Abstract
Background
Helicobacter pylori, a neutralophile, colonizes the acidic environment of the human stomach by employing acid acclimation mechanisms that regulate periplasmic and cytoplasmic pH. The regulation of urease activity is central to acid acclimation. Inactive urease apoenzyme, UreA/B, requires nickel for activation. Accessory proteins UreE, F, G and H are required for nickel insertion into apoenzyme. The ExbB/ExbD/TonB complex transfers energy from the inner to outer membrane, providing the driving force for nickel uptake. Therefore, the aim of this study was to determine the contribution of ExbD to pH homeostasis.
Materials and Methods
A nonpolar exbD knockout was constructed and survival, growth, urease activity, and membrane potential were determined in comparison to wildtype.
Results
Survival of the ΔexbD strain was significantly reduced at pH 3.0. Urease activity as a function of pH and UreI activation were similar to the wildtype strain, showing normal function of the proton-gated urea channel, UreI. The increase in total urease activity over time in acid seen in the wildtype strain was abolished in the ΔexbD strain, but recovered in the presence of supra-physiologic nickel concentrations, demonstrating that the effect of the ΔexbD mutant is due to loss of a necessary constant supply of nickel. In acid, ΔexbD also decreased its ability to maintain membrane potential and periplasmic buffering in the presence of urea.
Conclusions
ExbD is essential for maintenance of periplasmic buffering and membrane potential by transferring energy required for nickel uptake, making it a potential non-antibiotic target for H. pylori eradication.
Introduction
Helicobacter pylori, a Gram-negative neutralophile, is the only organism known to colonize the normal acid-secreting human stomach. The bacteria infects about 50% of the world’s population (1). Colonization is associated with several human gastric diseases, including gastritis, peptic and duodenal ulcers, gastric carcinoma and MALT lymphoma (2–5). Eradication of the bacteria involves a complex regimen of minimally two antibiotics and a proton pump inhibitor (6). Problems with patient compliance and emerging antibiotic resistance have made it progressively more difficult to successfully treat the infection (6). Targeting the mechanisms that allow the organism to survive the gastric environment, where the median luminal pH is 1.4 (7), offers potential for future non-antibiotic treatment regimens.
H. pylori is able to survive and grow in acid through the use of mechanisms collectively termed acid acclimation (8). This system is named to contrast it with the acid resistance or tolerance mechanisms seen in other neutralophiles that can only transit the stomach, such as Escherichia coli and Vibrio cholerae. In these organisms, cytoplasmic pH is maintained between pH 4–5 in an acidic environment via amino acid/amine counter transporters or proton export by the F1F0 ATPase, allowing survival but not growth (9). H. pylori instead employs mechanisms to maintain the periplasmic pH around pH 6.1 and the cytoplasmic pH near neutral in acidic medium, allowing both survival and growth (10).
At the center of acid acclimation is a neutral pH optimum urease enzyme, made up of structural subunits UreA and UreB, which form an apoenzyme complex that requires nickel for activation (11). Urease accounts for up to 7% of total protein expression in H. pylori (12). Cytoplasmic accessory protein pairs UreE/UreG and UreF/UreH are essential for nickel incorporation into apoenzyme and production of active enzyme (13). H. pylori express UreI, an inner membrane localized, proton gated urea channel with 50% open probability at pH 5.9, which provides urea to cytoplasmic urease (10, 14). Urease hydrolyzes urea into carbon dioxide and ammonia (15). Urease associates with UreI at the inside of the inner membrane, allowing for rapid hydrolysis of urea at the membrane and movement of carbon dioxide and ammonia back through the channel into the periplasm (13, 16). Once in the periplasm, ammonia is protonated to form ammonium. Carbon dioxide is converted to bicarbonate via a periplasmic localized a-carbonic anhydrase enzyme, and the periplasm is buffered to about pH 6.1, which allows for maintenance of cytoplasmic pH compatible with survival and growth despite low medium pH (8).
Finding other candidate proteins that may play a role in acid acclimation would lead to a better understanding of this complex system. Rain et al completed a protein-protein interaction map of H. pylori using a yeast two-hybrid system (17). On review of this data, an interaction was identified between UreI and ExbD (17). ExbD is the product of gene HP1340 in the 26695 strain and is an inner membrane protein that is part of a 3 protein complex (ExbB(HP1339)/ExbD/TonB(HP1341)) (18). The H. pylori genome possesses two additional homologues of both exbB (HP1445, HP1129) and exbD (HP1446, HP1130), genes of unknown function. The ExbB/ExbD/TonB protein complex is found in many Gram negative bacteria. In Gram negative bacteria, a proton motive force across the inner membrane maintains bacterial viability and powers active transport (19). The outer membrane forms a protective barrier. Small molecules are able to enter through porins (20). Larger molecules required by the bacteria, such as siderophore iron, need to cross the outer membrane via active transport, but the outer membrane cannot maintain an electrical potential (21). Import of these large molecules, required for survival, depends on the proton motive force maintained across the inner membrane (22, 23). The ExbB/ExbD/TonB complex, localized to the inner membrane, transfers energy to the outer membrane, in the form of conformational changes in TonB, allowing active transport of essential molecules such as iron and nickel (24, 25). TonB gathers energy from the inner membrane, transfers that energy to an outer membrane TonB-gated transporter, then re-energizes in a cyclic manner (26, 27). TonB and ExbD interact through their periplasmic domains, and ExbD aids with energy transfer and recycling of TonB (24, 28, 29).
Much of the work in other Gram negative bacteria has focused on iron transport. In H. pylori, iron is a required nutrient, but nickel plays a significant role in acid acclimation due to its required role in urease activity (30). In addition to urease, the [Fe-Ni]-hydrogenase requires nickel and is essential for colonization (31). One active urease molecule requires 24 nickel ions (32). Nickel moves through the inner membrane via NixA, a nickel transporter (33). More recent work has looked at movement of nickel through the outer membrane. FecA3 and FrpB4 are outer membrane proteins involved with movement of complexed or insoluble nickel through the outer membrane (34). Entry of nickel needs to be tightly regulated since too little would impair urease activity and acid survival and too much would lead to generation of reactive oxygen species and cell damage (35). NikR is involved with regulation of genes responsive to nickel, including ureA, ureB, nixA, frpB4, and fecA3 (34, 36, 37). In high nickel, NikR binds to the promoter regions for frpB4 and fec3A, leading to repression of these genes and decreased nickel uptake (34). Like other outer membrane proteins, FrpB4 and Fec3A are energized by the ExbB/ExbD/TonB system, and NikR regulates expression of exbB and exbD (34, 36). Schauer et al further demonstrated that the ExbB/ExbD/TonB system powers movement of nickel through FrpB4 (38). The authors proposed that the ExbB/ExbD/TonB system energizes iron uptake at neutral pH and nickel uptake at acidic pH, facilitating acid activation of urease and therefore acid survival (38).
The ExbB/ExbD/TonB system is clearly important for acid acclimation given its role in driving nickel transport. Since ExbD was shown to interact with UreI (17), this protein in particular may be important for survival and growth in the gastric environment. The aim of the present study was to further define the role of exbD in H. pylori bioenergetics and acid acclimation via characterization of an exbD deletion mutant.
