Abstract
Kaposi's sarcoma-associated herpesvirus (KSHV) maintains a latent infection in primary effusion lymphoma (PEL) cells, but treatment with tetradecanoyl phorbol acetate (TPA) can trigger the full lytic-cycle replication in some of these cells. During lytic-cycle replication, the KSHV-encoded replication and transcription activator (RTA or ORF50), the mRNA transport and accumulation protein (MTA), and the replication-associated protein (RAP) all play crucial roles in expression of downstream viral genes as well as in mediation of viral DNA replication. The cellular CCAAT/enhancer-binding protein alpha (C/EBPα) is induced in TPA-treated PEL cells and contributes to transactivation of the promoters for all of these genes through both direct binding and cooperative interactions with RTA and RAP targeted to upstream C/EBP sites. However, little is known about how RTA expression is triggered initially at the earliest stages after TPA induction when the C/EBPα levels are still limited. Treatment with TPA proved to significantly induce both AP1 DNA-binding activity and levels of activated phosphorylated cJUN in PEL cells and ectopic expression of cJUN-plus-cFOS-induced RTA protein expression in PEL cells. Cotransfected cJUN plus cFOS or TPA treatment transactivated the KSHV RTA, RAP, and MTA promoters in an AP1-binding site-dependent manner in all three promoters. Chromatin immunoprecipitation assays confirmed that cJUN associates with these KSHV target promoters in PEL cells as early as 4 h after TPA treatment. Furthermore, the KSHV RTA and RAP proteins both interact with cJUN or both cJUN and cFOS in vitro or by coimmunoprecipitation from induced PEL cells and enhance cJUN-plus-cFOS-mediated transactivation of these viral promoters. Both increased phosphorylated cJUN and AP1 DNA-binding activity was detected as early as 1 h after TPA treatment in PEL cells, suggesting that AP1 activity may be crucial for very early activation of the RAP, MTA, and RTA promoters during the KSHV lytic cycle. Finally, expression of RTA alone increased cJUN protein levels severalfold in DG75 cells but did not induce cJUN phosphorylation. Therefore, we suggest that the initiating effects of TPA via the AP1 pathway in PEL cells need to be amplified by RTA for full lytic-cycle induction.
Kaposi's sarcoma-associated herpesvirus (KSHV or human herpesvirus 8) is a gamma 2 class rhadinovirus (43) that establishes latent infection in vivo in both “spindloid” endothelial cells and plasmablast-like B cells and is associated with all forms of Kaposi's sarcoma (8) as well as with AIDS-associated multicentric Castleman's disease and primary effusion lymphoma (PEL) (3, 4, 50). In vitro culture of KSHV has been limited to several latently infected but inducible PEL cell lines harboring multicopy KSHV episomes and to primary or immortalized dermal microvascular endothelial cells (3, 4, 12). KSHV gene expression during latency is largely limited to the LANA, vCYC-D, and vFLIP proteins (5, 39). Disruption of KSHV latency and induction of full lytic-cycle progression can be triggered in some PEL cell lines by treatment with tetradecanoyl phorbol acetate (TPA) or sodium butyrate (37, 41). Upon viral reactivation, transcription of lytic genes carried by KSHV is extensively induced and the genes are expressed in a characteristic kinetic pattern. Traditional herpesvirus immediate-early (IE) genes encode nuclear regulatory proteins that are crucial to the switch from latency into the lytic cycle; their expression does not require new protein synthesis and is therefore resistant to the protein synthesis inhibitor cycloheximide (45, 63). The 110-kDa RTA (replication and transcription activator) protein, also known as ORF50, is the only KSHV IE gene product that can trigger the full lytic-cycle replication process when exogenously expressed in latently infected B lymphocytes (19, 35, 36, 38, 51).
KSHV RTA is a highly diverged homologue of the Epstein-Barr virus (EBV) DNA-binding transactivator RTA or BRLF1 (23, 40, 60), which can also trigger the lytic-cycle program in cells latently infected with EBV. Both are DNA-binding proteins with classical C-terminal acidic activation domains (7, 22, 24, 34, 46, 52). Several KSHV early lytic genes that have been identified so far as directly responding targets for RTA include the replication-associated protein (RAP or K8, a positional homologue of EBV ZTA), MTA (or ORF57, a homologue of EBV MTA), and PAN (or T1.1 polyadenylated nuclear RNA) genes, as well as the vIL6, K12 (Kaposin) and vOX2/GPCR genes (7, 11, 16, 34-36, 46, 49, 52). We have previously identified one or more functional CCAAT/enhancer-binding protein (C/EBP) binding sites in each of the first three promoters listed above (54, 55). At least two types of RTA-responsive elements (RREs) have been characterized, of which only the type I RRE found in the PAN promoter binds RTA directly with high affinity (7, 34, 48, 49, 54, 55). Although more-complex oligomeric interactions may occur (31), the defined type II RREs found in the RAP and MTA promoters themselves have low direct binding affinity for RTA in electrophoretic mobility shift assay (EMSA) experiments, but both encompass strong binding sites for C/EBPα, which mediates cooperative transactivation probably via DNA-bound C/EBPα-RTA protein complexes (54, 55). In the MTA promoter, RTA activates gene expression through interaction with the cellular CBF1 (CSL or RBP-Jκ) protein at a CBF1 binding site just downstream from the C/EBP binding site within the RRE (29, 30); however, no consensus CBF1 motif is present within the RAP promoter. Although RTA transactivates the PAN promoter through direct binding to the type I RRE, mutations in the C/EBP sites outside the originally defined RRE also lead to a partial decrease in RTA activation of the PAN promoter (55). It has been suggested that, besides C/EBPα and CBF1, cellular factors such as OCT, CREB, K-RBP, and SP1 play roles in RTA transactivation on certain target promoters (21, 44, 53, 61).
RTA has also been reported to positively autoregulate its own promoter (15, 44). However, in our experience this occurs only indirectly through the interaction of RTA with C/EBPα at one or more C/EBPα-responsive binding sites identified within the proximal RTA promoter (55). Therefore C/EBPα mediates RTA activation on all four of these early KSHV promoters during viral reactivation. Furthermore, strong induction of C/EBPα protein levels in those TPA-treated PEL cells that enter the lytic cycle is also observed, as judged by double-label indirect immunofluorescence assays (IFA) for RTA and RAP expression. This appears to create a self-reinforcing loop, because RTA and RAP, which also interact with each other (26, 54), both enhance C/EBPα-mediated transcriptional autoregulation of the C/EBPα promoter. They also both stabilize preexisting levels of short-half-life C/EBPα through protein-protein interactions (55, 59). In chromatin immunoprecipitation (ChIP) assays, C/EBPα, RTA, and RAP have all been detected in association with the C/EBPα, RTA, RAP, MTA, and PAN promoters in vivo in PEL cells, but not until at least 8 to 12 h after TPA treatment (54, 55, 59).
The bZIP (basic region leucine zipper) group of transcription factors include the JUN family, the FOS family, the CREB/ATF family, and the C/EBP family as well as the two gammaherpesvirus-encoded proteins EBV ZTA and KSHV RAP (9, 27, 32, 33, 47, 56). The most potent transcriptional activator in the JUN family is cJUN, and cFOS also contains a transcriptional activation domain. However, cFOS cannot homodimerize and instead forms stable heterodimers with JUN family proteins, a process which enhances their DNA-binding activity (47). Classic AP1 transcriptional activating complexes comprise cJUN and cFOS subunits that specifically bind to the DNA sequence 5′-TGA(G/C)TCA-3′, which is known as an AP1 site or a TPA-responsive element, known as TRE (2, 47). This same consensus AP1 motif is also a strong target recognition motif for EBV ZTA but is not bound by KSHV RAP (9, 59).
The phosphorylation and activation of cJUN are regulated by cJUN N-terminal kinase (JNK), which can be activated by TPA via protein kinase C (PKC) or mitogen-activated protein kinase pathways in numerous human cell lines (62). It has been previously observed that high levels of AP1 DNA-binding activity are induced upon TPA treatment of both EBV-positive Raji cells (32, 42) and KSHV-positive BCBL1 PEL cells (28). Therefore, in this study, we asked whether the induction of AP1-binding activity by TPA might contribute to KSHV lytic-cycle gene activation through AP1-mediated transcriptional regulation. The KSHV RTA and RAP proteins, which have both been shown to interact with and cooperate with C/EBPα in our previous work, also proved to functionally associate with cJUN-plus-cFOS transcriptional activating complexes and to enhance the AP1-mediated transactivation of KSHV early gene expression.
MATERIALS AND METHODS
Cells and plasmids.
HeLa cells were grown in Dulbecco's modified Eagle's medium (Invitrogen, Carlsbad, Calif.) containing 10% fetal bovine serum in a humidified 5% CO2 incubator at 37°C. KSHV-positive human PEL cell lines BCBL1 and BC3, as well as the KSHV-negative DG75 B-lymphoblast cell line and KSHV-negative U937 cells, were grown in RPMI 1640 medium (Invitrogen) containing 10% fetal bovine serum. TPA was added to the medium at a final concentration of 20 ng/ml for KSHV lytic-cycle induction in PEL cells and in DG75 cells, at 50 ng/ml for TPA treatment in U937 cells, and at 60 ng/ml for TPA treatment in HeLa cells.
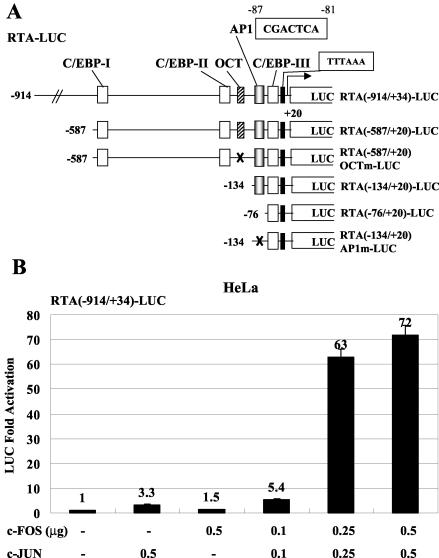
Plasmid pHC125C expresses Flag-tagged full-length human C/EBPα driven by the simian virus 40 (SV40) promoter-enhancer (SV2); plasmids pHC91A, pHC66, and pcJUN-DBD (kindly provided by Michael Birrer, National Cancer Institute) express SV2-driven wild-type cFOS, Flag-tagged wild-type cJUN, and a cJUN DNA-binding domain (DBD) mutant protein JUN(A→D265In265), respectively. The last contains an alanine-to-glutamic acid point mutation at amino acid 265 and an adjacent insertion of three acidic amino acids (2). Expression plasmids carrying full-length KSHV RTA(1-691) cDNA and Flag-tagged KSHV RAP(1-237) effector genes in pJX15 and pCJC581a, respectively, were described previously (54, 55). Plasmids pSEW-R21 and pFYW-07 express glutathione S-transferase (GST) fusion versions of the full-length RTA and RAP proteins. Plasmid pSEW-P01 contains the RAP(−190/+10)-LUC reporter gene as described previously (54). Three mutated reporter clones, RAP(PM1)-LUC (with TGT at positions −87 to −85 replaced with ATC), expressed by plasmid pSEW-P02, RAP(PM2)-LUC (with ACA at positions −71 to −69 replaced with TTC), expressed by pSEW-P03, and RAP(PM1+2)-LUC (with the two combined mutations), expressed by pSEW-P04, were generated from pSEW-P01. Plasmids pSEW-MP5 and pSEW-MP6 express reporter genes for KSHV MTA(−130/+10)-LUC and MTA(−130/+10)AP1m-LUC (in which TGAG at −121 to −118 has been changed to GATC), respectively. Reporter plasmids pGL3-FL(−914) (55), pSEW-RP1, pDH404, and pDH405 contain the KSHV RTA(−914/+34)-LUC, RTA(−587/+20)-LUC, RTA(−134/+20)-LUC, and RTA(−76/+20)-LUC reporter genes, respectively. Plasmid pSEW-RP8 contains the KSHV RTA(−587/+20 OCTm)-LUC reporter gene, in which ATCT at −214 to −211 has been changed to GGTA to destroy the OCT-binding site, and plasmid pSEW-RP9 contains the KSHV RTA(−134/+20)AP1m-LUC reporter gene in which ACTCA at −85 to −81 has been changed to GATCC to destroy the AP1-binding site.
