Abstract
The genome of the fish pathogen Aeromonas salmonicida subsp salmonicida harbors a large number of insertion sequences (ISs), many of which are located on plasmids. In the present study, we analyzed the small plasmid profile of A. salmonicida strains to identify evidences of plasmid alterations. Ten out of 78 strains analyzed displayed an unconventional plasmid profile. However the HER1104 strain was unique, having a positive PCR signal for pAsal1 plasmid despite not carrying this plasmid. Instead, HER1104 was bearing a plasmid at higher molecular weight than pAsal1. We characterized this new larger plasmid, which we called pAsal1B since it is a derivative of pAsal1 containing one more complete IS (ISAS5) than the parental plasmid. An additional 96 bp relic of ISAS5 was also present in pAsal1B. These results propose that ISAS5 is another active mobile genetic element in A. salmonicida subsp salmonicida and provided further proof of the genomic plasticity of this bacterium.
Keywords: furunculosis, Aeromonas salmonicida, plasmid, DNA rearrangement, insertion sequence
Introduction
Aeromonas salmonicida subsp salmonicida (A. salmonicida) is the Gram-negative bacterium responsible for furunculosis, a disease that mainly affects fish in the salmonid family.1,2
A. salmonicida strains usually contain many plasmids. Three small high-copy-number plasmids [pAsa1 (5.4 kb), pAsa2 (5.2 kb) and pAsa3 (5.6 kb)] have been found in the vast majority of the A. salmonicida strains that have been analyzed.3,4 These cryptic plasmids only bear genes for their own replication, mobilization and stability. pAsal1, another high-copy-number plasmid, is also commonly found in A. salmonicida strains.4,5 While this 6.4 kb plasmid is very similar to pAsa3, it also possesses the aopP gene that encodes a type three secretion system (TTSS) effector.5,6 The aopP gene is specific to pAsal1. This plasmid also possesses one copy of ISAS11, an insertion sequence (IS).7 These four plasmids represent the conventional set of plasmids seen in A. salmonicida strains and can easily be analyzed by EcoRI digestion followed by electrophoresis.4
A 155 kb low-copy-number plasmid (pAsa5) is also present in most A. salmonicida strains. It carries the vast majority of TTSS genes.8,9 Other plasmids such as pASOT, pASOT2, pASOT3, pRAS1, pRAS3.1, pRAS3.2, pAr-32, pAsa4 and pAsa6, which range in size from 11 to 167 kb, have also been identified in sub-groups of A. salmonicida strains.9-14 Most of these occasional plasmids carry antibiotic resistance genes.
The genomic plasticity of A. salmonicida is likely IS dependent given the involvement of ISs in gene inactivation and plasmid rearrangements15 and the fact that the A. salmonicida genome contains a large number of ISs.9 The vapA gene, which encodes the A protein involved in the A-layer at the surface of the bacterium, can be disrupted by the spontaneous transposition of ISAS1 and ISAS2.16,17 ISAS1 can also be inserted in abcA, a gene involved in O-polysaccharide synthesis and transport.18 Recently, it has been shown that three ISAS11 of pAsa5 trigger the rearrangement of this plasmid, resulting in the excision of the TTSS locus (loss profile 1) or the excision of this locus with an additional region of the plasmid (loss profile 2).7,8 These rearrangements of pAsa5 provoke the loss of virulence by A. salmonicida.8 More complex rearrangement profiles of pAsa5 has been also observed for some A. salmonicida strains, suggesting that ISs other than ISAS11 may be active in provoking multiple, sequential rearrangements of pAsa5.8,15
In the present study, we wanted to bring additional evidences supporting the hypothesis that the high content of mobile genetic elements in the genetic material of A. salmonicida might contribute to frequent rearrangements and rapid evolution of the bacterium’s genome. Therefore, we analyzed the small plasmids in A. salmonicida strains in order to identify unusual plasmid profiles that might provide evidence for the genomic plasticity of this bacterial species. This work led to the characterization of a new plasmid showing that ISAS5 is another active mobile genetic element in A. salmonicida.
Results
The small plasmid profile of 78 A. salmonicida strains was analyzed in order to identify those displaying unconventional plasmid profiles. After EcoRI digestion, the electrophoretic patterns of plasmids under 50 kb in size revealed that 68 of the 78 strains had bands corresponding to pAsa1, pAsa2, pAsa3 and pAsal1 (data not shown). This is considered the common profile for small plasmids in A. salmonicida.4 Moreover, all the strains with the common set of small plasmids also tested positive by PCR for the aopP gene, which is on the pAsal1 plasmid (data not shown).
On the other hand, 10 strains displayed unusual plasmid profiles (i.e., additional bands other than those of the common profile were present while others were absent) (Table 1). Five strains (HER1085, 2010-47K18, 2009-157K5, 2009-195K29 and 2009–144K3) displayed the bands for the complete set of common small plasmids but also had an additional band either at 14 or 11 kb. Two strains (HER1110 and A449) exhibited a smaller set of plasmids than expected. In the case of HER1110, only the pAsa3 plasmid was present, while A449 was missing the pAsal1 plasmid as previously reported.9 The band corresponding to pAsal1 was also missing in HER1084 but this strain had an additional band at 7.6 kb. The 2004–208 strain did not harbor the pAsa3 plasmid, but its plasmid profile included two additional bands at 3.0 and 7.6 kb. Lastly, HER1104 contained pAsa1, pAsa2, and pAsa3 but not pAsal1. However, HER1104 had a fourth band corresponding to a plasmid about 8.5 kb (Fig. 1).
