Abstract
We found human T-cell leukemia virus type 1- and simian T-cell leukemia virus type 1 (STLV-1)-related infections in 5 of 10 chimpanzees originating from three groups of wild chimpanzees. The new virus isolates showed a surprising heterogeneity not only in comparison to STLV-1 described previously in other primate species but also between the different chimpanzee groups, within a group, or even between strains isolated from an individual animal. The interdisciplinary combination of virology, molecular epidemiology, and long-term behavioral studies suggests that the primary route of infection might be interspecies transmission from other primates, such as red colobus monkeys, that are hunted and consumed by chimpanzees.
The potential for interspecies transmission of retroviruses through predatory relationships has often been evoked (5, 9, 17, 19, 31, 32), although direct evidence for this route of transmission between primate species has been elusive. The primate T-cell leukemia virus (PTLV) is reported to be endemic in numerous human (HTLV) and nonhuman primate (STLV) populations (8, 11, 13, 15, 21, 22, 28, 30). Indirect evidence suggests interspecies transmission of PTLV among various simian species and between nonhuman primates and humans (3, 7, 11, 14, 26, 29, 30); in support of this hypothesis, simian and human isolates sometimes show greater homology than isolates from within the same species (11). We demonstrate in the present study a high genetic diversity of STLV type 1 (STLV-1) strains in chimpanzees from the Taï National Park and link molecular epidemiological to ecological and behavioral data to substantiate the direct interspecies transmission of PTLV-1 from undomesticated western red colobus monkey (Piliocolobus badius) to chimpanzees.
In order to understand the behavior and ecology of wild chimpanzees, Boesch and Boesch-Achermann have been observing the chimpanzees of the Taï National Park (Côte d'Ivoire, Western Africa) for more than 20 years (1). These chimpanzees inhabit partially overlapping territories of three adjacent habitats (10), and individual chimpanzees could be distinguished. Various studies concerning diet and feeding behavior showed that these chimpanzees hunt and consume other nonhuman primates (1). Moreover, with the exception of the fur, the entire prey is ingested, including bones and skull, which may induce bleeding within the oral cavity or digestive tract. Respectively, adult male and female chimpanzees consume an average of 186 and 25 g of simian prey per day (1). Adolescent and younger chimpanzees consume considerably less than adults and generally nibble on small pieces discarded by adults or obtained from their mothers. Among the 10 different simian species hunted by the chimpanzees, the red colobus monkey represents by far the most frequent prey (1) (80% of simian prey) (Fig. 1).
FIG. 1.
After hunting and killing a red colobus monkey (P. badius), chimpanzees consume almost all parts of their prey.
Based on these observations and on the fact that chimpanzees share close physiological homologies with humans, conditions favoring a potential exchange and rapid dissemination of pathogens between these species, the transmission of PTLV in this “predator-prey system” (i.e., chimpanzee-red colobus) may serve as a novel model for studying the evolution of diseases resulting from interspecies transmission.