DNA transfection and LUC assays.
Transfection of HeLa cells was performed with 5 × 105 cells for each 1 μg of DNA sample in six-well plates using Lipofectamine (Invitrogen). The BCBL1 PEL cell line and DG75 B-lymphoblast cell line as well as U937 cells (107 cells/sample) were transfected with 5 to 10 μg of DNA by electroporation (Gene-pulser; Bio-Rad) at 300 V and 950 μF in 0.5 ml of RPMI medium as described previously (53, 54). In all cases, cells were harvested at 48 h posttransfection for LUC assays, which were carried out in triplicate using a LUC assay kit (Promega). Results are presented as means and ranges. All samples also received an added 0.2 μg of cytomegalovirus (CMV) β-galactosidase (β-Gal) expression plasmid DNA as an internal control for transfection variability. Activity of β-Gal in the cell lysate was measured with o-nitrophenol-β-d-galactopyranoside (ONPG) as the substrate. For TPA treatment of DNA-transfected HeLa cells, a final concentration of 60 ng/ml was added at 24 h posttransfection, followed by incubation for another 24 h before harvesting the cells. To normalize for TPA-induced cell apoptosis, protein assays were carried out to determine the total protein levels in TPA-treated and untreated cell lysates using bicinchoninic acid protein assay reagents (Pierce, Rockford, Ill.), although less than 20% divergence was observed before and after TPA treatment.
EMSA.
C/EBPα, cJUN, and cFOS protein samples used for EMSA were in vitro translated with the T7-TNT quick-coupled transcription-translation system (Promega) according to the manufacturer's procedures using corresponding expression plasmids pHC125C, pHC66, and pHC91A as templates. Annealed double-stranded oligonucleotides with 4-bp 5′ overhangs were radiolabeled with [α-32P]dCTP during a Klenow DNA polymerase fill-in reaction. For EMSA, approximately 50,000 cpm of the 32P-labeled probe and 2 μl of in vitro-translated proteins or of BCBL1 nuclear extract were incubated for 30 min at 20°C in a binding system containing 10 mM HEPES (pH 7.5), 50 mM KCl, 1 mM EDTA, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, 1% Triton X-100, 5% glycerol, and 2 μg of poly(dI/dC). For supershift experiments, 0.5 μl of mouse anti-Flag monoclonal antibody (MAb; catalog no. F-3165; Sigma, St. Louis, Mo.), rabbit anti-cJUN polyclonal antibody (PAb; catalog no. sc-1694; Santa Cruz Biotechnology, Santa Cruz, Calif.), or C/EBPα rabbit antiserum (54) was added to the mixture after 30 min and allowed to incubate for another 30 min before gel loading. Samples were separated on a 4.5% polyacrylamide gel in 1× HEE buffer (10 mM HEPES [pH 7.5], 1 mM EDTA, 0.5 mM EGTA) at 150 V and 4°C as described previously (10). The gels were dried and subjected to autoradiography with Kodak X-ray film.
All oligonucleotides used were purchased from Qiagen Operon (Alameda, Calif.). Nucleotide sequences of the annealed DNA probes for AP1 consensus, RAP-PWT, RAP-PM1, RAP-PM2, RAP-PM1+2, MTA-AP1, MTA-AP1m, RTA-AP1, and RTA-AP1m are indicated in the figures.
Recombinant protein expression and in vitro GST affinity binding assay.
GST fusion protein expression was induced in Escherichia coli (BL21 strain) with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 4 h at 30°C. Bacterial pellets were resuspended in ice-cold phosphate-buffered saline (PBS; 150 mM NaCl, 16 mM Na2HPO4, 4 mM NaH2PO4, pH 7.3) and sonicated for 30 s. After centrifugation, clarified lysates were immobilized on the glutathione-Sepharose 4B beads (Amersham Pharmacia). Input [35S]methionine-labeled proteins were synthesized in vitro using the T7-TNT quick-coupled transcription-translation system as described above. Recombinant GST fusion proteins immobilized on beads were pretreated with 0.2 U of DNase I and 0.2 μg of RNase A per μl for 30 min at 20°C in pretreating buffer (50 mM Tris-HCl [pH 8.0], 5 mM MgCl2, 2.5 mM CaCl2, 100 mM NaCl, 5% glycerol, 1 mM dithiothreitol). The beads were washed twice with binding buffer (20 mM Tris-Cl [pH 7.5], 100 mM NaCl, 1 mM EDTA, 0.5% Nonidet P-40, 1 mM dithiothreitol) and blocked in the same buffer containing 10 mg of bovine serum albumin (BSA)/ml for 30 min at 4°C. After being blocked, the bead-immobilized GST fusion protein was resuspended in binding buffer containing 1 mg of BSA/ml, and labeled proteins were subsequently added and allowed to incubate for 1 h at 4°C. The beads were then washed five times in binding buffer at 10-min intervals, resuspended in 20 μl of 2× sodium dodecyl sulfate (SDS)-gel loading buffer, and boiled for 5 min before being loaded onto SDS-polyacrylamide gel electrophoresis (PAGE) gels. After electrophoresis, the gels were fixed in 50% methanol-40% H2O-10% acetic acid for 30 min and dried for X-ray autoradiography.
In vivo coimmunoprecipitation from PEL cell lysates.
Nuclear extracts of BCBL1 cells were prepared for coimmunoprecipitation by procedures similar to those described previously (54, 57). Approximately 108 cells were harvested at 30 h or at various time points after TPA induction (as indicated in Fig. 1B8, 9A9A, 9C, and 12), washed once with PBS, and gently resuspended in 10 ml of cold hypotonic buffer A (10 mM HEPES [pH 7.9], 10 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA, 1 mM dithiothreitol, 0.5 mM phenylmethylsulfonyl fluoride, 1 mM sodium orthovanadate). The mixture was allowed to swell on ice for 30 min, and NP-40 was then added to a final concentration of 0.62%. The mixture was vortexed vigorously for 10 s, and the nuclear pellet was collected after centrifuging for 3 min at 1,200 × g at 4°C. The nuclear pellet was resuspended in 3 ml of cold immunoprecipitation buffer (50 mM Tris-Cl [pH 7.9], 50 mM NaCl, 0.1 mM EDTA, 1% glycerol, 0.2% NP-40, 1 mM dithiothreitol, 0.5 mM phenylmethylsulfonyl fluoride, 1 mM sodium orthovanadate) and sonicated for 2 s at the minimal setting. The samples were then centrifuged at 15,000 × g for 1 min at 4°C, and the supernatant was collected and stored at −70°C. For coimmunoprecipitation, 400 μl of prepared nuclear extract was pretreated with 0.2 U of DNase I and 0.2 μg of RNase A per μl and precleared with 5 μl of rabbit preimmune serum and 100 μl of a 50% slurry of protein A/G (20%:80%)-Sepharose beads for 1 h. After preclearing, 3 μg of anti-RAP or anti-RTA rabbit PAb or rabbit preimmune serum was added to the precleared nuclear extracts and incubated for 1 h at 4°C; then 100 μl of a 50% slurry of protein A/G (20%:80%)-Sepharose beads blocked with 5% BSA in PBS was added to the nuclear extracts and incubated for 1 h at 4°C. The beads were then washed three times with cold immunoprecipitation buffer at 15-min intervals, resuspended in 2× SDS-gel loading buffer, and boiled for 5 min before being loaded onto SDS-PAGE gel.
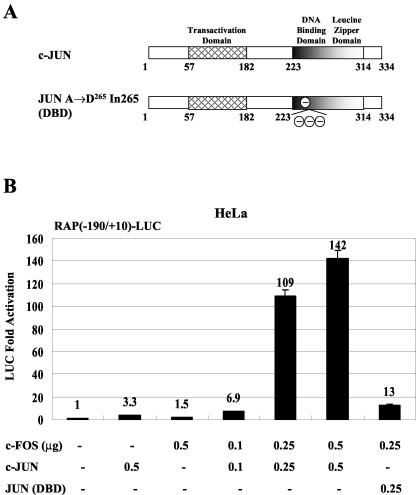
FIG. 1.
cJUN and cFOS activate RAP-LUC reporter gene expression in cotransfected cells. (A) Schematic diagram of both the wild-type cJUN protein and the DBD mutant protein cJUN(A→D265In265). (B) HeLa cells were transfected with 0.2 μg of target plasmid DNA encoding RAP(−190/+10)-LUC and the indicated amounts of effector plasmid DNA expressing either SV2-cJUN, SV2-cFOS, or SV2-cJUN(DBD). The total amount of effector plasmid DNA used in each transfection was normalized to 1 μg by adding empty SV2 promoter vector (pSG5) DNA. Activation of LUC activity relative to that for the basal-level control obtained using 1 μg of empty SV2 promoter vector DNA as the effector was calculated.
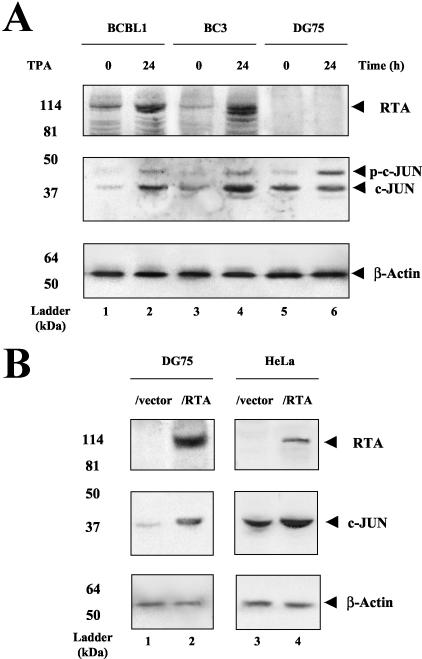
FIG. 9.
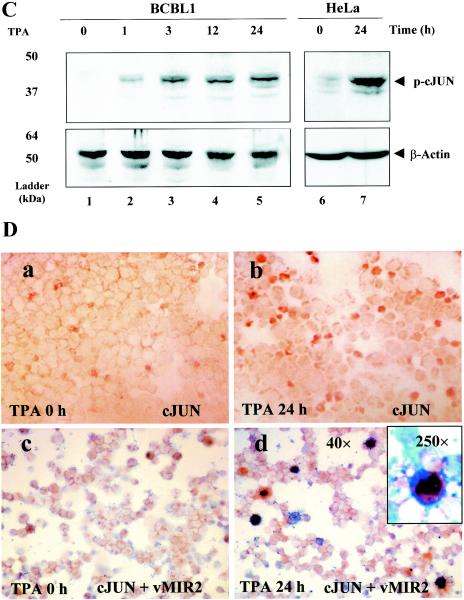
Induction of the phosphorylated form of cJUN after TPA treatment in various cell lines. (A) Protein samples were isolated from BCBL1 (lanes 1 and 2), BC3 (lanes 3 and 4), or DG75 cells (lanes 5 and 6) at either 0 or 24 h after TPA treatment. Western immunoblotting was performed with either the anti-RTA PAb (top) or anti-cJUN PAb (middle). An antibody against β-actin was used as a loading control (bottom). The positions of protein size markers are indicated at the left of each panel. (B) Western immunoblotting with protein samples isolated from DG75 cells (lanes 1 and 2) or HeLa cells (lanes 3 and 4) transfected with empty vector DNA (lanes 1 and 3) or with the SV2-RTA expression plasmid (lanes 2 and 4). (C) BCBL1 cells were harvested at 0, 1, 3, 12, and 24 h after TPA treatment (lanes 1 to 5), or HeLa cells were isolated at 0 and 24 h after TPA treatment (lanes 6 and 7). The cell lysates were then analyzed by Western immunoblotting with a PAb specific for phosphorylated cJUN (top) or a β-actin MAb (bottom) as a loading control. (D) Immunohistochemical staining showing the induction of phosphorylated cJUN in the nuclei of BCBL1 cells undergoing KSHV lytic-cycle replication. BCBL1 cells were harvested at either 0 (a and c) or 24 h (b and d) after TPA treatment. A 3,3′-diaminobenzidine peroxidase (DAB)-labeled antibody (brown) was used to detect the presence of phosphorylated cJUN (a to d). For double staining, True Blue reagent-labeled antibody (blue) was used to detect the presence of the KSHV vMIR2 lytic-cycle protein in the cytoplasm (c and d).