Materials and Methods
Bacterial strains and culture conditions
H. pylori strain ATCC 43504 was obtained from the American Type Culture Collection. A non-polar ATCC 43504 exbD/HP1340 deletion mutant was constructed by allelic exchange using a kanamycin resistance gene as described below. Bacteria were grown under microaerobic conditions (5% O2, 10% CO2, 85% N2) either on Trypticase Soy Agar (TSA) plates supplemented with 5% sheep blood (Gibco BRL-Life Technologies) or in brain heart infusion (BHI) medium (Difco Laboratories) supplemented with 7% horse serum (Gibco BRL-Life Technologies) and 0.25% yeast extract (Difco Laboratories). All bacteria grown in media were in the presence of Dent selective supplement (Oxoid Limited) and the ΔexbD was grown in the presence of 20 μg/mL kanamycin (Sigma).
Construction of H. pylori ΔexbD strain by allelic exchange mutagenesis
A genomic knockout of HP1340 was constructed by homologous recombination. pBluescript (Stratagene) containing a kanamycin resistance gene in the multi-cloning site flanked by SalI (5′) and BglII (3′) was used to generate the knockout plasmid. Primers were designed to flank the regions approximately 400 base pairs upstream of the 5′ end of the gene and 600 base pairs downstream from the 3′ end. The construct was designed for insertion into the pBluescript multi-cloning site between the XbaI and KpnI restriction sites, the intervening restriction sites were removed in the process of knockout construction. The 400 base pair upstream segment was amplified with a 5′ primer containing a site for digestion by XbaI (5′-cgtttctagattatgtggacatatttg-3′) and a 3′ primer containing a site for digestion by SalI (5′-gatggtcgacattttttcatgatcctg-3′). The 600 base pair downstream segment was amplified with a 5′ primer containing a site for digestion by BglII (5′-cagcattatggagatcttaaaagagcataatcatg-3′) and a 3′ primer containing a site for digestion by Acc65I (5′-ccgtaacgctcggtaccgcattgatcgtaaagctcacc-3′). The purified PCR products were sequentially ligated into pBluescript around the kanamycin resistance gene. The construct was introduced into H. pylori strain 43504 by natural transformation and colonies were selected in the presence of 40 μg/mL kanamycin on plates (once mutants were selected, colonies were expanded in liquid media as described above, in the presence of 20 μg/mL kanamycin). Knockouts were confirmed by a series of PCR reactions with primers to the up and downstream genes and the antibiotic resistance marker (data not shown). The exbD gene is in the middle of a 3-gene operon, and the products of these 3 genes have interrelated functions. Reverse transcriptase (RT)-PCR was used to confirm not only the presence, but also the transcription of tonB, the downstream gene. RNA was isolated from wildtype H. pylori and ΔexbD and subjected to RT-PCR using primers to both the exbD and tonB genes. As expected, a transcript for the exbD gene was seen in the wildtype bacteria but not in the ΔexbD strain, but the downstream tonB gene was expressed in both strains, confirming the exbD deletion is nonpolar.
RNA extraction and RT-PCR
Total RNA was isolated from H. pylori strain 43504 and 43504 ΔexbD using TRIzol® reagent (Invitrogen) combined with RNeasy columns (Qiagen). The bacterial pellet was resuspended in 500 μl of TRIzol® reagent and lysed at room temperature for 5 min before 100 μl of chloroform was added. After centrifugation at 12,000 × g for 10 min at 4°C, the supernatant was mixed with 250 μl of ethanol and applied to an RNeasy spin column (Qiagen), and RNA purification was performed following the manufacturer’s instructions (beginning with the application to the column). RT-PCR was then carried out with RNA from both strains, with 6 μg of RNA as the starting material. RT reaction was completed using RT omniscript (Qiagen) according to the manufacturer’s instructions. PCR on the generated cDNA was done using primers to HP1340 and HP1341 (tonB). Primers for the exbD gene were 5′-ggcgatgggctgaatgttgtccc-3′ (sense) and 5′-gtctataagggttttagggtc-3′ (antisense). Primers for the tonB gene were 5′-taaaatggcgcagattaggg-3′ (sense) and 5′-gatgcacggtttcttctggt-3′ (antisense). All PCRs were done with an annealing temperature of 55°C on a PTC-200 DNA engine PCR machine (MJ Research). PCR products were detected by capillary electrophoresis using an Agilent Bioanalyzer (Agilent Biosystems).
Measurement of growth at neutral pH
H. pylori strain 43504 wt and ΔexbD were grown overnight on TSA plates. 75 mL BHI (Difco) with 7% horse serum (Gibco) was added to each of three 250 mL flasks. 40 μg/mL kanamycin was added to one flask. Bacteria were added at an OD600 of 0.1 (1 flask with wt, 1 with ΔexbD, 1 with ΔexbD + kanamycin 40 μg/mL). 3 mL was removed immediately from each flask and the OD600 checked as the zero time point. The flasks were incubated under microaerobic conditions and aliquots were removed regularly to record the OD600 for a total of 30 hours. The log10 of the OD600 was plotted as a function of time for each culture.
Measurement of survival at acidic and neutral pH
H. pylori 43504 wt and ΔexbD were grown overnight on TSA plates. BHI was prepared at pH 7.4 and pH 3.0. The media at pH 3.0 was made with 50 mM homopipes and the pH was lowered with HCl. 5 mM urea was added to the pH 3.0 media for each strain. Each strain was resuspended in 600 μL BHI and 220 μL was added to 10 mL at each pH. Bacteria were incubated for 1hr in microaerobic conditions. 1 mL of each condition was added to 9 mL BHI, and serial dilutions were done using 1 mL into 9 mL BHI. 100 μL was plated in duplicate for each condition. Colonies were counted after 3 days. Data was expressed as percent survival at pH 3.0 compared to pH 7.4 for each strain.
Urease activity of intact bacteria
Urease activity was measured radiometrically (39, 40). Bacteria were added to 100 mM sodium phosphate buffer containing 5 mM KCl, 138 mM NaCl, 0.5 mM MgCl2, 1 mM CaCl2, 10 mM glucose, 1 mM glutamine, and 5 mM [14C]urea with a specific activity of 10 mCi/mmol. The range of pH of the buffer used was between pH 3.0 and 8.0. The pH of the buffer between 4.5 and 8.0 was achieved by mixing various amounts of 100 mM sodium phosphate monobasic and 100 mM sodium phosphate dibasic to the desired pH. Below a pH of 4.5 the desired pH was achieved by the addition of HCl. The pH of the buffer during the course of the experiment did not change by more than 0.1 pH units. Plastic wells containing 500 mM KOH soaked filter paper hung from rubber stoppers were used to collect the liberated 14CO2 that resulted from the hydrolysis of urea by urease. Urease activity was measured for 30 min at 37°C with constant agitation. The reaction was terminated by the addition of 5N H2SO4 and incubated 30 min at 37°C. The wells were placed in scintillation cocktail (HiIonicFluor; Packard Instruments), and the radioactivity was measured by scintillation counting (1216 RackBeta; LKB Institute). Protein concentration was determined by the BCA method (Pierce). Results were expressed in μmol/min/mg protein.