Table 1. Small plasmid profiles for the A. salmonicida strains displaying an unconventional plasmid profile following EcoRI digestion.
| Name | Origina | Source and/or referenceb | Plasmid profilec | aopP by PCRd | Tetracycline resistancee |
|---|---|---|---|---|---|
| 01-B526 |
Brook trout (QC, Canada) |
FMVUM19 |
Cryptic + pAsal1 |
P |
S |
| HER1110 |
*(Japan) |
FHRC20 |
pAsa3 only |
N |
S |
| HER1104 |
*(France) |
FHRC |
Cryptic + 8.5 kb |
P |
S |
| HER1085 |
* |
FHRC |
Cryptic + pAsal1 + 14 kb |
P |
S |
| HER1084 |
*(France) |
FHRC20 |
Cryptic + 7.6 kb |
N |
S |
| A449 |
Brown trout (France) |
21 |
Cryptic |
N |
R |
| 2009–157K5 |
Brook trout (NB, Canada) |
FOC |
Cryptic + pAsal1 + 11 kb |
P |
R |
| 2010–47K18 |
Brook trout (NB, Canada) |
FOC |
Cryptic + pAsal1 + 11 kb |
P |
R |
| 2009–195K29 |
Brook trout (NB, Canada) |
FOC |
Cryptic + pAsal1 + 11 kb |
P |
R |
| 2009–144K3 |
Brook trout (NB, Canada) |
FOC |
Cryptic + pAsal1 + 11 kb |
P |
R |
| 2004–208 | * | FOC | pAsa1 + pAsa2 + pAsal1 +3.0 kb + 7.6 kb | P | S |
The 01-B526 strain is shown as an example of strain displaying the conventional profile. a*Further information not known or not traceable. bFMVUM: Laboratoire de bactériologie clinique, Faculté de médecine vétérinaire, Université de Montréal (Montreal, QC, Canada); FHRC: Félix d’Hérelle Reference Center, Département de biochimie, de microbiologie et de bio-informatique, Université Laval (Quebec City, QC, Canada); MAPAQ: Ministère de l'Agriculture, des Pêcheries et de l'Alimentation du Québec; FOC: Aquatic Animal Health Department, Fisheries and Oceans Canada (NB, Canada); NB: New Brunswick; NS: Nova Scotia; QC: Quebec. cCryptic means that the bands for the pAsa1, pAsa2 and pAsa3 plasmids were visible on the gel. The sizes of bands other than pAsa1, pAsa2, pAsa3, or pAsal1 are indicated in kb. dP: positive PCR; N: negative PCR. eR: resistant; S: sensitive
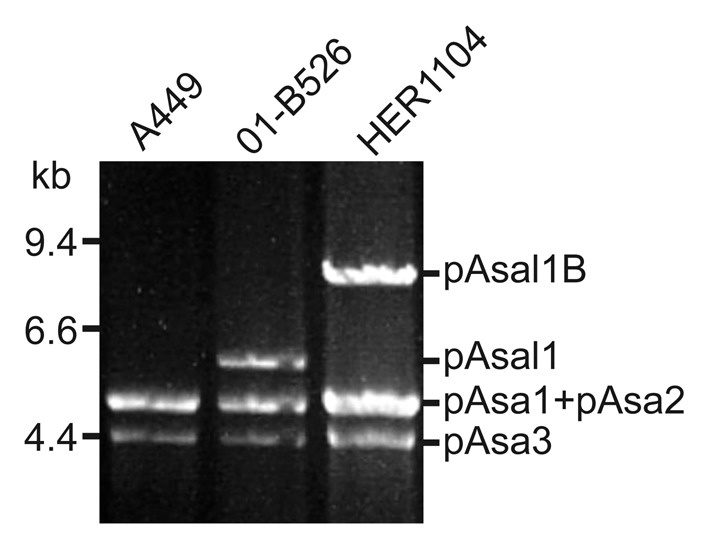
Figure 1. HER1104 displays an uncommon small plasmid profile. Plasmids under 50 kb in size were extracted from A. salmonicida A449, 01-B526, and HER1104 and were digested with EcoRI. The digested DNA samples were separated by pulsed field gel electrophoresis. The three strains tested contained pAsa1, pAsa2, and pAsa3. It was already known that A449 does not possess pAsal1 while this plasmid is present in 01-B526. 4,7 EcoRI-digested HER1104 DNA does not contain a band corresponding to pAsal1 but does contain a band corresponding to an unknown plasmid with a higher molecular weight, which was named pAsal1B after further analyses.