FIG. 12.
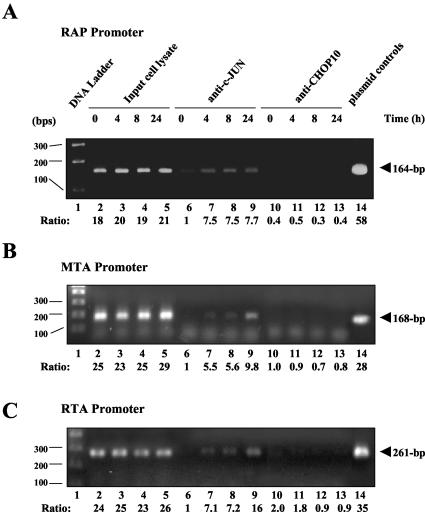
Time course ChIP assays showing the association of cJUN protein with three KSHV target viral promoters during early-lytic-cycle reactivation in PEL cells. At different time points after TPA treatment, aliquots of sonicated chromatin from the BCBL1 cell lysates were precipitated with antibodies against either cJUN or CHOP10. Levels of protein association with the target promoters were measured by DNA PCR using primers specific for the promoter regions of KSHV RAP (A), MTA (B), and RTA (C). Lane 1, DNA size marker; lanes 2 to 5, input lysate controls at 0, 4, 8, and 24 h, respectively; lanes 6 to 13, ChIP assay PCR products from either uninduced input BCBL1 cell lysates (0 h, lanes 6 and 10) or those at 4 (lanes 7 and 11), 8 (lanes 8 and 12), and 24 h (lanes 9 and 13) after TPA induction; lanes 6 to 9, PCR products from immunoprecipitates obtained with the PAb against cJUN; lanes 10 to 13, PCR products from immunoprecipitates obtained with the PAb against CHOP10; lane 14, size control PCR amplification products from the corresponding promoter-driven LUC reporter plasmids encoding RAP(−190/+10)-LUC, MTA(−160/+10)-LUC, and RTA(−914/+34)-LUC.
Protein isolation and Western immunoblot assay.
Approximately 107 BCBL1, BC3, or DG75 cells were harvested at different time points after TPA induction or at 30 h posttransfection, washed once with PBS, gently resuspended in 0.2 ml of ice-cold immunoprecipitation buffer, and sonicated for 30 s. The samples were then centrifuged at 15,000 × g for 5 min at 4°C, and the supernatants were collected. Western blot analysis was performed after SDS-PAGE using 10 μl of each protein sample or coimmunoprecipitated sample. The proteins were detected by immunoblotting with a rabbit antipeptide PAb against KSHV RTA recognizing an epitope mapping between amino acids 527 and 539, a rabbit PAb against cJUN (Santa Cruz Biotechnology; catalog no. sc-1694), or a rabbit PAb against phospho-JUN(Ser73) (catalog no. 06-659; Upstate Biotechnology, Lake Placid, N.Y.) as described previously (54).
Immunohistochemical staining.
Immunohistochemical staining was performed with TPA-induced or uninduced BCBL1 cells. Cells were washed three times in 1× PBS and fixed in 4% paraformaldehyde in 1× PBS for 10 min, followed by boiling in 10 mM sodium/citrate buffer (pH 6.0) for 10 min. After cells were quenched in freshly prepared ethanol-PBS-H2O2 (77:20:3) for 1 h in blocking solution (1% BSA and 10% normal goat serum in 1× PBS with 1% Tween 20), the primary rabbit phospho-JUN(Ser73) PAb was applied first at 1:200 dilution in blocking solution for 12 h at 20°C and detected with a biotinylated goat anti-rabbit immunoglobulin (Ig) antibody plus the streptavidin-peroxidase-ABC complex (Dako Corporation, Carpinteria, Calif.), followed by 3,3′-diaminobenzidine peroxidase substrate (Sigma). For double staining, after cells were washed in 1× PBS and treated with an avidin-biotin blocking kit (Vector Labs, Burlingame, Calif.), the secondary rabbit PAb against the KSHV vMIR2 (ZMP-A or K5) protein (11, 12) was applied at a 1:500 dilution for 1 h at 20°C and then detected with the biotinylated goat anti-rabbit Ig antibody plus the streptavidin-peroxidase-ABC complex, followed by True Blue peroxidase substrate (Kirkegaard & Perry Laboratories, Gaithersburg, Md.).
IFA.
IFA was performed 40 h after BCBL1 cells were transfected by the acetone-methanol (1:1) fixation method (54, 55). Primary antibodies included rabbit antipeptide antisera against KSHV RTA, KSHV vMIR2, and KSHV vIL6 (K2) (1:200 dilution) (3, 12) and a mouse anti-Flag MAb (Sigma; 1:100 dilution). Secondary donkey- or goat-derived rhodamine- or fluorescein isothiocyanate (FITC)-conjugated anti-rabbit or anti-mouse IgG (Jackson Pharmaceuticals, West Grove, Pa.; both at a 1:100 dilution) was used to detect the primary antibodies.
ChIP.
BCBL1 PEL cells were treated with TPA, and 106 cells were harvested at different time points for ChIP assays using a ChIP assay kit (Upstate Biotechnology) in accordance with the manufacturer's protocol. Histones were cross-linked to DNA by adding formaldehyde directly to the culture medium to a final concentration of 1% and incubating for 10 min at 37°C. Cells were then washed twice with ice-cold PBS containing protease inhibitors (1 mM phenylmethylsulfonyl fluoride, 1 μg of aprotinin/ml, and 1 μg of pepstatin A/ml), resuspended in 200 μl of SDS lysis buffer containing the protease inhibitors, and incubated for 10 min on ice. DNA was sheared by sonicating samples on ice, and samples were centrifuged to collect the supernatants. The supernatants were then diluted in 1,800 μl of ChIP dilution buffer before 80 μl of salmon sperm DNA-protein A agarose slurry was added, followed by agitation for 30 min at 4°C. After the agarose beads were spun down, precleared samples were aliquoted, antibodies against cJUN or CHOP10 (Santa Cruz Biotechnology; catalog no. sc-7351) were added to each aliquot of 500 μl, and samples were rotated for 2 h at 4°C. Sixty microliters of salmon sperm DNA-protein A agarose slurry was then added to each sample, and samples were rotated for another hour at 4°C. The beads were subsequently washed in low-salt immune complex wash buffer, high-salt immune complex wash buffer, and LiCl immune complex wash buffer and then washed twice in Tris-EDTA (TE) buffer. After the washing steps, 250 μl of freshly prepared elution buffer (1% SDS, 0.1 M NaHCO3) was added to the beads and samples were vortexed and incubated for 15 min at 20°C by rotation. The supernatants were collected by centrifugation, and the elution was repeated with another 250 μl of elution buffer. Then 20 μl of 5 M NaCl was added to each sample, and samples were incubated at 65°C for 12 h before proteinase K treatment, phenol-chloroform extraction, and ethanol precipitation. DNA pellets were resolved in 20 μl of TE buffer, and 2 μl was used as the template for PCR. Primers LGH3641 and LGH4362 (54), specific for a 164-bp region in the KSHV RAP promoter from −160 to +4, primers LGH4980 and LGH4987 (55), specific for a 168-bp region in the KSHV MTA promoter from −160 to +8, and primers LGH4354 and LGH4355 (55), specific for a 261-bp region in the KSHV RTA promoter from −241 to +20, were used for PCR detection (30 cycles). The PCR products were fractionated on a 2% agarose gel and detected by ethidium bromide staining.
RESULTS
JUN and FOS transactivate the KSHV RAP promoter through an AP1-binding site mapping at −87 to −81.
Studies on EBV gene regulation have defined several AP1-binding elements in various EBV lytic gene promoters; these AP1 elements respond to both AP1 and the EBV transactivator ZTA and therefore play important roles in the regulation of these promoters (32). AP1 DNA-binding activity was observed to be dramatically induced upon TPA treatment of EBV-immortalized B cells (32, 42) and in KSHV-positive PEL cells (28). This is usually considered to be a result of TPA-dependent induction of PKC and JNK, which regulates cJUN activity though phosphorylation (62). Based on these observations, we asked whether the TPA-induced JNK pathway may also play a role in KSHV reactivation by activating viral gene expression through the cJUN-plus-cFOS-containing heterodimeric transcription factor AP1.
We first investigated the effect of AP1 on the KSHV RAP gene promoter by cotransfecting cJUN and cFOS expression plasmids either alone or in combination with the RAP(−190/+10)-LUC target reporter gene. Transfected HeLa cells expressing either cJUN or cFOS alone showed little increase in LUC expression compared to control cells transfected with the empty vector DNA, whereas cells cotransfected with equal amounts of cJUN and cFOS produced up to 142-fold activation of RAP-LUC activity in a dose-responsive manner (Fig. 1B). A cJUN mutant DBD with a disrupted basic region (Fig. 1A) that is unable to recognize consensus AP1 DNA-binding sites (2) was also used as a specificity control in place of the wild-type cJUN in some cotransfection experiments. Activation of the RAP promoter by a combination of JUN(DBD) and cFOS reduced transactivation from 109-fold (0.25 μg each) to 13-fold (Fig. 1B), suggesting that direct binding to specific DNA sites by the AP1 transcription factor complex rather than potential alternative indirect effects is crucial here.
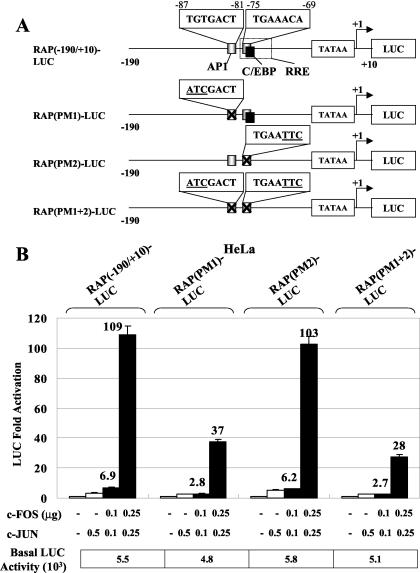
The proximal upstream RAP(−190/+10)-LUC promoter contains two potential AP1-like sequences, located at positions −87 to −81 (5′-TGTGACT-3′) and −75 to −69 (5′-TGAAACA-3′), that diverge from the 7-bp AP1 consensus binding sequence 5′-TGA(G/C)TCA-3′ at one or two positions, respectively (Fig. 2A). The sequence at −75 to −69 overlaps with the previously identified strong C/EBP site and the RRE. To test the potential of these AP1-like sequences to mediate AP1 transactivation, we introduced point mutations into the RAP(−190/+10)-LUC reporter gene to destroy either one or both of the two sites (Fig. 2A). Mutation at −87 to −81 in RAP(PM1)-LUC led to a 3-fold decrease in cJUN-plus-cFOS-mediated activation from 109-fold down to 37-fold (Fig. 2B). However, mutation of the other site at −75 to −69 in RAP(PM2)-LUC, which abolished most of the C/EBPα and the RTA responsiveness (55), had little effect on AP1 responsiveness, although activation of the double-mutant gene fell further to 28-fold. None of the mutations had any significant effect on the basal level of expression. These results indicated that the sequence at positions −87 to −81 may be one of possibly several AP1-responsive elements within the RAP promoter.
