Abstract
Replication forks formed at bacterial origins often encounter template roadblocks in the form of DNA adducts and frozen protein–DNA complexes, leading to replication-fork stalling and inactivation. Subsequent correction of the corrupting template lesion and origin-independent assembly of a new replisome therefore are required for survival of the bacterium. A number of models for replication-fork restart under these conditions posit that nascent strand regression at the stalled fork generates a Holliday junction that is a substrate for subsequent processing by recombination and repair enzymes. We show here that early replication intermediates containing replication forks stalled in vitro by the accumulation of excess positive supercoils could be cleaved by the Holliday junction resolvases RusA and RuvC. Cleavage by RusA was inhibited by the presence of RuvA and was stimulated by RecG, confirming the presence of Holliday junctions in the replication intermediate and supporting the previous proposal that RecG could catalyze nascent strand regression at stalled replication forks. Furthermore, RecG promoted Holliday junction formation when replication intermediates in which the replisome had been inactivated were negatively supercoiled, suggesting that under intracellular conditions, the action of RecG, or helicases with similar activities, is necessary for the catalysis of nascent strand regression.
The picture of how DNA replication proceeds around the bacterial chromosome has changed over the last decade as a result of research in many laboratories (1). Even though the two replication forks that form at oriC have a sufficiently high enough inherent processivity to complete replication of the chromosome, it is clear that this is generally not what happens. Instead, the replication forks formed at the origin become inactivated at high frequency as a result of an encounter with roadblocks either in or on the template strands. These roadblocks can take many forms: a nick in one of the template strands, a DNA adduct formed as a result of endogenous damage, secondary structure in the template, and frozen proteins on the DNA. Survival then depends on both correction of the damage and reactivation of DNA replication.
Although there is a large body of both genetic and biochemical data informing the mechanisms that act to repair damaged nucleotides in DNA, except in one instance, the mechanisms of replication-fork restart are less well defined. Replication-fork restart after an encounter with a template nick, leading to double-strand break (DSB) generation and detachment from the growing fork of one of the nascent sister duplexes—sometimes termed replication fork collapse (2)—is effected by a marriage of homologous recombination and DNA replication proteins (3). Here, the DSB generated is processed by RecBCD to generate a recombinogenic 3′ single-stranded tail that is used for RecA-catalyzed strand invasion with the intact sister duplex, creating a D loop. This structure is recognized by PriA (4, 5), which then directs the assembly of a new replication fork at the site through the loading of a primosome (6). The strand crossover initiating D loop formation is resolved subsequently.
Both the DNA structures formed and the mechanism of replication restart in other cases are less clear. Insight to the problem has been acquired through the examination of the consequences of stalling replication forks in vivo by various means. Placement of Ter sequences outside of the usual configuration at the terminus region of the chromosome generated strains that required RecA and RecBCD for survival if the replication-fork arrest protein Tus also was present (7, 8). The models developed to explain these observations suggested that DSB formation was occurring at the stalled replication fork. Replication-fork restart then presumably could proceed via the pathway discussed above for restart after an encounter with a template nick.
Michel and colleagues have studied the consequences of stalling replication forks by interfering with DNA helicase action. They demonstrated an increased frequency of DSB formation in rep recBC mutant strains (9). These researchers argued that the absence of Rep, a 3′ → 5′ DNA helicase (10) known to be able to displace some bound proteins from DNA (11), caused forks to pause more often because of poor clearing of protein obstacles from the template. Interestingly, DSB formation depended on RuvABC (12), the homologous recombination combination branch migration/Holliday junction resolvase machine (13). Thus, this observation suggested that Holliday junctions were forming at stalled replication forks as a result of pairing of the nascent strands and fork regression. RuvAB could either be catalyzing nascent strand regression to form the Holliday junction or be acting subsequent to its formation. Similar observations were made at the nonpermissive temperature in strains carrying conditional-lethal mutations in the replication fork helicase, DnaB (9). Additional processing of the Holliday junction presumably leads to the generation of substrates for replication-fork restart.
More recent genetic and biochemical data from McGlynn and Lloyd (14) argues that it is RecG, another branch migration DNA helicase (15, 16), that is responsible for catalyzing nascent strand regression at stalled forks. In this study, the presence of transcribing RNA polymerase on UV-irradiated chromosomes was shown to promote replication-fork stalling and lethality in ΔruvAC mutant strains. Survival of UV-irradiated Δruv strains was found to be enhanced in derivatives carrying either spoT1 [which increases the steady-state level of the RNA polymerase modulator (p)ppGpp] or rpo* [mutations in rpoB that mimic the effect of (p)ppGpp binding to RNA polymerase]. This ruv-independent recovery relied heavily on UvrABC-mediated excision repair and on the activities of PriA, RecG, and RecA. The need for PriA implied that replication forks had stalled and that the replication machinery had to be reassembled at sites remote from oriC. RecBCD was not required, and the typical UV sensitivity of recB mutations could only be observed in the presence of the Holliday junction resolvase RusA. These observations led us to propose that RecG-catalyzed replication fork regression occurred at forks stalled at UV lesions in the template and that replication restart proceeded via a pathway that did not involve D loop formation by RecBCD-mediated recombination. This finding was supported by the demonstration that RecG, but not RuvAB, could convert three-way junctions to Holliday junctions that could be cleaved by a resolvase such as RusA or RuvC.