Four strains (2010-47K18, 2009-157K5, 2009-195K29 and 2009-144K3) from New Brunswick (Canada) displayed the same profile, with an additional band at about 11 kb detected following the EcoRI digestion (Table 1). These four strains were resistant to tetracycline (Table 1). The pRAS3 plasmid (11.8 kb) is known to bear a TetC gene that confers resistance to tetracycline.11 We confirmed by PCR analyses that 2010-47K18, 2009-157K5, 2009-195K29 and 2009-144K3 bear this plasmid (data not shown), which is likely the one detected at 11 kb after electrophoresis. The A449 strain also displayed a resistance to tetracycline, but in this case this is due to the presence of a tetracycline resistance gene found on its pAsa4 plasmid.9
Strains without pAsal1 were negative by PCR for the presence of the aopP gene, except for HER1104 (Table 1). In theory, it was expected that the aopP gene was present only in strains displaying pAsal1. This discrepancy between the plasmid profile and the PCR result for HER1104 set this strain apart from all others we analyzed, and suggested that elements from pAsal1 are still present in the genome of HER1104. Since pAsal1 contains ISAS11, which is known to be active in genome rearrangements,7 we suspected that the additional 8.5 kb band in HER1104 may be the result of modifications to pAsal1. The 8.5 kb band was thus isolated and characterized.
DNA restriction fragments encompassing the entire 8.5 kb plasmid were cloned and sequenced. The sequence analysis revealed a high level of identity with the sequence of pAsal1 and ISAS5. This unexpected result suggested that the additional plasmid in HER1104 might be a combination of pAsal1 and ISAS5. This 2614 bp IS belongs to the IS21 family.9,22 A449 contains 12 complete and three partial versions of ISAS5 in its genome.9
To obtain the complete sequence of the additional plasmid in HER1104, primers were designed from the sequences of the clones. The sequences obtained with these primers made it possible to identify the insertion site of ISAS5 in the plasmid. A second set of primers was designed to fill in the gaps and to get a complete circular sequence. Due to its high degree of similarity with pAsal1, the new plasmid (8989 bp) was named pAsal1B. The nucleotide sequence of pAsal1B was deposited in DDBJ/EMBL/GenBank under accession number KC686700.
The linear maps of pAsal1 and pAsal1B are shown in Figure 2. While the two plasmids were nearly identical, pAsal1B contained two ISAS5 elements. One was complete and was identical to other ISAS5 in the genome of A. salmonicida. It was inserted in mobA, resulting in the interruption of this gene. A fragment of ISAS5 was also found in the non-coding region between the repA and the mobA genes in pAsal1B (Fig. 2). This 96 bp fragment corresponded to part of the non-coding region upstream from the istA gene in ISAS5.22 This ISAS5 fragment was associated with the absence of a 121 bp sequence ranging from position 603 through 723 of pAsal1, which was replaced by the ISAS5 portion.
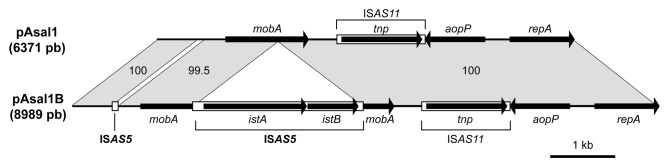
Figure 2. Maps of the pAsal1 and pAsal1B plasmids. Genes are illustrated by arrows while IS elements are illustrated by rectangles. The numbers between the two plasmids indicate the identity (in %) for the plasmid regions indicated. As shown by the maps, pAsal1B contains all the elements found in pAsal1 except that a 121 bp fragment was replaced by the insertion of the 96 bp ISAS5 fragment in the first third of the plasmid. In addition, pAsal1B bears a complete ISAS5 inserted in the mobA gene.
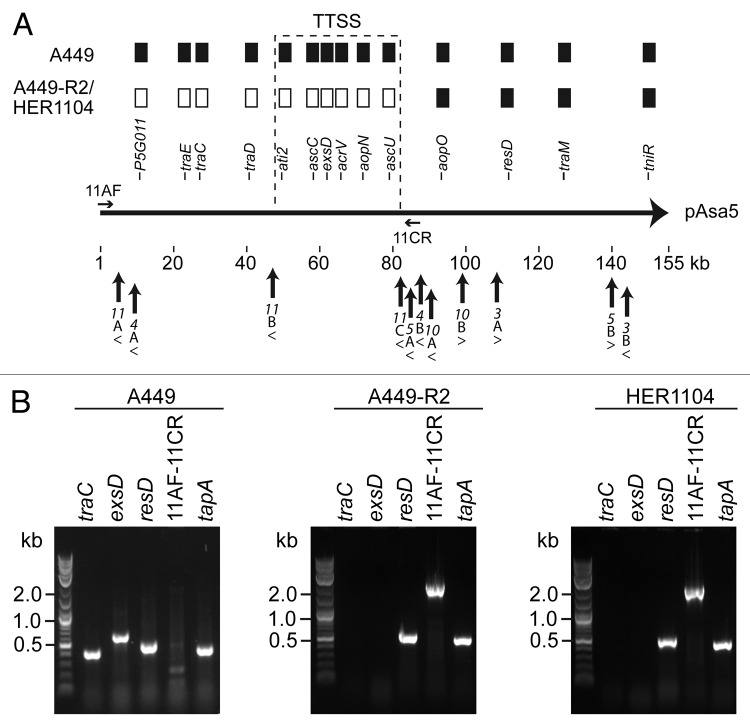
It has already been reported that pAsa5 in HER1104 is altered.8 Under stressful conditions, two typical pAsa5 alterations are most likely produced, that is, loss profile 1 and loss profile 2.7,8 As already mentioned, loss profile 1 results from the deletion of the TTSS locus, while loss profile 2 results from an additional 40 kb deletion upstream from this locus.7 The alteration of pAsa5 in HER1104 fits the loss profile 2 category (Fig. 3A). Loss profile 2 was observed less frequently than loss profile 1. However, until now, all loss profile 2 plasmids have been explained by homologous recombination of flanking ISAS11 (A and C).7
Figure 3. Analysis of pAsa5 rearrangements in HER1104. (A) Map of pAsa5 with rearrangements in A449-R2 and HER1104. As already shown 7,8, both strains displayed a loss profile 2 based on PCR analyses of 14 pAsa5 genes shown as filled rectangles on this figure when present. The positions of the primers (11AF and 11CR) used to study the pAsa5 rearrangements are also shown by horizontal arrows. The primers shown are not to scale. Upright arrows indicate IS positions on pAsa5 while arrowheads indicate the orientation of the ISs. (B) PCR analyses. Strains tested and primers used are indicated above the images of the gels. The primer pairs traC, exsD and resD targeted the corresponding genes in pAsa5, while the tapA primers were used as positive chromosome hybridization controls.
