Abstract
The primary cilia are microtubule-based organelles that protrude from most of the eukaryotic cells. Recognized as the cell’s antenna, primary cilia function as a signaling hub for many physiologically and developmentally important signaling cascades. Ciliary dysfunction causes a wide spectrum of syndromic human genetic diseases collectively termed "ciliopathies”. Mounting evidences have shown that various small GTPases have been implicated in the context of cilia as well as human ciliopathies. However, how these small GTPases affect cilia formation and function remains poorly understood. Here we review and discuss the ciliary role of three Arf-like small GTPases (Arls), Arl3, Arl6, and Arl13b.
Introduction
Cilia are micrometer-scale hair-like organelles that protrude from the surface of most eukaryotic cells. Based on their function, cilia are divided into two types: motile cilia and non-motile cilia (or primary cilia). Motile cilium has a rhythmic waving or beating motion that promotes cell motility or fluid flow over the cell surface. Primary cilium is recognized as the cell’s antenna, which functions as a signaling hub for many physiologically and developmentally important signaling cascades, including polycystins, Sonic hedgehog, Wnt, and various GPCR signaling pathways [1–3]. Recently, positional cloning and pathogenesis studies have identified that mutations in more than 50 human genes, most of which encode exclusively and highly conserved ciliary proteins across species, cause various syndromic human genetic diseases, which are now collectively termed "ciliopathies". Consistent with the ubiquitous existence of cilia, most ciliopathies exhibit disrupted developmental homeostasis in many organs, including the kidneys, central nervous system (CNS), cardiovascular system, liver, bones, eyes, ears, nose, and fat storage tissue [4, 5]. Despite the clinical relevance of cilia, we know little about how cilia form and function as well as the mechanistic insights into the ciliopathy genes. In this regard, the greatest challenges ahead are to explore how cilia form and function; determine the pathogenesis underlying ciliopathies; and design therapies to prevent, delay, or halt disease progression.
Cilia are microtubule-based structure that tightly covered by the plasma membrane. No ribosome exists inside the cilium and, thus, the protein needed for cilia biogenesis and function need to be synthesized in the cytoplasm and then transported into the cilium. A phylogenetically conserved cellular process, termed Intraflagellar Transport (IFT), is then responsible for the bidirectional transport of ciliary structural and signaling proteins inside cilia. The IFT complex, which contains more than 20 proteins, is composed of IFT-A and IFT-B subcomplex that cooperate together to mediate the movement of IFT cargos along the axoneme [6].
Small GTPases are key molecular switches that bind and hydrolyze GTP in diverse membrane- and cytoskeleton-related cellular processes. The unique enzymatical characteristic of small GTPases has made this group of proteins favorite and viable drug targets in many human diseases [7, 8]. Recently, emerging evidences have highlighted the roles of various small GTPases, including the members in Arf, Arl (Arf-like), Rab, and Ran subfamilies, in cilia formation and function [9, 10]. Among all small GTPases involved in the context of cilia, Arls are particularly intriguing due to two facts: First, the whole Arl family contains over 20 members. However, all Arls lack the biochemical or genetic activities characteristic of Arfs, which makes Arls the least studied group compared with other small GTPases [11]; Second, Arl6 and Arl13b are the only two small GTPases identified so far to be mutated in human ciliopathy patients. Thus, understanding the role of Arls in cilia would have imminent impact on either basic science in regarding to how the Arls act in cells or translational study in regarding to the pathogenesis of ciliopathies and the potential to identify ciliary switch(es) that can be used as therapeutic target(s).
Studies of Ras/Rho small GTPases have demonstrated that numerous proteins bind to small GTPases to control their localization, activity, and downstream signaling pathways. The interactors include enzymes involved in posttranslational modifications, guanine nucleotide exchange factors (GEF), GTPase-activating factors (GAP), and effectors (such as adaptors, motors, kinases, and phosphatases) that bind specifically to the active form of small GTPases [12–14]. This mini-review summarizes recent developments in the molecular mechanisms underlying the role of poorly understood ciliopathy Arls in the context of cilia, with particular emphasis on their ciliary effectors and the coordinated action.
Arl3 acts negatively in ciliogenesis but positively in cilia signaling
The implication of Arl3 in the context of cilia was first documented in a study on LdARL-3A gene (the ortholog of the human Arl3 gene in Leishmania donovani) that showed overexpression of the constitutively active form of LdARL-3(Q70L) leads to shortened flagellum [15]. Later, immunofluorescence study on human retina photoreceptor cells showed that Arl3 predominantly localizes to the connecting cilium [16]. A comparative genomics study also indicated that Arl3, along with Arl6, is exclusively found in the genome of ciliated organisms during evolution [17]. Although Arl3 has not been identified as a causal locus in any human ciliopathy, Arl3 knockout mice do exhibit typical ciliopathy manifestations characterized by cysts in the kidney, liver, and pancreas and impaired photoreceptor development [18]. Similar to many other cilia-defective knockout mouse models, Arl3−/− mice are very sick and die within 3 weeks after birth [18].
In C. elegans, a functional GFP-tagged ARL-3 exhibits a strong ciliary localization [19]. Arl-3−/− worms still possess properly formed cilia. However, similar to the observations made in Leishmania, the overexpression of a dominant active ARL-3(Q72L) caused ciliogenesis defects, suggesting that ARL-3 is a negative regulator of ciliogenesis [19]. This also explains why depletion of Arl3 protein by RNAi treatment in mammalian systems does not affect ciliogenesis [20]. The fact that depletion of ARL-3 can partially rescue the ciliogenesis defects in arl-13−/− worms further strengthens the conclusion that ARL-3 negatively regulates ciliogenesis [19]. Our studies in C. elegans indicated that ARL-3 plays a role in regulating the integrity of IFT complex through Histone deacetylatase 6 (HDAC6)- dependent pathway [19]. Nevertheless, the exact molecular mechanism underlying the function of ARL-3 in ciliogenesis is still poorly understood.