FIG. 2.
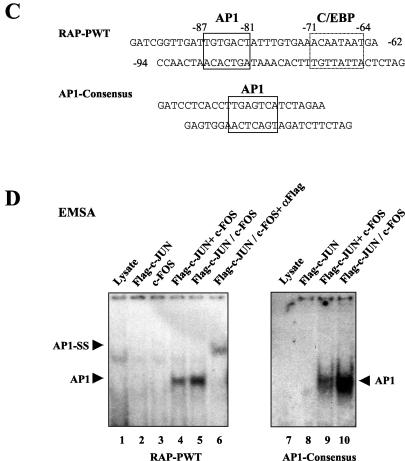
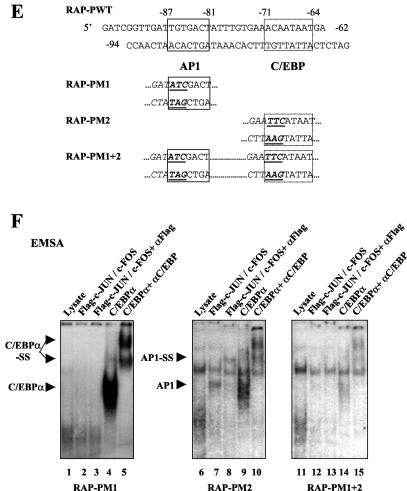
Identification of an AP1-binding site and response element in the KSHV RAP promoter. (A) Schematic diagram of the RAP promoter region between positions −190 and +10 and the positions of the two potential AP1-like sites relative to the known C/EBP and RRE sites. The three specific RAP promoter point mutant LUC reporter genes used in cotransfection experiments are also shown. (B) Responsiveness of wild-type and point mutant RAP-LUC derivatives to AP1 transactivation in cotransfected HeLa cells. Transfections were carried out with 0.2 μg of target reporter promoter DNA and the indicated amounts of SV2-cJUN and SV2-cFOS expression plasmid DNA. The basal activity of each reporter gene cotransfected with vector DNA is also indicated. (C) Sequences of the RAP promoter and AP1 consensus oligonucleotide probes used in EMSA experiments. The potential AP1 sites as well as the previously identified C/EBP site are boxed. (D) Results of an EMSA experiment showing the relative AP1-binding ability of the AP1 motif in the RAP promoter (lanes 1 to 6) compared to that of a consensus AP1 probe used as a positive control (lanes 7 to 10). Lane 1, unprogrammed reticulocyte lysate; lane 2, in vitro-translated Flag-tagged cJUN; lane 3, cFOS alone; lane 4, mixed Flag-cJUN and cFOS; lane 5, cotranslated Flag-cJUN and cFOS. Protein samples were incubated with the 32P-labeled RAP-PWT or AP1 consensus probes. A rabbit anti-Flag antibody was used to generate a specific supershifted band (lane 6). AP1, AP1-specific shifted bands; SS, antibody supershift. (E) Sequences of the wild-type and mutated RAP promoter oligonucleotide probes used for EMSA. Underlining, boldface, and italics indicate mutated positions. (F) EMSA experiment showing binding of in vitro-translated AP1 or C/EBPα to the three 32P-labeled mutated RAP promoter probes. Either the cotranslated Flag-cJUN and cFOS proteins (lanes 2, 3, 7, 8, 12, and 13) or the C/EBPα protein (lanes 4, 5, 9, 10, 14, and 15) was incubated with the probes. Anti-Flag (lanes 3, 8, and 13) or anti-C/EBPα antibodies (lanes 5, 10, and 15) were used to generate specific supershifted bands. Arrowheads, C/EBPα- and AP1-specific shifted bands and their supershifted bands (SS).
To test the ability of this AP1 site to bind to JUN-FOS complexes, we performed EMSA with in vitro-translated Flag-tagged cJUN and untagged cFOS proteins and a 32P-labeled oligonucleotide probe (RAP-PWT) that contains both the identified AP1 and C/EBP sites (Fig. 2C). Neither cJUN nor cFOS alone showed any binding affinity for the probe; however, when the two proteins were either mixed or cotranslated, the heterodimers formed gave a typical AP1 shifted band, which could be further supershifted by the anti-Flag antibody (Fig. 2D). A probe containing a consensus high-affinity AP1-binding motif was also used in the EMSA experiments as a positive control (Fig. 2C and D). When probes corresponding to all three mutant RAP promoter reporters used in the transfection above were tested in the same EMSA experiments (Fig. 2E), RAP-PM1 failed to bind to AP1 but still bound to C/EBPα as expected, whereas RAP-PM2 retained AP1-binding ability but lost most of the C/EBPα binding (Fig. 2F). The double-mutant probe RAP-PM1+2 failed to bind to either of the two proteins. Therefore, these EMSA results are consistent with our cotransfection data and indicate that cJUN plus cFOS transactivate the KSHV RAP gene promoter by a mechanism that includes direct binding to the moderate-affinity AP1 site at the position −87 to −81.
JUN and FOS transactivate the KSHV MTA promoter through an AP1-binding site mapping at −121 to −115.
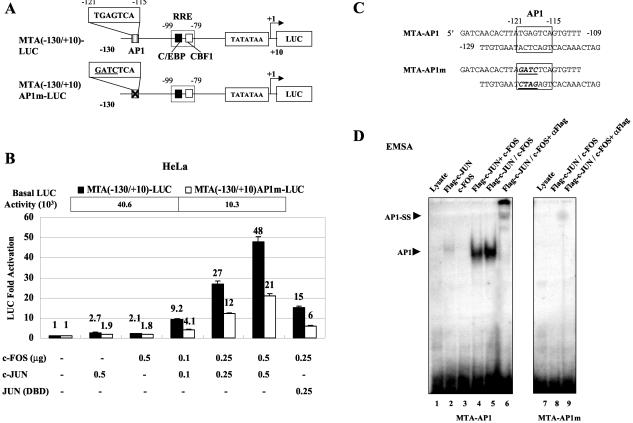
The KSHV MTA promoter is also induced during the early stages of viral reactivation and responds to transactivation by KSHV RTA through a type I RRE, which is partially homologous to that in the RAP promoter and which also does not directly interact with RTA but encompasses a strong functional C/EBP site as well as an adjacent CBF1 site (55). Therefore, we were interested in whether or not AP1 also transactivates the MTA promoter. We recognized only one potential AP1 site within the first 500 bp upstream of the MTA TATA box, represented by the consensus sequence 5′-TGAGTCA-3′ mapping at positions −121 to −115, adjacent to the previously identified RRE and C/EBP sites. To test the contribution of this potential high-affinity AP1 site in the MTA promoter, we cotransfected HeLa cells with either a wild-type MTA(−130/+10)-LUC reporter gene or with a mutated version [MTA(−130/+10)AP1m-LUC] that was designed to destroy the AP1 site (Fig. 3A). Plasmids expressing cJUN or cFOS proved to have little effect on MTA promoter activity when transfected alone but in combination were again able to activate wild-type MTA-LUC by up to 48-fold (Fig. 3B). The point mutant MTA reporter gene showed a 2.3-fold decrease in responsiveness to cJUN plus cFOS (down to 21-fold) but also had a 4-fold reduction in basal-level expression. In addition, when the cJUN(DBD) mutant protein was used in the cotransfection to replace wild-type cJUN, the activation level was reduced to 15-fold (Fig. 3B). Therefore, direct DNA binding by AP1 at this site contributes to both basal and cJUN-plus-cFOS-mediated activation of the MTA promoter, but again there are likely to be additional weaker target sites.
FIG. 3.
cJUN and cFOS transactivate the MTA promoter through an AP1-binding site. (A) Schematic diagram of the wild-type and mutated MTA promoter region between positions −130 and +10 used in transfection experiments. The locations of the potential AP1 site relative to the C/EBP and CBF1 sites and the RRE are indicated. (B) HeLa cells were cotransfected with 0.2 μg of target reporter plasmid DNA encoding either wild-type MTA(−130/+10)-LUC (solid bars) or the point mutant version MTA(−130/+10)AP1m-LUC (open bars) and the indicated amounts of effector plasmid DNA expressing SV2-cJUN, SV2-cFOS, or SV2-cJUN(DBD). The total amount of effector plasmid DNA used in each transfection was normalized to 1 μg by adding empty SV2 promoter vector (pSG5) DNA. The basal activity of each reporter gene cotransfected with vector DNA is also indicated. (C) Sequences of the wild-type MTA-AP1 and mutated MTA-AP1m probes used. The potential AP1 sites are boxed. Underlining, boldface, and italics indicate mutated positions. (D) EMSA experiment showing the binding of in vitro-translated cJUN-plus-cFOS complexes to the wild-type and mutated AP1 site probes from the MTA promoter. Positions of AP1-specific shifted bands are indicated. SS, antibody supershift.
The binding ability of both the wild-type and mutated versions of the AP1 site from the MTA promoter was confirmed in EMSA experiments with the in vitro-translated cJUN plus cFOS proteins. Only the mixture of cJUN and cFOS or the cotranslated samples were able to bind to the wild-type probe MTA-AP1, but the same samples showed no affinity for the mutant MTA-AP1m probe (Fig. 3C and D).
JUN and FOS transactivate the KSHV RTA promoter through an AP1-binding site mapping at −87 to −81.
In the RTA promoter, three C/EBP sites that both contribute to C/EBPα transactivation and mediate enhanced positive autoregulation by RTA, apparently through cooperative formation of RTA protein complexes with C/EBPα bound at these sites, have been identified (55). Although addition of C/EBPα alone can activate endogenous RTA expression, the highest levels of induction in PEL cells appear to require cooperative interaction by C/EBPα and RTA activating both the cellular C/EBPα and viral RTA promoters, as well as a stabilization effect on C/EBPα protein through physical interactions with both RTA and RAP (55, 59). We were interested in addressing whether TPA-triggered AP1 induction contributes to the rapid accumulation of the RTA IE protein at even earlier stages during viral reactivation, prior to initiation of the self-regulating positive-feedback loop mediated by C/EBPα and RTA on both the C/EBPα and RTA promoters. An OCT site located at −220 to −213 has also been reported to contribute to RTA induction (44), and we included a promoter with a mutation at this site in our studies (Fig. 4A).
FIG. 4.