Here we have examined the fate of nascent DNA formed in vitro on plasmid templates carrying oriC where replication forks have been stalled as a result of the absence of a topoisomerase. These early θ-type replication intermediates could be cleaved by either RusA or RuvC. RusA cleavage was inhibited by RuvA and, in intermediates where the replisome had been inactivated, by negative supercoiling, suggesting that Holliday junctions had indeed formed spontaneously in the positively supercoiled early replication intermediate (ERI). RecG stimulated RusA cleavage of ERIs where the replisome had been inactivated and promoted cleavage when the inactivated ERI was negatively supercoiled. These results suggest that under normal conditions in the cell, when the replicating chromosome is expected to be maintained at a net negative linking difference, the action of RecG, or enzymes with similar activities, catalyzes nascent strand regression at stalled replication forks.
Materials and Methods
Replication and Recombination Proteins.
Proteins for oriC replication—DnaA, DnaB, DnaC, DnaG, HU, the single-stranded DNA-binding protein (SSB), the DNA polymerase III holoenzyme (Pol III HE) (reconstituted from preparations of Pol III* and the β subunit), topoisomerase IV (Topo IV), and DNA gyrase—were purified as described (17, 18). RusA (19), RuvA and RuvB (20), RuvC (21), and RecG (22) were purified as indicated.
oriC DNA Replication.
Standard oriC DNA replication reaction mixtures (20 μl) were as described by Hiasa and Marians (17) except that no topoisomerase was added so that ERI accumulated. Template DNA was plasmid pBROTB535 type I, a 6-kb-long molecule (23). Gel electrophoretic analyses of replication products were performed as described (17).
Results
Holliday Junctions Form in Early Replication Intermediates Containing Paused Replication Forks.
We used the oriC DNA replication system to generate paused replication forks that could then be probed to detect Holliday junction formation. We took advantage of the fact that the plasmid DNA templates used in the replication system are negatively supercoiled. Thus, initiation can occur in the absence of a topoisomerase. Replication fork progression then occurs until excess positive windings accumulate, typically as positive supercoils. We have demonstrated that this intermediate (the ERI) is a true kinetic intermediate in the replication pathway (22). In the oriC replication system, the ERI contains a nascent leading strand that is about 1 kb in length (Fig. 2). Although at first glance this seems longer than expected, we have shown recently that under these conditions, only one of the two forks that are formed at oriC is released to form the ERI (N. Smelkova and K.J.M., unpublished data). And, as shown here, origin-proximal regression of the nascent DNA in the ERI allows more fork progression than one would have predicted based on the superhelical density of the starting template DNA. Further replication fork progression in the ERI requires relief of the accumulated topological strain by either adding a topoisomerase or cutting one of the template strands (24).
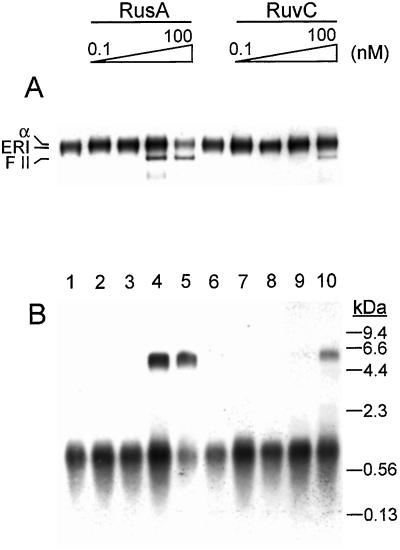
Figure 2.
Both RusA and RuvC can cleave ERIs. Standard oriC replication reactions containing either RusA (lanes 2–5, concentration varied by a factor of 10 in each lane, left to right), RuvC (lanes 7–10, concentration varied by a factor of 10 in each lane, left to right), or having no addition (lanes 1 and 6) were incubated at 37°C for 5 min. The reactions then were terminated by the addition of EDTA, and the DNA products then were analyzed by electrophoresis through either a neutral 1% agarose gel (A) or a denaturing 1% alkaline agarose gel (B) as described in Materials and Methods. α, α structure; FII, form II (nicked, circular DNA). The mobility difference between the ERI and the α structure is subtle and can be seen more clearly in Figs. 5–7.