Except for the role in cilia formation, Arl3 also regulates ciliary signaling through, probably, regulating the proper ciliary targeting of various key signaling receptors/molecules. For example, the worm polycystins LOV-1/Polycystin-1 and PKD-2/Polycystin-2 localize to the cilia of male-specific sensory neurons and are required for the two cilia-mediated male mechanosensory behaviors: response (Rsp) and location of vulva (Lov) [21, 22]. Arl-3−/− worms show compromised ciliary targeting of polycystins and defective Rsp and Lov behaviors (unpublished data in the Hu lab). Arl3 was implicated in regulating rhodopsin localization as well as the membrane association and traffic of heterotrimeric G-protein transducin in photoreceptor cells [18, 23]. Knockdown of Arl3 in mammalian cells also disrupts the transportation of Gli3, the key signaling molecule in Hedgehog pathway [20].
So far, several interactors/effectors, including Golgin-245, CCDC104, C20orf194, Binder of Arl Two (BART), HDAC6, Phosphodiesterase 6 delta subunit (PDE6δ), Uncoordinated 119 protein (UNC-119), and one GAP protein, Retinitis Pigmentosa 2 (RP2), have been implicated in Arl3 pathway [19, 24–27]. Among these candidates, Golgin-245, CCDC104, C20orf194, and BART were only biochemically confirmed as interactors of ARL3, with no clear biological significance proven yet [26, 27]. HDAC6 is a microtubule-associated deacetylase that can deacetylates substrates, such as tubulin, heat shock protein 90, and cortactin [28]. We found that ARL-3 regulates the association between IFT subcomplex B and KIF17 motor in an HDAC-6-dependent manner in C. elegans, but the detailed mechanism still remains undefined [19]. Reminiscent of the ciliary role of Arl3, activation of HDAC6 promotes cilia disassembly and loss of HDAC6 activity selectively stabilizes cilia in human retinal epithelial cells [29]. Intriguingly, HDAC6 is highly expressed in cholangiocarcinoma cells and HDAC6 inhibition not only restores ciliogenesis but also slows tumor growth in a cilia-dependent way [30]. Combining with the observation that ARL3 protein is highly expressed in various human tumor cell lines [31], this certainly raise an interesting question about the functional correlation among ARL3/HDAC6, cilia and malignant transformation.
The other two Arl3 effectors, PDE6δ and UNC119, share extensive sequence homology. These proteins have similar hydrophobic lipid binding pockets and were shown to work together with GTP-bound ARL3 and its GAP protein RP2 to regulate the proper ciliary targeting of various lipid-modified proteins [23]. PDE6δ was originally identified as a prenyl-binding protein [32]. In mammalian system, GTP-bound ARL3 binds with PDE6δ to release the farnesylated cargo protein from PDE6δ [33, 34]. Compared with PDE6δ, UNC119 binds to myristoylated proteins, such as Nephronophthisis 3 (NPHP3) and transducin α subunit [27, 35]. In a working model, UNC119 binds and transports the myristoylated NPHP3 to the cilium, then the association between the ciliary GTP-bound ARL3 and UNC119 results in the release of myristoylated NPHP3 protein. RP2, which acts as the GAP for ARL3, converts GTP-bound ARL3 to GDP-bound ARL3 and consequently releases UNC119 from ARL3 [27]. Recently, it was even proposed that the UNC119, PDE6δ, and RP2 might all function together as effectors of GTP-bound ARL3 in regulating the assembly and membrane targeting of transducin, in which UNC119 binds to acylated transducin α subunit Gα1, RP2 binds to transducin β subunit Gβ1, PDE6δ binds to prenylated transducin γ subunit Gγ1 [23]. Given these data, ARL3 probably acts in a GTP-dependent manner as the allosteric release factor for lipid-modified cargo proteins from its effectors or GAP protein.
Arl6 is indispensable in cilia signaling but dispensable in ciliogenesis
Arl6 is the first known Arl to be associated with an inherited ciliopathy [36]. Bardet-Biedl syndrome (BBS) is an autosomal recessive inherited disease characterized by six major defects including obesity, mental retardation, renal anomalies, polydactyly, retinal degeneration, and hypogenitalism [37]. Seventeen genes (BBS1, BBS2, BBS3 (ARL6), BBS4, BBS5, BBS6 (MKKS), BBS7, BBS8 (TTC8), BBS9, BBS10, BBS11 (TRIM32), BBS12, MKS1, CEP290, C2ORF86, SDCCAG8, and LZTFL1) have been implicated in causing BBS so far [37–40]. When mapping the causal gene in BBS3 patients, the authors reasoned that the ortholog of BBS3 should also exist in other ciliated organisms. By using ciliated organism’s genome as references, two groups were able to identify ARL6 as the causal locus for BBS3 [36, 41]. Further analyses with the determination of the crystal structure of GTP-bound human ARL6 protein suggested that mutations identified in BBS3 patients locate around ARL6 GTP binding motif, suggesting the importance of small GTPase activity in ciliopathy etiology [42].
The role of Arl6 in the context of cilia is largely unknown due to the lack of the knowledge on its ciliary effectors. Arl6 knockdown zebrafishes and Arl6−/− mice both show the common BBS associated phenotypes, suggesting that the ciliary function of Arl6 is highly conserved [43, 44]. In C. elegans, Arl-6, the ortholog of human ARL6, localizes predominantly to the basal body and the axoneme of the cilium, and occasionally moves along the axoneme [36]. In ciliated IMCD3 cells, ARL6 exhibits a unique ring-like localization pattern at the distal end of basal body [42]. In RPE1-hTERT cells, ARL6 colocalizes with the BBSome in punctate at the ciliary membrane [45].