Tissue and blood samples were collected from 10 chimpanzees belonging to the three study groups and nine western red colobus monkeys that had all died of natural causes in the research area of Taï National Park between 1997 and 2002 (Table 1). All samples were screened for the presence of PTLV-1 by using serological assays, as well as by molecular genetic analysis. Five of ten chimpanzees and five of nine red colobus monkeys tested positive for PTLV-1 antibodies; these animals showed a surprisingly high PTLV prevalence (Table 1). Because the long terminal repeat (LTR) and env are among the most variable regions of the HTLV and STLV genomes, these regions are frequently used for phylogenetic tree analyses. Primers SK43 (12) and LTR R6 (4) derived from the prototype HTLV-1 sequence ATK (23) were used in the primary PCR to amplify a 1.4-kb fragment from the 3′ end of the PTLV-provirus genome. The internal primers X (CAGATGACAATGACCATGAG) and H (TTCAGGAGGCACCACAGGC) were used for nested or seminested PCR analyses combining primers X with LTR R6 and SK43 with H. PCR conditions for first-round PCR and for seminested PCRs were used as previously described (13). From all serologically positive chimpanzees and from two red colobus monkeys a PCR product was obtained (Table 1). The env region from three chimpanzees and one red colobus was successfully amplified by using the primers S3 (CCTCAAGCGAGCTGCATGCCC) and SK44, followed by a nested PCR with the primers S4 (CTCCCTTCTAGTCGACGCTCCAGG) and R4 (GGAGTCCTTGGAGGCTGAACGG). DNA preparations from spleen, ganglion, and blood were performed for all chimpanzees and for two red colobus monkeys (animals 2/02 and 3/02) at the Robert Koch-Insitut on separate days, and only samples from one animal were treated at a time. DNA was stored in aliquots. DNA preparation from the remaining seven red colobus monkeys was performed at the University of Montpellier. Initially, PCRs were performed only on the chimpanzee-derived DNA samples. After a >2-month delay, the red colobus samples were analyzed. PCRs were performed individually for one animal at a time, and appropriate negative controls were always included. All PCR products were sequenced in both orientations by using the PCR primers, as well as internal primers. Once the initial PCR products were obtained and sequenced, a PCR was repeated at least two times on previously untouched aliquots of DNA from the same extraction (all individuals) and on different tissues, as well as on newly prepared DNA from tissue aliquots untouched since necropsy (all chimpanzees and red colobus monkeys 2/02 and 3/02); these results confirmed the previously obtained PCR results. All PCR products from positive samples were used for sequencing and confirmed the initially obtained sequence information. The sequences obtained were submitted to the public databases (accession numbers AY267829 to AY267838 and AY333376). Direct sequencing of the PCR product obtained for the LTR region from one of the chimpanzees (designated Rafiki) suggested a relatively high heterogeneity. Therefore, the PCR product was cloned into a PCR cloning vector, and several clones were sequenced. Two distinct sequences (Ptr-Rafiki-I and Ptr-Rafiki-II) were obtained. The comparison with published HTLV-1 and STLV-1 sequences showed that all PTLV sequences isolated from the five chimpanzees were significantly different from published chimpanzee and other PTLV-1 sequences (≥4% divergence). Interestingly, these six chimpanzee sequences also differed among each other, the divergence ranging between 2 and 5%. Also, the LTR sequences isolated from the two red colobus monkeys showed an unexpectedly high divergence among each other (3% comparing 547 bp) and were only distantly related to other PTLV-1 sequences. Surprisingly, the colobus monkey-derived LTR sequence (STLVwrc) was almost identical to that of Ptr-Dorry (0.2% divergence). Within the 1.4-kb tax/LTR sequence, only three nucleotide changes were identified. These differences were localized in the coding region of the second exon of tax but did not alter the amino acid sequence. In the 1-kb env fragments of Ptr-Dorry and STLVwrc, five nucleotide changes were identified (0.6% divergence), only one of them resulted in an amino acid substitution from asparagine to serine. One of the two STLV-1 sequences isolated from chimpanzee Rafiki (Ptr-Rafiki-I) was closely related to a sequence found in red colobus monkey 14/97. In a 422-bp fragment of the LTR region there was only a three-nucleotide difference between these two isolates (0.8%). Cross-contamination between chimpanzee tissues and red colobus tissues is highly unlikely. Necropsy of the red colobus (animal 02/02) occurred 6 months later than the necropsy of chimpanzee Dorry. Tissue samples were kept separate at any time. Tissues from the red colobus monkey 14/97 and chimpanzee Rafiki were collected almost 3 years apart (see Table 1) during different research projects. Although DNA from the red colobus monkey was extracted at the University of Montpellier, DNA from the chimpanzee was extracted at the Robert Koch-Insitut in Berlin. In addition, extensive precautions were taken to avoid cross-contamination during DNA preparations and PCR analysis.
TABLE 1.