To confirm that pAsa5 loss profile 2 in HER1104 was due to the recombination of ISAS11A with ISAS11C, 11AF-11CR PCR was performed to amplify the recombined ISAS11A/C as well as the flanking sequences on either side of the reclosed plasmid. 11AF-11CR PCR analyses of the A449 and A449-R2 strains served as controls. The A449-R2 strain, which was derived from A449 and displayed a pAsa5 loss profile 2 due to the recombination of ISAS11A with ISAS11C,7 was used as a positive control. HER1104 generated the same PCR products as A449-R2, confirming that the loss profile 2 seen in HER1104 is also caused by an ISAS11 recombination (Fig. 3B).
Discussion
The goal of the present study was to discover additional evidence of the genomic plasticity of A. salmonicida. We thus analyzed the small plasmid profiles of 78 strains from different parts of the world, but mainly from Canada. The vast majority of the strains harbored only the cryptic plasmids pAsa1, pAsa2 and pAsa3 in addition to pAsal1. Strains with an unusual profile can be classified into two categories: (1) strains grown in stressful conditions and (2) strains possessing antibiotic resistance related to the presence of a small plasmid bearing an antibiotic resistance gene.
The plasmid rearrangements and plasmid losses seen in HER1110, HER1104, HER1085 and HER1084 were the result of growing the parental strain in stressful conditions (i.e., grown repeatedly at temperatures above 22°C).7,8 The absence of pAsal1 in A449 can also be explained by being grown in stressful conditions since this strain was isolated a long time ago and was commonly grown at 25°C before it has been included in this study, which is not suitable for preserving the integrity of plasmids in A. salmonicida.7,8,23 On the other hand, no unconventional plasmid profile was detected for the 57 bacterial strains isolated in Province of Quebec in the last five years. In the same period, clear instructions have been given to veterinarians and veterinary microbiologists to manipulate and grow these isolates under appropriate conditions (i.e., at temperatures below 20°C), which was not the case before. All together, these observations support the idea that stressful growth conditions promote genomic plasticity in A. salmonicida.
The present study is the first to describe the mobility of IS from the IS21 family (ISAS5 in this case) in A. salmonicida. Moreover, the presence of a partial ISAS5 on pAsal1B compared with pAsal1 provides support for the idea that ISs in A. salmonicida can generate incomplete and complex integrations and rearrangements as already suggested.7,15 If the two ISAS5-related sequences are not taken into consideration, the sequence identity between pAsal1 and pAsal1B is higher than 99.9%, indicating that pAsal1B is a direct derivative of pAsal1 in which two ISAS5 (one complete and one partial) have been integrated.
A cluster of four different ISs located downstream from the TTSS locus in pAsa5 (Fig. 3A) may be the source of the ISAS5 observed in pAsal1B. In this cluster, ISAS11C is directly followed by an ISAS5 (ISAS5A). It is thus possible that, due to its relative proximity, the ISAS11 recombination detected in pAsa5 promoted the mobility of ISAS5A and its activation and transposition in pAsal1B. However, since the 11CR primer hybridizes in ISAS5A, we cannot infer that this IS copy is the source of the transposition event into pAsal1 that generated pAsal1B. If ISAS5A is the source of the ISAS5 in pAsal1B, this event occurred by replicative transposition, a mechanism already observed in the IS21 family.22 In fact, the presence of mobile genetic elements of the IS21 family such as ISAS5 has been reported in numerous bacterial22 and even Archaea24 genomes. These ISs have been shown to be involved in different types of genetic rearrangements, including recombinations, the creation of new composite transposons, replicon fusion, interplasmid recombinations and deletions by intramolecular transposition.22,25-27
The transposition of ISAS5 in the pAsal1 replicon reported here is another illustration of the genetic activity of these IS and their impact on the genome of their host. IS21-related genetic events can occasionally leave remnants of the IS in the “donor” DNA when the transposition mechanism is defective. These sequence relics have been reported in every genome bearing these transposons, including A. salmonicida.9,22,24,28,29 The 96 bp portion of the ISAS5 sequence detected in pAsal1B is an example of an aborted transposition. The fact that two transposition events by ISAS5 occurred in the pAsal1 replicon confirms the involvement of this IS in A. salmonicida genome plasticity and reinforces the idea that the high content of mobile genetic elements in the genetic material of this species might indicate that A. salmonicida is subject to frequent rearrangements and rapid evolution.
