples of total RNA were used as template for cDNA synthesis using Superscript III First Strand kit (Invitrogen). cDNA was diluted in RT buffer and amounts corresponding to 100 ng of original RNA was used for quantitative gene expression by RT-PCR. RT-PCR used TaqMan® Gene Expression Assay probes and primers (Applied Biosystems) and the ABI Prism 7900 RT-PCR system at the University of Connecticut Health Center Gene Array Core Facility. 3 most upregulated genes (CCL20, LTF and TCL1) and 3 most downregulated genes (BEST4, PYY, GCG) were measured by RT-PCR to validate our array data. Additional genes relevant to one-carbon metabolism (TYMS, DHFR) and inflammation (IL6, OLFM4) were also measured. Expression quantification used the delta method. 18S mRNA and/or GAPDH endogenous controls were used. Three independent RT-PCR reactions were used for each sample to calculate results.
Gene Array Analyses
Expression data were analyzed by Genespring software (Agilent Technologies, Inc, Santa Clara, CA) after normalization. Quality control was performed analyzing gene expression correlation coefficients. Arrays were normalized to 50th percentile per chip and median per gene, expression values below noise level were set to the minimum detection level. Probes with missing values for at least one experimental condition were excluded. For the actual analysis biological duplicate samples were averaged. The differences in gene expression were determined using repeated measures ANOVA or paired t-test, multiple hypothesis testing adjustment were made using Benjamini-Hochberg method at FDR<0.05. If no adjustments were made it is directly stated in the results. Genes differentially expressed following folic acid administration were subjected to Gene Ontology analysis using the Hypergeometric method (http://www.geneontology.org) corrected by Benjamini-Yekuteili method at FDR-q<0.05. Subsequently, Gene Set Enrichment Analysis (GSEA) was used. GSEA is a computational method that determines whether an a priori defined set of genes shows statistically significant differences between two phenotypes. We ranked gene expression differences between folate intervention time-points and baseline to identify gene sets that were significantly enriched after depletion/repletion or supplementation. (http://www.broadinstitute.org/gsea). Both curated and computationally derived data sets were used for result interpretation. False discovery rate (FDR-q) was used to rank the enrichment results.
Statistics
Statistical analysis of gene arrays is described above. Biological duplicates were used to generate array data except for the supplementation study, where at the baseline and 8 weeks time points, technical replicates were added as well. A non-parametric Repeat Measures Friedman’s Test with Dunn’s Multiple Comparisons Test was used to compare endpoints before and after folic acid depletion and repletion or supplementation. Significance was set at a two-tailed value of p<0.05. For analysis of promoter methylation, a repeat measures analysis with post-hoc testing was applied to identify those genes that underwent significant changes over the course of the four time points, and a Benjamini-Hochberg test was used to adjust for multiple observations (FDR<0.05).
RESULTS
All 10 depletion-repletion subjects completed the run-in and experimental periods without development of anemia. None of the study subjects complained of significant adverse effects. Based upon pill counts, subjects consumed an average of 99 percent of their prescribed supplements.
Folate depletion-repletion study
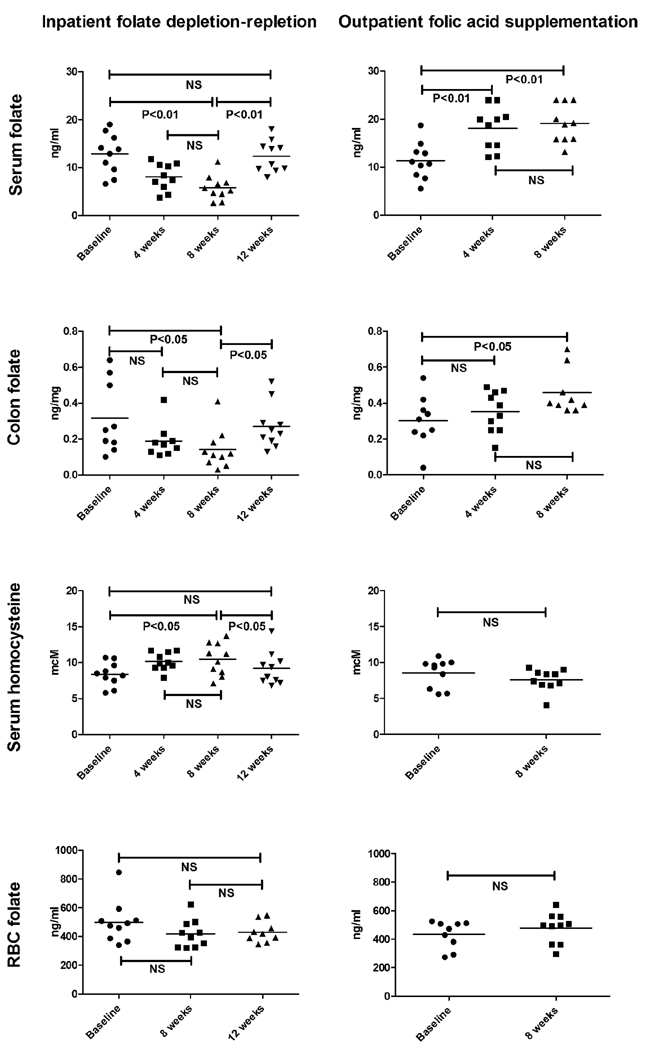
Mean serum and rectosigmoid folate concentrations each decreased by 50–80% during the 8 weeks of folate depletion compared to baseline (P<0.01 and <0.05, respectively, Figure 2). In addition, a nonsignificant decline of 16% was observed in RBC folate after 8 weeks of depletion. In response to folate depletion serum homocysteine levels rose (by 13%, P<0.05). The folate repletion was also effective: although most indicators of folate status fell just short of reverting to baseline levels, they demonstrated a significant restoration towards baseline levels compared to the values observed after 8 weeks of depletion: significant increases in serum and colonic folate, and a decrease in serum homocysteine levels were each observed (all P<0.05). No statistically significant increase in red blood cell (RBC) folate levels occurred between the end of the depletion phase and after supplementation (increase of 2%, P=0.33). No statistically significant differences in serum or colonic folate or serum homocysteine were found between baseline values and those observed at the end of the study.