Characteristics of the primates analyzed
| Primatea | Group or territory | Age (yr) or age level | Date of death (mo yr) | Sex | PTLV-1 serologyb | Sequence |
|---|---|---|---|---|---|---|
| Chimpanzees | ||||||
| Dorry | North | 10 | Oct 01 | F | + | Ptr-Dorry |
| Loukoum | North | ∼27 | May 99 | F | + | Ptr-Loukoum |
| Lefkas | North | 8 | May 99 | M | − | —d |
| Leonardo | North | 2 | June 99 | M | − | — |
| Leo | Middle | ∼29 | Feb 02 | M | + | Ptr-Leo |
| Kady | Middle | ∼30 | Dec 01 | F | + | Ptr-Kady |
| Noah | Middle | 7 | Feb 02 | M | − | — |
| Rafiki | South | ∼19 | Sept 00 | M | + | Ptr-Rafiki-I and II |
| Olduvai | South | 8 | June 02 | M | − | — |
| Tita | South | ∼25 | Aug 98 | F | − | — |
| Red colobus monkeys | ||||||
| 9/96 | North | Adult | Nov 96 | F | +* | — |
| 6/96 | North | Adult | Oct 96 | F | −* | NDc |
| 14/97 | Between South/North | Adult | Oct 97 | F | +* | STLVwrc-14/97 |
| 2/02 | Middle | Adult | Feb 02 | M | + | STLVwrc |
| 15/98 | Middle | Adult | Oct 98 | F | −* | ND |
| 9/97 | Middle/South | Adult | Aug 98 | M | −* | ND |
| 3/02 | South | Juvenile | March 02 | M | − | — |
| 4/98 | South | Adult | April 98 | M | +* | — |
| 04/00 | Adjacent territory | Adult | Unknown | M | + | ND |
Lefkas and Leonardo are sons of Loukoum.
Animals were tested by enzyme-linked immunosorbent assay and Western blotting. *, animals tested only by enzyme-linked immunosorbent assay.
ND, not determined (only serum was available).
—, negative PCR results.
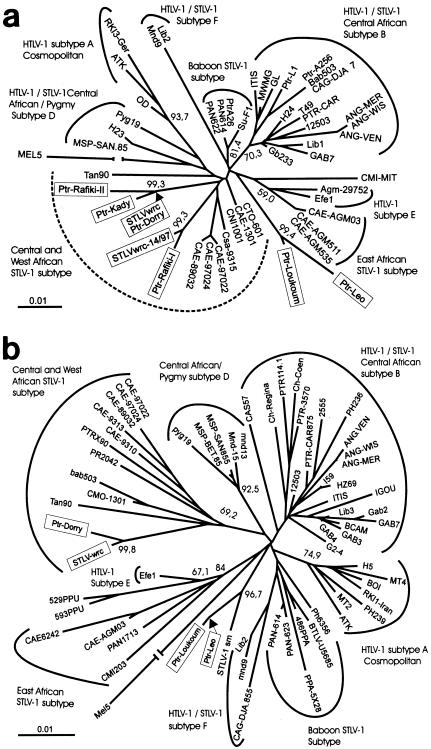
In contrast, the second STLV-1 sequence found in chimpanzee Rafiki (Ptr-Rafiki-II) was only distantly related to Ptr-Rafiki-I (4.3% divergence), as well as to the red colobus monkey 14/97 isolate. In addition to these differences, the Rafiki-I sequence showed a 48-bp duplication in the LTR region. However, in order to avoid a distortion of the results by calculating a distance matrix for phylogenetic tree analyses, this duplication was valued as only a 1-bp difference since it most likely represents a single mutation step. Phylogenetic analyses were performed with a 422-bp LTR fragment corresponding to nucleotides 8331 to 8745 of HTLV-1 ATK sequence (J02029) and for the env region by using a 522-bp fragment corresponding to nucleotides 6046 to 6567 of the ATK sequence. The sequences were aligned with the CLUSTAL program (27) and a DNA distance matrix was calculated. Using the neighbor-joining (24) (Fig. 2) and maximum-likelihood (data not shown) methods, we constructed phylogenetic trees. For the isolates from the Taï National Park, three distinct groups were identified in the phylogenetic tree. The Taï-I group consisted of Ptr-Loukoum clustering with Ptr-Leo, an isolate that was recently identified in a dead chimpanzee that had lived in a neighboring community in an adjacent, partially overlapping territory of the Taï forest (10). This group also clustered with an isolate detected previously in a sooty mangabey (5) (only the env sequence was available). The second group (Taï-II) included isolates from red colobus monkey 2/02 and chimpanzees Dorry and Kady, as well as the Ptr-Rafiki-II sequence. The Taï-III group included the sequence from red colobus monkey 14/97 and the second sequence obtained from chimpanzee Rafiki (Ptr-Rafiki-I).