The discovery and characterization of pAsal1B revealed for the first time the mobile nature of ISAS5 in A. salmonicida. It is the fourth IS, after ISAS1, ISAS2 and ISAS1115 for which experimental evidences indicate the involvement of an IS in gene inactivation or plasmid rearrangements through its transposition in this bacterium. The present study also provided additional proofs demonstrating that the genomic plasticity of A. salmonicida is complex. As already mentioned, stressful conditions lead to the activation of plasmid rearrangements in A. salmonicida.8 In the future, it would be interesting to determine which conditions specifically provoke the mobilization of ISAS5.
Material and Methods
Bacterial strains and growth conditions
The A. salmonicida strains used in our study are listed in Table S1. In addition to these strains, A449-R2 was also used.7 The strains were grown on furunculosis agar medium (10 g of bacto-tryptone, 5 g of yeast extract, 1 g of l-tyrosine, 2.5 g of NaCl and 15 g of agar in one liter of water) at 18°C for 3 d. Escherichia coli MC1061 was used for cloning.
Plasmid DNA isolation and restriction enzyme profiles
Plasmid miniprep kits (Feldan, 9K-006-0010) were used as recommended by the manufacturer to extract < 50 kb plasmids from the strains listed in Table 1. The extracts (25 µl) were digested with EcoRI [New England Biolabs (NEB), R3101S]. The digested DNA samples were separated either by regular gel electrophoresis or by pulsed field gel electrophoresis with the following parameters: a pulse time ranging from to 0.2 to 13.0 sec, a run time of 16 h at 6 V/cm and a 120° angle. The agarose gels were stained with ethidium bromide to visualize the DNA bands under UV illumination.
Tetracycline resistance analyses
Strains listed in Table 1 were analyzed for their resistance to tetracycline. A homogeneous bacterial lawn of 108 bacteria was made on Mueller-Hinton agar medium (Oxoid, CM0337). The discs containing 5 µg tetracycline (BD, B31343) were put on the petri dish. Afterwards, the plates were incubated at 18°C. The results were taken an incubation of 24 h and were interpreted depending on the presence or absence of a growth inhibition zone.
PCR analyses
PCR analyses were performed to test the presence of the aopP gene in all the A. salmonicida strains analyzed, to confirm the presence of pRAS3 in some strains, and to verify the rearrangement status of pAsa5 in HER1104 strain. The PCR primers used are listed in Table 2. DNA templates were prepared by lysing one bacterial colony of every A. salmonicida strain in 25 µl of SWL buffer (50 mM KCl, 10 mM Tris, pH 8.3, 2.5 mM MgCl2, 0.45% NP-40 and 0.45% Tween 20).31 The lysates were heated at 95°C for 5 min. The PCR mixture contained 4 µl of 5 × Go-Taq buffer (Promega, M791A), 1.6 µl of 2 mM dNTP, 1.3 µl of forward and reverse primers (100 ng/µl each), 0.1 µl of GoTaq [5 U, (Promega, M3005)], 10.7 µl of H2O and 1 µl of DNA template. The PCR program was as follows: 2 min 30 sec at 95°C, 30 cycles of 30 sec at 95°C, 30 sec at 60°C and 2 min at 68°C, followed by a final extension for 5 min at 68°C. The samples were separated on 1% agarose gels, which were stained with 0.5 µg/ml ethidium bromide. The PCR reactions were performed at least twice.
Table 2. Primers used in this study.