It recently has been demonstrated, by using intermediates made with the oriC replication system (25), that the ERI, in fact, migrates during electrophoresis through neutral agarose gels as if it were relaxed, rather than containing supercoils of any type. This finding suggested that the positive supercoiling generated during replication actually promoted nascent strand regression. This result is because the preferred resting state of closed circular DNA is the relaxed form, negatively supercoiled molecules thus tend to favor unwinding of the duplex turns, whereas positively supercoiled molecules favor rewinding of the duplex turns. Either process results in the removal of supercoils from the respective molecule. Thus, in a positively supercoiled ERI, the tendency to rewind the template strands actually drives nascent strand regression. Postow et al. (25) were able to observe the reversed fork in purified replication intermediates by using scanning force microscopy.
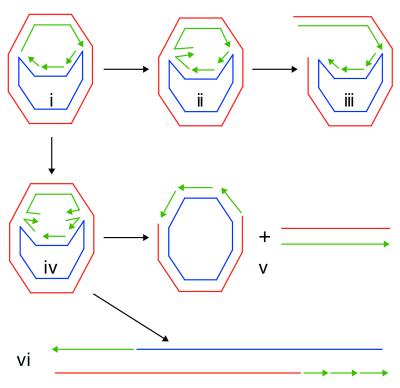
We wanted to examine whether fork regression was occurring during the replication reaction, thus the use of analytical techniques such as scanning force microscopy were not applicable. Instead, we reasoned that if nascent strand regression occurred in the ERI, the Holliday junction formed should be susceptible to cleavage by the Holliday junction-specific resolvases RusA and RuvC. Cleavage of ERIs containing Holliday junctions can generate three major species (Fig. 1). Presumably, either the origin-proximal, origin-distal, or both ends of the nascent DNA can pair to generate four-way junctions. Cleavage at only one of these points generates an α structure (Fig. 1iii). This DNA form will migrate slightly slower than the ERI in neutral agarose gels. If both ends of the nascent DNA regress and both Holliday junctions in the same molecule are cut, two products will be generated, distinguished by the orientation of resolution. In one case, a nicked circular (form II) molecule and a small duplex fragment corresponding to the distance along the nascent DNA between the resolution points will arise (Fig. 1v). In the other case, a nicked linear DNA will arise that is longer by the remaining regions of nascent DNA than the linear form (form III) of the original plasmid template (Fig, 1vi). This DNA form (labeled as form III′ in the figures) will migrate with a mobility that is intermediate between that of form II and form III DNAs on neutral agarose gels.
Figure 1.
Resolution of Holliday junctions in early replication intermediates can generate a number of different products. (i) The ERI. (ii) An ERI in which one end (the origin-proximal end) of the nascent DNA has regressed. This could happen with equal probability at the other end as well. (iii) Cleavage of the Holliday junction in ii generates an α structure. (iv) An ERI in which both ends of the nascent DNA have regressed. Because resolution of each Holliday junction can occur in one of two ways, two sets of products are generated: a nicked circle (form II) and a short duplex DNA corresponding to the distance on the replicated portion of the template that is between the sites of resolution (which will vary depending on the amount of nascent DNA that has regressed) (v); and a linear molecule that is longer than the original template by the distance on the template that is between the sites of resolution (vi). Red, the leading-strand template; blue, the lagging-strand template; green, nascent DNA.
To examine whether Holliday junctions formed in ERIs where replication forks had paused because of accumulated topological strain, RusA and RuvC were included in oriC replication reactions that contained DnaA, DnaB, DnaC, DnaG, HU, the single-stranded DNA-binding protein (SSB), and the DNA polymerase III holoenzyme (Pol III HE), but no topoisomerase. Analysis of the products of reaction by neutral agarose gel electrophoresis revealed that, indeed, the ERI could be cleaved by the Holliday junction resolvases, generating at least α structures and form II DNA (Fig. 2A). Cleavage could be detected at lower concentrations of RusA than RuvC. As expected from previous studies (26), cleavage of the parental strands removed the topological constraint and thus released the paused replication fork. This release allowed extension of the nascent leading strand to full length (Fig. 2B) and suggested that, under these conditions, nascent strand regression and subsequent cleavage occurred primarily at the origin-proximal end of the nascent DNA (Fig. 1iii). Presumably, the presence of the replisome inhibited fork regression at the origin-distal end of the nascent DNA.