FIG. 2.
Rooted phylogenetic tree of PTLV sequences. The tree was generated by the neighbor-joining method based on a 422-bp fragment that corresponds to the LTR (a) and a 522-bp fragment that corresponds to a region of env (b). The HTLV-1 subtype isolate Mel5 was used as an outgroup. The trees were statistically evaluated with a bootstrap analysis with 1,000 bootstraps. Bootstrap values for the major branch points are given as percentages. The different PTLV-1 subtypes are indicated as previously described (7, 10, 14, 15, 16, 21, 25). The strains identified in the chimpanzees and the red colobus monkey of the Taï Forest are highlighted by shaded boxes.
In general, HTLV-1 and STLV-1 isolates from the same region and species display close sequence homologies, reflecting the geographic origin of the isolates (25). Based on these observations, phylogenetic analyses are frequently used to determine the geographic origin of infections (4). The remarkable heterogeneity of PTLV-1 sequences detected in wild chimpanzees that had lived in three partially overlapping territories was highly unexpected. It is unlikely that the heterogeneity of the PTLV isolates found in the chimpanzees reflects sexual or mother-child dissemination among the chimpanzees, but rather this heterogeneity suggests multiple virus transmission from other sources. The data obtained here favor the hypothesis that red colobus monkeys are one of these sources. This hypothesis is supported by six observations: (i) the close homology between PTLV sequences identified (PTR-Dorry and PTR-Rafiki-I with STLVwrc and STLVwrc14/97, respectively); (ii) the close genetic relationship between env sequences from Ptr-Leo and a sooty mangabey isolate (U94516) (although uncommon, predation of sooty mangabeys by the Taï chimpanzees has been observed [1]); and (iii) the fact that Rafiki was infected with two divergent strains of PTLV-1, suggesting that each virus was transmitted separately to the chimpanzee. (Indeed, one of the isolates, Ptr-Rafiki-I, clustered with the PTLV-1 isolated from red colobus monkey 14/97 [STLVwrc-14/97], and the second sequence obtained from Rafiki with red colobus monkey 2/02 [STLVwrc], Ptr-Dorry, and Ptr-Kady, both supported by high bootstrap values.) (iv) The closest STLV-1 strain published so far was isolated from chimpanzees originating from other areas and displayed 4% divergence from the sequences identified in the present study. Thus, the STLV-1 isolates from the Taï National Park are at least as divergent from other STLV-1 sequences as from human isolates. These other chimpanzee STLV-1 strains mainly cluster with HTLV-1/STLV-1 Central African subtype B. (v) Within the study group, all samples from chimpanzees that died at an age younger than 8 years were negative for PTLV-1, including chimpanzees Leonardo and Lefkas, whose mother, chimpanzee Loukoum, was PTLV positive. This might be explained by the fact that young chimpanzees consume only small amounts of prey (1) that are less likely to cause injury when ingested. Furthermore, a study conducted on chimpanzees in captivity showed that mother-child transmission of PTLV has not been observed over a period of 20 years (18) in contrast to humans, among whom mother-child transmission, especially via breast-feeding, is a major route of HTLV transmission (16). (vi) Finally, four of five chimpanzees older than 10 years of age were PTLV-1 positive. This fact correlates infection profiles with behavioral patterns, demonstrating that adult chimpanzees are at a higher risk of becoming infected due to their direct predatory relationship with the red colobus monkeys, their consumption of larger amounts of prey (which are almost completely consumed), and the fact that masticated bones may induce cuts and bleeding within the oral cavity and digestive tract, thereby increasing the risk of infection. Even though transmission from one chimpanzee to another by sexual contact or injuries due to aggression cannot be excluded, no evidence for these routes of transmission was obtained in the present study.