Figure 2.
Changes in serum, rectosigmoid, and RBC folate concentrations and serum homocysteine concentrations after dietary folate depletion followed by repletion and after folic acid supplementation. Data is shown as individual data points for all 10 subjects. P values are shown in the figure, horizontal lines indicate mean values. Parallel changes in serum and rectosigmoid folate concentrations occurred as a result of depletion and supplementation and were accompanied by reciprocal changes in serum homocysteine, validating the biochemical effects of folate depletion, repletion, and supplementation.
Supplementation study
Serum and rectosigmoid folate concentrations each increased by 50–80% during the 8 weeks of folic acid supplementation compared to baseline (P<0.05, Figure 2). No statistically significant changes in red blood cell (RBC) folate or homocysteine levels occurred between baseline and the end of the study although numerically, an 11% decrease in homocysteine was observed (mean decrease from 8.54 to 7.6 µmol/L) and RBC folate increased from a mean of 472 ng/ml to 498 ng/ml (Figure 2).
p53 strands breaks
DNA strand breaks contained within exon 6 and exon 8 of the p53 gene were independently examined in rectosigmoid biopsies. In the inpatient depletion-repletion protocol, a gradual increase in exon 6 strand breaks of p53 gene were observed over the course of the entire study, although this did not achieve statistical significance until the 12 week time point. Similarly, an increasing trend was observed in exon 8 strand breaks although the mean values never achieved statistical significance (Supplement Figure). In the outpatient supplementation arm no changes in strand breaks were observed in the colon over the course of the study.
Genomic DNA methylation
Genomic DNA methylation was studied at 4 and 8 weeks following dietary folate depletion, at 12 weeks following 4 weeks of folic acid repletion, as well as during outpatient supplementation. No statistically significant changes were detected.
Promoter methylation
Due to the limited amount of DNA extracted from biopsies, promoter methylation could not be studied in the outpatient supplementation protocol. In 3 out of 10 subjects from the inpatient depletion-repletion protocol, complete methylation data was available for all 4 time points of the study (inadequate quantities of colonic DNA were available at one or more time points for the remaining subjects). After adjusting for multiple observations by Benjamini-Hochberg Test, no promoter was found to be differentially methylated to a significant degree. These data suggest that relatively short-term modest changes in folate status do not result in significant changes in promoter methylation in the colonic mucosa.
Gene Array Analysis
Quality assurance analysis was performed by analyzing gene expression correlations between biological and/or technical duplicates. All samples from inpatient depletion-repletion study showed correlation coefficient of ≥0.99 so that all array data were included in the analysis. In the supplementation study, 2 subjects (subject 23 and 24) showed correlation coefficient of 0.95 or less and thus were excluded from the gene array analysis. Unsupervised hierarchical condition clustering analysis showed that samples from the 18 subjects divided into 18 distinct clusters according to their similarity measures. Each of these 18 clusters corresponded to samples from a single subject suggesting that gene expression for the 24,000 interrogated genes maintained similarity within the rectosigmoid mucosa during the length of the study. Similar data were observed in a previous study (21).
Depletion-repletion study
Using Repeat Measures ANOVA Test and Benjamini Hochberg Multiple Testing Correction 1585 genes were identified as being differentially expressed compared to the baseline (Table 2), all significant genes are listed in Supplement S1. Adjusted Gene Ontology (GO) analysis showed that genes that were downregulated at 4 and 8 weeks were related to cellular metabolic processes (P=0.029), nucleus (P=0.048), immune system process (P=0.069) and immune response (P=0.076). There were no significantly enriched upregulated GO categories in adjusted analysis but unadjusted analysis revealed that genes upregulated at 4 and 8 weeks of depletion were most significantly related to GABA signaling and receptor activity (P=0.001). Next we examined changes in gene expression after folic acid repletion and observed that repletion phase of the study generally produced reciprocal changes to those observed with depletion, although exceptions did exist (top part of Figure GSEA). There were 281 genes whose expression was downregulated by folate depletion and re-upregulated by folate repletion (vs 8 week depletion time point). Adjusted analysis showed that immune response (P=0.027) and immune system process (P=0.027) GO categories were enriched. There were 689 genes upregulated by folate depletion and re-downregulated after repletion. Adjusted analysis showed no significant GO categories but top enriched GO groups in unadjusted analysis were again GABA receptor signaling and activity (P=0.001). Because of the folate intervention the methionine (‘1-carbon’) metabolic pathway was analyzed it was not significantly enriched during folate depletion-repletion.
TABLE 2.