| Primer name | Sequence (5′-3′) | Ref. |
|---|---|---|
| Primers used for the walk on pAsal1B | ||
| MT5-R1 |
AAGTCCTCGATCTCGTCCCTGATA |
This study |
| MT7-R1 |
TTGTGGAGCGTGTTTCTCGTAGCC |
This study |
| MT9-R1 |
AGGACCGACAACCAGAACTTTGCC |
This study |
| MT11-F1 |
TTCATCTACTGGCACCATGAGCCT |
This study |
| MT13-R1 |
GTTTGATACATCCAATGGGAAGGG |
This study |
| MT15-F1 |
TGGCCTATCGCCATATCCAACACA |
This study |
| MT18-R2 |
TGTTGGATATGGCGATAGGCCAGT |
This study |
| MT19-R1 |
CTCTATGTGCTTGCACCTGGCTAA |
This study |
| MT21-F1 |
CTCACGTCATCTCGTTTCTAACT |
This study |
| MT22-R1 |
GTCACTGAAGGGAGAATCGATGAG |
This study |
| MT23-F1 |
CGGTCAGTTTCAGGTTTCTCAGG |
This study |
| MT24-R1 |
GTCCTTACAAGCCCAAGGACAAG |
This study |
| MT25-F1 |
TGCGTTAAGGATGATTCTGGTGAA |
This study |
| MT26-R1 |
GACCAATGTGAACGCTCATTCC |
This study |
| MT27-F1 |
CTTGCTCGATCTGATCCCATATCC |
This study |
| MT28-R1 |
GATACAGCAGGCTTCAAAGAAGT |
This study |
| MT29-F1 |
TCTTTAGTTGCCTCTTGGATTG |
This study |
| MT30-R1 |
CGGAATGAAGTGTGATGTTCTTG |
This study |
| MT31-F1 |
TTGTGTTGGATATGGCGATAGG |
This study |
| MT32-R1 |
TTAGAGAGTACTGGCGGCCTAA |
This study |
| MT33-F1 |
CTTCCAGCAGTTCCCGATTTAT |
This study |
| MT33-F2 |
TAACGTTACTCTCCTGCTGTTTG |
This study |
| MT34-F1 |
CTATCGATGCCAATCAGCAAAC |
This study |
| MT34-F2 |
ATCGATGCCAATCAGCAAAC |
This study |
| MT35-R1 |
CATGGATGATCGGTATGGGAAATA |
This study |
| MT35-R2 |
GGTCAACTGGCAAGGATAGAG |
This study |
| MT36-R1 |
CAAACCAGCTTCCCATCAGTAA |
This study |
| MT36-R2 |
CATATCCAGCTCATGCCAAAC |
This study |
| Primers used for the PCR analyses | ||
| To test the presence of the aopP gene | ||
| pAsal1 F |
TAACATGGGTGAGTCAGGA |
4 |
| pAsal1 R |
TGCATGTTTGTAAAAAGTAGGTG |
4 |
| To test the presence of pRAS3 | ||
| pRAS3deb-F |
CATGAGCATTGCGGTAGCACTCAA |
This study |
| pRAS3deb-R |
TCGCTTGCGGGAACTTCTCATACT |
This study |
| pRAS3mil-F |
GCGGGTCGAACAAATCTGGTTCTT |
This study |
| pRAS3mil-R |
ACTGTTGGCTGTGCTCAAACATCC |
This study |
| To test the rearrangement status of pAsa5 | ||
|
tapA F |
ACATGAAGAAGCAATCAGGC |
30 |
|
tapA R |
AGAGGTCATGCGTTAGCAG |
30 |
|
traC F |
TGCACTATCCCCAGCTATCC |
8 |
|
traC R |
TCGGTAATCGCGGTCTTGTC |
8 |
|
exsD F |
AGAAGTGATCCTGACCCAAGGCAA |
8 |
|
exsD R |
TTGCAACGACTGTTGCCAAGAACC |
8 |
|
resD F |
TCAGAAACTTGGCCATCGCTCACA |
8 |
|
resD R |
TGATGTGCAGATTTCCCTGGAGACA |
8 |
| 11AF |
AATAGGTGTCGCAAGCTGGGTTGA |
7 |
| 11CR | AACTGGCAAGGATAGAGCTGCTGA | 7 |
Cloning and sequencing of pAsal1B
EcoRI-digested plasmid DNA from HER1104 was deposited in 18 wells of a 0.4% agarose gel prestained with ethidium bromide and was migrated for 5 h at 70 V. Bands in the 8 kb region were excised under UV illumination and were extracted using gel extraction kits (Feldan, 9K-006-0001). Purified DNA (30 µl) was digested with DpnI (NEB, R0176S), which generated multiple bands, with major bands between 300 and 1500 bp (data not shown). The pSP73 plasmid (Promega, P2221) was also digested with SmaI (NEB, R0141S), and the digest was dephosphorylated with Antarctic phosphatase (NEB, M0289S). The digested DNA from pSP73 and HER1104 was purified using PureLink PCR purification kits (Invitrogen, K3100-01). T4 ligase (NEB, M02025) was used to insert pAsal1B fragments in pSP73. The ligation products were introduced into E. coli MC1061 by heat shock. Twenty clones obtained by growing the bacteria on LB agar + ampicillin (50 µg/ml) were selected and amplified. Plasmid DNA from the clones was extracted using plasmid miniprep kits (Feldan, 9K-006-0010). Plasmid DNA samples were digested with EcoRI and XhoI. The sizes of the inserts were verified on agarose gels. The DNA was sent to the genome analysis platform (IBIS, Université Laval, Quebec City, QC, Canada) for sequencing. Nucleotide BLAST analyses of the sequences were performed using tools available on the National Center for Biotechnology Information website (www.ncbi.nlm.nih.gov). To complete the assembly of pAsalB, the DNA was sequenced by primer walking. The primers used are listed in Table 2. Primers were designed using primer design software available online on the Integrated DNA Technologies website (www.idtdna.com). The sequences of pAsal1 and pAsa5 were used to design the required primers (GenBank accession numbers: CP000646 and AJ508382).5,9
Supplementary Material
Acknowledgments
We thank the Laboratoire de bactériologie clinique, Faculté de médecine vétérinaire, Université de Montréal (Montreal, QC, Canada), the Félix d’Hérelle Reference Center (Département de biochimie, de microbiologie et de bio-informatique, Université Laval, Quebec City, QC, Canada), the Ministère de l'Agriculture, des Pêcheries et de l’Alimentation du Québec, and the Aquatic Animal Health Department, Fisheries and Oceans Canada (NB, Canada) for the A. salmonicida strains. We also thank Laurence Nadeau, Valérie Paquet and Stéphanie Dallaire-Dufresne for technical support.