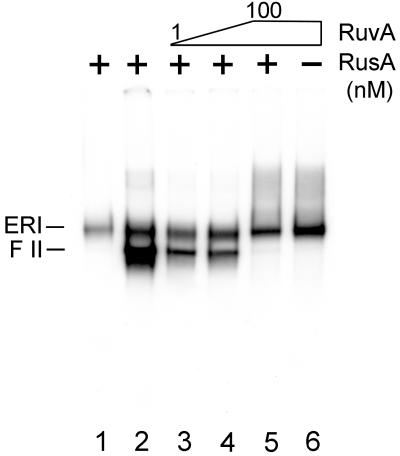
To confirm that cleavage was occurring at Holliday junctions, RuvA, which specifically recognizes and binds to Holliday junctions (13) and prevents their resolution by either RusA or RuvC (27, 28), was included in replication reactions that also contained RusA (Fig. 3). The presence of RuvA alone in the replication reaction had no effect (Fig. 3, lane 6) and the presence of RusA alone led to the expected cleavage (Fig. 3, lane 2). Increasing concentrations of RuvA inhibited RusA cleavage (Fig. 3, lanes 3–5), suggesting that the enzymes were competing for the same site on the DNA, a Holliday junction. We therefore conclude that Holliday junctions form as a result of nascent strand regression in the ERI.
Figure 3.
RuvA inhibits cleavage of the ERI by RusA. Standard oriC replication reactions containing either 10 nM RusA (lane 2), 10 nM RusA and RuvA (lanes 3–5, RuvA concentration varied by a factor of 10 in each lane from left to right), 100 nM RuvA (lane 6), or having no addition (lane 1) were incubated first for 2 min at 37°C after RuvA addition and then for an additional 5 min at 37°C after RusA addition. The reactions were terminated by the addition of EDTA, and the DNA products then were analyzed by electrophoresis through a neutral 1% agarose gel.
Nascent Strand Regression Is Favored by Positive Supercoiling.
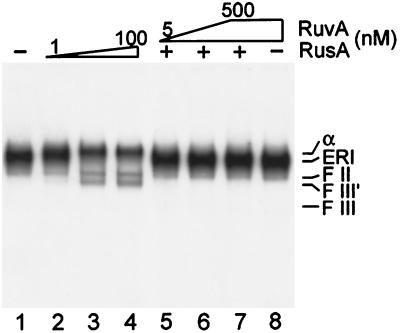
To investigate the topological requirements for Holliday junction formation in the ERI, we adopted a slightly different approach. Because inclusion of topoisomerases in the reaction mixture suppresses the formation of the ERI and promotes completion of replication, we formed the ERI first, heat-inactivated the proteins in the reaction mixture, and then probed for Holliday junction formation. ERI treated in this fashion still contained Holliday junctions, as evinced by RusA-catalyzed cleavage that could be inhibited by the presence of RuvA (Fig. 4). The pattern of cleavage was slightly different from that shown in Fig. 2A, which presumably reflects the fact that either one or both of the forked structures could be cleaved once the replication proteins were inactivated.
Figure 4.
Holliday junctions form in inactivated ERI. Inactivated ERI was formed as follows. Nine standard reactions for ERI formation as described in Materials and Methods were incubated at 37°C for 10 min. The reactions then were terminated by heating to 65°C for 5 min. After cooling, the reactions were pooled together. Nine and one-half microliters of the inactivated ERI pool was incubated in new reaction mixtures (10 μl) containing either no RuvA or RusA (lane 1), RusA (lanes 2–7, concentration in lanes 2–4 varied by a factor of 10 from left to right and was 100 nM in lanes 5–7), and RuvA (lanes 5–8, concentration varied by a factor of 10 in lanes 5–7 from left to right and was 500 nM in lane 8) and incubated for 2 min at 37°C after RuvA addition and for an additional 5 min at 37°C after RusA addition. The reactions then were terminated by the addition of EDTA, and the DNA products were analyzed by neutral 1% agarose gel electrophoresis. F III′, form III′ (see text for definition); F III, form III (full length, linear form).
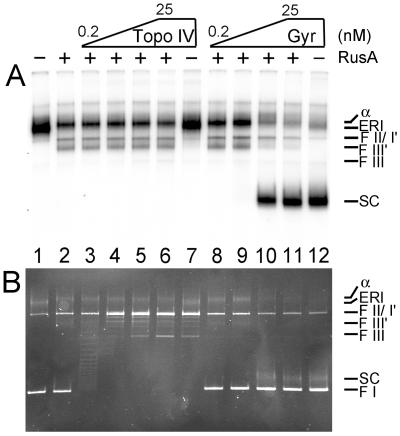
RusA cleavage of the inactivated ERI was unaffected by prior treatment with Topo IV (Fig. 5). The electrophoretic mobility of the ERI was unaffected as well (Fig. 5A, compare lanes 1 and 7), even though the topoisomerase was clearly active, as demonstrated by relaxation of the form I DNA that had not been replicated and was present in the reaction mixture (Fig. 5B, lanes 1–7). Because Topo IV can remove both positive and negative supercoils (29, 30), this observation indicates that the ERI has no net supercoiling and is effectively relaxed, as noted by Postow et al. (25).
Figure 5.