The BBSome is composed of eight BBS proteins, including BBS1, BBS2, BBS4, BBS5, BBS7, BBS8, BBS9, and BBIP10 [46, 47]. The BBSome components are highly conserved during evolution and, as a whole, shares common structural features with COPI, COPII and clathrin coats [45]. Loss of Arl6 does not affect BBSome formation but disrupts proper localization of Melanin concentrating hormone receptor 1 (MCHR1) to ciliary membranes in mouse model [44]. Because MCHR1 is involved in the regulation of feeding behavior and BBS is associated with hyperphagia-induced obesity, these results suggest that altered signaling caused by the mislocalization of ciliary sensory receptors underlies the manifestations in BBS patients and provides a potential molecular mechanism to link cilia defects with obesity [44, 48]. Additionally, ARL6 and BBSome are required for the ciliary entry of Somatostatin receptor 3 in mammalian cells [45]. It was proposed that GTP-bound ARL6 recruits the BBSome and BBSome-associated cargo complex to membrane and assembles a polymerized coat-cargo complex that can be dragged through the periciliary diffusion barrier [45]. Leroux’s lab also proved that ARL6/BBS3 GTPases activity is required for the ciliary Wnt signaling in mammalian cells [42]. Interestingly, other than the ciliary entry of membrane receptors, loss of Arl6 can affect the retrograde transport of sensory receptor Smoothened inside cilia in mammalian cells, which leads to modest decrease in ciliary Shh signaling [44]. Studies in our lab also confirmed that various sensory receptors consistently mislocalize in arl-6 knockout worms (unpublished data in the Hu lab). Taken together, these evidences suggest that the role of Arl6 in the context of cilia is highly conserved and the variety of symptoms found in BBS patients is likely due to the compromised ciliary targeting or removal of various sensory receptors and/or signaling molecules.
In drastic contrast to its essential role in cilia signaling, Arl6 appears to be dispensable in the process of cilia formation. In C. elegans, all other bbs mutants exhibited disrupted IFT integrity and slightly truncated cilia [49], whereas arl-6−/− animals show completely normal IFT and cilia morphology (unpublished data in the Hu lab). Similarly, no difference could be found in primary cilia numbers and length between WT and Arl6−/− mice [44]. Interestingly, it was reported that the abrogation of ARL6 activity leads to increased portion of ciliated cells with longer cilia in IMCD3 cells [42], suggesting that Arl6 may function similarly as Arl3 to negatively regulate ciliogenesis. Thus, it would be interesting to examine the potential functional crosstalk between Arl3 and Arl6, which both limit the ciliogenesis.
Arl13, the all-rounder in cilia formation and signaling
Arl13b was initially identified as a candidate that potentially plays a role in multiple ciliated organs in zebrafish [50, 51]. Arl13bhnn mouse embryos show ciliopathy defects in neural tube patterning, limbs and eyes [52]. In 2008, ARL13B mutations were identified in two families with the classical form of Joubert Syndrome (JBTS), which is a rapidly expanding group of ciliopathies characterized by abnormalities in the central nervous system as well as other variable common-shared ciliopathy manifestations such as cystic kidney, blindness, and polydactyly [53, 54]. ARL13B proteins specifically enrich inside cilia in different organisms [19, 51, 52]. For example, when overexpressing a functional GFP-tagged ARL-13 in C. elegans, almost all ARL-13 are transported into the cilium, with little to none protein detected in the cytoplasm [19]. This observation suggests that ARL-13 probably functions exclusively in the context of cilia. Ultrastructurally, the cilia in arl-13−/− worms and Arl13bhnn mice are surprisingly similar, with truncated axoneme plus defective B-tubule closing [19, 52], which suggesting the highly conserved function for ARL-13/Arl13b in cilia.
Using C. elegans as a model, others and we previously demonstrated that ARL-13 regulates cilia formation via maintaining the integrity of IFT particles [19, 55]. Our studies indicated that worm ARL-13 regulates cilia formation by enhancing the binding force between IFT subcomplex A and subcomplex B. In arl-13 mutant worms, the dissociation between IFT-A and IFT-B leads to disrupted IFT integrity and compromised ciliogenesis. ARL-13(R83Q), which resembles the R79Q mutation identified in patients and only retains half of the GTPase activity of the wide-type protein, could not fully rescue the ciliogenesis defect of arl-13−/− worms, which further suggests that the requirement of the GTPase activity of ARL-13 in ciliogenesis [19]. However, until now, no GEF and GAP protein for Arl13b was identified yet.
Except for the defects in cilia structure, abnormalities in cilia signaling pathways, including Sonic Hedgehog (Shh) signaling pathway, Bone Morphogenetic Protein (BMP) signaling pathway, and abnormally expressed Wnt ligands were also reported in Arl13bhnn mouse mutant [52, 56]. It has been proved that the abnormal Shh signaling pathway is a result from the mislocalization of Smoothened (Smo) and Shh signaling components [57]. Several ciliary chemo- or mechano-sensory receptors, like PKD-2, ODR-10, TAX-2, and OSM-9, also mislocalize or accumulate in C. elegans arl-13 mutants [55].
In C. elegans, it is known that PKD-2 does not bind to the moving IFT particles [58], thus, the reported role of ARL-13 in IFT assembly could not explain why sensory receptors, at least for some of them, mislocalize in arl-13 mutant cilia. We proposed that ARL-13 might have different effectors in cilia formation and signaling. Recently, this hypothesis was proved to be true by our discoveries that the SUMOylation modification of ARl-13/ARL13B specifically regulates the proper ciliary targeting of various sensory receptors across species [59]. We found that ARL13B associates with, and as a consequence, is SUMOylated by UBC9 (the sole E2 small ubiquitin-like modifier (SUMO)-conjugating enzyme) both in C. elegans and mammalian cells. Mutations that totally abolish the SUMOylation of ARL-13/ARL13B do not affect the role of ARL-13/ARL13B in ciliogenesis, but only compromise the ciliary targeting of sensory receptors, which indicates that UBC-9 is a specific ARL-13/ARL-13B effector in cilia signaling. Considering the fact that more and more critical signaling pathways have been identified as using primary cilium as a central hub and that JBTS patients exhibit diverse phenotypes in different organs, it would be interesting to verify whether ARL13B SUMOylation also generally regulates cilia sensory functions in every affected organs by determining the proper ciliary targeting of the corresponding sensory receptors [59].