Measurement of total urease activity over time
H. pylori 43504 wt and ΔexbD grown on TSA plates were harvested and suspended in 500 μL of BHI media, pH 7.3. For some experiments with the ΔexbD strain, 250 μM nickel chloride (Sigma) was added to the media to bypass the requirement for ExbD. A total of 220 μL of the cell suspension was added to 15 mL of BHI at pH 7.3 or to 15 mL BHI adjusted to pH 5.5 by the addition of HCl and was incubated at 37°C under microaerobic conditions. 1.5 mL aliquots were obtained at 0, 30, 90, and 180 minutes and immediately centrifuged at 17,900 × g for 5 minutes at 4°C. The pellets were resuspended in 25 mM ice-cold phosphate buffer (PB), pH 7.0, and centrifuged at 17,900 × g for 5 minutes at 4°C. The pellet was resuspended in 150 μL of 25 mM phosphate buffer, pH 7.0 (PB) containing 0.01% polyoxyethylene-8-lauryl ether (C12E8; Sigma). C12E8 was used to permeabilize the bacterial membranes without disruption of the bacteria, as visualized by acridine orange fluorescence in a confocal microscope (14).
Urease activity was measured radiometrically as outlined above using 10 μL of the C12E8-treated bacterial suspension.
Measurement of membrane potential
Membrane potential was determined as previously described (41). Briefly, the bacteria were harvested from plates into 300 μl HP medium (5 mM KCl, 138 mM NaCl, 0.5 mM MgCl2, 1 mM CaCl2, 10 mM glucose, 1 mM glutamine) buffered with 100 mM phosphate. The fluorescent membrane potential sensitive dye, 3,3′ - dipropylthiadicarbocyanine iodide (DiSC3(5)), was dissolved in dimethyl sulfoxide (DMSO) and 3 μl added to 3 ml of appropriate buffer to give a final concentration of 1 μM. The bacterial suspension was then added to 3 mL of the dye solution at different pHout in a fluorimeter cuvette to reach an OD of 0.160 at 600 nm (usually 15–20 μl).
Fluorescence quenching due to potential dependent uptake of the dye was measured in a fluorimeter set at an excitation wavelength of 600 nm and emission wavelength of 665 nm. Data points were collected every 0.050 seconds. The dye solution was added 5 minutes before adding the bacteria to allow temperature equilibration. With addition of the bacteria, the fluorescence quenched due to dye uptake dependent on the interior negative potential. Urea was added once the fluorescence stabilized.
The calibration of the membrane potential was carried out as previously described by the addition of valinomycin followed by the addition of K+ until no further change in fluorescence was observed (40). This enables calculation of the K+ equilibrium potential found with the addition of the K+ selective ionophore, valinomycin, using the Nernst equation:
where [K+]in is equal to the external K+ concentration at which the potential difference becomes zero in the presence of valinomycin, i.e. where [K+]out = [K+]in, and 5 mM is medium concentration when valinomycin is added. The membrane potential in the absence of valinomycin can then be calculated. No change in medium pH was found in these strong buffers before or after the addition of urea over the time course of measurement. Membrane potential is displayed based on the calibration once the dye reaches equilibrium. The initial fluorescence quench depends on accumulation of the dye inside the bacteria and the fluorescence present prior to equilibrium does not necessarily imply a positive interior potential. Membrane potential was determined at pH 3.0, 4.5 and 7.4 in the wt and ΔexbD strains with the addition of 5 mM urea. An increase in membrane potential following urea addition in acid is due to reduction of the ΔpH, thus elevating ΔΨ.
Results
Growth of H. pylori wildtype and ΔexbD strains at pH 7.4
Growth was measured at pH 7.4 in the wildtype and the ΔexbD strains. Growth was also measured in the ΔexbD strain in the presence of kanamycin to control for any effect of kanamycin on growth independent of the gene deletion. The OD600 was measured at time zero, every 2 hours for the first 10 hours (data not shown), and again every 2 hours between 24–30 hours. Growth leveled off in all conditions between 24–30 hours (data not shown). Data was plotted as log of the OD600 as a function of time. There was no significant difference in the slope of the growth curves over time between the three conditions, suggesting that ExbD is not required for normal growth at neutral pH. The presence of kanamycin also did not have a deleterious effect on bacterial growth (data not shown).
Survival of ΔexbD and wildtype H. pylori at pH 3.0 and pH 7.4
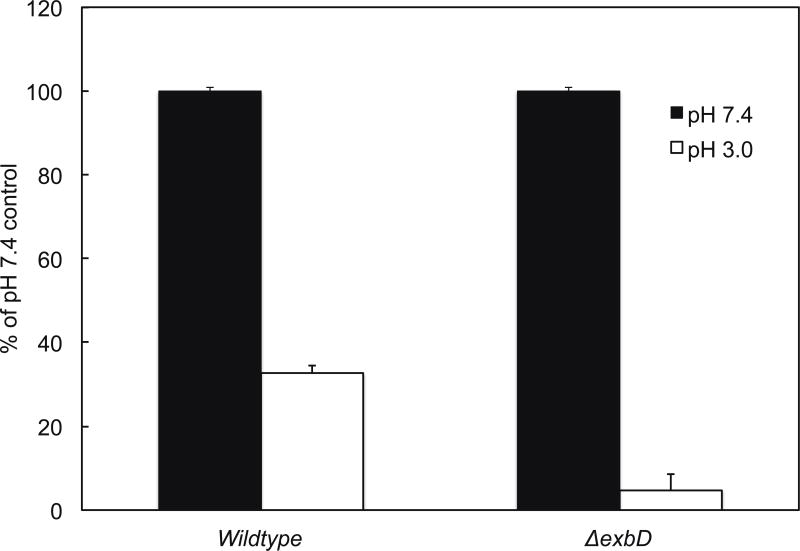
Wildtype H. pylori and ΔexbD strains were incubated for 1 hour at pH 3.0 or 7.4, with 5 mM urea added to the pH 3.0 conditions. Bacteria were then serially diluted, plated, and colonies counted. Data were expressed as percent surviving at pH 3.0 compared to pH 7.4 (100%) for each strain (figure 1). The wildtype bacteria showed 32.6 ± 2.0 % survival at pH 3.0 (mean ± s.e.m, n=3). The ΔexbD mutant showed only 4.9 ± 3.7 % survival (mean ± s.e.m, n=3). In the absence of exbD, there is decreased survival at acidic pH in the presence of urea as compared to wildtype.
Figure 1.
Survival of wildtype H. pylori and ΔexbD at pH 3.0 vs pH 7.4. Bacteria were incubated for 1 hour at pH 3.0 + 5mM urea or pH 7.4, then serially diluted and plated. Colony counts showed a decrease in survival in the ΔexbD at acidic pH compared to wildtype (mean ± s.e.m., n=3).