Negative supercoiling inhibits RusA cleavage of the ERI. Inactivated ERI was formed as in the legend to Fig. 4 except that 12 reaction mixtures were pooled. Fresh ATP was added to this pool to a concentration of 2 mM. Nine microliters of the inactivated ERI pool was incubated in new reaction mixtures (10 μl) containing either no RusA (lanes 1, 7, and 12) or 100 nM RusA (all other lanes), and either Topo IV (lanes 3–7, concentration varied by a factor of 5 in lanes 3 to 6 from left to right and was 25 nM in lane 7) or DNA gyrase (lanes 8–12, concentration varied by a factor of 5 in lanes 7–11 from left to right and was 25 nM in lane 12) for 5 min at 37°C. The reactions were terminated by the addition of EDTA, and the DNA products were analyzed by neutral 1% agarose gel electrophoresis. The autoradiogram of the gel is shown in A and a photograph of the ethidium bromide stain gel is shown in B. FI, negatively supercoiled template DNA; F I′, form I′ (intact template DNA with no supercoils); SC, supercoiled ERI.
On the other hand, prior treatment with DNA gyrase inhibited cleavage by RusA (Fig. 5A, lanes 8–12). In this instance as well, it was clear that the topoisomerase was working because the ERI became negatively supercoiled (Fig. 5B, compare lane 1 to lanes 10–12). Because of the presence of a replication bubble, the supercoiled ERI (labeled SC in Fig. 5) had a mobility that was slightly reduced compared with the form I template DNA. Thus, just as positive supercoiling drives nascent strand regression, negative supercoiling favors unwinding of the template duplex turns by rewinding the nascent DNA with its parental strand. Note, because the nascent DNA is not covalently closed, only windings of the parental strands about themselves affect the topology of the template DNA.
RecG Promotes Holliday Junction Formation in ERIs That Are Negatively Supercoiled.
The observations described in the previous section raised a conundrum: Although positive supercoiling drives fork regression, a replicating chromosome is expected to be negatively supercoiled in vivo because of the presence of DNA gyrase in the cell. If Holliday junction formation plays a role in the processing of stalled replication forks in vivo, one would then predict that there must be some agent that overcomes the inhibitory effect of the negative supercoils and catalyzes fork regression. Based on our previous studies (14), we suspected that this agent was RecG. We therefore investigated the effect of RecG on RusA-catalyzed cleavage of the ERI.
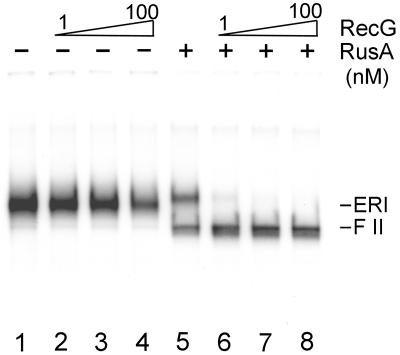
RusA-catalyzed cleavage of inactivated ERI was stimulated by the addition of RecG (Fig. 6, lanes 5–8). RecG alone had no effect on the electrophoretic mobility of the ERI even at concentrations 100-fold higher than necessary to observe the stimulation of RusA cleavage (Fig. 6, lanes 1–4). This stimulation presumably reflects our previous demonstration that RecG could branch-migrate three-way junctions into four-way junctions (14).
Figure 6.
RecG stimulates RusA-catalyzed cleavage of the ERI. Inactivated ERI was formed as in the legend to Fig. 4 except that six reaction mixtures were pooled. Fresh ATP was added to this pool to a concentration of 2 mM. Six and three-tenths microliters of the inactivated ERI pool was then incubated in new reaction mixtures (7 μl) containing either no RecG or RusA (lane 1), RecG (lanes 2–4 and 6–8, concentration varied by a factor of 10 in lanes 2–4 and 6–8, left to right), and 10 nM RusA (lanes 5–8) for 5 min at 37°C. The reactions were terminated by the addition of EDTA, and the DNA products were analyzed by neutral 1% agarose gel electrophoresis.