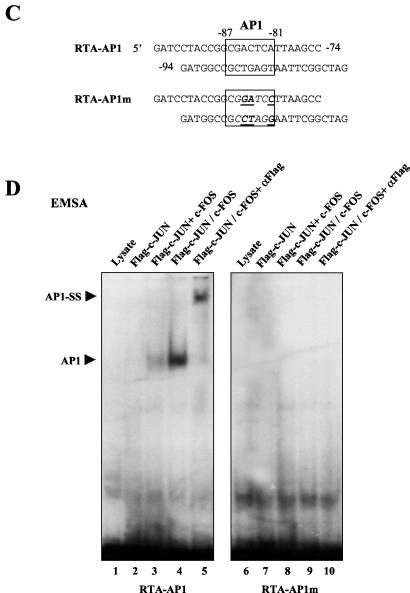
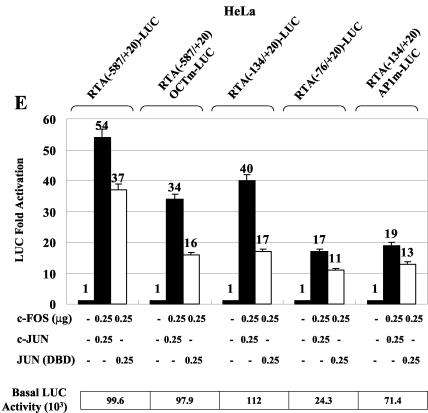
cJUN and cFOS transactivate the RTA promoter through an AP1-binding site. (A) Schematic diagram of the RTA promoter region between positions −914 and +34 showing the relative locations of the potential AP1 site, as well as the structures of wild-type and mutated RTA promoter reporter derivatives used in cotransfection experiments. (B) HeLa cells were transfected with 0.2 μg of reporter plasmid DNA encoding RTA(−914/+34)-LUC and the indicated amounts of effector plasmid DNA expressing SV2-cJUN or SV2-cFOS. The total amount of effector plasmid DNA used in each transfection was normalized to 1 μg by adding empty SV2 promoter vector (pSG5) DNA. (C) Sequences of the wild-type RTA-AP1 and mutated RTA-AP1m probes used. The AP1 site is boxed. Underlining, boldface, and italics indicate mutated positions. (D) EMSA experiment showing binding of in vitro-translated Flag-cJUN and cFOS complexes to the wild-type or mutated AP1 site in the RTA promoter. AP1-specific shifted bands are indicated. SS, antibody supershift. (E) Responsiveness of different RTA-LUC mutation derivatives to cJUN-plus-cFOS transactivation in cotransfected HeLa cells. Transfections were carried out with 0.2 μg of target reporter promoter DNA and the indicated amounts of SV2-cJUN, SV2-cFOS, and SV2-cJUN(DBD) effector plasmid DNA. The basal activity measured for each reporter gene cotransfected with empty vector DNA is also indicated.
HeLa cells were first cotransfected with the intact wild-type RTA(−914/+34)-LUC reporter gene (Fig. 4A) together with expression plasmids for the two AP1 subunits. The combined level of transactivation by cotransfected cJUN and cFOS proteins on the RTA promoter reached 72-fold in a dose-responsive manner (Fig. 4B), indicating that the AP1 transcription factor complex indeed affects the RTA promoter very significantly. The most likely potential AP1-binding site in the 1,000-bp RTA promoter was 5′-CGACTCA-3′ located at position −87 to −81. In subsequent EMSA experiments, we also tested the ability of JUN-FOS complexes to bind to both the wild-type AP1 site and a mutated version of it from the RTA promoter (Fig. 4C) and confirmed that the wild-type probe but not the mutated probe interacted directly with the in vitro-translated AP1 protein complex (Fig. 4D).
To test the contribution by this site to AP1-mediated activation of the RTA promoter, as well to evaluate the possibility of other potential AP1 sites, we examined the AP1 responsiveness of five RTA reporter genes with deletions or mutations, namely, the RTA(−587/+20)-LUC, RTA(−587/+20)OCTm-LUC, RTA(−134/+20)-LUC, RTA(−76/+20)-LUC, and RTA(−134/+20)AP1m-LUC genes, in HeLa cells (Fig. 4A). The RTA(−587/+20)-LUC gene was activated by cotransfected cJUN plus cFOS at a level similar to that of the full-length RTA (RTA(−914/+34)-LUC) promoter, whereas the RTA(−587/+20)OCTm-LUC and RTA(−134/+20)-LUC genes each gave a 20 to 40% reduction in AP1 responsiveness without reduced basal activity (Fig. 4E). This suggests that the OCT site at −220 to −213 may have a small indirect contribution to AP1 activation of the RTA promoter in HeLa cells. In contrast, either mutation of the confirmed AP1-binding site at −87 to −81 in the RTA(−134/+20)AP1m-LUC gene or its deletion in the RTA(−76/+20)-LUC gene gave a twofold loss of responsiveness compared to that for the wild-type RTA(−134/+20)-LUC gene (Fig. 4E). This deletion also lowered the basal activity fourfold compared to only a small effect for the AP1 site point mutation, although there are no other obvious recognizable motifs within this region. Use of the DBD mutant form of cJUN in combination with cFOS reduced residual transactivation of the latter two AP1 site-negative targets by only 35%, whereas for the wild-type RTA(−134/+20)-LUC target gene, which retained the AP1 site, the effect was more than twofold (Fig. 4E). Therefore, we infer from these results that what causes the rather surprisingly high residual cJUN-plus-cFOS responsiveness may be somewhat indirect.
Exogenously introduced JUN plus FOS trigger endogenous RTA and vIL6 protein expression in PEL cells.
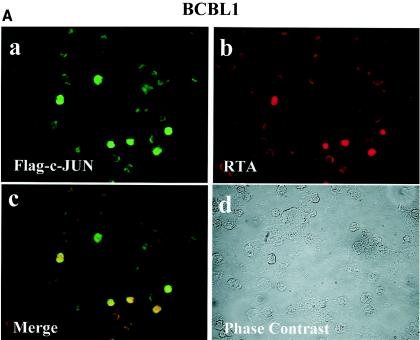
Cotransfected cJUN plus cFOS strongly transactivate the KSHV RTA promoter in transient reporter gene assays in HeLa cells. To investigate whether activation of the RTA promoter also occurs in KSHV-infected PEL cells, resulting in an induction of RTA protein levels, double-label IFA experiments were performed on BCBL1 cells cotransfected with SV2-Flag-cJUN plus SV2-cFOS. Spontaneous expression of the endogenous RTA protein was highly limited in latently infected BCBL1 cells and could be detected in less than 1% of the cells (55). However, in cJUN-plus-cFOS-cotransfected cells, 20% of the cells expressed exogenous Flag-tagged cJUN in the nucleus and 83% of these cJUN-positive cells were also positive for RTA protein expression in the nucleus (Fig. 5A). Therefore, expression of exogenous cJUN plus cFOS can activate RTA expression in PEL cells from a latent state, and because RTA alone is sufficient to trigger progression of the entire lytic cycle, cJUN plus cFOS are potentially capable of fully reactivating PEL cells from latency into the lytic cycle.
FIG. 5.
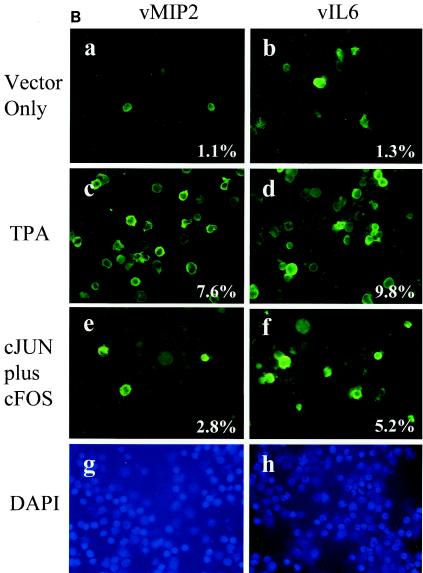
Exogenously introduced cJUN plus cFOS trigger endogenous RTA, vMIP2, and vIL6 protein expression in PEL cells. (A) BCBL1 cells were transfected by electroporation with SV2-Flag-cJUN plus SV2-cFOS, and the expression of Flag-cJUN (FITC, green) and endogenous RTA (rhodamine, red) proteins was detected by double-label IFA. (a and b) Single images. (c) Merged image (yellow). (d) Phase-contrast image. Spontaneous background RTA expression occurred in 1.2% of the untreated BCBL1 cells. (B) BCBL1 cells were transfected by electroporation with exogenous cJUN-plus-cFOS expression plasmids or with an empty vector control in the presence or absence of TPA treatment for 40 h and were then examined for expression of endogenous vMIR2 or vIL6 by IFA with specific FITC-labeled rabbit PAbs. (a, c, and e) vMIR2; (b, d, and f) vIL6; (a and b) control vector without TPA; (c and d) control vector plus TPA; (e and f) cJUN plus cFOS; (g and h) DAPI (4′,6′-diamidino-2-phenylindole) nuclear staining shown in panels e and f.
In addition to examining the induction of RTA protein by exogenous introduction of cJUN plus cFOS, we also examined their effects on two other KSHV-encoded early lytic proteins by single-label IFA, namely, the cytoplasmic vMIR2 (K5 or ZMP-A) and vIL6 (K2) proteins. In this experiment, although cJUN plus cFOS increased the number of vMIR2-positive BCBL1 cells by 2.5-fold from 1.1% spontaneous to 2.8%, this was considerably less than the 7-fold effect (to 7.6%) of parallel treatment with TPA (Fig. 5B, a, c, e, and g). For vIL6, both the introduction of cJUN plus cFOS and treatment with TPA led to somewhat higher levels of induction, which were measured at fourfold and eightfold, respectively (Fig. 5B, b, d, f, and h). Additional double-label IFA experiments confirmed that the increased vIL6 and vMIR2 expression occurred only in AP1-transfected cells expressing the exogenous Flag-tagged cJUN protein (data not shown). These results demonstrate that increased AP1 protein levels can induce more than just the viral RTA lytic-cycle trigger protein, but just as with TPA the possibility of direct effects on the endogenous viral vMIR2 and vIL6 promoters instead of or in addition to an RTA-mediated downstream effect cannot be ruled out.
Both RTA and RAP enhance JUN-plus-FOS-mediated transactivation of KSHV promoters.
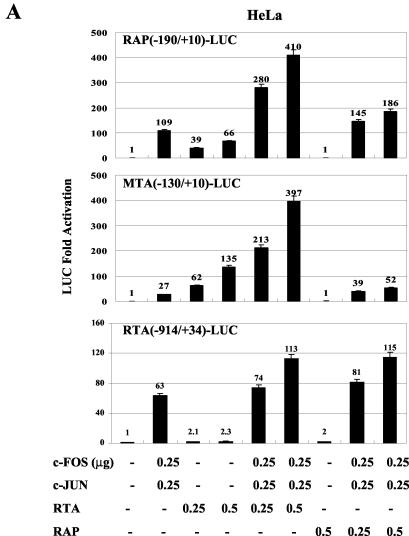
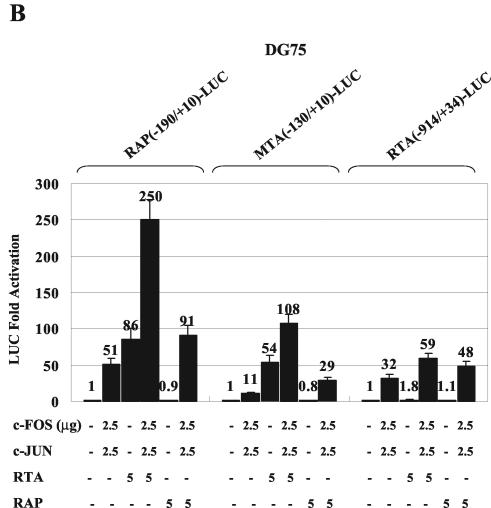
KSHV RTA activates expression of the RTA, RAP, MTA, and PAN promoters either directly through binding to type II RREs or indirectly by associating with other transcription factors, such as C/EBPα and CBF1, that bind to type I RREs (29, 54, 55). Similarly, KSHV RAP, although it does not itself bind to or transactivate any of these promoters, can also enhance the transactivation by RTA and C/EBPα on both their own and other target promoters severalfold, apparently through strong protein-protein interactions with DNA-bound C/EBPα (54, 55, 58, 59). To ask whether the AP1 transactivator complex behaves similarly to C/EBPα, we performed cotransfection experiments together with RTA and RAP using the same three wild-type KSHV promoter driven reporter genes that had been shown to respond to cJUN-plus-cFOS transactivation. On all three promoters investigated, both RTA and RAP were able to cooperate with cJUN and cFOS to significantly stimulate transactivation, and the effects were more than additive compared to transactivation in samples that were transfected with equivalent amounts of either the cJUN-plus-cFOS, RTA, or RAP expression plasmid alone (Fig. 6). Essentially equivalent results were obtained with transfected HeLa cells (Fig. 6A) and electroporated DG75 B lymphocytes (Fig. 6B), except that 20 times as many cells and 10 times as much DNA were used in the latter case.