Intact urease activity of the ΔexbD strain
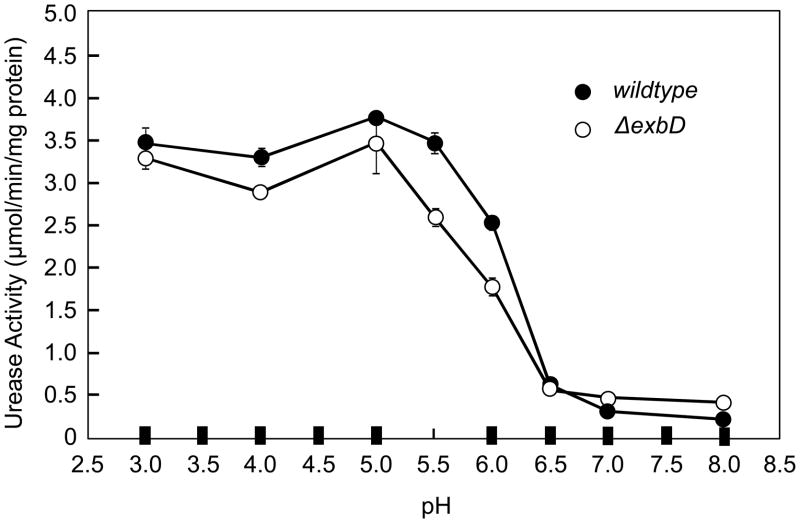
Intact urease activity was measured radiometrically in the wildtype H. pylori and ΔexbD strains over a pH range from 3–8. Urease activity was expressed in μmol/min/mg protein (figure 2). As expected, and as shown previously (40), in intact wildtype H. pylori, urease activity is maximal between pH 3–5.5, with a sharp decline to minimal activity at pH 7.0 and 8.0. In the intact organism, urea access to urease is accelerated by UreI, an inner membrane localized, H+ gated urea channel, with a pKa of ~6 (10, 14). The enzyme activity curve of the ΔexbD mutant is similar to the wildtype, suggesting that ExbD does not play a direct role in UreI function. The decrease in acid survival in the mutant is not attributable to inhibition of UreI opening.
Figure 2.
Intact urease activity curve of wildtype H. pylori and ΔexbD. Urease activity was measured in the intact bacteria by measuring release of [14C]O2 from [14C]urea at various media pHs. In the wildtype bacteria, there is low activity at neutral pH, with a steep increase in activity between pH 6.5–5.5 and maximal activity below pH 5.5. This is due to opening of the proton gated urea channel, UreI, allowing urea access to cytoplasmic urease. The shape of the urease activity curve is similar in the ΔexbD, demonstrating that absence of ExbD does not affect channel function itself.
Variation in total urease activities of H. pylori wildtype and ΔexbD strains incubated at pH 5.5 and 7.4
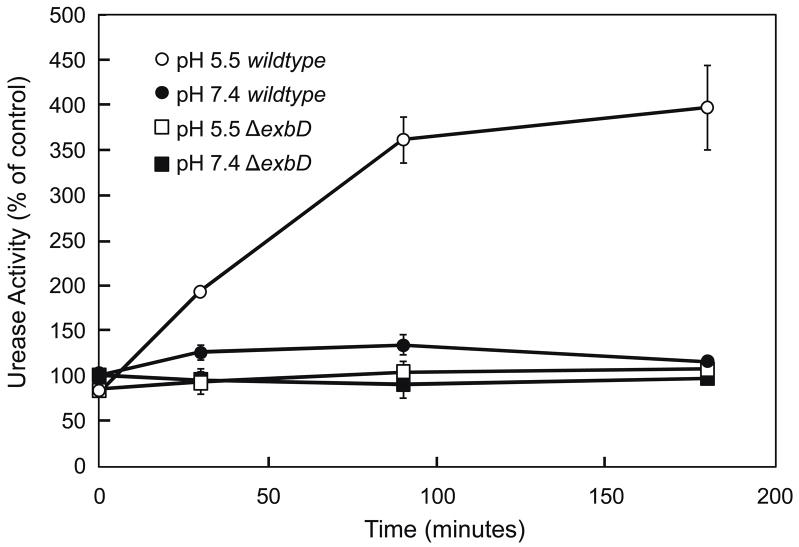
H. pylori wildtype and the ΔexbD mutant were incubated for 180 minutes and aliquots were removed (at 0, 30, 90, 180 minutes) for measurement of total urease activity. Total urease activity was measured in the presence of C12E8 in order to bypass UreI. Urease activity was expressed as percent of control (wildtype pH 7.4) as a function of time (figure 3). There was no increase in urease activity over time at neutral pH in either strain. The wildtype showed a rapid and sustained increase in urease activity over time in acid, as shown previously (42). In the wildtype, at pH 5.5, total urease activity was 175% of control within 30 minutes and ultimately reached a maximal level of 3.5-4x control. This rapid, sustained increase in urease activity over time was abolished in the ΔexbD mutant. This suggests a role for ExbD in activation of apourease in acid.
Figure 3.
Time course of urease activation of H. pylori. H. pylori wildtype and ΔexbD strains were incubated in BHI, pH 5.5 or 7.4, for 30 to 180 min, and total urease activity was measured in the presence of the nonionic detergent C12E8 to bypass UreI. In the wildtype strain, there was a >3-fold increase in urease activity at pH 5.5 compared to that at pH 7.4 after 30 min, and it remained elevated for 3 h. The absence of ExbD had no effect on urease activity at pH 7.4 but abolished the increase in urease activity in acidic pH seen in the wildtype strain.
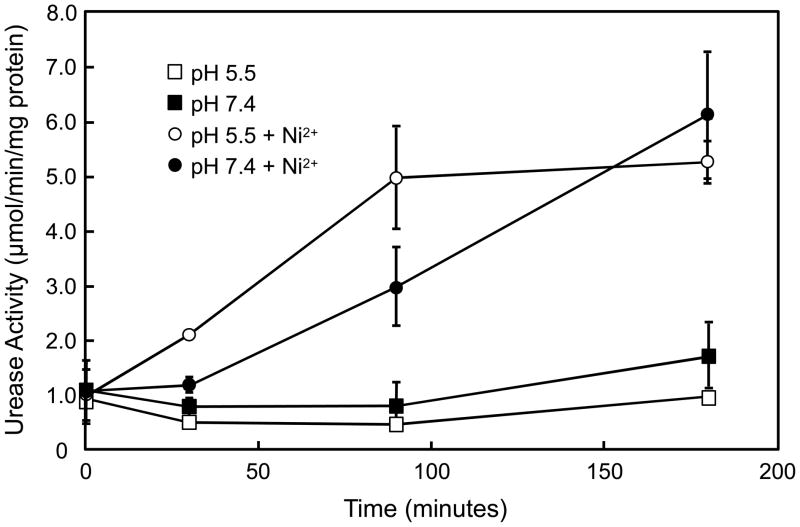
Total urease activity over time was measured in the ΔexbD strain in the presence of very high nickel in the media (250 μM NiCl2) to see if ExbD regulation of NixA could be overcome. As above, the acid induced increase in total urease activity is abolished in the ΔexbD, but urease activity is restored to wildtype levels in the presence of high nickel (figure 4). This confirms the singular role for ExbD in regulating nickel for urease activation.
Figure 4.
Time course of urease activation in the ΔexbD in the presence of high nickel. H. pylori ΔexbD strain was incubated in BHI, pH 5.5 or 7.4, in the presence of 250 μM NiCl2 for 30 to 180 min, and total urease activity was measured in the presence of the nonionic detergent C12E8 to bypass UreI. In the presence of supraphysiologic levels of nickel, the acid-induced activation of urease activity over time is restored in the ΔexbD.