Gene expression changes detected by statistical methods
| REPEATED MEASURES ANOVA# | |||
| FOLATE STATUS | DEPLETION | DEPLETION | REPLETION |
| Time-point | 4 weeks | 8 weeks |
12 weeks (vs 8 wks) |
| No of Genes | 1585 | ||
| UPREGULATED | 806 | 999 | 365 |
| 720* | |||
| DOWNREGULATED | 779 | 584 | 1209 |
| 500* | |||
| REPEATED MEASURES ANOVA$ | |||
| FOLATE STATUS | SUPPLEMENTATION | ||
| Time-point | 4 weeks | 8 weeks | |
| No of Genes | 1001 | ||
| UPREGULATED | 522 | 477 | |
| 306* | |||
| DOWNREGULATED | 479 | 524 | |
| 308* | |||
| PAIRED T-TEST BASELINE VS 8 WEEKS SUPPLEMENTATION$ | |||
| No of Genes | 2073 | ||
| UPREGULATED | 642 | ||
| DOWNREGULATED | 1431 | ||
Modulated at both 4 AND 8 weeks
Adjusted by Benjamini-Hochberg Multiple Correction Test
Unadjusted analysis
Folic acid supplementation
Using Repeat Measures ANOVA Test and Benjamini Hochberg Multiple Testing Correction no genes were identified as being differentially expressed compared to the baseline. Unadjusted test showed 1001 genes were differentially expressed at P<0.05. At both 4 weeks and 8 weeks of supplementation 306 genes were upregulated and 308 genes were downregulated (Table 2). Most upregulated GO categories were immune system process (P<0.001) and immune response (P=0.001) and most downregulated categories were regulation of transcription (P=0.001) and regulation of nucleotide and nucleic acid metabolic process (P=0.001, all unadjusted analyses). We then compared baseline to 8 weeks of supplementation by paired t-test using additional technical replicates. 2073 genes were differentially expressed at P<0.05 (unadjusted paired t-test, Table 2). More than 80% of genes down- or up-regulated at 8 weeks of supplementation showed the same changes at 4 weeks. 642 upregulated genes were consistently related to immune and inflammatory processes such as immune system process and immune response (P<0.001), inflammatory response (P=0.001), chemokine activity (P=0.004) and cytokine receptor binding (P<0.001, all adjusted analyses, Table 3). 1431 downregulated genes were related to cellular metabolic processes (P<0.001), nucleus (P<0.001), regulation of nucleotide and nucleic acid metabolic process (P<0.001), cell cycle (P<0.001), mitosis (P=0.002, all adjusted analyses); all significant genes are listed in the Supplement S2.
TABLE 3.
List of genes involved in selected inflammatory and immune-related GO categories that were significantly up-regulated following folic acid supplementation
| CYTOKINE AND CHEMOKINE ACTIVITY |
| CCL14, CCL19, CCL20, CCL21, CCL22, CCL23, CSH2, CXCL1, CXCL13, EBI3, ECGF1, IL17F, IL27, IL29, IL6, LTB, PF4V1, TNF, TNFSF11 |
| IMMUNE RESPONSE |
| APS, BATF, BF, C3, C4A, CCL14, CCL20, CCL21, CCL22, CCR6, CCR7, CD1C, CD22, CD72, CFHL1, CRLF1, CTLA4, DEFA5, DEFA6, EBI3, EDG6, FADD, FCGR3A, G1P2, G1P3, GPSM3, ICOS, IKBKE, IL12RB1, IL6, IL6ST, INDO, ISGF3G, KIR2DS2, LTB, LTF, MHC2TA, MX1, MX2, OAS2, PF4V1, PFC, PSMB9, RNF125, SCAP1, TLR10, TNF, TNFRSF9, TNFSF11, TNFSF13, UBD |
| INFLAMMATORY RESPONSE |
| ALOX5AP, BF, C3, C4A, CCL19, CCL20, CCL21, CCL22, CCL23, CCL23, CCR7, CFHL1, CXCL1, CXCL13, FN1, IL17F, IL6, INS, NCR3, PFC, SERPINF2, TLR10 |
Analysis of the methionine (‘1-carbon’) metabolic pathway showed significant downregulation during folic acid supplementation (P=0.02). The supplementation regimen modulated expression of several enzymes in the colonic mucosa important in nucleotide synthesis and biological methylation (Figure 3). Additional downregulated genes linked to 1-carbon metabolism that are not shown in the figure include the folate receptor 1 (FOLR1), and gamma glutamyl hydrolase (conjugase, GGH).
Figure 3.
Intracellular 1-carbon metabolism (simplified), emphasizing pathways relevant to the text. Multiple significant alterations in gene expression in this metabolic pathway were observed. Enzyme gene names are shown in capital letters, upregulated genes are shown in green and downregulated genes red, genes whose expression did not change are shown in black. Major end-products relevant to the discussion are in bold italics. Methionine adenosyltransferase 2b (MAT2b) is upregulated but all other genes whose expression were altered were downregulated. Notably thymidylate synthase (TYMS),dihydrofolate reductase (DHFR) and 2 de novo DNA methyltransferases 3A and 3B (DNMT3A and DNMT3B) are downregulated. CTH (cystathionine lyase) is reduced as well. The function of the gene product of AHCYL1 (S-adenosylhomocysteine hydrolase-like 1) is not known with certainty but a highly homologous protein catalyzes the reversible hydrolysis of S-adenosylhomocysteine (S-Adohcy) to adenosine and L-homocysteine. Additional genes that were downregulated include folate receptor 1 (FOLR1); gamma glutamyl hydrolase (‘conjugase’,GGH), an enzyme that cleaves the polyglutamyl tail of folates; methylenetetrahydrofolate dehydrogenease (MTHFD1), which is involved in the interconversion of 1-carbon derivatives of tetrahydrofolate; 5-methyltetrahydrofolate-homocysteine methyltransferase reductase (MTRR), which regenerates an active form of methionine synthase (MTR) via reductive methylation with vitamin B12 as cofactor; and phosphoribosylglycinamide formyltransferase (GART), which is pivotal in purine biosynthesis. THF = tetrahydrofolate; S-AdoMet = S-adenosylmethionine; S-AdoHcy = S-adenosylhomocysteine.