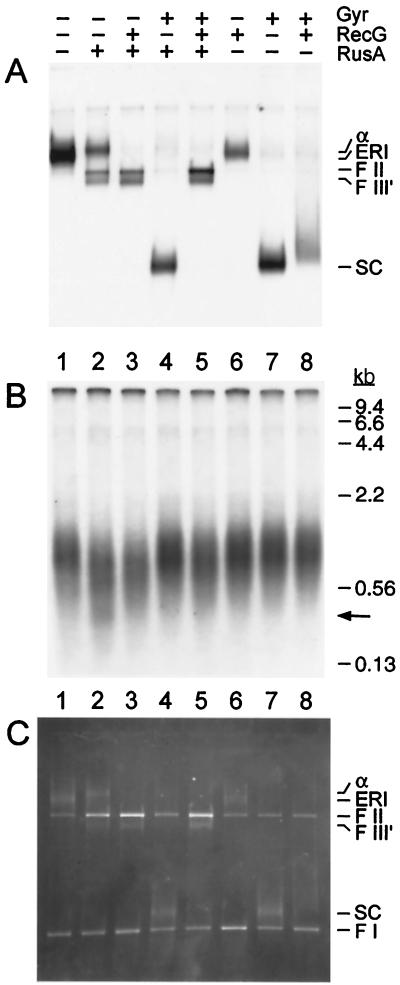
It seemed reasonable therefore to expect that the ATP-dependent helicase activity of RecG that was responsible for branch migration could drive nascent strand regression in negatively supercoiled ERI molecules. This expectation proved to be the case (Fig. 7). Consistent with the data displayed in Fig. 6, RusA-catalyzed cleavage of the inactivated ERI was stimulated by RecG (Fig. 7A, compare lanes 2 and 3), but inhibited by the addition of DNA gyrase to supercoil the DNA (Fig. 7, compare lanes 2 and 4). Remarkably, RusA-catalyzed cleavage of the ERI could be effected in the presence of gyrase if RecG was present as well (Fig. 7, compare lanes 4 and 5). Gyrase was clearly active in the reaction, as indicated by the shift in mobility of the ERI in its presence (Fig. 7 A and C, compare lanes 1 and 7). Thus, we conclude that RecG is necessary in order for nascent strand regression to occur in these negatively supercoiled replication intermediates.
Figure 7.
RecG promotes nascent strand regression in negatively supercoiled ERI. Inactivated ERI was formed as in the legend to Fig. 4 except that 12 reaction mixtures were pooled. Fresh ATP was added to this pool to a concentration of 2 mM. Twelve and three-quarter microliters of the inactivated ERI pool was then incubated in new reaction mixtures (15 μl) containing DNA gyrase (25 nM), RecG (10 nM), and RusA (100 nM) as indicated for 5 min at 37°C. The reactions were then terminated by the addition of EDTA. One-half of the reaction mixture was analyzed by neutral 1% agarose gel electrophoresis [autoradiogram (A); photograph of the ethidium bromide-stained gel (C)] and the other half was analyzed by denaturing 1% alkaline agarose gel electrophoresis [autoradiogram (B)]. The arrow on the right-hand side of B marks a faint band that may correspond to the linear fragment formed by cleavage of the ERI at both ends of the nascent DNA (see Fig. 1v).
The experiment displayed in Fig. 7 also shows clearly that RusA is cleaving the nascent DNA in the ERI. The population of nascent DNA centered around 1 kb in length is decreased in size on the denaturing alkaline agarose gel as a result of RusA treatment (Fig. 7B, compare lanes 1 and 2) and a band of about 0.4 kb in length is just visible in lane 2. This cleavage by RusA provides direct evidence for the formation of a Holliday junction by nascent strand regression in the ERI (Fig. 1). The ≈1-kb population of nascent strands also is shortened in the presence of RecG, but the 0.4-kb band is not observed (Fig. 7, compare lanes 3 and 5), which implies that RecG not only stimulates fork regression, but also alters the pattern of cleavage by altering the distribution of the junctions in the regressed molecules.
Discussion
It is now clear that replication forks stall with significant frequency during normal bacterial growth, as well as, of course, under conditions where the chromosome has been damaged by exogenous insult. Mechanisms that preserve genomic integrity are in place that work essentially as both housekeeping functions and inducible systems that respond to heavy DNA damage loads. Central to the survival of the organism is both repair of the damaged DNA template and reactivation of the stalled replication fork. The recognition of replication-fork stalling as a common occurrence during each cell cycle has led to important questions focused on how the cell processes the stalled fork: What is the fate of the replication proteins? What are the enzymatic pathways of restart? What are the structures formed at the stalled fork? And how do these structures influence restart? In this paper, we have focused on assessing the type of DNA structure assumed by the nascent DNA of a stalled fork.
Replication forks were stalled during θ-type DNA replication on plasmid templates carrying Escherichia coli oriC as a result of the accumulation of positive supercoils. These ERI molecules were found to be substrates for the Holliday junction-resolving enzymes RusA and RuvC. This observation suggested that a four-way junction had been generated in the ERI as a result of rewinding of the replicated parental template and pairing of the nascent DNA. The existence of a four-way junction was confirmed by the fact that RuvA, which recognizes Holliday junctions with high specificity (13), could inhibit cleavage of the ERI by RusA.
Nascent strand regression should be equally likely at either end of the nascent DNA in the ERI, i.e., at either the origin-proximal region or near the replication fork. When RusA was present in reaction mixtures where replication was ongoing, it appeared that the majority of cleavage occurred near the origin, away from the replication fork. This observation is consonant with our previous observations that the paused replication fork in the ERI can resume replication if the accumulated topological strain is relieved (24). Therefore, the replication proteins are likely to remain at the replication fork in an enzymatically competent state for a certain period. By using pulse–chase analysis, we have found that the functional half-life of these forks is 5–7 min (26).