Alteration of membrane potential in the H. pylori ΔexbD mutant at acidic pH
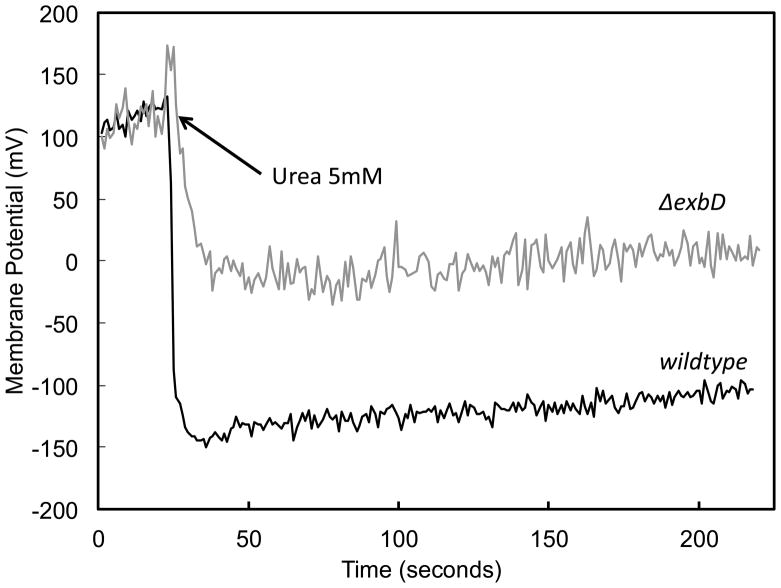
Membrane potential was measured in the wildtype H. pylori and ΔexbD strains using the potential sensitive fluorescent cationic dye DiSC3(5). The dye accumulates on the membrane and is translocated into the lipid bilayer. Aggregation within the cytoplasmic membrane leads to a decrease in fluorescence, corresponding to an increase in membrane potential (larger negative number with respect to zero in mV). Wildtype H. pylori maintains a constant proton motive force (PMF) across the inner membrane between pH 3.0 and 8.0. The PMF is the algebraic sum of the pH difference (ΔpH) and the potential difference (ΔΨ) across the inner membrane. The PMF is maintained at a relatively constant level via reciprocal changes in ΔpH and ΔΨ (43). The cytoplasmic pH is about 7.8, so at acidic pH, the ΔpH is the major contributor to the PMF. In order to grow at pH 4.0 with a neutral cytoplasmic pH, H. pylori needs to maintain the periplasmic pH above the medium pH to prevent an inward proton gradient from driving the membrane potential close to 0 mV, incompatible with many required cellular functions (43, 44). In the wildtype, at pH 4.5, and more significantly at pH 3.0, addition of urea results in an increase in ΔΨ as the periplasm becomes less acidic since the ΔpH across the inner membrane decreases. This increase in ΔΨ was abolished in the ΔexbD mutant at pH 4.5 (data not shown) and 3.0 (figure 5). This failure to increase ΔΨ in the ΔexbD strain suggests that ExbD is necessary for periplasmic buffering by increasing urease activity via regulation of cytoplasmic Ni2+ uptake.
Figure 5.
Membrane potential measurement in the presence and absence of ExbD. DiSC3(5) was used to measure membrane potential at pH 3.0. In the wildtype (black line), with the addition of urea, there is a sustained increase in membrane potential, allowing maintenance of the proton motive force in the face of acidic medium pH. The ΔexbD (grey line) is unable to sustain an increase in membrane potential under the same conditions.
Discussion
H. pylori has developed the mechanism of acid acclimation to combat the acidity at its site of colonization, the human gastric mucosa. As a neutralophile, it relies on periplasmic pH homeostasis in order to grow in the stomach. As the periplasmic pH falls in acidic media, UreI, the inner membrane proton gated urea channel, opens and urea moves into the cytoplasm (14). Urease and its accessory proteins move to the membrane at acidic pH, allowing for rapid breakdown of urea in close proximity to the periplasm (13, 16). The products of urea hydrolysis move back through UreI into the periplasm, again adding to the efficiency of periplasmic buffering (16). Nickel is required for activation of urease and is found in trace levels in the blood (45). Nickel incorporation into apourease, leading to enzyme activation, is increased at acidic pH, and is independent of protein synthesis (42). The availability of nickel therefore provides one of several levels of regulation of urease activity and aids survival in acid.
Nickel crosses the outer membrane before it enters the cytoplasm via the inner membrane nickel transporter NixA (33). The outer membrane of Gram negative bacteria contains porins that allow passive entry of small molecules (46). Many compounds essential to bacterial survival are often either in too low abundance and/or too large to pass through porins in sufficient amounts, and therefore require active transport (47). The ExbB/ExbD/TonB system transfers energy to the outer membrane in order to drive active transport of critical molecules (47). This system is most extensively studied in the context of iron and cobalamin transport in E. coli (21). Given the presence of this complex of genes in H. pylori and the requirement for nickel, a molecule in relatively low abundance, investigation has illustrated a role for ExbB/ExbD/TonB in nickel uptake (38).
Fur, an iron responsive regulator, and NikR, a nickel responsive regulator, have an effect on the exbB/exbD/tonB operon (36, 48–51). Based on this finding, Schauer et al studied the dual role of ExbB/ExbD/TonB in iron and nickel uptake (38). The protein complex was necessary for iron uptake at neutral pH, where Fe2+ is oxidized to the insoluble Fe3+ form (38, 52). At acidic pH, the complex was also responsible for nickel accumulation, an effect that was abolished when the operon was deleted (38). This work also identified FrpB4 as the outer membrane, TonB dependent receptor involved with nickel transport (38).
The ExbB/ExbD/TonB complex is clearly important in regulation and energizing of nickel movement through the outer membrane. Given that ExbD was shown to interact with UreI (17) and that additional components of acid acclimation have been identified in recent years, the work presented here clarifies the specific role of ExbD in the context of gastric colonization.
In E. coli, knockout of exbD leads to TonB protein degradation (53). This illustrates the importance of ExbD for function of the complex. Survival of the nonpolar ΔexbD was normal at neutral pH but survival was diminished compared to wildtype at pH 3.0, suggesting a role for this protein in the gastric environment. exbD is upregulated in acid in only 1 of 4 reported in vivo and in vitro acid transcriptome studies (48, 51, 54, 55), likely reflecting its dual role in iron and nickel import rather than diminishing its importance at acidic pH. exbD is part of the regulon controlled by the HP0165/HP0166 TCS (56), which is activated in response to low pH. Although ExbD interacts with UreI (17), it is not required for acid activation of channel opening. Increase in total urease activity with time is dependent on the presence of UreI, as demonstrated by comparison with a ΔureI strain (42), and is also dependent on the presence of ExbD, underscoring the significance of nickel uptake for enhanced urease activity.