In addition to the statistical analysis we analyzed the top 1% or 5% regulated genes in our supplementation trial and we observed similar results to those from unadjusted ANOVA or t-tests (Supplement S3)
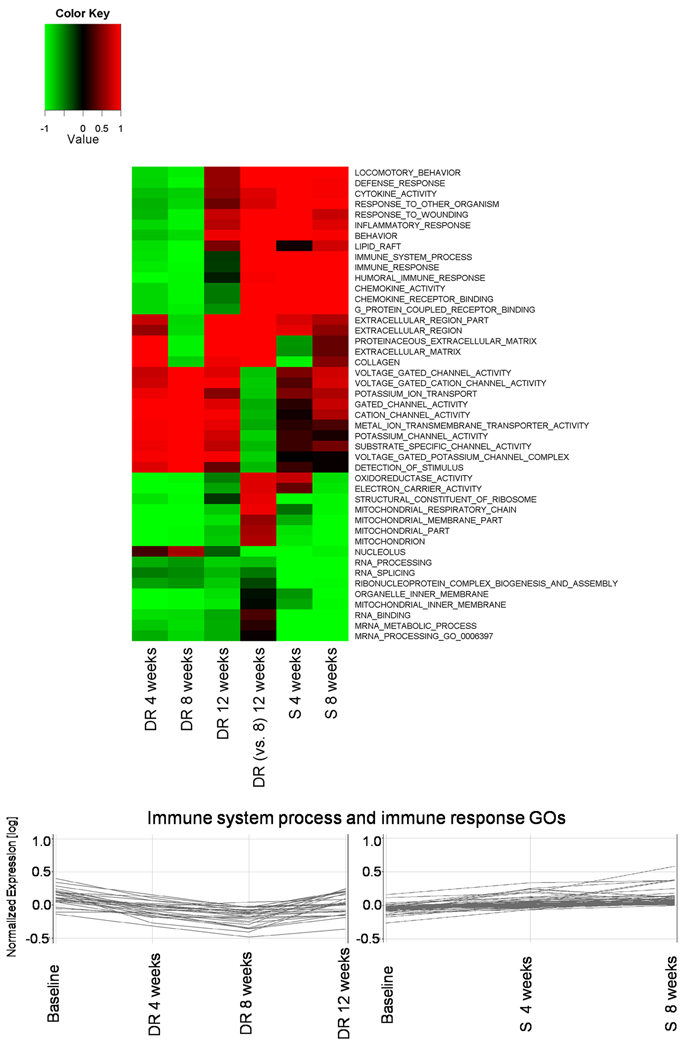
We observed that changes in immune, inflammatory and other pathways were associated with changes in folate status. To confirm these finding and to directly compare changes across both studies we used Gene Set Enrichment Analysis. Figure 4 shows GO categories most significantly and consistently modulated by folate status were indeed related to immune and inflammatory pathways. Individual genes contributing to enrichment of immune system process and immune response pathways are listed in the Supplement S4. Of interest is that another group of genes regulated by folate status is related to neuronal channel activities that also includes GABA activity. Full list of significantly enriched pathways across both studies is listed in the Supplement S5.
Figure 4.
Heatmap of Gene Set Enrichment Analysis for Gene Ontology across folate depletion-repletion study and folic acid supplementation study. Only categories with FDR-q-value of less than 0.001 in at least one condition are shown in the figure. Colors indicate down-regulation (green) or up-regulation (red) and values are 1 - P values (with down-regulation given negative values). DR indicate depletion-repletion study and S indicates folate supplementation study. Figure includes enrichment results for comparison of nadir of depletion (8 weeks) vs 4 week repletion timepoints. Note that categories related to immune functions and inflammation are strongly associated with folate status. To demonstrate these changes on individual gene level, lower part of the figure shows general relative changes of significantly expressed genes that belong to immune response process and immune response GOs and whose expression is linked to folate status, left part for depletion-repletion and right panel for supplementation study. Individual contributing genes are listed in the Supplement S4.
Quantitative RT-PCR Validation of Differentially Expressed Genes
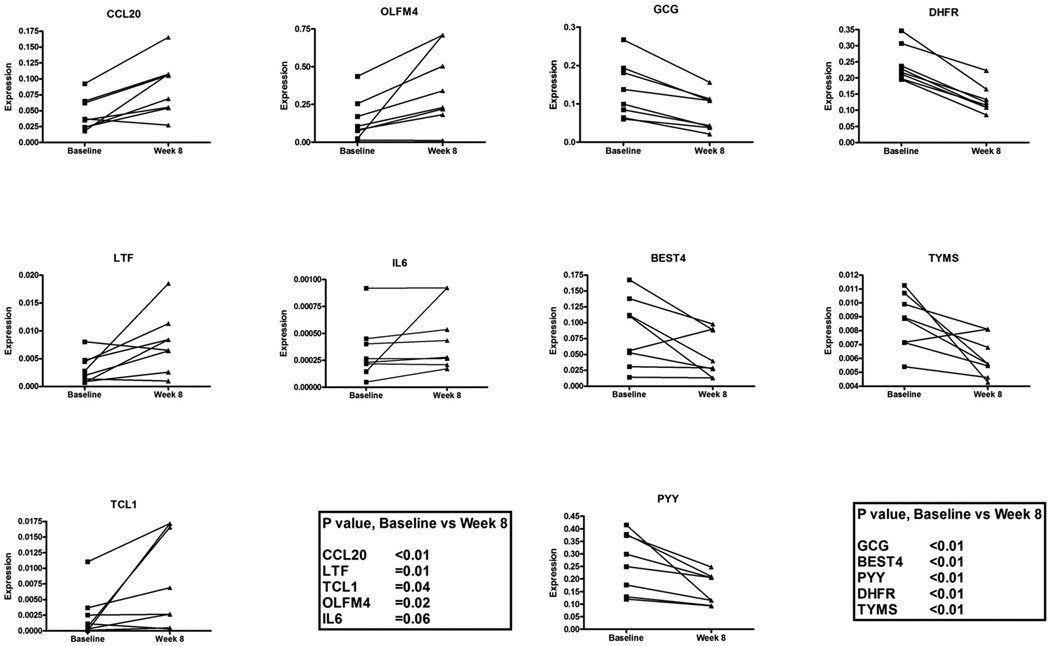
To validate array data, the three most upregulated genes (LTF, CCL20 and TLC1) and the 3 most downregulated genes (BEST4, GCG and PYY) were selected from outpatient folic acid supplementation study for RT-PCR from a list of significant genes identified by paired t test. To validate the changes in inflammatory genes and genes involved in 1-carbon metabolism (Figure 3) OLFM4, IL-6, DHFR and TYMS were also measured. RT-PCR results confirmed upregulation of TCL1 (P=0.04) and upregulation of inflammatory markers OLFM4, CCL20, LTF and IL6 by RT-PCR (P=0.02, P<0.01, P=0.01 and P=0.06, respectively) and dowregulation of BEST4, GCG and PYY (P<0.01 for all 3 genes). Downregulation of DHFR and TYMS, 2 genes involved in 1-carbon metabolism, was also confirmed (P<0.01 for both genes, Figure 5).