Besides targeting the ciliary sensory receptors, ARL13B also facilitates the ciliary targeting of inositol polyphosphate-5-phosphatase E (INPP5E), another causal protein of JBTS, through a distinct function network formed by ARL13B, INPP5E, phosphodiesterase 6δ (PDE6δ) and centrosomal protein 164 (CEP164) in mammalian cells [60]. This observation raises a couple of interesting questions. First, does ARL13B also regulate the ciliary entry of other ciliopathy proteins, especially JBTS proteins? Second, is this regulation depending on the SUMOylation of ARL13B? Third, since it was found that PDE6δ, one of known ARL3 effector, probably acts upstream of ARL13B in facilitating the ciliary targeting of INPP5E [60], it would be intriguing to examine the functional crosstalk between ARL13B and ARL3 in this cellular process. Although the authors did not observe the mislocalization of INPP5E after knockdown ARL3 [60], it does not exclude the possibility that ARL3 and ARL13B may function redundantly in regulating the ciliary entry of INPP5E. At least in C. elegans, the defects in the ciliary entry of polycystins as well as their ciliary signaling are more severe in arl-3; arl-13 double knockout animals than in arl-3 or arl-13 single knockout animals (unpublished data in the Hu lab).
Concluding remarks
In C. elegans, ARL-3 and ARL-13 both act to stabilize the integrity of IFT complex, but through different mechanisms. Comparing to our understanding that ARL-3 acts through HDAC6 to regulate the binding between IFT-B and KIF17 motor, little is known about how ARL-13 regulates the association between IFT-A and IFT-B [19]. Interestingly, HDAC6 also interacts with BBIP10, a core subunit of the BBSome in mammalian cells [47]. Similar to ARL-13, the C. elegans BBSome also play an important role in stabilizing the association between IFT-A and IFT-B [49]. On the other hand, GTP-bound ARL6/BBS3 recruits the BBSome onto the ciliary membrane in mammalian cells [45]. All these facts suggest unexpectedly sophisticated crosstalks, synergistically and/or antagonistically, exist for the three ciliary Arls in regarding to their roles in cilia biogenesis and signaling.
One key to deduce the coordinated function of Arls in the context of cilia is to identify the ciliary GEFs, GAPs, and effectors of Arls. Small GTPases use GDP/GTP alternation to act as a variety of functional switches critical for cells’ homeostasis. The GTPase switch is turned on by GEFs, which stimulate the release of GDP to allow the binding of GTP, and turned off by GAPs, which accelerate the hydrolysis of GTP. Once activated, small GTPases can bind to various effectors to fulfill the switching role. However, no GEF and only one GAP protein (RP2 for Arl3) was identified for the ciliary Arls so far. Although Arf GEF/GAP proteins are defined by the conserved signature domains and this facilitated the identification process for other Arf GEF/GAP proteins from yeast to human, no evidence demonstrated that the Arf GEF/GAP proteins also work on Arls proteins [11]. Functional identification and characterization of Arl regulators in the context of cilia will undoubtedly provide seminal clues to their ciliary roles. To this end, combining the power of simple genetic models and traditional biochemical toolkits, such as tandem affinity purification, can definitely accelerate the discovery process.
In the past few years, the function of the ciliary Arls has been investigated in greater detail than ever before, and we now have a much better molecular understanding of multiple, yet complicated, roles that these three Arls play in seemingly every biological process inside cilia. No doubt the ciliary Arls have been well accepted to be critical switches in cilia and ciliopathies. However, key questions will keep biologists busy for a while due to our poor understanding of the molecular mechanisms, especially the identity of Arl regulators/effectors. The fact that Arls as well as many of the identified effectors of small GTPases are enzymatic proteins promises the potential that further exploration in this field could lead to the identification of novel therapeutic targets for certain ciliopathies.
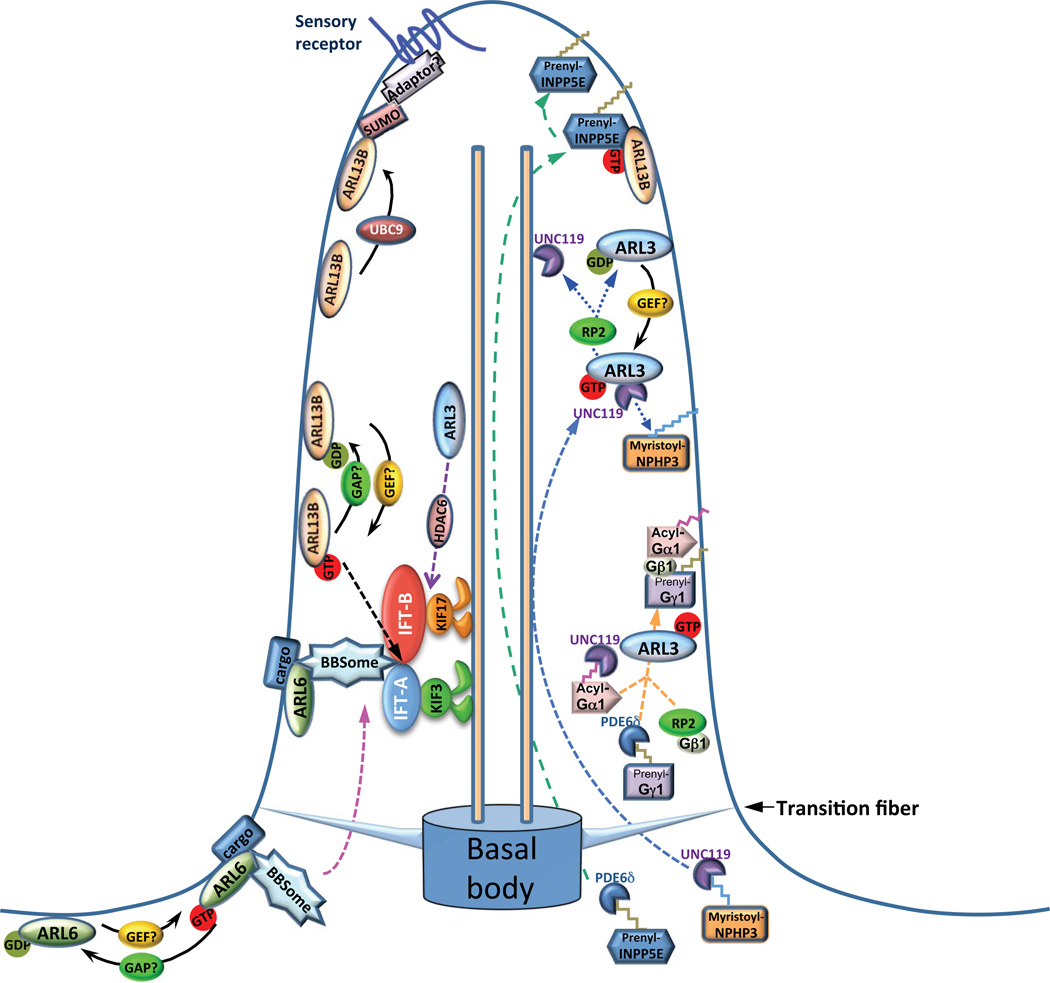
Fig. 1. A working model for the role of ciliary Arls.