The effect of the loss of ExbD on the time course of urease activation can be overcome in the presence of high levels of nickel, demonstrating its role in nickel uptake. Over the three hour time course of the experiment, at acidic pH, the total urease activity in the presence of nickel increased about 4 fold over the baseline at zero time. With supra-physiologic levels of nickel present, there is likely sufficient passive movement through outer membrane porins to obviate the need for active transport. The total activity in the ΔexbD strain at neutral pH with high nickel reaches the levels seen at acidic pH after 3 hours, although the rate of rise is slower. This likely reflects such an excess of nickel that a higher proportion of apoenzyme is activated than is possible under physiologic conditions in the ΔexbD mutant.
Wildtype H. pylori is unable to maintain a membrane potential below pH 4.0 in the absence of urea, with more physiologic potential levels seen between pH 6–8 (41). In the absence of ExbD, at acidic medium pH, in contrast to wildtype, the bacteria are unable to generate a membrane potential, diminishing survival, despite the addition of urea. This demonstrates the importance of ExbD in urease dependent periplasmic buffering. Without energy transfer for sufficient nickel entry to activate enough urease to buffer the periplasm in acid, ΔpH increases and proton motive force will approach zero.
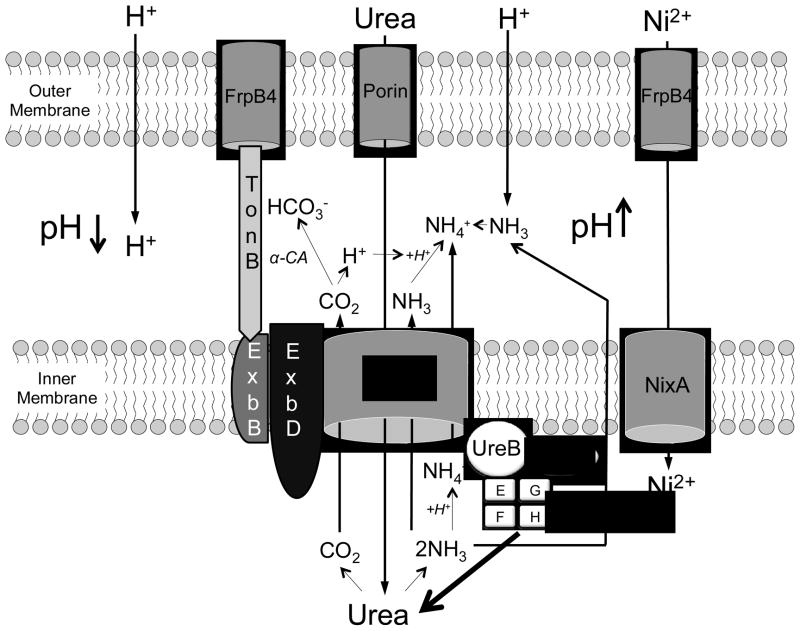
ExbD is an inner membrane protein, putting it in close proximity to the site of urea hydrolysis. ArsS, the inner membrane localized histidine kinase that senses acidity, has exbD in its regulon (56). As mentioned above, ArsS is involved with trafficking of cytoplasmic urease and its accessory proteins to the inner membrane, in association with UreI (57). The role of ExbD in acid acclimation is illustrated by the model shown in figure 6. Since ExbD interacts with UreI, it likely adds an additional level of control to ensure that urease is activated and breaks down urea in as close proximity to the periplasm as possible, maintaining cytoplasmic and periplasmic pH in acceptable ranges.
Figure 6.
The role of ExbD in the acid acclimation response in H. pylori. In acidic media, the periplasmic pH is maintained at ~6.1 by acid acclimation. At this pH, the membrane potential is ~−100 mV, about 100 mV less negative then at pH 7.4. The ExbB/ExbD/TonB complex energizes the uptake of nickel through the TonB-dependent transporter FrpB4 by responding to the change in membrane potential and increasing the flux of nickel through NixA into the cytoplasm. Once in the cytoplasm, nickel incorporation, required for urease activation, is aided by cytoplasmic urease accessory proteins UreE-H. Simultaneously, protonation of UreI leads to its opening. Decreased periplasmic pH is also sensed by HP0165 and subsequent activation of its cognate response regulator, HP0166 (not shown in the model), leading to mobilization of urease and its nickel insertion accessory proteins at the inner membrane. Urea moves through UreI, where it is hydrolyzed at the inner membrane to CO2 and 2NH3. The products of urea hydrolysis move back through UreI into the periplasm. CO2 is converted to bicarbonate, catalyzed by a periplasmic, inner membrane localized carbonic anhydrase. The periplasm is then buffered by the products of urea hydrolysis, allowing H. pylori, a neutralophile, to survive and grow in an acidic environment.
ExbD is required for stability of the ExbB/ExbD/TonB complex (53) that is also shown here to be required for acid survival and periplasmic buffering in H. pylori. Periplasmic pH regulation is required for gastric survival of H. pylori. ExbD is required for acute response to acidic media for maintenance of membrane potential and activation of urease activity, as a component of the acid acclimation mechanism. Given the critical role for ExbD in acid survival, this protein represents a potential non-antibiotic drug target for eradication of infection.
Acknowledgments
This work was supported by NIH and USVA grants 5K12 HD034610 (EAM), P30 DK41301 (EAM), DK053642 (GS), 1I01BX001006 (GS).