Phylogenetic analyses of HTLV-1 and STLV-1 sequences suggest that interspecies transmission plays a role in the evolution of these retroviruses (3, 7, 11, 14, 26, 28, 30). We demonstrate here the presence of highly divergent STLV-1 strains in free-living chimpanzees and, for the first time, the presence of STLV-1 in Piliocolobus badius. Furthermore, behavioral and ecological observations and molecular epidemiological analysis demonstrated that interspecies transmission of PTLV-1 to chimpanzees most likely occurs as a result of predation and consumption of different monkey species. Multiple infections with different STLV-1 strains (as seen for chimpanzee Rafiki) could facilitate recombination between viruses and potentially lead to the selection of new variants, as described for human immunodeficiency virus (6, 20). It could be speculated that such mechanisms can alter the host range and the pathological potential of such viruses. The observed heterogeneity of the two red colobus monkey-derived STLV-1 sequences requires further investigation since red colobus monkeys, unlike the chimpanzees, are herbivores. Humans hunt numerous primate species in this part of Africa, including chimpanzees, colobus monkeys, and sooty mangabeys (2). Therefore, humans may be at increased risk for interspecies transmission of retroviruses (5, 9, 17, 19, 31, 32), just as suggested for the chimpanzees.
Acknowledgments
We thank the Ivorian authorities for long-term support, especially the Ministry of the Environment and Forests, as well as the Ministry of Research, the directorship of the Taï National Park, and the Swiss Research Center in Abidjan. We thank S. Neumann, H. von Spreckelsen, S. Pociuli, and H. Emmel for skillful technical support, L. Vigilant and F. Kim for helpful discussions, and U. Erikli for copy editing.
For the fieldwork F. H. Leendertz was awarded a grant from the German Academic Exchange Service.
REFERENCES
- 1.Boesch, C., and H. Boesch-Achermann. 2000. The chimpanzees of the Taï forest: behavioural ecology and evolution. Oxford University Press, Oxford, United Kingdom.
- 2.Caspary, H. U., I. Koné, C. Prouot, and M. de Pauw. 2001. La chasse et la filière viande de brousse dans l'espace Taï, Côte d'Ivoire. Tropenbos-Côte d'Ivoire Série 2. Tropenbos International, Wageningen, The Netherlands.
- 3.Crandall, K. A. 1996. Multiple interspecies transmissions of human and simian T-cell leukemia/lymphoma virus type I sequences. Mol. Biol. Evol. 13:115-131. [DOI] [PubMed] [Google Scholar]
- 4.Ellerbrok, H., C. Fleischer, M. Salemi, P. Reinhardt, W. D. Ludwig, A. M. Vandamme, and G. Pauli. 1998. Sequence analysis of the first HTLV-1 infection in Germany without relations to endemic areas. AIDS Res. Hum. Retrovir. 14:1199-1203. [DOI] [PubMed] [Google Scholar]
- 5.Fultz, P. N., L. Su, P. May, and J. T. West. 1997. Isolation of sooty mangabey simian T-cell leukemia virus type I [STLV-I(sm)] and characterization of a mangabey T-cell line coinfected with STLV-I(sm) and simian immunodeficiency virus SIVsmmPBj14. Virology 235:271-285. [DOI] [PubMed] [Google Scholar]
- 6.Gao, F., E. Bailes, D. L. Robertson, Y. Chen, C. M. Rodenburg, S. F. Michael, L. B. Cummins, L. O. Arthur, M. Peeters, G. M. Shaw, P. M. Sharp, and B. H. Hahn. 1999. Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature 397:436-441. [DOI] [PubMed] [Google Scholar]