Figure 5.
Changes in gene expression in rectosigmoid mucosa by quantitative RT-PCR. Gene symbols are listed on a top of each graph. Data is shown as individual paired expression data points for all 8 subjects at baseline and after 8 weeks of folate supplementation. To validate the arrays three most upregulated genes (LTF, CCL20 and TLC1) and the 3 most downregulated genes (BEST4, GCG and PYY) were measured. To validate the changes in inflammatory genes and genes involved in 1-carbon metabolism OLFM4, IL-6, DHFR and TYMS were also measured. P values are listed in the graph.
DISCUSSION
This study examined the effects of dietary folate depletion/repletion on gene expression-and select other molecular events-in the human colon, as well as changes occurring after one month of folate repletion; and 2) the effects of two months of folic acid supplementation in folate-replete subjects. Subjects who by history were at modestly increased risk of colon cancer were studied since the underlying concept was to define which cancer-relevant pathways are altered by varying levels of folate intake in individuals whose colons are susceptible to neoplastic transformation.
Although folate depletion is difficult to achieve in the present era of mandatory folic acid fortification, the first study successfully induced a significant degree of depletion, as evidenced by substantial declines in serum and colonic folate, and an increase in serum homocysteine (Figure 2). In fact, homocysteine rises when intracellular folate concentrations can no longer adequately support biological re-methylation (22), so the degree of depletion achieved in the study is indicative of a true intracellular depletion of the vitamin. Nevertheless, the degree of depletion at the end of the study was rather modest: since after depletion the mean serum folate concentration (=5.8 ng/mL) did not fall below the threshold that is conventionally considered to define the lower limit of normality (i.e: 5 ng/mL). This is clinically relevant since enhancement of colorectal cancer risk due to folate inadequacy often occurs in segments of the population whose low folate status is at the lower end of the normal range rather than in the zone of frank deficiency (23). Furthermore repletion of folate also was successful since indicators of folate status, both systemic and colonic, returned nearly to baseline status. RBC folate status did not similarly increase to a significant degree presumably since the half-life of red cell folate parallels the half-life of those cells. These data agree with previous studies which have demonstrated that serum folate is a more accurate proxy measure of human colonic folate status than RBC folate (14). In the second study, in which folic acid supplementation was examined, serum and colonic folate concentrations rose by 50–60%, confirming successful augmentation of both systemic and colon-specific folate status.
A seminal observation in the supplementation protocol is increased expression of genes involved in immune and pro-inflammatory processes. Moreover, the gene expression data from the depletion/repletion protocol are highly complementary to those of the supplementation study: all three phases (i.e.: depletion, repletion, and supplementation) indicate that higher folate status increases expression of pro-inflammatory and immune pathways (Figure 4). In particular, chemokine and cytokine genes including those of the complement and coagulation systems and immune recognition system were downregulated by folate depletion and this effect was reversed with folate repletion and supplementation (Supplement S1–3). Although repleting folate-deplete subjects with folic acid supplementation is physiologically distinct from supplementing folate-replete subjects, the observed effects on the expression of pro-inflammatory and immune-related genes were very similar, suggesting that the effect occurs incrementally over the entire range of folate status in these two protocols. Along these lines, a recent human study showed that administration of 1.2 mg folic acid for 12 weeks resulted in an increase of serum proteins involved in regulation and activation of immune function and complement cascade (24). Interestingly, these results might in part explain the mechanistic basis for recent observations emerging from the human Aspirin/Folate Polyp Prevention trial, which suggested that folic acid administration antagonizes the suppressive effects of aspirin on circulating inflammatory markers (10). Furthermore, in preclinical studies, supraphysiologic levels of dietary folic acid also led to enhanced inflammation in a liver tissue injury model (25).
Our data also show that neuronal receptor pathways were also linked to folate status. Expression of genes such as GABA receptors or other G-protein coupled receptors such as taste transduction receptors were increased during folate depletion and reciprocally decreased during folate repletion. The potential association between these genes and colon carcinogenesis is unclear but the GABA may be over-expressed in colon cancer and GABA receptor signaling was linked to metastatic behavior of colon cancer cells in experimental models (26).
One-carbon metabolism is complex, making it difficult to accurately predict the net metabolic effects of the observed changes in gene expression. However, some of the observed changes warrant speculation: thymidylate synthase expression was suppressed by supplementation (confirmed by qRT-PCR, Figure 5), which could diminish thymidylate availability for DNA synthesis and thereby enhance uracil incorporation into DNA, a potentially mutagenic event. Thus, our observations are consistent with a report in which subjects receiving 5 mg of folic acid per day developed increased levels of uracil in their colonic mucosa (27).