Footnotes
Disclosures
None
References
- 1.Suerbaum S, Michetti P. Helicobacter pylori infection. The New England journal of medicine. 2002 Oct 10;347(15):1175–86. doi: 10.1056/NEJMra020542. [DOI] [PubMed] [Google Scholar]
- 2.Blaser MJ. Hypotheses on the pathogenesis and natural history of Helicobacter pylori-induced inflammation. Gastroenterology. 1992 Feb;102(2):720–7. doi: 10.1016/0016-5085(92)90126-j. [DOI] [PubMed] [Google Scholar]
- 3.Nomura A, Stemmermann GN, Chyou PH, Kato I, Perez-Perez GI, Blaser MJ. Helicobacter pylori infection and gastric carcinoma among Japanese Americans in Hawaii. The New England journal of medicine. 1991 Oct 17;325(16):1132–6. doi: 10.1056/NEJM199110173251604. [DOI] [PubMed] [Google Scholar]
- 4.Parsonnet J. Gastric adenocarcinoma and Helicobacter pylori infection. The Western journal of medicine. 1994 Jul;161(1):60. [PMC free article] [PubMed] [Google Scholar]
- 5.Parsonnet J, Friedman GD, Vandersteen DP, Chang Y, Vogelman JH, Orentreich N, et al. Helicobacter pylori infection and the risk of gastric carcinoma. The New England journal of medicine. 1991 Oct 17;325(16):1127–31. doi: 10.1056/NEJM199110173251603. [DOI] [PubMed] [Google Scholar]
- 6.Malfertheiner P, Megraud F, O’Morain C, Bazzoli F, El-Omar E, Graham D, et al. Current concepts in the management of Helicobacter pylori infection: the Maastricht III Consensus Report. Gut. 2007 Jun;56(6):772–81. doi: 10.1136/gut.2006.101634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schade C, Flemstrom G, Holm L. Hydrogen ion concentration in the mucus layer on top of acid-stimulated and -inhibited rat gastric mucosa. Gastroenterology. 1994 Jul;107(1):180–8. doi: 10.1016/0016-5085(94)90075-2. [DOI] [PubMed] [Google Scholar]
- 8.Marcus EA, Moshfegh AP, Sachs G, Scott DR. The periplasmic alpha-carbonic anhydrase activity of Helicobacter pylori is essential for acid acclimation. Journal of bacteriology. 2005 Jan;187(2):729–38. doi: 10.1128/JB.187.2.729-738.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Foster JW. Escherichia coli acid resistance: tales of an amateur acidophile. Nature reviews. 2004 Nov;2(11):898–907. doi: 10.1038/nrmicro1021. [DOI] [PubMed] [Google Scholar]
- 10.Scott DR, Marcus EA, Weeks DL, Lee A, Melchers K, Sachs G. Expression of the Helicobacter pylori ureI gene is required for acidic pH activation of cytoplasmic urease. Infection and immunity. 2000 Feb;68(2):470–7. doi: 10.1128/iai.68.2.470-477.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferrero RL, Cussac V, Courcoux P, Labigne A. Construction of isogenic urease-negative mutants of Helicobacter pylori by allelic exchange. Journal of bacteriology. 1992 Jul;174(13):4212–7. doi: 10.1128/jb.174.13.4212-4217.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bauerfeind P, Garner R, Dunn BE, Mobley HL. Synthesis and activity of Helicobacter pylori urease and catalase at low pH. Gut. 1997 Jan;40(1):25–30. doi: 10.1136/gut.40.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Voland P, Weeks DL, Marcus EA, Prinz C, Sachs G, Scott D. Interactions among the seven Helicobacter pylori proteins encoded by the urease gene cluster. American journal of physiology. 2003 Jan;284(1):G96–G106. doi: 10.1152/ajpgi.00160.2002. [DOI] [PubMed] [Google Scholar]
- 14.Weeks DL, Eskandari S, Scott DR, Sachs G. A H+-gated urea channel: the link between Helicobacter pylori urease and gastric colonization. Science. 2000 Jan 21;287(5452):482–5. doi: 10.1126/science.287.5452.482. [DOI] [PubMed] [Google Scholar]
- 15.Bury-Mone S, Mendz GL, Ball GE, Thibonnier M, Stingl K, Ecobichon C, et al. Roles of alpha and beta carbonic anhydrases of Helicobacter pylori in the urease-dependent response to acidity and in colonization of the murine gastric mucosa. Infection and immunity. 2008 Feb;76(2):497–509. doi: 10.1128/IAI.00993-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scott DR, Marcus EA, Wen Y, Singh S, Feng J, Sachs G. Cytoplasmic histidine kinase (HP0244)-regulated assembly of urease with UreI, a channel for urea and its metabolites, CO2, NH3, and NH4+, is necessary for acid survival of Helicobacter pylori. Journal of bacteriology. 2010 Jan;192(1):94–103. doi: 10.1128/JB.00848-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rain JC, Selig L, De Reuse H, Battaglia V, Reverdy C, Simon S, et al. The protein-protein interaction map of Helicobacter pylori. Nature. 2001 Jan 11;409(6817):211–5. doi: 10.1038/35051615. [DOI] [PubMed] [Google Scholar]
- 18.Tomb JF, White O, Kerlavage AR, Clayton RA, Sutton GG, Fleischmann RD, et al. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997 Aug 7;388(6642):539–47. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 19.Mitchell P. Chemiosmotic coupling in oxidative and photosynthetic phosphorylation. Biological reviews of the Cambridge Philosophical Society. 1966 Aug;41(3):445–502. doi: 10.1111/j.1469-185x.1966.tb01501.x. [DOI] [PubMed] [Google Scholar]
- 20.Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev. 2003 Dec;67(4):593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ferguson AD, Deisenhofer J. Metal import through microbial membranes. Cell. 2004 Jan 9;116(1):15–24. doi: 10.1016/s0092-8674(03)01030-4. [DOI] [PubMed] [Google Scholar]
- 22.Bradbeer C. The proton motive force drives the outer membrane transport of cobalamin in Escherichia coli. Journal of bacteriology. 1993 May;175(10):3146–50. doi: 10.1128/jb.175.10.3146-3150.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sen K, Hellman J, Nikaido H. Porin channels in intact cells of Escherichia coli are not affected by Donnan potentials across the outer membrane. The Journal of biological chemistry. 1988 Jan 25;263(3):1182–7. [PubMed] [Google Scholar]
- 24.Ollis AA, Manning M, Held KG, Postle K. Cytoplasmic membrane protonmotive force energizes periplasmic interactions between ExbD and TonB. Molecular microbiology. 2009 Aug;73(3):466–81. doi: 10.1111/j.1365-2958.2009.06785.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Postle K, Larsen RA. TonB-dependent energy transduction between outer and cytoplasmic membranes. Biometals. 2007 Jun;20(3–4):453–65. doi: 10.1007/s10534-006-9071-6. [DOI] [PubMed] [Google Scholar]
- 26.Fischer E, Gunter K, Braun V. Involvement of ExbB and TonB in transport across the outer membrane of Escherichia coli: phenotypic complementation of exbB mutants by overexpressed tonB and physical stabilization of TonB by ExbB. Journal of bacteriology. 1989 Sep;171(9):5127–34. doi: 10.1128/jb.171.9.5127-5134.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Larsen RA, Thomas MG, Postle K. Protonmotive force, ExbB and ligand-bound FepA drive conformational changes in TonB. Molecular microbiology. 1999 Mar;31(6):1809–24. doi: 10.1046/j.1365-2958.1999.01317.x. [DOI] [PubMed] [Google Scholar]