- 7.Georges-Courbot, M. C., P. Moisson, E. Leroy, A. M. Pingard, E. Nerrienet, G. Dubreuil, E. J. Wickings, F. Debels, I. Bedjabaga, V. Poaty-Mavoungou, N. T. Hahn, and A. J. Georges. 1996. Occurrence and frequency of transmission of naturally occurring simian retroviral infections (SIV, STLV, and SRV) at the CIRMF Primate Center, Gabon. J. Med. Primatol. 25:313-326. [DOI] [PubMed] [Google Scholar]
- 8.Gessain, A., and R. Mahieux. 2000. Épidemiologie, origine et diversité génétique du rétrovirus HTLV-1 et des rétrovirus simiens apparentés à STLV-1. Bull. Soc. Pathol. Exot. 93:163-171. [PubMed] [Google Scholar]
- 9.Hahn, B. H., G. M. Shaw, K. M. De Cock, and P. M. Sharp. 2000. AIDS as a zoonosis: scientific and public health implications. Science 287:607-614. [DOI] [PubMed] [Google Scholar]
- 10.Herbinger, I., C. Boesch, and H. Rothe. 2001. Territory characteristics among three neighboring chimpanzee communities in the Taï National Park, Côte d'Ivoire. Int. J. Primatol. 22:143-167. [Google Scholar]
- 11.Koralnik, I. J., E. Boeri, W. C. Saxinger, A. L. Monico, J. Fullen, A. Gessain, H. G. Guo, R. C. Gallo, P. Markham, and V. Kalyanaraman. 1994. Phylogenetic associations of human and simian T-cell leukemia/lymphotropic virus type I strains: evidence for interspecies transmission. J. Virol. 68:2693-2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kwok, S., G. Ehrlich, B. Poiesz, R. Kalish, and J. J. Sninsky. 1988. Enzymatic amplification of HTLV-1 viral sequences from peripheral blood mononuclear cells and infected tissues. Blood 72:1117-1123. [PubMed] [Google Scholar]
- 13.Leendertz, F. H., C. Boesch, S. Junglen, G. Pauli, and H. Ellerbrok. 2003. Characterisation of a new simian T-lymphocyte virus type 1 in a wild living chimpanzee (Pan troglodytes verus) from Ivory Coast: evidence for a new STLV-1 group? AIDS Res. Hum. Retrovir. 19:255-258. [DOI] [PubMed] [Google Scholar]
- 14.Liu, H. F., P. Goubau, M. Van Brussel, K. Van Laethem, Y. C. Chen, J. Desmyter, and A. M. Vandamme. 1996. The three human T-lymphotropic virus type I subtypes arose from three geographically distinct simian reservoirs. J. Gen. Virol. 77:359-368. [DOI] [PubMed] [Google Scholar]
- 15.Mahieux, R., C. Chappey, L. Meertens, P. Mauclere, J. Lewis, and A. Gessain. 2000. Molecular characterization and phylogenetic analyses of a new simian T-cell lymphotropic virus type 1 in a wild-caught African baboon (Papio anubis) with an indeterminate STLV type 2-like serology. AIDS Res. Hum. Retrovir. 16:2043-2048. [DOI] [PubMed] [Google Scholar]
- 16.Miyoshi, I., H. Muneishi, and Y. Tanaka. 1992. Vertical transmission of HTLV-1. Lancet 339:305-306. [DOI] [PubMed] [Google Scholar]
- 17.Nerrienet, E., L. Meertens, A. Kfutwah, Y. Foupouapouognigni, and A. Gessain. 2001. Molecular epidemiology of simian T-lymphotropic virus (STLV) in wild-caught monkeys and apes from Cameroon: a new STLV-1, related to human T-lymphotropic virus subtype F, in a Cercocebus agilis. J. Gen. Virol. 82:2973-2977. [DOI] [PubMed] [Google Scholar]
- 18.Niphuis, H., E. J. Verschoor, I. Bontjer, M. Peeters, and J. L. Heeney. 2003. Reduced transmission and prevalence of simian T-cell lymphotropic virus in a breeding colony of chimpanzees (Pan troglodytes verus). J. Gen. Virol. 84:615-620. [DOI] [PubMed] [Google Scholar]