Arl3 stabilize the association between IFT-B and the KIF17 motor in an HDAC-6-dependent manner; GTP-bound ARL3 binds to UNC119/myristoylated-NPHP3 complex and release NPHP3 into the cilium. GTP-bound ARL3 works together with different effectors to mediate the proper ciliary trafficking/targeting of transducin. GTP-bound Arl6 recruits the BBSome and sensory receptors into cilia. ARL-13 enhances the association between IFT-A and IFT-B subcomplexes; The SUMOylation of Arl13b by UBC9 is required for the ciliary targeting of sensory receptors; GTP-bound Arl13b interacts with and facilitates the ciliary targeting of INPP5E. Dashed lines indicate the underlying molecular mechanisms remain unclear.
Table 1.
The ciliary Arls and their interactors.
| Name | organism | Gene name | Function | GEF | GAP | Effector |
|---|---|---|---|---|---|---|
| Arl3 | C. elegans | arl-3 | ciliogenesis[19] | HDAC-6[19] | ||
| Mouse | Arl3 | cilia signaling[18, 20] | ||||
| Human | ARL3 | RP2[24] | Golgin-245[26], BART[26], UNC119[27], CCDC104[27], C20orf194[27], PDEδ[25] | |||
| Arl6 | C. elegans | arl-6/bbs-3 | ||||
| Mouse | Arl6/Bbs-3 | cilia signaling[44] | ||||
| Human | ARL6/BBS3 | cilia signaling[45] | The BBSome[45] | |||
| Arl13b | C. elegans | arl-13 | ciliogenesis[19, 55]; sensory receptor targeting[59] | UBC-9[59]; | ||
| Zebrafish | arl13b/scorpion | ciliogenesis[51] | ||||
| Mouse | Arl13b | ciliogenesis[52], cilia signaling[52, 56, 57] | ||||
| Human | ARL13B | UBC-9[59]; INPP5E[60] |
Acknowledgements
We are sorry for not including all the references due to the space limitation. J.H. and coworkers were supported by the National Institutes of Health research grant 1R01DK090038 and P30 center grant P30DK90728, a Pilot and Feasibility Award from the Mayo Clinic Center for Cell Signaling in Gastroenterology (P30DK084567), and the PKD Foundation Young Investigator Award 04YI09a. K.L. was supported by National Cancer Institute (NCI; 1R01CA149039-01A1), Susan G. Komen for the Cure (KG100902).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Goetz SC, Anderson KV. The primary cilium: a signalling centre during vertebrate development. Nat Rev Genet. 2010;11:331–344. doi: 10.1038/nrg2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harris PC. Molecular basis of polycystic kidney disease: PKD1, PKD2 and PKHD1. Curr Opin Nephrol Hypertens. 2002;11:309–314. doi: 10.1097/00041552-200205000-00007. [DOI] [PubMed] [Google Scholar]
- 3.Singla V, Reiter JF. The primary cilium as the cell's antenna: signaling at a sensory organelle. Science. 2006;313:629–633. doi: 10.1126/science.1124534. [DOI] [PubMed] [Google Scholar]
- 4.Davis EE, Katsanis N. The ciliopathies: a transitional model into systems biology of human genetic disease. Curr Opin Genet Dev. 2012;22:290–303. doi: 10.1016/j.gde.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Badano JL, Mitsuma N, Beales PL, Katsanis N. The ciliopathies: an emerging class of human genetic disorders. Annu Rev Genomics Hum Genet. 2006;7:125–148. doi: 10.1146/annurev.genom.7.080505.115610. [DOI] [PubMed] [Google Scholar]
- 6.Rosenbaum JL, Witman GB. Intraflagellar transport. Nat Rev Mol Cell Biol. 2002;3:813–825. doi: 10.1038/nrm952. [DOI] [PubMed] [Google Scholar]
- 7.Benard V, Bokoch GM, Diebold BA. Potential drug targets: small GTPases that regulate leukocyte function. Trends Pharmacol Sci. 1999;20:365–370. doi: 10.1016/s0165-6147(99)01367-x. [DOI] [PubMed] [Google Scholar]
- 8.Vigil D, Cherfils J, Rossman KL, Der CJ. Ras superfamily GEFs and GAPs: validated and tractable targets for cancer therapy? Nat Rev Cancer. 2010;10:842–857. doi: 10.1038/nrc2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Y, Hu J. Small GTPases and cilia. Protein Cell. 2011;2:13–25. doi: 10.1007/s13238-011-1004-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Y, Ling K, Hu J. The emerging role of Arf/Arl small GTPases in cilia and ciliopathies. J Cell Biochem. 2012;113:2201–2207. doi: 10.1002/jcb.24116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burd CG, Strochlic TI, Gangi Setty SR. Arf-like GTPases: not so Arf-like after all. Trends Cell Biol. 2004;14:687–694. doi: 10.1016/j.tcb.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 12.Cherfils J, Chardin P. GEFs: structural basis for their activation of small GTP-binding proteins. Trends Biochem Sci. 1999;24:306–311. doi: 10.1016/s0968-0004(99)01429-2. [DOI] [PubMed] [Google Scholar]
- 13.Donovan S, Shannon KM, Bollag G. GTPase activating proteins: critical regulators of intracellular signaling. Biochim Biophys Acta. 2002;1602:23–45. doi: 10.1016/s0304-419x(01)00041-5. [DOI] [PubMed] [Google Scholar]