- 28.Brinkman KK, Larsen RA. Interactions of the energy transducer TonB with noncognate energy-harvesting complexes. Journal of bacteriology. 2008 Jan;190(1):421–7. doi: 10.1128/JB.01093-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Larsen RA, Deckert GE, Kastead KA, Devanathan S, Keller KL, Postle K. His(20) provides the sole functionally significant side chain in the essential TonB transmembrane domain. Journal of bacteriology. 2007 Apr;189(7):2825–33. doi: 10.1128/JB.01925-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eaton KA, Gilbert JV, Joyce EA, Wanken AE, Thevenot T, Baker P, et al. In vivo complementation of ureB restores the ability of Helicobacter pylori to colonize. Infection and immunity. 2002 Feb;70(2):771–8. doi: 10.1128/iai.70.2.771-778.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Olson JW, Mehta NS, Maier RJ. Requirement of nickel metabolism proteins HypA and HypB for full activity of both hydrogenase and urease. Helicobacter pylori. 2001;39:176–82. doi: 10.1046/j.1365-2958.2001.02244.x. [DOI] [PubMed] [Google Scholar]
- 32.Ha NC, Oh ST, Sung JY, Cha KA, Lee MH, Oh BH. Supramolecular assembly and acid resistance of Helicobacter pylori urease. Nature structural biology. 2001 Jun;8(6):505–9. doi: 10.1038/88563. [DOI] [PubMed] [Google Scholar]
- 33.Mobley HL, Island MD, Hausinger RP. Molecular biology of microbial ureases. Microbiological reviews. 1995 Sep;59(3):451–80. doi: 10.1128/mr.59.3.451-480.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ernst FD, Stoof J, Horrevoets WM, Kuipers EJ, Kusters JG, van Vliet AH. NikR mediates nickel-responsive transcriptional repression of the Helicobacter pylori outer membrane proteins FecA3 (HP1400) and FrpB4 (HP1512) Infection and immunity. 2006 Dec;74(12):6821–8. doi: 10.1128/IAI.01196-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mulrooney SB, Hausinger RP. Nickel uptake and utilization by microorganisms. FEMS microbiology reviews. 2003 Jun;27(2–3):239–61. doi: 10.1016/S0168-6445(03)00042-1. [DOI] [PubMed] [Google Scholar]
- 36.Contreras M, Thiberge JM, Mandrand-Berthelot MA, Labigne A. Characterization of the roles of NikR, a nickel-responsive pleiotropic autoregulator of Helicobacter pylori. Molecular microbiology. 2003 Aug;49(4):947–63. doi: 10.1046/j.1365-2958.2003.03621.x. [DOI] [PubMed] [Google Scholar]
- 37.van Vliet AH, Poppelaars SW, Davies BJ, Stoof J, Bereswill S, Kist M, et al. NikR mediates nickel-responsive transcriptional induction of urease expression in Helicobacter pylori. Infection and immunity. 2002 Jun;70(6):2846–52. doi: 10.1128/IAI.70.6.2846-2852.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schauer K, Gouget B, Carriere M, Labigne A, de Reuse H. Novel nickel transport mechanism across the bacterial outer membrane energized by the TonB/ExbB/ExbD machinery. Molecular microbiology. 2007 Feb;63(4):1054–68. doi: 10.1111/j.1365-2958.2006.05578.x. [DOI] [PubMed] [Google Scholar]
- 39.McDonald JA, Speeg KVJ, Campbell JW. Urease: a sensitive and specific radiometric assay. Enzymologia. 1972;42(1):1–9. [PubMed] [Google Scholar]
- 40.Scott DR, Weeks D, Hong C, Postius S, Melchers K, Sachs G. The role of internal urease in acid resistance of Helicobacter pylori. Gastroenterology. 1998 Jan;114(1):58–70. doi: 10.1016/s0016-5085(98)70633-x. [DOI] [PubMed] [Google Scholar]
- 41.Meyer-Rosberg K, Scott DR, Rex D, Melchers K, Sachs G. The effect of environmental pH on the proton motive force of Helicobacter pylori. Gastroenterology. 1996 Oct;111(4):886–900. doi: 10.1016/s0016-5085(96)70056-2. [DOI] [PubMed] [Google Scholar]
- 42.Scott DR, Marcus EA, Weeks DL, Sachs G. Mechanisms of acid resistance due to the urease system of Helicobacter pylori. Gastroenterology. 2002 Jul;123(1):187–95. doi: 10.1053/gast.2002.34218. [DOI] [PubMed] [Google Scholar]
- 43.Kashket ER, Wong PT. The intracellular pH of Escherichia coli. Biochimica et biophysica acta. 1969 Oct 14;193(1):212–4. doi: 10.1016/0005-2736(69)90074-1. [DOI] [PubMed] [Google Scholar]
- 44.von Heijne G. Membrane protein assembly: rules of the game. Bioessays. 1995 Jan;17(1):25–30. doi: 10.1002/bies.950170107. [DOI] [PubMed] [Google Scholar]
- 45.Templeton DM, Sunderman FW, Jr, Herber RF. Tentative reference values for nickel concentrations in human serum, plasma, blood, and urine: evaluation according to the TRACY protocol. The Science of the total environment. 1994 Jun 6;148(2–3):243–51. doi: 10.1016/0048-9697(94)90400-6. [DOI] [PubMed] [Google Scholar]
- 46.Nikaido H. Porins and specific diffusion channels in bacterial outer membranes. The Journal of biological chemistry. 1994 Feb 11;269(6):3905–8. [PubMed] [Google Scholar]
- 47.Postle K, Kadner RJ. Touch and go: tying TonB to transport. Molecular microbiology. 2003 Aug;49(4):869–82. doi: 10.1046/j.1365-2958.2003.03629.x. [DOI] [PubMed] [Google Scholar]
- 48.Bury-Mone S, Thiberge JM, Contreras M, Maitournam A, Labigne A, De Reuse H. Responsiveness to acidity via metal ion regulators mediates virulence in the gastric pathogen Helicobacter pylori. Molecular microbiology. 2004 Jul;53(2):623–38. doi: 10.1111/j.1365-2958.2004.04137.x. [DOI] [PubMed] [Google Scholar]
- 49.Ernst FD, Bereswill S, Waidner B, Stoof J, Mader U, Kusters JG, et al. Transcriptional profiling of Helicobacter pylori Fur- and iron-regulated gene expression. Microbiology (Reading, England) 2005 Feb;151(Pt 2):533–46. doi: 10.1099/mic.0.27404-0. [DOI] [PubMed] [Google Scholar]
- 50.Ernst FD, Kuipers EJ, Heijens A, Sarwari R, Stoof J, Penn CW, et al. The nickel-responsive regulator NikR controls activation and repression of gene transcription in Helicobacter pylori. Infection and immunity. 2005 Nov;73(11):7252–8. doi: 10.1128/IAI.73.11.7252-7258.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Merrell DS, Goodrich ML, Otto G, Tompkins LS, Falkow S. pH-regulated gene expression of the gastric pathogen Helicobacter pylori. Infection and immunity. 2003 Jun;71(6):3529–39. doi: 10.1128/IAI.71.6.3529-3539.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wandersman C, Delepelaire P. Bacterial iron sources: from siderophores to hemophores. Annual review of microbiology. 2004;58:611–47. doi: 10.1146/annurev.micro.58.030603.123811. [DOI] [PubMed] [Google Scholar]
- 53.Ahmer BM, Thomas MG, Larsen RA, Postle K. Characterization of the exbBD operon of Escherichia coli and the role of ExbB and ExbD in TonB function and stability. Journal of bacteriology. 1995 Aug;177(16):4742–7. doi: 10.1128/jb.177.16.4742-4747.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Scott DR, Marcus EA, Wen Y, Oh J, Sachs G. Gene expression in vivo shows that Helicobacter pylori colonizes an acidic niche on the gastric surface. Proceedings of the National Academy of Sciences of the United States of America. 2007 Apr 24;104(17):7235–40. doi: 10.1073/pnas.0702300104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wen Y, Marcus EA, Matrubutham U, Gleeson MA, Scott DR, Sachs G. Acid-adaptive genes of Helicobacter pylori. Infection and immunity. 2003 Oct;71(10):5921–39. doi: 10.1128/IAI.71.10.5921-5939.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pflock M, Finsterer N, Joseph B, Mollenkopf H, Meyer TF, Beier D. Characterization of the ArsRS regulon of Helicobacter pylori, involved in acid adaptation. Journal of bacteriology. 2006 May;188(10):3449–62. doi: 10.1128/JB.188.10.3449-3462.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Marcus EA, Sachs G, Wen Y, Feng J, Scott DR. The role of the Helicobacter pylori sensor kinase ArsS in protein trafficking and acid acclimation. Journal of bacteriology. 2012 Aug 3; doi: 10.1128/JB.01263-12. [DOI] [PMC free article] [PubMed] [Google Scholar]