In the experiments described here, the presence of the replication proteins at the stalled replication fork are likely to have stabilized the nascent DNA, preventing it from regressing. How should this observation affect our thinking about what happens in the cell? Clearly, regression of the origin-proximal end of the replicated chromosome arm will provide little benefit for restarting a stalled fork. Currently there is no information extent that addresses the fate of the proteins at a stalled fork in vivo. If the half-life that we have measured in vitro reflects what happens in the cell, then it seems reasonable to suggest that there may be active mechanisms that act to displace the replisome from a stalled replication fork to allow regression. Perhaps there are helicases that target stalled forks to displace the bound proteins.
Spontaneous nascent strand regression required that the ERI be positively supercoiled. The positive supercoiling is an impressive driving force for this reaction in that all of the ERI existed as topologically relaxed DNA as a result of nascent strand regression. If the ERI was negatively supercoiled by the action of gyrase, fork regression was severely inhibited. Can fork regression therefore occur spontaneously in vivo? This is a difficult question to answer. Previous studies on the decatenating activity of Topo IV and DNA gyrase in vitro (29) and in vivo (31) suggested that these enzymes lacked the catalytic turnover necessary to keep pace with advancing replication forks that generate nearly 200 excess positive windings per second. If this scenario holds, then the replicating chromosome would probably be positively supercoiled. However, recent single enzyme studies (32) with Topo IV show that the number average turnover values calculated previously are gross underestimates. If the same holds for DNA gyrase, then it is more reasonable to expect that the replicating chromosome is, in fact, negatively supercoiled.
Thus, if nascent strand regression plays a significant role at stalled replication forks in vivo, it would seem that there is a need for enzymes that act to facilitate formation of the Holliday junction. We have shown here that RecG fulfills this requirement. RecG stimulated RusA cleavage of the ERI and was required to observe any significant cleavage when the ERI was negatively supercoiled. Thus, the RecG branch-migration helicase activity can overcome the inhibitory effect of negative supercoiling and cause nascent strands to pair and regress enough that the Holliday junction that forms can be recognized and cleaved by RusA. We do not know whether the extent of RecG-catalyzed strand regression is as extensive as the regression that occurs spontaneously in the ERI.
These findings suggest that RecG plays a central role in processing of stalled replication forks in the cell. In an accompanying paper in this colloquium (33), McGlynn and Lloyd show that RecG unwinds forked DNA by translocating simultaneously along both the leading- and lagging-strand templates. This unique helicase activity explains how RecG may promote formation of a Holliday junction. Elucidating how this enzyme cooperates with all of the others that are competing for the various DNA ends and single-stranded gaps that are present at the site of fork stalling will likely keep many laboratories occupied for some time to come.
Acknowledgments
We thank Ed Bolt for providing RusA protein. These studies were supported by National Institutes of Health Grant GM34557 (to K.J.M.), a Medical Research Council Career Establishment grant (to P.M.), and a Medical Research Council Program grant (to R.G.L.). P.M. is a Lister Institute–Jenner Research Fellow.
Abbreviations
- DSB
double-strand break
- ERI
early replication intermediate
- Topo IV
topoisomerase IV
Footnotes
This paper results from the National Academy of Sciences colloquium, “Links Between Recombination and Replication: Vital Roles of Recombination,” held November 10–12, 2000, in Irvine, CA.