- 14.Bernards A, Settleman J. GAP control: regulating the regulators of small GTPases. Trends Cell Biol. 2004;14:377–385. doi: 10.1016/j.tcb.2004.05.003. [DOI] [PubMed] [Google Scholar]
- 15.Cuvillier A, Redon F, Antoine JC, Chardin P, DeVos T, Merlin G. LdARL-3A, a Leishmania promastigote-specific ADP-ribosylation factor-like protein, is essential for flagellum integrity. J Cell Sci. 2000;113(Pt 11):2065–2074. doi: 10.1242/jcs.113.11.2065. [DOI] [PubMed] [Google Scholar]
- 16.Grayson C, Bartolini F, Chapple JP, Willison KR, Bhamidipati A, Lewis SA, Luthert PJ, Hardcastle AJ, Cowan NJ, Cheetham ME. Localization in the human retina of the X-linked retinitis pigmentosa protein RP2, its homologue cofactor C and the RP2 interacting protein Arl3. Hum Mol Genet. 2002;11:3065–3074. doi: 10.1093/hmg/11.24.3065. [DOI] [PubMed] [Google Scholar]
- 17.Avidor-Reiss T, Maer AM, Koundakjian E, Polyanovsky A, Keil T, Subramaniam S, Zuker CS. Decoding cilia function: defining specialized genes required for compartmentalized cilia biogenesis. Cell. 2004;117:527–539. doi: 10.1016/s0092-8674(04)00412-x. [DOI] [PubMed] [Google Scholar]
- 18.Schrick JJ, Vogel P, Abuin A, Hampton B, Rice DS. ADP-ribosylation factor-like 3 is involved in kidney and photoreceptor development. Am J Pathol. 2006;168:1288–1298. doi: 10.2353/ajpath.2006.050941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Y, Wei Q, Zhang Y, Ling K, Hu J. The small GTPases ARL-13 and ARL-3 coordinate intraflagellar transport and ciliogenesis. J Cell Biol. 2010;189:1039–1051. doi: 10.1083/jcb.200912001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lai CK, Gupta N, Wen X, Rangell L, Chih B, Peterson AS, Bazan JF, Li L, Scales SJ. Functional characterization of putative cilia genes by high-content analysis. Mol Biol Cell. 2011;22:1104–1119. doi: 10.1091/mbc.E10-07-0596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barr MM, DeModena J, Braun D, Nguyen CQ, Hall DH, Sternberg PW. The Caenorhabditis elegans autosomal dominant polycystic kidney disease gene homologs lov-1 and pkd-2 act in the same pathway. Curr Biol. 2001;11:1341–1346. doi: 10.1016/s0960-9822(01)00423-7. [DOI] [PubMed] [Google Scholar]
- 22.Barr MM, Sternberg PW. A polycystic kidney-disease gene homologue required for male mating behaviour in C. elegans. Nature. 1999;401:386–389. doi: 10.1038/43913. [DOI] [PubMed] [Google Scholar]
- 23.Schwarz N, Hardcastle AJ, Cheetham ME. Arl3 and RP2 mediated assembly and traffic of membrane associated cilia proteins. Vision Res. 2012;75:2–4. doi: 10.1016/j.visres.2012.07.016. [DOI] [PubMed] [Google Scholar]
- 24.Veltel S, Gasper R, Eisenacher E, Wittinghofer A. The retinitis pigmentosa 2 gene product is a GTPase-activating protein for Arf-like 3. Nat Struct Mol Biol. 2008;15:373–380. doi: 10.1038/nsmb.1396. [DOI] [PubMed] [Google Scholar]
- 25.Linari M, Hanzal-Bayer M, Becker J. The delta subunit of rod specific cyclic GMP phosphodiesterase, PDE delta, interacts with the Arf-like protein Arl3 in a GTP specific manner. FEBS letters. 1999;458:55–59. doi: 10.1016/s0014-5793(99)01117-5. [DOI] [PubMed] [Google Scholar]
- 26.Van Valkenburgh H, Shern JF, Sharer JD, Zhu X, Kahn RA. ADP-ribosylation factors (ARFs) and ARF-like 1 (ARL1) have both specific and shared effectors: characterizing ARL1-binding proteins. The Journal of biological chemistry. 2001;276:22826–22837. doi: 10.1074/jbc.M102359200. [DOI] [PubMed] [Google Scholar]
- 27.Wright KJ, Baye LM, Olivier-Mason A, Mukhopadhyay S, Sang L, Kwong M, Wang W, Pretorius PR, Sheffield VC, Sengupta P, Slusarski DC, Jackson PK. An ARL3-UNC119-RP2 GTPase cycle targets myristoylated NPHP3 to the primary cilium. Genes & development. 2011;25:2347–2360. doi: 10.1101/gad.173443.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Valenzuela-Fernandez A, Cabrero JR, Serrador JM, Sanchez-Madrid F. HDAC6: a key regulator of cytoskeleton, cell migration and cell-cell interactions. Trends Cell Biol. 2008;18:291–297. doi: 10.1016/j.tcb.2008.04.003. [DOI] [PubMed] [Google Scholar]
- 29.Pugacheva EN, Jablonski SA, Hartman TR, Henske EP, Golemis EA. HEF1-dependent Aurora A activation induces disassembly of the primary cilium. Cell. 2007;129:1351–1363. doi: 10.1016/j.cell.2007.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gradilone SA, Radtke BN, Bogert PS, Huang BQ, Gajdos GB, Larusso NF. HDAC6 inhibition restores ciliary expression and decreases tumor growth. Cancer Res. 2013 doi: 10.1158/0008-5472.CAN-12-2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cavenagh MM, Breiner M, Schurmann A, Rosenwald AG, Terui T, Zhang C, Randazzo PA, Adams M, Joost HG, Kahn RA. ADP-ribosylation factor (ARF)-like 3, a new member of the ARF family of GTP-binding proteins cloned from human and rat tissues. J Biol Chem. 1994;269:18937–18942. [PubMed] [Google Scholar]