FIG. 6.
Cooperative activation of KSHV promoters by cJUN plus cFOS with RTA or RAP in HeLa and DG75 cells. (A) HeLa cell cotransfections were carried out with 0.2 μg of target reporter plasmid DNAs encoding RAP(−190/+10)-LUC (top), MTA(−130/+10)-LUC (middle), and RTA(−914/+34)-LUC (bottom) and the indicated amounts of SV40 promoter-enhancer-driven effector plasmid DNAs encoding cJUN, cFOS, RTA, or RAP. The total amount of effector plasmid DNA used in each transfection was normalized to 1 μg by adding empty SV2 promoter vector (pSG5) DNA. (B) DG75 cells were electroporated with 2 μg of target LUC reporter plasmid DNAs and the indicated amounts of effector plasmid DNAs. The total amount of effector plasmid DNA used in each transfection was normalized to 10 μg by adding pSG5 DNA.
In both cell types, cJUN plus cFOS alone gave stronger effects on the RAP and RTA promoters than on the MTA promoter and RTA alone had strong effects on both the RAP and MTA promoters but, as expected, had minimal effect on the RTA promoter (55), whereas, also as expected, RAP alone had essentially no effect on any of the three target promoters (54, 55, 59). Similarly, in both cell types, the level of cooperative enhancement between cJUN plus cFOS and RTA together reached 4-fold for both the RAP and RTA promoters, giving up to 400-fold overall transactivation. In comparison, in the situations where either RTA alone or RAP alone had minimal effects, the cooperative effects of either RTA or RAP with cJUN plus cFOS were all approximately twofold. Therefore, similar to the cooperativity between RTA or RAP and C/EBPα, there may also be interactions between cJUN or cFOS and RTA or RAP that lead to further recruitment of viral and cellular transcription factors at the AP1 sites in these early-lytic-cycle promoters.
RTA and RAP interact with the JUN or FOS proteins both in vitro and intracellularly.
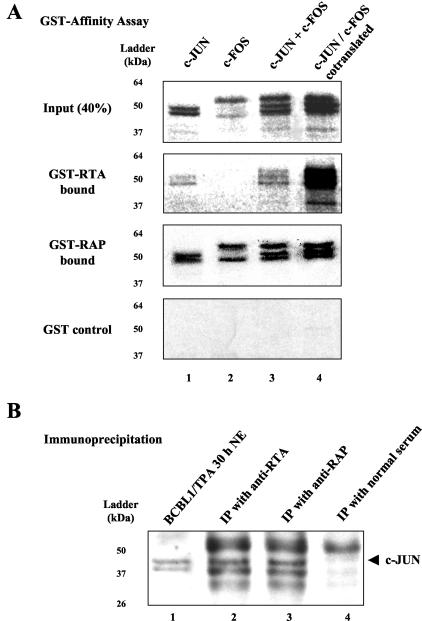
To detect a potential physical association between the individual AP1 protein subunits and RTA or RAP, we carried out GST affinity-binding assays using bacterially expressed GST-RTA or GST-RAP recombinant proteins. In vitro-translated cJUN and cFOS proteins were labeled with [35S]methionine and incubated with the GST fusion proteins immobilized on glutathione-Sepharose beads. These experiments revealed that GST-RTA was able to interact efficiently with cJUN, but had little affinity for cFOS (Fig. 7A, second panel from the top, lanes 1 and 2). However, when cJUN and cFOS were either admixed together or cotranslated before addition to the system, the interaction between cJUN and cFOS evidently allowed GST-RTA to retain both proteins on the beads (Fig. 7A, second panel, lanes 3 and 4). In contrast, in the same experiments GST-RAP displayed a high affinity for both cJUN and cFOS individually, as well as after the proteins were mixed or cotranslated (Fig. 7A, third panel). None of the labeled cJUN or cFOS samples bound to GST alone (Fig. 7A, fourth panel).
FIG. 7.
The KSHV RTA and RAP proteins bind to AP1 subunit proteins. (A) In vitro GST affinity binding assay. cJUN (lane 1) and cFOS (lane 2) were in vitro translated alone and then admixed (lane 3) or both were cotranslated (lane 4) before addition to the GST binding assay mixtures. The autoradiographs show PAGE fractionation of 32S-labeled input samples (first [top] panel) compared to recovered labeled proteins bound to GST-RTA(1-691) (second panel), GST-RAP(1-237) (third panel), or GST alone (fourth panel). Positions of protein size markers are indicated to the left of each panel. (B) Coimmunoprecipitation of cJUN with RTA or RAP in KSHV-positive BCBL1 cells undergoing the lytic cycle after TPA induction. Western immunoblotting was carried out with an anti-cJUN PAb. Lane 1, positive control of cJUN protein in the input cell lysate (10 μl, 10% of input sample); lane 2, recovery of cJUN by immunoprecipitation with the anti-RTA PAb; lane 3, recovery of cJUN by immunoprecipitation with the anti-RAP PAb; lane 4, negative control for the immunoprecipitation with normal preimmune rabbit serum.
To evaluate the interactions of RTA and RAP with AP1 complexes in vivo in virus-infected cells, we performed coimmunoprecipitation experiments with a BCBL1 nuclear extract prepared after 30 h of TPA treatment using either RTA or RAP antiserum and analyzed the precipitated proteins by SDS-PAGE followed by immunoblotting with a rabbit anti-cJUN PAb. The results revealed that cJUN could be detected in both the RTA- and RAP-immunoprecipitated samples but was absent in control samples precipitated with normal rabbit serum (Fig. 7B), indicating that cJUN associates with both KSHV RTA and RAP in vivo in TPA-induced PEL cells undergoing lytic-cycle progression.
Induction of AP1 DNA-binding activity early after TPA treatment in PEL cells.
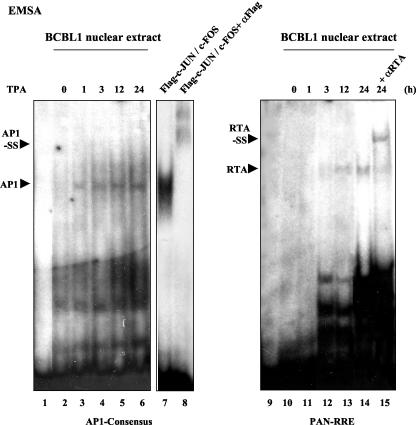
The upregulation of several KSHV lytic-cycle promoters by AP1 in cotransfection assays suggests that AP1 may have an important role in reactivating the viral lytic replication cycle from latency in vivo through targeting to the promoters of viral regulatory genes such as the RTA, RAP, and MTA genes. Cultured PEL cell lines that carry multicopy latent KSHV episomes can be induced to undergo lytic-cycle infection and viral DNA replication in response to triggering by adding TPA to the medium. To evaluate how early induced AP1 DNA-binding ability can be detected after TPA treatment in PEL cells, we prepared nuclear extracts from BCBL1 cells at 0, 1, 3, 12, and 24 h after TPA treatment and performed EMSA experiments using these nuclear extracts with the 32P-labeled consensus AP1 oligonucleotide probe. Shifted bands migrating at the same positions as in vitro-translated AP1-bound DNA complexes as a positive control were detected as early as 1 h after TPA treatment and continued to increase in abundance up to at least 24 h (Fig. 8, left). In comparison, a parallel experiment using the PAN-RRE probe showed that RTA did not start to associate with its target DNA-binding site until later than 3 h (Fig. 8, right), suggesting that AP1 DNA-binding activity may be induced significantly earlier than when RTA DNA-binding activity can be first detected.
FIG. 8.
Induction of AP1 and RTA DNA-binding activities in TPA-treated BCBL1 cells. BCBL1 cells were harvested at 0, 1, 3, 12, and 24 h (lanes 2 to 6 and lanes 9 to 13, as indicated) after TPA treatment, and the nuclear extracts were prepared and tested in an EMSA experiment using either the AP1 consensus probe (lanes 1 to 6) or the PAN-RRE probe for RTA binding activity (lanes 9 to 15). Lanes 1 and 9, no nuclear extract; lanes 2 to 6 and 10 to 14, nuclear extract samples from the different time points; lanes 7 and 8, positive mobility control of in vitro-cotranslated Flag-cJUN plus cFOS and its anti-Flag supershifted forms from the same gel; lane 15, 24-h sample plus RTA PAb. Positions of the AP1-DNA or RTA-DNA complexes and the supershifted (SS) bands are indicated.
Induction of phosphorylation of JUN by TPA treatment in PEL cells.
TPA is known to effectively induce AP1 transcriptional activity through activating the JNK pathway that phosphorylates and activates cJUN in various cell lines (28). To study the induction of activated AP1 in two latently KSHV- infected PEL cell lines (BCBL1 and BC3) and in a control KSHV-negative B-lymphoblast cell line (DG75), we performed immunoblotting using a cJUN PAb that recognizes both hypo- and hyperphosphorylated forms of cJUN. As expected, the phosphorylation of cJUN was induced in all three cell lines at 24 h after TPA treatment, and an increased level of hypophosphorylated cJUN was also observed in TPA-treated BCBL1 and BC3 cells but not in the DG75 cells (Fig. 9A, middle, lanes 1 to 6).
To examine the time course of cJUN activation in BCBL1 cells at different time points, we examined protein extracts harvested at 0, 1, 3, 12, and 24 h after TPA treatment for the levels of phosphorylated cJUN by immunoblotting with a PAb that detects only the phosphorylated form of cJUN. Phosphorylated cJUN was barely detectable at 0 h but started to appear as early as 1 h after TPA treatment and continued to accumulate up to 24 h (Fig. 9C, top, lanes 1 to 5). A large increase in phosphorylated cJUN was also evident at 24 h after TPA induction in HeLa cells (lanes 6 and 7). Therefore, induction of a phosphorylated form of cJUN that is active for DNA binding correlates with the rapid appearance of the AP1 DNA-binding activity in EMSA experiments, and both assays detected induction within 1 h upon TPA treatment in PEL cells.
We subsequently performed immunocytochemical staining using the PAb against the phosphorylated form of cJUN. Before TPA treatment 5% of the BCBL1 cells showed positive staining in the nucleus (Fig. 9D, a), whereas after TPA treatment for 24 h, 15% of the cells were positive and had significantly stronger staining (b). To further address the question of whether the phosphorylation and activation of cJUN observed in both Western immunoblotting and immunocytochemical staining occur in PEL cells undergoing KSHV lytic-cycle replication, we carried out a double-label immunohistochemical procedure with BCBL1 cells at either 0 or 24 h after TPA treatment using a PAb against vMIR2 as well as the phosphorylated cJUN antibody. As expected, more of the cells (10%) expressed cytoplasmic vMIR2 (blue) after TPA treatment (Fig. 9D, d) than before TPA treatment (1%; c). However, among the TPA-treated BCBL1 cells, at least 60 to 70% of the vMIR2-positive cells were also positive for nuclear phosphorylated cJUN (brown) by double-stain immunohistochemical procedures (d; see inset). Therefore, most cells undergoing KSHV lytic-cycle replication were also positive for phosphorylated cJUN staining in the nucleus, suggesting that the phosphorylation of cJUN is highly related to lytic-cycle replication in these cells.