- 19.Peeters, M., V. Courgnaud, B. Abela, P. Auzel, X. Pourrut, F. Bibollet-Ruche, S. Loul, F. Liegeois, C. Butel, D. Koulagna, E. Mpoudi-Ngole, G. M. Shaw, B. H. Hahn, and E. Delaporte. 2002. Risk to human health from a plethora of simian immunodeficiency viruses in primate bushmeat. Emerg. Infect. Dis. 8:451-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robertson, D. L., B. H. Hahn, and P. M. Sharp. 1995. Recombination in AIDS viruses. J. Mol. Evol. 40:249-259. [DOI] [PubMed] [Google Scholar]
- 21.Saksena, N. K., V. Herve, J. P. Durand, B. Leguenno, O. M. Diop, J. P. Digouette, C. Mathiot, M. C. Muller, J. L. Love, and S. Dube. 1994. Seroepidemiologic, molecular, and phylogenetic analyses of simian T-cell leukemia viruses (STLV-I) from various naturally infected monkey species from central and western Africa. Virology 198:297-310. [DOI] [PubMed] [Google Scholar]
- 22.Salemi, M., J. Desmyter, and A. M. Vandamme. 2000. Tempo and mode of human and simian T-lymphotropic virus (HTLV/STLV) evolution revealed by analyses of full-genome sequences. Mol. Biol. Evol. 17:374-386. [DOI] [PubMed] [Google Scholar]
- 23.Seiki, M., S. Hattori, Y. Hirayama, and M. Yoshida. 1983. Human adult T-cell leukemia virus: complete nucleotide sequence of the provirus genome integrated in leukemia cell DNA. Proc. Natl. Acad. Sci. USA 80:3618-3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 25.Slattery, J. P., G. Franchini, and A. Gessain. 1999. Genomic evolution, patterns of global dissemination, and interspecies transmission of human and simian T-cell leukemia/lymphotropic viruses. Genome Res. 9:525-540. [PubMed] [Google Scholar]
- 26.Song, K. J., V. R. Nerurkar, N. Saitou, A. Lazo, J. R. Blakeslee, I. Miyoshi, and R. Yanagihara. 1994. Genetic analyses and molecular phylogeny of simian T-cell lymphotropic virus type I: evidence for independent virus evolution in Asia and Africa. Virology 199:56-66. [DOI] [PubMed] [Google Scholar]
- 27.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vandamme, A. M., H. F. Liu, M. Van Brussel, W. De Meurichy, J. Desmyter, and P. Goubau. 1996. The presence of a divergent T-lymphotropic virus in a wild-caught pygmy chimpanzee (Pan paniscus) supports an African origin for the human T-lymphotropic/simian T-lymphotropic group of viruses. J. Gen. Virol. 77:1089-1099. [DOI] [PubMed] [Google Scholar]
- 29.Voevodin, A., E. Samilchuk, H. Schatzl, E. Boeri, and G. Franchini. 1996. Interspecies transmission of macaque simian T-cell leukemia/lymphoma virus type 1 in baboons resulted in an outbreak of malignant lymphoma. J. Virol. 70:1633-1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Voevodin, A. F., B. K. Johnson, E. I. Samilchuk, G. A. Stone, R. Druilhet, W. J. Greer, and C. J. Gibbs. 1997. Phylogenetic analysis of simian T-lymphotropic virus type I (STLV-1) in common chimpanzees (Pan troglodytes): evidence for interspecies transmission of the virus between chimpanzees and humans in Central Africa. Virology 238:212-220. [DOI] [PubMed] [Google Scholar]
- 31.Weiss, R. A., and R. W. Wrangham. 1999. From Pan to pandemic. Nature 397:385-386. [DOI] [PubMed] [Google Scholar]
- 32.Wolfe, N. D., A. A. Escalante, W. B. Karesh, A. Kilbourn, A. Spielman, and A. A. Lal. 1998. Wild primate populations in emerging infectious disease research: the missing link? Emerg. Infect. Dis. 4:149-159. [DOI] [PMC free article] [PubMed] [Google Scholar]