Our results suggest that altered expression in 1-carbon enzymes may represent a protective mechanism against an oversupply of folate co-factor. By down-regulating DHFR, folic acid would be less capable of entering into the 1-carbon metabolic network (Figure 3). Moreover, the terminal enzymes for two critical functions of 1-carbon metabolism--thymidylate synthesis and biological methylation of DNA--were both downregulated, an effect which might limit the synthesis of end-products in the face of excess co-factor. This result is consistent with those of Basten et al, who observed that supplemental folic acid administered to healthy volunteers in a similar dose and duration as in our study diminished DNA excision repair (28). They postulated that the surfeit of nucleotides provided for by excess folic acid might downregulate pathways related to DNA repair, a concept that was borne out by our observation that supplementation led to a downregulation of the pathways integral to both purine and thymidine synthesis. Feasibly these changes also may alter cell cycle dynamics since we observed significant down-regulation of genes that are involved in regulation of cell cycle and DNA replication including cyclins, cyclin-dependent kinases and PCNA (Supplement S2).
Both genomic DNA methylation and promoter methylation were examined as well. No significant changes in genomic DNA methylation were observed in folate depletion-repletion study or in the parallel supplementation study. The observation that the colon is relatively resistant to changes in genomic methylation due to altered folate status has also been noted in animal studies (29), as well as some (30), but not all (31) clinical trials.
Probably more relevant to mechanistic issues than genomic methylation were the observations pertaining to loci-specific methylation. We found no significant changes that occurred in promoter methylation in response to folate depletion, or in the subsequent phase of repletion. Although modification of promoter methylation has been observed with folate depletion in some cell culture studies (32), the severity of deficiency in such pre-clinical models is rather profound and it is entirely feasible that the magnitude of depletion that is encountered in clinical situations is simply not robust enough to alter promoter methylation. Additional caveat is that small shifts in methylation (<5%) may not have been reliably detected by the technology used in this study. A case-control study has explored this issue previously and the results suggested that low folate intake may be related to hypermethylation of the p16 gene (33), but this is the first time that any study has directly tested whether folate depletion in the human alters promoter methylation in the colon. The fact that many substantial shifts in gene expression were observed in the depletion/repletion protocol in the absence of changes in gene methylation suggest that the alterations in expression, as well as the changes in DNA strand breaks, were mediated by processes other than changes promoter methylation. This is consistent with a recent review of the topic, which proposed that other molecular anomalies that arise in the setting of folate depletion, such as impaired DNA synthesis and repair, are the primary drivers of enhanced carcinogenesis (34). Our observations regarding gene-specific methylation are nevertheless limited to the folate depletion-repletion protocol, by the number of subjects with a complete set of samples, and by the marked stringency of the multiple comparisons test we imposed on our data, so we cannot exclude the possibility that some modest changes in promoter methylation occurred that went undetected or that supplementation of folate-replete subjects might induce changes.
We also assessed DNA strand breaks in two exonic loci within the colonic p53 gene. Whereas supplementation produced no apparent changes (data not shown), an increase in exon 6 breaks was observed at week 12 of the depletion protocol; strand breaks in exon 8 followed a nearly identical course but the changes in the latter never achieved statistical significance. This is consistent with an earlier study in rodents, where instability of the so-called ‘hypermutable region’ of the p53 gene was noted to increase progressively with increasing severity of folate depletion (19). Interestingly, this was one molecular marker that did not reverse at all with repletion; indeed, numerically it continued to increase during the repletion phase, suggesting that the effect is either irreversible or that it takes longer than 4 weeks to return to its former state. The ramifications of single- and double-stranded DNA breaks in the exons of the p53 gene are not at all well understood. On the one hand, they may be a marker of instability of that gene and indicate an increased risk of mutations, which is consistent with the very high rate of mutations found at these loci (35). On the other hand, base excision DNA repair transiently creates strand breaks (36), and therefore higher levels of strand breaks may instead indicate exceptionally robust and effective DNA repair activity. Consistent with the last statement was the observed significant increase in gene expression of 2 genes in DNA ligase IV complex at 8 weeks of the study (Supplement S1, genes XRCC4 and LIG4), although changes in DNA repair pathways overall fell short of statistical significance.
In conclusion, these two studies demonstrate that mild dietary folate depletion over eight weeks decreases the expression of genes involved in pro-inflammatory and immune-related pathways, and that repletion of deplete individuals or supplementation of replete individuals produces changes in expression of these pathways that are reciprocal to those observed with depletion. All of these altered profiles of gene expression occurred in the absence of significant changes in promoter methylation, thereby implicating other mechanisms that would mediate the changes in expression. Over the entire course of the depletion/repletion protocol there was a progressive increase in p53 DNA strand breaks within the hypermutable region of the gene, although the increase did not achieve significance until after the repletion phase, thereby obscuring the phase of the study that was actually responsible for the increase. These data suggest that folate status modulates mucosal inflammation, and are therefore consistent with recent observations in two clinical trials (10) (24). Although the functional ramifications of what we have observed remain to be fully defined, our observations make it quite clear that alterations in folate availability significantly impact on the molecular milieu of the human colonic mucosa in ways that may modulate the risk of carcinogenesis.
Supplementary Material
ACKNOWLEDGMENTS
Supported by the GCRC and 5UL1RR024143 from the NCRR and the NIH Roadmap for Medical Research to Rockefeller University, NIH grant NO1-CN35111 (PI: JM), CA-29502-24 (PI: PP), Carol and Ray Neag Colon Cancer Prevention Program research funds at University of Connecticut Health Center to PP; K05 CA100048 (JBM).
Footnotes
ClinicalTrials.gov: NCT00220012.