- 32.Zhang H, Liu XH, Zhang K, Chen CK, Frederick JM, Prestwich GD, Baehr W. Photoreceptor cGMP phosphodiesterase delta subunit (PDEdelta) functions as a prenyl-binding protein. J Biol Chem. 2004;279:407–413. doi: 10.1074/jbc.M306559200. [DOI] [PubMed] [Google Scholar]
- 33.Zhang H, Li S, Doan T, Rieke F, Detwiler PB, Frederick JM, Baehr W. Deletion of PrBP/delta impedes transport of GRK1 and PDE6 catalytic subunits to photoreceptor outer segments. Proc Natl Acad Sci U S A. 2007;104:8857–8862. doi: 10.1073/pnas.0701681104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ismail SA, Chen YX, Rusinova A, Chandra A, Bierbaum M, Gremer L, Triola G, Waldmann H, Bastiaens PI, Wittinghofer A. Arl2-GTP and Arl3-GTP regulate a GDI-like transport system for farnesylated cargo. Nat Chem Biol. 2011;7:942–949. doi: 10.1038/nchembio.686. [DOI] [PubMed] [Google Scholar]
- 35.Zhang H, Constantine R, Vorobiev S, Chen Y, Seetharaman J, Huang YJ, Xiao R, Montelione GT, Gerstner CD, Davis MW, Inana G, Whitby FG, Jorgensen EM, Hill CP, Tong L, Baehr W. UNC119 is required for G protein trafficking in sensory neurons. Nat Neurosci. 2011;14:874–880. doi: 10.1038/nn.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fan Y, Esmail MA, Ansley SJ, Blacque OE, Boroevich K, Ross AJ, Moore SJ, Badano JL, May-Simera H, Compton DS, Green JS, Lewis RA, van Haelst MM, Parfrey PS, Baillie DL, Beales PL, Katsanis N, Davidson WS, Leroux MR. Mutations in a member of the Ras superfamily of small GTP-binding proteins causes Bardet-Biedl syndrome. Nat Genet. 2004;36:989–993. doi: 10.1038/ng1414. [DOI] [PubMed] [Google Scholar]
- 37.Zaghloul NA, Katsanis N. Mechanistic insights into Bardet-Biedl syndrome, a model ciliopathy. J Clin Invest. 2009;119:428–437. doi: 10.1172/JCI37041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim SK, Shindo A, Park TJ, Oh EC, Ghosh S, Gray RS, Lewis RA, Johnson CA, Attie-Bittach T, Katsanis N, Wallingford JB. Planar cell polarity acts through septins to control collective cell movement and ciliogenesis. Science. 2010;329:1337–1340. doi: 10.1126/science.1191184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marion V, Stutzmann F, Gerard M, De Melo C, Schaefer E, Claussmann A, Helle S, Delague V, Souied E, Barrey C, Verloes A, Stoetzel C, Dollfus H. Exome sequencing identifies mutations in LZTFL1, a BBSome and smoothened trafficking regulator, in a family with Bardet--Biedl syndrome with situs inversus and insertional polydactyly. J Med Genet. 2012;49:317–321. doi: 10.1136/jmedgenet-2012-100737. [DOI] [PubMed] [Google Scholar]
- 40.Otto EA, Hurd TW, Airik R, Chaki M, Zhou W, Stoetzel C, Patil SB, Levy S, Ghosh AK, Murga-Zamalloa CA, van Reeuwijk J, Letteboer SJ, Sang L, Giles RH, Liu Q, Coene KL, Estrada-Cuzcano A, Collin RW, McLaughlin HM, Held S, Kasanuki JM, Ramaswami G, Conte J, Lopez I, Washburn J, Macdonald J, Hu J, Yamashita Y, Maher ER, Guay-Woodford LM, Neumann HP, Obermuller N, Koenekoop RK, Bergmann C, Bei X, Lewis RA, Katsanis N, Lopes V, Williams DS, Lyons RH, Dang CV, Brito DA, Dias MB, Zhang X, Cavalcoli JD, Nurnberg G, Nurnberg P, Pierce EA, Jackson PK, Antignac C, Saunier S, Roepman R, Dollfus H, Khanna H, Hildebrandt F. Candidate exome capture identifies mutation of SDCCAG8 as the cause of a retinal-renal ciliopathy. Nature genetics. 2010;42:840–850. doi: 10.1038/ng.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chiang AP, Nishimura D, Searby C, Elbedour K, Carmi R, Ferguson AL, Secrist J, Braun T, Casavant T, Stone EM, Sheffield VC. Comparative genomic analysis identifies an ADP-ribosylation factor-like gene as the cause of Bardet-Biedl syndrome (BBS3) Am J Hum Genet. 2004;75:475–484. doi: 10.1086/423903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wiens CJ, Tong Y, Esmail MA, Oh E, Gerdes JM, Wang J, Tempel W, Rattner JB, Katsanis N, Park HW, Leroux MR. Bardet-Biedl syndrome-associated small GTPase ARL6 (BBS3) functions at or near the ciliary gate and modulates Wnt signaling. J Biol Chem. 2010;285:16218–16230. doi: 10.1074/jbc.M109.070953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yen HJ, Tayeh MK, Mullins RF, Stone EM, Sheffield VC, Slusarski DC. Bardet-Biedl syndrome genes are important in retrograde intracellular trafficking and Kupffer's vesicle cilia function. Hum Mol Genet. 2006;15:667–677. doi: 10.1093/hmg/ddi468. [DOI] [PubMed] [Google Scholar]