Addition of RTA alone induces the levels of endogenous unphosphorylated JUN protein.
Our evidence above that RTA physically interacts with cJUN-cFOS complexes raised the possibility that, similar to the situation with C/EBPα, this may result in increased overall levels of AP1 components in RTA-expressing cells. To examine this question, we first carried out Western blotting for total cJUN protein levels in Flag-RTA-transfected DG75 and HeLa cells. Indeed, as shown in Fig. 9B, the addition of RTA alone induced a large increase in the level of unphosphorylated cJUN (middle, lanes 2 to 4), although there was no increase in the slower-migrating phosphorylated form.
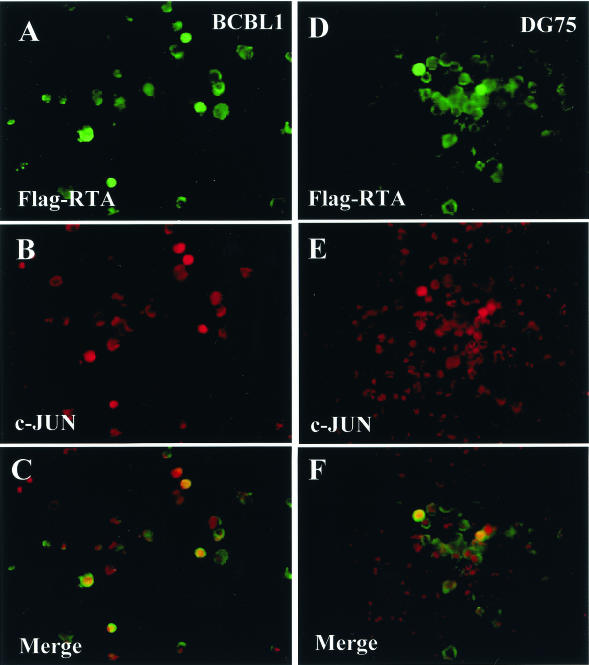
To further support these results, we also carried out double-label IFA experiments after transfection of BCBL1 and DG75 cells with Flag-RTA by electroporation (Fig. 10). The results confirmed that, although a small fraction (1.2%) of the untreated BCBL1 cells were spontaneously positive for nuclear cJUN, virtually all of the 10% of cells in the transfected culture that expressed exogenous nuclear Flag-RTA (Fig. 10A, FITC, green) also became positive for endogenous nuclear cJUN protein expression (Fig. 10B, rhodamine, red, and C, merge, yellow). In the KSHV-negative DG75 culture, again almost all of the 3% of transfected cells that expressed either endogenous cJUN or Flag-RTA were positive for both proteins (Fig. 10D to F). These results suggest that, once RTA is induced early in the KSHV lytic cycle, it is capable of in turn activating cJUN expression either directly or indirectly by an as-yet-unknown mechanism.
FIG. 10.
Addition of exogenous RTA induces endogenous cJUN expression in both KSHV-infected PEL cells and uninfected B lymphoblasts. The Flag-RTA expression vector was introduced into either BCBL1 cells (A to C) or DG75 cells (D to F), and expression of both exogenous Flag-RTA and endogenous cJUN was detected by double-label IFA. (A and B) Anti-Flag MAb (FITC, green); (C and D) anti-cJUN PAb (rhodamine, red); (E and F) merge (yellow).
TPA synergistically stimulates transactivation by JUN and FOS but not by C/EBPα on the RAP, MTA, and RTA promoters.
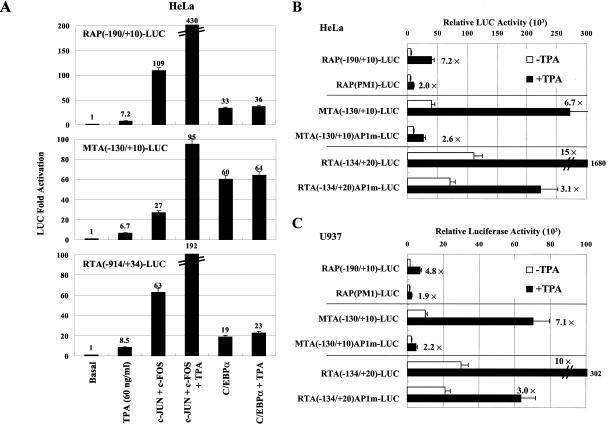
AP1 is thought to be one of the most important pathways through which TPA stimulates target gene promoters. When transfected into HeLa cells, all three KSHV lytic-cycle-promoter-driven reporter genes studied in this work, the RAP(−190/+10)-LUC, MTA(−130/+10)-LUC, and RTA(−914/+34)-LUC genes, were responsive to TPA treatment after 24 h, showing 7.2-, 6.7-, and 8.5-fold-increased LUC activity, respectively (Fig. 11A). When these promoters were both cotransfected with the cJUN plus cFOS plasmids and then treated with TPA to analyze the dual effect by AP1 and TPA, activation by AP1 was stimulated three- to fourfold by TPA compared to that for untreated AP1-transfected samples (Fig. 11A). However, in parallel experiments TPA treatment had no added effect on the transactivation levels achieved by C/EBPα on these same promoters.
FIG. 11.
TPA synergistically enhances AP1-binding site-mediated cJUN-plus-cFOS activation but not C/EBPα-mediated activation of the RAP, MTA, and RTA promoters. (A) HeLa cells were cotransfected with 0.2 μg of target reporter plasmid DNA encoding RAP(−190/+10)-LUC (top), MTA(−130/+10)-LUC (middle), or RTA(−914/+34)-LUC (bottom) and 0.25 μg each of SV2-cJUN and SV2-cFOS or 0.5 μg of SV2-C/EBPα effector plasmid DNAs as indicated. The total amount of effector plasmid DNA used in each transfection was normalized to 1 μg by adding empty SV2 promoter vector (pSG5) DNA. TPA was added into the medium, and the cells were incubated for 24 h before harvesting. (B and C) Mutations of the identified AP1-binding sites in the RAP, MTA, and RTA promoters all impair their responsiveness to TPA stimulation. Either Lipofectamine-transfected HeLa cells (B) or electroporated U937 cells (C) were transfected with 0.2 μg or 2 μg, respectively, of target reporter plasmid DNA encoding either RAP(−190/+10)-LUC, RAP(PM1)-LUC, MTA(−130/+10)-LUC, MTA(−130/+10)AP1m-LUC, RTA(−134/+20)-LUC, or RTA(−134/+20)AP1m-LUC. Cells were harvested at 48 h posttransfection, either with or without TPA treatment for 24 h, and lysed for LUC assays.
The AP1 sites in the RAP, MTA, and RTA promoters are crucial for the responsiveness to TPA treatment.
To further test whether the AP1 sites identified in this study mediate TPA stimulation on the three KSHV promoters, we tested the TPA responsiveness of both the wild-type promoters and the AP1 site-mutated versions by transfecting the corresponding reporter genes into HeLa cells. The point mutant RAP(PM1)-LUC reporter gene that retained only one-third of the cJUN-plus-cFOS activation level compared to the wild-type promoter (Fig. 2B) also showed a 3.6-fold decrease in TPA responsiveness (from 7.2-fold to 2.0-fold) (Fig. 11B). Similarly, a mutation in the AP1 site in the MTA promoter that led to a 2.1-fold loss of cJUN-plus cFOS responsiveness (Fig. 3B) led to a 2.5-fold reduction in TPA responsiveness (from 6.7-fold to 2.6-fold). Finally, mutation of the identified AP1 site in the RTA promoter that led to a 2.1-fold loss of cJUN-plus-cFOS responsiveness (Fig. 4E) also gave a 5-fold loss in TPA responsiveness (from 15-fold to 3.1-fold) (Fig. 11B).
Interestingly, in both sets of experiments we observed that, compared to the wild-type promoters, the promoters with AP1 site mutations (especially the mutation in the MTA promoter) also gave some reductions in basal activity, even without exogenous cJUN-plus-cFOS coexpression. This may result from relatively high endogenous AP1 expression in HeLa cells, giving basal activation of the AP1 sites of the wild-type promoters but not of the mutated promoters. To address the possibility of cell type-specific effects, we also performed the same transfection and TPA treatment experiments with the U937 premonocytic cell line, used successfully previously for measuring TPA responsiveness in the CMV MIE promoter (6) (Fig. 11C). However, essentially identical effects on TPA responsiveness as well as on the basal activities of these reporter genes by mutations in the AP1 sites were also observed in U937 cells.
Association of the JUN protein with KSHV target promoters by ChIP assay during early-lytic-cycle reactivation.
The above experiments indicated that cotransfected cJUN and cFOS can transactivate the expression of the KSHV RAP, MTA, and RTA promoters primarily through the AP1-binding sites that we identified above and that the phosphorylated form of cJUN and AP1 DNA-binding activity itself are both induced as early as 1 h after TPA treatment in PEL cells. To investigate whether and when AP1 protein complexes can be detected in association with the three viral promoters during the early stages of lytic-cycle reactivation in PEL cells, we carried out a time course ChIP assay using BCBL1 cell lysates prepared at 0, 4, 8, and 24 h after TPA treatment. Antibodies against either cJUN or CHOP10 (as a negative control) were used to immunoprecipitate sonicated chromatin and any associated DNA fragments (Fig. 12). Following PCR amplification, DNA regions representing all three promoters (RAP, MTA, and RTA promoters) were detected in the cJUN immunoprecipitates as early as 4 h after TPA treatment (Fig. 12, lanes 7 to 9) and for MTA and RTA the levels increased greatly by 24 h. Importantly, none of these promoter DNAs were detected at any time point in parallel immunoprecipitates obtained with the CHOP10 antibody (lanes 10 to 13). Furthermore, as expected, little or no cJUN-bound promoter DNA was detected in the uninduced (0-h) BCBL1 cell extracts (Fig. 12, lanes 6).
DISCUSSION
In previous studies, we found that the cellular transcription factor C/EBPα interacts with two important KSHV-encoded lytic-cycle regulatory proteins RTA and RAP (54, 58, 59) and mediates KSHV early-lytic-cycle gene regulation by directly binding to and transactivating at least four viral promoters, including those for RTA, RAP, MTA, and PAN (54, 55). Both the RTA and RAP proteins cooperate with C/EBPα through these protein-protein interactions to augment target gene expression, including having positive autoregulatory effects on their own promoters. The expression of C/EBPα itself is also strongly induced in TPA-treated PEL cells, and both RTA and RAP reciprocally contribute to this effect by enhancing positive autoregulation of the C/EBPα promoter as well as by stabilizing the C/EBPα protein (54, 58, 59). As an IE protein, RTA is one of the first viral proteins expressed upon primary infection or viral reactivation, and the initial induction of RTA mRNA does not depend on new protein synthesis (45, 51, 63). Despite the fact that ectopic expression of C/EBPα induces RTA and RAP expression in latently infected PEL cells, we knew that C/EBPα protein levels are not induced by TPA treatment in the absence of RTA or RAP. As shown here, C/EBPα transactivation is also not induced or enhanced by TPA. Therefore, some other cellular factors that are regulated through signal transduction pathways and by protein modification at the posttranslational level must be responsible for the initial induction of RTA mRNA expression by TPA, which is then subsequently boosted by the RTA and RAP transcriptional autoregulatory loop mediated by C/EBPα.