Complete expression array data and other supporting material can be accessed at: http://protivalab.com/REVIEWERS/FOLATE_SUPP/data.htm; User Name: AACR, Password: ?q2w3e4
Competing Interest: None to declare for all authors
Reference List
- 1.Mason JB. Folate, cancer risk, and the Greek god, Proteus: a tale of two chameleons. Nutr Rev. 2009 Apr;67(4):206–212. doi: 10.1111/j.1753-4887.2009.00190.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ulrich CM, Potter JD. Folate and cancer--timing is everything. JAMA. 2007 Jun 6;297(21):2408–2409. doi: 10.1001/jama.297.21.2408. [DOI] [PubMed] [Google Scholar]
- 3.Giovannucci E. Epidemiologic studies of folate and colorectal neoplasia: a review. J Nutr. 2002 Aug;132(8 Suppl):2350S–2355S. doi: 10.1093/jn/132.8.2350S. [DOI] [PubMed] [Google Scholar]
- 4.Harnack L, Jacobs DR, Jr, Nicodemus K, Lazovich D, Anderson K, Folsom AR. Relationship of folate, vitamin B-6, vitamin B-12, and methionine intake to incidence of colorectal cancers. Nutr Cancer. 2002;43(2):152–158. doi: 10.1207/S15327914NC432_5. [DOI] [PubMed] [Google Scholar]
- 5.Sanjoaquin MA, Allen N, Couto E, Roddam AW, Key TJ. Folate intake and colorectal cancer risk: a meta-analytical approach. Int J Cancer. 2005 Feb 20;113(5):825–828. doi: 10.1002/ijc.20648. [DOI] [PubMed] [Google Scholar]
- 6.Liu Z, Choi SW, Crott JW, Keyes MK, Jang H, Smith DE, et al. Mild depletion of dietary folate combined with other B vitamins alters multiple components of the Wnt pathway in mouse colon. J Nutr. 2007 Dec;137(12):2701–2708. doi: 10.1093/jn/137.12.2701. [DOI] [PubMed] [Google Scholar]
- 7.Liu Z, Choi SW, Crott JW, Smith DE, Mason JB. Multiple B-vitamin inadequacy amplifies alterations induced by folate depletion in p53 expression and its downstream effector MDM2. Int J Cancer. 2008 Aug 1;123(3):519–525. doi: 10.1002/ijc.23599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shpitz B, Bomstein Y, Mekori Y, Cohen R, Kaufman Z, Grankin M, et al. Proliferating cell nuclear antigen as a marker of cell kinetics in aberrant crypt foci, hyperplastic polyps, adenomas, and adenocarcinomas of the human colon. Am J Surg. 1997 Oct;174(4):425–430. doi: 10.1016/s0002-9610(97)00122-0. [DOI] [PubMed] [Google Scholar]
- 9.Stover P. Folate Biochemical Pathways and Their Regulation. In: Bailey LB, editor. Folate in Health and Disease. 2nd ed. Boca Raton: Taylor and Francis; 2010. pp. 49–74. [Google Scholar]
- 10.Ho GY, Xue X, Cushman M, Keown-Eyssen G, Sandler RS, Ahnen DJ, et al. Antagonistic effects of aspirin and folic acid on inflammation markers and subsequent risk of recurrent colorectal adenomas. J Natl Cancer Inst. 2009 Dec 2;101(23):1650–1654. doi: 10.1093/jnci/djp346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim YI, Baik HW, Fawaz K, Knox T, Lee YM, Norton R, et al. Effects of folate supplementation on two provisional molecular markers of colon cancer: a prospective, randomized trial. Am J Gastroenterol. 2001 Jan;96(1):184–195. doi: 10.1111/j.1572-0241.2001.03474.x. [DOI] [PubMed] [Google Scholar]
- 12.Yeung L, Yang Q, Berry RJ. Contributions of total daily intake of folic acid to serum folate concentrations. JAMA. 2008 Dec 3;300(21):2486–2487. doi: 10.1001/jama.2008.742. [DOI] [PubMed] [Google Scholar]
- 13.Kim YI, Fawaz K, Knox T, Lee YM, Norton R, Arora S, et al. Colonic mucosal concentrations of folate correlate well with blood measurements of folate status in persons with colorectal polyps. Am J Clin Nutr. 1998 Oct;68(4):866–872. doi: 10.1093/ajcn/68.4.866. [DOI] [PubMed] [Google Scholar]
- 14.Kim YI, Fawaz K, Knox T, Lee YM, Norton R, Libby E, et al. Colonic mucosal concentrations of folate are accurately predicted by blood measurements of folate status among individuals ingesting physiologic quantities of folate. Cancer Epidemiol Biomarkers Prev. 2001 Jun;10(6):715–719. [PubMed] [Google Scholar]
- 15.Varela-Moreiras G, Selhub J. Long-term folate deficiency alters folate content and distribution differentially in rat tissues. J Nutr. 1992 Apr;122(4):986–991. doi: 10.1093/jn/122.4.986. [DOI] [PubMed] [Google Scholar]
- 16.Friso S, Choi SW, Dolnikowski GG, Selhub J. A method to assess genomic DNA methylation using high-performance liquid chromatography/electrospray ionization mass spectrometry. Anal Chem. 2002 Sep 1;74(17):4526–4531. doi: 10.1021/ac020050h. [DOI] [PubMed] [Google Scholar]
- 17.Bibikova M, Lin Z, Zhou L, Chudin E, Garcia EW, Wu B, et al. High-throughput DNA methylation profiling using universal bead arrays. Genome Res. 2006 Mar;16(3):383–393. doi: 10.1101/gr.4410706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fan JB, Gunderson KL, Bibikova M, Yeakley JM, Chen J, Wickham GE, et al. Illumina universal bead arrays. Methods Enzymol. 2006;410:57–73. doi: 10.1016/S0076-6879(06)10003-8. [DOI] [PubMed] [Google Scholar]
- 19.Kim YI, Shirwadkar S, Choi SW, Puchyr M, Wang Y, Mason JB. Effects of dietary folate on DNA strand breaks within mutation-prone exons of the p53 gene in rat colon. Gastroenterology. 2000 Jul;119(1):151–161. doi: 10.1053/gast.2000.8518. [DOI] [PubMed] [Google Scholar]