- 44.Zhang Q, Nishimura D, Seo S, Vogel T, Morgan DA, Searby C, Bugge K, Stone EM, Rahmouni K, Sheffield VC. Bardet-Biedl syndrome 3 (Bbs3) knockout mouse model reveals common BBS-associated phenotypes and Bbs3 unique phenotypes. Proc Natl Acad Sci U S A. 2011;108:20678–20683. doi: 10.1073/pnas.1113220108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jin H, White SR, Shida T, Schulz S, Aguiar M, Gygi SP, Bazan JF, Nachury MV. The conserved Bardet-Biedl syndrome proteins assemble a coat that traffics membrane proteins to cilia. Cell. 2010;141:1208–1219. doi: 10.1016/j.cell.2010.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nachury MV, Loktev AV, Zhang Q, Westlake CJ, Peranen J, Merdes A, Slusarski DC, Scheller RH, Bazan JF, Sheffield VC, Jackson PK. A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell. 2007;129:1201–1213. doi: 10.1016/j.cell.2007.03.053. [DOI] [PubMed] [Google Scholar]
- 47.Loktev AV, Zhang Q, Beck JS, Searby CC, Scheetz TE, Bazan JF, Slusarski DC, Sheffield VC, Jackson PK, Nachury MV. A BBSome subunit links ciliogenesis, microtubule stability, and acetylation. Dev Cell. 2008;15:854–865. doi: 10.1016/j.devcel.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 48.Berbari NF, Lewis JS, Bishop GA, Askwith CC, Mykytyn K. Bardet-Biedl syndrome proteins are required for the localization of G protein-coupled receptors to primary cilia. Proc Natl Acad Sci U S A. 2008;105:4242–4246. doi: 10.1073/pnas.0711027105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ou G, Blacque OE, Snow JJ, Leroux MR, Scholey JM. Functional coordination of intraflagellar transport motors. Nature. 2005;436:583–587. doi: 10.1038/nature03818. [DOI] [PubMed] [Google Scholar]
- 50.Sun Z, Amsterdam A, Pazour GJ, Cole DG, Miller MS, Hopkins N. A genetic screen in zebrafish identifies cilia genes as a principal cause of cystic kidney. Development. 2004;131:4085–4093. doi: 10.1242/dev.01240. [DOI] [PubMed] [Google Scholar]
- 51.Duldulao NA, Lee S, Sun Z. Cilia localization is essential for in vivo functions of the Joubert syndrome protein Arl13b/Scorpion. Development. 2009;136:4033–4042. doi: 10.1242/dev.036350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Caspary T, Larkins CE, Anderson KV. The graded response to Sonic Hedgehog depends on cilia architecture. Dev Cell. 2007;12:767–778. doi: 10.1016/j.devcel.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 53.Cantagrel V, Silhavy JL, Bielas SL, Swistun D, Marsh SE, Bertrand JY, Audollent S, Attie-Bitach T, Holden KR, Dobyns WB, Traver D, Al-Gazali L, Ali BR, Lindner TH, Caspary T, Otto EA, Hildebrandt F, Glass IA, Logan CV, Johnson CA, Bennett C, Brancati F, Valente EM, Woods CG, Gleeson JG. Mutations in the cilia gene ARL13B lead to the classical form of Joubert syndrome. Am J Hum Genet. 2008;83:170–179. doi: 10.1016/j.ajhg.2008.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Parisi MA. Clinical and molecular features of Joubert syndrome and related disorders. Am J Med Genet C Semin Med Genet. 2009;151C:326–340. doi: 10.1002/ajmg.c.30229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cevik S, Hori Y, Kaplan OI, Kida K, Toivenon T, Foley-Fisher C, Cottell D, Katada T, Kontani K, Blacque OE. Joubert syndrome Arl13b functions at ciliary membranes and stabilizes protein transport in Caenorhabditis elegans. J Cell Biol. 2010;188:953–969. doi: 10.1083/jcb.200908133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Horner VL, Caspary T. Disrupted dorsal neural tube BMP signaling in the cilia mutant Arl13b hnn stems from abnormal Shh signaling. Dev Biol. 2011;355:43–54. doi: 10.1016/j.ydbio.2011.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Larkins CE, Aviles GD, East MP, Kahn RA, Caspary T. Arl13b regulates ciliogenesis and the dynamic localization of Shh signaling proteins. Mol Biol Cell. 2011;22:4694–4703. doi: 10.1091/mbc.E10-12-0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Qin H, Burnette DT, Bae YK, Forscher P, Barr MM, Rosenbaum JL. Intraflagellar transport is required for the vectorial movement of TRPV channels in the ciliary membrane. Curr Biol. 2005;15:1695–1699. doi: 10.1016/j.cub.2005.08.047. [DOI] [PubMed] [Google Scholar]
- 59.Li Y, Zhang Q, Wei Q, Zhang Y, Ling K, Hu J. SUMOylation of the small GTPase ARL-13 promotes ciliary targeting of sensory receptors. J Cell Biol. 2012;199:589–598. doi: 10.1083/jcb.201203150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Humbert MC, Weihbrecht K, Searby CC, Li Y, Pope RM, Sheffield VC, Seo S. ARL13B, PDE6D, and CEP164 form a functional network for INPP5E ciliary targeting. Proc Natl Acad Sci U S A. 2012;109:19691–19696. doi: 10.1073/pnas.1210916109. [DOI] [PMC free article] [PubMed] [Google Scholar]