M.V.T. received a Bourse FONCER scholarship from Ressources Aquatiques Québec (RAQ). K.H.T. received a scholarship from the Natural Sciences and Engineering Research Council of Canada (NSERC). R.K.D. received a scholarship from the Programme canadien de bourses de la francophonie (PCBF). This project was funded by an NSERC Discovery grant to S.J.C.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Materials
Supplemental materials may be found here: www.landesbioscience.com/journals/mge/article/25640
Footnotes
Previously published online: www.landesbioscience.com/journals/mge/article/25640
References
- 1.Wiklund T, Dalsgaard I. Occurrence and significance of atypical Aeromonas salmonicida in non-salmonid and salmonid fish species: a review. Dis Aquat Organ. 1998;32:49–69. doi: 10.3354/dao032049. [DOI] [PubMed] [Google Scholar]
- 2.Hiney M, Olivier G. Furunculosis (Aeromonas salmonicida). In: Woo P, Bruno D, eds. Fish Diseases and Disorders III: Viral, Bacterial and Fungal Infections: Oxford: CAB Publishing, 1999:341-425 [Google Scholar]
- 3.Belland RJ, Trust TJ. Aeromonas salmonicida plasmids: plasmid-directed synthesis of proteins in vitro and in Escherichia coli minicells. J Gen Microbiol. 1989;135:513–24. [Google Scholar]
- 4.Boyd J, Williams J, Curtis B, Kozera C, Singh R, Reith M. Three small, cryptic plasmids from Aeromonas salmonicida subsp. salmonicida A449. Plasmid. 2003;50:131–44. doi: 10.1016/S0147-619X(03)00058-1. [DOI] [PubMed] [Google Scholar]
- 5.Fehr D, Casanova C, Liverman A, Blazkova H, Orth K, Dobbelaere D, et al. AopP, a type III effector protein of Aeromonas salmonicida, inhibits the NF-kappaB signalling pathway. Microbiology. 2006;152:2809–18. doi: 10.1099/mic.0.28889-0. [DOI] [PubMed] [Google Scholar]
- 6.Charette SJ, Brochu F, Boyle B, Filion G, Tanaka KH, Derome N. Draft genome sequence of the virulent strain 01-B526 of the fish pathogen Aeromonas salmonicida. J Bacteriol. 2012;194:722–3. doi: 10.1128/JB.06276-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tanaka KH, Dallaire-Dufresne S, Daher RK, Frenette M, Charette SJ. An insertion sequence-dependent plasmid rearrangement in Aeromonas salmonicida causes the loss of the type three secretion system. PLoS One. 2012;7:e33725. doi: 10.1371/journal.pone.0033725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daher RK, Filion G, Tan SG, Dallaire-Dufresne S, Paquet VE, Charette SJ. Alteration of virulence factors and rearrangement of pAsa5 plasmid caused by the growth of Aeromonas salmonicida in stressful conditions. Vet Microbiol. 2011;152:353–60. doi: 10.1016/j.vetmic.2011.04.034. [DOI] [PubMed] [Google Scholar]
- 9.Reith ME, Singh RK, Curtis B, Boyd JM, Bouevitch A, Kimball J, et al. The genome of Aeromonas salmonicida subsp. salmonicida A449: insights into the evolution of a fish pathogen. BMC Genomics. 2008;9:427. doi: 10.1186/1471-2164-9-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adams CA, Austin B, Meaden PG, McIntosh D. Molecular characterization of plasmid-mediated oxytetracycline resistance in Aeromonas salmonicida. Appl Environ Microbiol. 1998;64:4194–201. doi: 10.1128/aem.64.11.4194-4201.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.L’Abée-Lund TM, Sørum H. A global non-conjugative Tet C plasmid, pRAS3, from Aeromonas salmonicida. Plasmid. 2002;47:172–81. doi: 10.1016/S0147-619X(02)00001-X. [DOI] [PubMed] [Google Scholar]
- 12.Najimi M, Balado M, Lemos ML, Osorio CR. Genetic characterization of pAsa6, a new plasmid from Aeromonas salmonicida subsp. salmonicida that encodes a type III effector protein AopH homolog. Plasmid. 2009;61:176–81. doi: 10.1016/j.plasmid.2009.01.001. [DOI] [PubMed] [Google Scholar]
- 13.Sandaa RA, Enger O. Transfer in Marine Sediments of the Naturally Occurring Plasmid pRAS1 Encoding Multiple Antibiotic Resistance. Appl Environ Microbiol. 1994;60:4234–8. doi: 10.1128/aem.60.12.4234-4238.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sørum H, L’Abée-Lund TM, Solberg A, Wold A. Integron-containing IncU R plasmids pRAS1 and pAr-32 from the fish pathogen Aeromonas salmonicida. Antimicrob Agents Chemother. 2003;47:1285–90. doi: 10.1128/AAC.47.4.1285-1290.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tanaka KH, Frenette M, Charette SJ. IS-mediated loss of virulence by Aeromonas salmonicida: A tangible piece of an evolutionary puzzle. Mobile Genet Elements. 2013;3:1–4. doi: 10.4161/mge.23498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gustafson CE, Chu S, Trust TJ. Mutagenesis of the paracrystalline surface protein array of Aeromonas salmonicida by endogenous insertion elements. J Mol Biol. 1994;237:452–63. doi: 10.1006/jmbi.1994.1247. [DOI] [PubMed] [Google Scholar]