References
- 1.Cox M M, Goodman M F, Kreuzer K N, Sherratt D J, Sandler S J, Marians K J. Nature (London) 2000;404:37–41. doi: 10.1038/35003501. [DOI] [PubMed] [Google Scholar]
- 2.Kuzminov A. Mol Microbiol. 1995;16:373–384. doi: 10.1111/j.1365-2958.1995.tb02403.x. [DOI] [PubMed] [Google Scholar]
- 3.Kogoma T. Cell. 1996;85:625–627. doi: 10.1016/s0092-8674(00)81229-5. [DOI] [PubMed] [Google Scholar]
- 4.McGlynn P, Al-Deib A A, Liu J, Marians K J, Lloyd R G. J Mol Biol. 1997;270:212–221. doi: 10.1006/jmbi.1997.1120. [DOI] [PubMed] [Google Scholar]
- 5.Nurse P, Liu J, Marians K J. J Biol Chem. 1999;274:25026–25032. doi: 10.1074/jbc.274.35.25026. [DOI] [PubMed] [Google Scholar]
- 6.Liu J, Xu L, Sandler S J, Marians K J. Proc Natl Acad Sci USA. 1999;96:3552–3555. doi: 10.1073/pnas.96.7.3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Horiuchi T, Fujimura Y. J Bacteriol. 1995;177:783–791. doi: 10.1128/jb.177.3.783-791.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sharma B, Hill T M. Mol Microbiol. 1995;18:45–61. doi: 10.1111/j.1365-2958.1995.mmi_18010045.x. [DOI] [PubMed] [Google Scholar]
- 9.Michel B, Ehrlich S D, Uzest M. EMBO J. 1997;16:430–438. doi: 10.1093/emboj/16.2.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scott J F, Eisenberg S, Bertsch L L, Kornberg A. Proc Natl Acad Sci USA. 1977;74:193–197. doi: 10.1073/pnas.74.1.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yancey-Wrona J E, Matson S W. Nucleic Acids Res. 1992;20:6713–6721. doi: 10.1093/nar/20.24.6713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seigneur M, Bidnenko V, Ehrlich S D, Michel B. Cell. 1998;95:419–430. doi: 10.1016/s0092-8674(00)81772-9. [DOI] [PubMed] [Google Scholar]
- 13.West S C. Annu Rev Genet. 1997;31:213–244. doi: 10.1146/annurev.genet.31.1.213. [DOI] [PubMed] [Google Scholar]
- 14.McGlynn P, Lloyd R G. Cell. 2000;101:35–45. doi: 10.1016/S0092-8674(00)80621-2. [DOI] [PubMed] [Google Scholar]
- 15.Lloyd R G, Sharples G J. EMBO J. 1993;12:17–22. doi: 10.1002/j.1460-2075.1993.tb05627.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Whitby M C, Vincent S, Lloyd R G. EMBO J. 1994;13:5220–5228. doi: 10.1002/j.1460-2075.1994.tb06853.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hiasa H, Marians K J. J Biol Chem. 1994;269:6058–6063. [PubMed] [Google Scholar]
- 18.Marians K J. Methods Enzymol. 1995;262:507–521. doi: 10.1016/0076-6879(95)62042-7. [DOI] [PubMed] [Google Scholar]
- 19.Bolt E L, Sharples G J, Lloyd R G. J Mol Biol. 1999;286:403–415. doi: 10.1006/jmbi.1998.2499. [DOI] [PubMed] [Google Scholar]
- 20.Tsaneva I R, Illing G T, Lloyd R G, West S C. Mol Gen Genet. 1992;235:1–10. doi: 10.1007/BF00286175. [DOI] [PubMed] [Google Scholar]
- 21.Dunderdale H J, Sharples G J, Lloyd R G, West S C. J Biol Chem. 1994;269:5187–5194. [PubMed] [Google Scholar]
- 22.McGlynn P, Lloyd R G. Nucleic Acids Res. 1999;27:3049–3056. doi: 10.1093/nar/27.15.3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hiasa H, Marians K J. J Biol Chem. 1994;269:26959–26968. [PubMed] [Google Scholar]
- 24.Hiasa H, Marians K J. J Biol Chem. 1994;269:16371–16375. [PubMed] [Google Scholar]
- 25.Postow L, Ulsperger C, Keller R W, Bustamante C, Vologodski A V, Cozzarelli N R. J Biol Chem. 2001;276:2790–2796. doi: 10.1074/jbc.M006736200. [DOI] [PubMed] [Google Scholar]
- 26.Marians K J, Hiasa H, Kim D R, McHenry C S. J Biol Chem. 1998;273:2452–2457. doi: 10.1074/jbc.273.4.2452. [DOI] [PubMed] [Google Scholar]
- 27.Whitby M C, Bolt E L, Chan S N, Lloyd R G. J Mol Biol. 1996;264:878–890. doi: 10.1006/jmbi.1996.0684. [DOI] [PubMed] [Google Scholar]
- 28.Chan S N, Harris L, Bolt E L, Whitby M C, Lloyd R G. J Biol Chem. 1997;272:14873–14882. doi: 10.1074/jbc.272.23.14873. [DOI] [PubMed] [Google Scholar]
- 29.Hiasa H, Marians K J. J Biol Chem. 1996;271:21529–21535. doi: 10.1074/jbc.271.35.21529. [DOI] [PubMed] [Google Scholar]
- 30.Ulsperger C, Cozzarelli N R. J Biol Chem. 1996;271:31549–31555. doi: 10.1074/jbc.271.49.31549. [DOI] [PubMed] [Google Scholar]
- 31.Zecheidrich E L, Cozzarelli N R. Genes Dev. 1995;9:2859–2869. doi: 10.1101/gad.9.22.2859. [DOI] [PubMed] [Google Scholar]
- 32.Crisona N J, Strick T R, Bensimon D, Croquette V, Cozzarelli N R. Genes Dev. 2000;14:2881–2892. doi: 10.1101/gad.838900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McGlynn P, Lloyd R G. Proc Natl Acad Sci USA. 2001;98:8227–8234. doi: 10.1073/pnas.111008698. [DOI] [PMC free article] [PubMed] [Google Scholar]