- 20.Crott JW, Liu Z, Choi SW, Mason JB. Folate depletion in human lymphocytes up-regulates p53 expression despite marked induction of strand breaks in exons 5–8 of the gene. Mutat Res. 2007 Jan 10;626(1–2):171–179. doi: 10.1016/j.mrgentox.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 21.Protiva P, Cross HS, Hopkins ME, Kallay E, Bises G, Dreyhaupt E, et al. Chemoprevention of colorectal neoplasia by estrogen: potential role of vitamin D activity. Cancer Prev Res (Phila Pa) 2009 Jan;2(1):43–51. doi: 10.1158/1940-6207.CAPR-08-0103. [DOI] [PubMed] [Google Scholar]
- 22.Kohlmeier M, da Costa KA, Fischer LM, Zeisel SH. Genetic variation of folate-mediated one-carbon transfer pathway predicts susceptibility to choline deficiency in humans. Proc Natl Acad Sci U S A. 2005 Nov 1;102(44):16025–16030. doi: 10.1073/pnas.0504285102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fuchs CS, Willett WC, Colditz GA, Hunter DJ, Stampfer MJ, Speizer FE, et al. The influence of folate and multivitamin use on the familial risk of colon cancer in women. Cancer Epidemiol Biomarkers Prev. 2002 Mar;11(3):227–234. [PubMed] [Google Scholar]
- 24.Duthie SJ, Horgan G, de RB, Rucklidge G, Reid M, Duncan G, et al. Blood folate status and expression of proteins involved in immune function, inflammation, and coagulation: biochemical and proteomic changes in the plasma of humans in response to long-term synthetic folic acid supplementation. J Proteome Res. 2010 Apr 5;9(4):1941–1950. doi: 10.1021/pr901103n. [DOI] [PubMed] [Google Scholar]
- 25.Marsillach J, Ferre N, Camps J, Riu F, Rull A, Joven J. Moderately high folic acid supplementation exacerbates experimentally induced liver fibrosis in rats. Exp Biol Med (Maywood) 2008 Jan;233(1):38–47. doi: 10.3181/0703-RM-59. [DOI] [PubMed] [Google Scholar]
- 26.Thaker PH, Yokoi K, Jennings NB, Li Y, Rebhun RB, Rousseau DL, Jr, et al. Inhibition of experimental colon cancer metastasis by the GABA-receptor agonist nembutal. Cancer Biol Ther. 2005 Jul;4(7):753–758. doi: 10.4161/cbt.4.7.1827. [DOI] [PubMed] [Google Scholar]
- 27.van den DM, Pellis L, Crott JW, van EM, Friederich P, Nagengast FM, et al. Folic acid and vitamin B-12 supplementation does not favorably influence uracil incorporation and promoter methylation in rectal mucosa DNA of subjects with previous colorectal adenomas. J Nutr. 2007 Sep;137(9):2114–2120. doi: 10.1093/jn/137.9.2114. [DOI] [PubMed] [Google Scholar]
- 28.Basten GP, Duthie SJ, Pirie L, Vaughan N, Hill MH, Powers HJ. Sensitivity of markers of DNA stability and DNA repair activity to folate supplementation in healthy volunteers. Br J Cancer. 2006 Jun 19;94(12):1942–1947. doi: 10.1038/sj.bjc.6603197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Choi SW, Friso S, Dolnikowski GG, Bagley PJ, Edmondson AN, Smith DE, et al. Biochemical and molecular aberrations in the rat colon due to folate depletion are age-specific. J Nutr. 2003 Apr;133(4):1206–1212. doi: 10.1093/jn/133.4.1206. [DOI] [PubMed] [Google Scholar]
- 30.Figueiredo JC, Grau MV, Wallace K, Levine AJ, Shen L, Hamdan R, et al. Global DNA hypomethylation (LINE-1) in the normal colon and lifestyle characteristics and dietary and genetic factors. Cancer Epidemiol Biomarkers Prev. 2009 Apr;18(4):1041–1049. doi: 10.1158/1055-9965.EPI-08-0926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pufulete M, Al-Ghnaniem R, Khushal A, Appleby P, Harris N, Gout S, et al. Effect of folic acid supplementation on genomic DNA methylation in patients with colorectal adenoma. Gut. 2005 May;54(5):648–653. doi: 10.1136/gut.2004.054718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jhaveri MS, Wagner C, Trepel JB. Impact of extracellular folate levels on global gene expression. Mol Pharmacol. 2001 Dec;60(6):1288–1295. doi: 10.1124/mol.60.6.1288. [DOI] [PubMed] [Google Scholar]
- 33.Kraunz KS, Hsiung D, McClean MD, Liu M, Osanyingbemi J, Nelson HH, et al. Dietary folate is associated with p16(INK4A) methylation in head and neck squamous cell carcinoma. Int J Cancer. 2006 Oct 1;119(7):1553–1557. doi: 10.1002/ijc.22013. [DOI] [PubMed] [Google Scholar]
- 34.Ciappio E, Mason JB. Folate and Carcinogenesis. Basic Mechanisms. In: Bailey LB, editor. Folate in Health and Disease. 2nd ed. Boca Raton: Taylor and Francis; 2010. pp. 235–262. [Google Scholar]
- 35.Walker DR, Bond JP, Tarone RE, Harris CC, Makalowski W, Boguski MS, et al. Evolutionary conservation and somatic mutation hotspot maps of p53: correlation with p53 protein structural and functional features. Oncogene. 1999 Jan 7;18(1):211–218. doi: 10.1038/sj.onc.1202298. [DOI] [PubMed] [Google Scholar]
- 36.Barnes DE, Lindahl T. Repair and genetic consequences of endogenous DNA base damage in mammalian cells. Annu Rev Genet. 2004;38:445–476. doi: 10.1146/annurev.genet.38.072902.092448. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.