- 17.Belland RJ, Trust TJ. Cloning of the gene for the surface array protein of Aeromonas salmonicida and evidence linking loss of expression with genetic deletion. J Bacteriol. 1987;169:4086–91. doi: 10.1128/jb.169.9.4086-4091.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chu S, Noonan B, Cavaignac S, Trust TJ. Endogenous mutagenesis by an insertion sequence element identifies Aeromonas salmonicida AbcA as an ATP-binding cassette transport protein required for biogenesis of smooth lipopolysaccharide. Proc Natl Acad Sci U S A. 1995;92:5754–8. doi: 10.1073/pnas.92.12.5754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dautremepuits C, Fortier M, Croisetiere S, Belhumeur P, Fournier M. Modulation of juvenile brook trout (Salvelinus fontinalis) cellular immune system after Aeromonas salmonicida challenge. Vet Immunol Immunopathol. 2006;110:27–36. doi: 10.1016/j.vetimm.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 20.Popoff M. Etude sur les Aeromonas salmonicida. II. Caractérisation des bactériophages actifs sur les Aeromonas salmonicida et lysotypie. Ann Rech Vet. 1971;2:33–45. [Google Scholar]
- 21.Dacanay A, Knickle L, Solanky KS, Boyd JM, Walter JA, Brown LL, et al. Contribution of the type III secretion system (TTSS) to virulence of Aeromonas salmonicida subsp. salmonicida. Microbiology. 2006;152:1847–56. doi: 10.1099/mic.0.28768-0. [DOI] [PubMed] [Google Scholar]
- 22.Berger B, Haas D. Transposase and cointegrase: specialized transposition proteins of the bacterial insertion sequence IS21 and related elements. Cell Mol Life Sci. 2001;58:403–19. doi: 10.1007/PL00000866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stuber K, Burr SE, Braun M, Wahli T, Frey J. Type III secretion genes in Aeromonas salmonicida subsp salmonicida are located on a large thermolabile virulence plasmid. J Clin Microbiol. 2003;41:3854–6. doi: 10.1128/JCM.41.8.3854-3856.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Filée J, Siguier P, Chandler M. Insertion sequence diversity in archaea. Microbiol Mol Biol Rev. 2007;71:121–57. doi: 10.1128/MMBR.00031-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kehrenberg C, Schwarz S. Distribution of florfenicol resistance genes fexA and cfr among chloramphenicol-resistant Staphylococcus isolates. Antimicrob Agents Chemother. 2006;50:1156–63. doi: 10.1128/AAC.50.4.1156-1163.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Franco AA. The Bacteroides fragilis pathogenicity island is contained in a putative novel conjugative transposon. J Bacteriol. 2004;186:6077–92. doi: 10.1128/JB.186.18.6077-6092.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Szeverényi I, Nagy Z, Farkas T, Olasz F, Kiss J. Detection and analysis of transpositionally active head-to-tail dimers in three additional Escherichia coli IS elements. Microbiology. 2003;149:1297–310. doi: 10.1099/mic.0.26121-0. [DOI] [PubMed] [Google Scholar]
- 28.Dziewit L, Baj J, Szuplewska M, Maj A, Tabin M, Czyzkowska A, et al. Insights into the transposable mobilome of Paracoccus spp. (Alphaproteobacteria) PLoS One. 2012;7:e32277. doi: 10.1371/journal.pone.0032277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shore AC, Brennan OM, Ehricht R, Monecke S, Schwarz S, Slickers P, et al. Identification and characterization of the multidrug resistance gene cfr in a Panton-Valentine leukocidin-positive sequence type 8 methicillin-resistant Staphylococcus aureus IVa (USA300) isolate. Antimicrob Agents Chemother. 2010;54:4978–84. doi: 10.1128/AAC.01113-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ebanks RO, Knickle LC, Goguen M, Boyd JM, Pinto DM, Reith M, et al. Expression of and secretion through the Aeromonas salmonicida type III secretion system. Microbiology. 2006;152:1275–86. doi: 10.1099/mic.0.28485-0. [DOI] [PubMed] [Google Scholar]
- 31.Charette SJ, Cosson P. Preparation of genomic DNA from Dictyostelium discoideum for PCR analysis. Biotechniques. 2004;36:574–5. doi: 10.2144/04364BM01. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.