The activity of the AP1 transcriptional activating complexes consisting of cJUN-plus-cFOS heterodimers is known to be significantly induced by TPA in various cell lines through induction of the JNK pathway that activates cJUN by phosphorylation. In this study, we have shown that TPA indeed induces AP1 DNA-binding activity and phosphorylated cJUN in PEL cells and that the three KSHV lytic-cycle promoters tested, including the RTA promoter itself, are all responsive to both TPA and cJUN-plus-cFOS transactivation. We also identified specific AP1-binding sites that contributed to these effects in all three promoters. The strongest of these sites (the classical AP1 motif in the MTA promoter) also contributed to basal-level expression. Even after mutation of the identified AP1 sites, each promoter still retained significant additional cJUN-plus-cFOS responsiveness, suggesting the presence of other unidentified target sites. However, some of this residual activity, including that involving the OCT site in the RTA promoter, appears to be relatively indirect, because it was less dependent on the presence of the DBD of cJUN than was the responsiveness mediated by the AP1 sites themselves.
Addition of exogenous RTA or Flag-tagged RTA increased overall cJUN protein levels detectable by Western blotting and double-label IFA of PEL cells, as well as of KSHV-negative DG75 and HeLa cells, but did not increase phosphorylated cJUN levels. Although this effect closely resembles that for KSHV RTA and RAP as well as for EBV ZTA on c/EBPα and p21 protein levels (55, 57, 58, 59), we do not know at present whether RTA itself has direct effects on the cJUN promoter or whether this is a consequence of protein stabilization associated with the binding of RTA to cJUN and cFOS protein complexes or both. In contrast, TPA alone induced just the phosphorylation of cJUN in an RTA-independent manner in DG75 and HeLa cells but had no effect on overall levels of unphosphorylated cJUN protein.
Nevertheless, in TPA-treated KSHV-positive PEL cells, increases in total cJUN protein levels were detected at 24 h, cJUN association with the viral promoters was detectable by ChIP assay within 4 h, and both increased activated phosphorylated cJUN and increased AP1 DNA-binding activity occurred within 1 h. Furthermore, introduction of exogenous Flag-tagged cJUN plus cFOS into PEL cells (in the absence of TPA) induced endogenous RTA protein expression in the majority of the cells (83%) that expressed Flag-cJUN, as detected by double-label IFA. Therefore, although activation of the JNK pathway and phosphorylation of cJUN to activate AP1 DNA-binding activity by TPA might be responsible for (or at least participate in) the initial activation of the RTA promoter, subsequent positive amplification of cJUN-plus-cFOS levels by RTA may also be critical. This may be accomplished by the interaction of newly synthesized RTA with cJUN-plus-cFOS complexes bound to AP1 sites, which results in a cooperative stimulation of further RTA promoter activity and enhances the levels of both the RTA and cJUN proteins, followed by the switching on of the RAP promoter and other early promoters. Later, when RTA also stimulates C/EBPα induction, all of these promoters, which are independently responsive to both C/EBPα and to cJUN-plus-cFOS activation, are also further stimulated by the newly synthesized RAP protein, in turn cooperatively enhancing transactivation of both its own promoter and those for the RTA gene and the other early genes by a combination of interactions with RTA, C/EBPα, and cJUN plus cFOS.
We have shown previously that RTA and RAP both reciprocally enhance transcriptional autoregulation of the cellular C/EBPα promoter and that both also stabilize the C/EBPα protein through direct protein-protein contacts. Although we demonstrate here that RTA and RAP both coimmunoprecipitate cJUN and bind to either or both cJUN and cFOS in vitro, we have yet to address whether either of those transcriptional or posttranslational mechanisms also applies to cJUN or cFOS.
The effects of TPA treatment on all three viral early promoters in HeLa cells ranged from only 6.7- to 8.5-fold compared to 27- to 109-fold for cotransfected cJUN plus cFOS, but, importantly, the identified AP1 sites proved to play more significant roles in the TPA responsiveness than they did in activation by cJUN-plus-cFOS cotransfection, with point mutations in each giving 3- to 5-fold reductions in cJUN-plus-cFOS responsiveness and 8- to 10-fold reductions in overall activity for the TPA-treated MTA and RTA promoters. Interestingly, addition of TPA together with cotransfection by cJUN plus cFOS increased overall activation of all three wild-type promoters by 3- to 4-fold to between 95- and 430-fold over basal levels. Curiously, addition of TPA also boosted RTA responsiveness of the wild-type promoters three- to fourfold in HeLa cells (data not shown) but had no effect on C/EBPα responsiveness of the same promoters in parallel experiments in either the presence or absence of RTA. The synergistic effects of TPA on basal-level activity, on cJUN-plus-cFOS-induced activity, and on RTA-induced activity can all be reasonably well explained by the observed increase in phosphorylation of cJUN over the preexisting basal or activated levels, but we have no explanation at present for the apparent ability of C/EBPα to block, prevent, or counteract the added effects of TPA in the transient reporter gene assays.
AP1 sites were originally defined in the SV40 T-antigen promoter-enhancer region, where they were also recognized as a subset of TREs (13). They also occur in the human CMV MIE enhancer (25) and in the herpes simplex virus type 1 IE110 first intron (20). However, there are also other targets of TPA responsiveness, such as the ZI and ZII sites in the upstream EBV ZTA promoter (17) and the classical CRE, NF-κB, and overlapping SRE/ETS sites within the CMV MIE enhancer (6). Therefore, it is not particularly surprising that cJUN and cFOS and activated AP1 DNA-binding activity play a role in TPA induction in B lymphocytes, but few studies have addressed the JUN/FOS/AP1 pathway even in the EBV system, and no previous studies have addressed these issues specifically for the KSHV lytic cycle in PEL cell lines. There was also a question about whether this pathway would apply only to the RTA promoter rather than also acting independently on several or perhaps many early-lytic-cycle promoters, as our results imply. Although we show here that both cJUN-plus-cFOS complexes alone (as well as C/EBPα) can clearly contribute to activation of the downstream viral promoters, their full level of activity is still likely to be highly dependent on probably both RTA and RAP in lytically infected cells in vivo. Other cellular proteins that have been reported to activate the RTA promoter or downstream promoters used here, especially OCT1 or OCT2 and CBF1 (RBP-Jκ), may also contribute in similar ways (29, 44), but in neither case have the levels of those proteins or their functional activation in KSHV lytically infected cells yet been addressed. In the EBV system, there is evidence that TPA and PKC also act at the posttranslational level by phosphorylation of S186 in the ZTA protein, which appears to play a key role in lytic-cycle reactivation (1, 18), although the exact mechanism and consequences of this phosphorylation event are not fully understood.
Several other interesting questions about the role of the cJUN-plus-cFOS pathway remain to be resolved. In particular, unlike what was found for C/EBPα, we could expect activated phosphorylated cJUN to be induced in perhaps all PEL cells by TPA in the absence of viral gene products. However, our initial immunocytochemistry results suggest that, just like for the overall C/EBPα and cJUN levels, enhanced levels of phosphorylated cJUN may be detectable only at the single-cell level in those TPA-treated cells that are expressing viral lytic-cycle gene products. Unfortunately, we do not yet have either good enough phosho-cJUN or -cFOS antibodies to carry out these kinds of studies as thoroughly as we have been able to do for C/EBPα, but the question arises as to why the detectable increased AP1 complexes and DNA-binding activity are apparently limited to just those cells undergoing the lytic cycle. Two obvious points would be that the positive-amplification loops mediated by RTA and RAP, perhaps including protein stabilization events, are needed for a sustained effect and that TPA treatment alone might be expected to give just a transient effect on AP1 expression. As we have shown here, RTA increases the levels of unphosphorylated cJUN in the absence of TPA, and perhaps that may also apply to the levels of activated phosphorylated cJUN in the presence of TPA. Other factors, such as inhibitory effects of latent-state functions or potential apoptosis in cells that are not protected from it by viral lytic-cycle gene products, might all play a role here. But if indeed TPA initially induces activated AP1-binding activity in all cells, why do the cells not all go on into the lytic cycle and produce the conditions that sustain and enhance the AP1 pathway? Could it instead be that only a subset of the PEL cells initially induce activated AP1 in response to TPA and that these are the only ones that go on into the lytic cycle? And if so are there cell cycle-related factors, for example, that modulate the ability of the individual cells to activate AP1 or are some other conditional factors also needed to trigger sustained lytic-cycle induction? It has been suggested that both promoter CpG methylation and methotrexate (an anti-inflammatory agent) treatment are complicating factors that may negatively regulate RTA promoter expression in PEL cell lines (10a, 14).
In addition to the strong AP1 site at −121 in the MTA promoter, the KSHV genome contains classic 5′-TGA(C/G)TCA-3′ AP1 motifs at positions −10 in the PAN promoter, −20 in the vIL6 promoter, −20 in the ORF9 (POL) promoter, −55 in the ORF10 promoter, and −150 in the ORF37 (EXO) promoter, as well as three each within a 360-bp stretch of the duplicated ORI-LYT domains known as DS-B and DS-E (59). The latter have also been shown elsewhere to contribute to both the basal and TPA responsiveness of the powerful DSBR and DSEL promoters associated with ORI-LYTR and ORI-LYTL (C.-J. Chiou et al., unpublished data). Therefore, the roles of cJUN and cFOS in both early-lytic-cycle transcription and KSHV lytic-cycle DNA replication are likely to be much more extensive than just those demonstrated here.
.
Acknowledgments
These studies were funded by National Cancer Institute Research grants (R01 CA73585 and P01 CA81400) to G.S.H. from the National Institutes of Health. S.E.W. was a postdoctoral fellow in the Viral Oncology Program at the Sidney Kimmel Comprehensive Cancer Center at Johns Hopkins School of Medicine.
We thank all members of the G. S. Hayward and S. D. Hayward laboratories for valuable discussions.
REFERENCES
- 1.Baumann, M., H. Mischak, S. Dammeier, W. Kolch, O. Gires, D. Pich, R. Zeidler, H. J. Delecluse, and W. Hammerschmidt. 1998. Activation of the Epstein-Barr virus transcription factor BZLF1 by 12-O-tetradecanoylphorbol-13-acetate-induced phosphorylation. J. Virol. 72:8105-8114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown, P. H., S.-H. Kim, S. C. Wise, A. L. Sabichi, and M. J. Birrer. 1996. Dominant-negative mutants of cJUN inhibit AP-1 activity through multiple mechanisms and with different potencies. Cell Growth Differ. 7:1013-1021. [PubMed] [Google Scholar]
- 3.Cannon, J. S., D. Ciufo, A. L. Hawkins, C. A. Griffin, M. J. Borowitz, G. S. Hayward, and R. F. Ambinder. 2000. A new primary effusion lymphoma-derived cell line yields a highly infectious Kaposi's sarcoma herpesvirus-containing supernatant. J. Virol. 74:10187-10193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cesarman, E., P. S. Moore, P. H. Rao, G. Inghirami, D. M. Knowles, and Y. Chang. 1995. In vitro establishment and characterization of two acquired immunodeficiency syndrome-related lymphoma cell lines (BC-1 and BC-2) containing Kaposi's sarcoma-associated herpesvirus-like (KSHV) DNA sequences. Blood 86:2708-2714. [PubMed] [Google Scholar]
- 5.Cesarman, E., R. G. Nador, F. Bai, R. A. Bohenzky, J. J. Russo, P. S. Moore, Y. Chang, and D. M. Knowles. 1996. Kaposi's sarcoma-associated herpesvirus contains G protein-coupled receptor and cyclin D homologs which are expressed in Kaposi's sarcoma and malignant lymphoma. J. Virol. 70:8218-8223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan, Y.-J., C.-J. Chiou, Q. Huang, and G. S. Hayward. 1996. Synergistic interactions between overlapping binding sites for the serum response factor and ELK-1 proteins mediate both basal enhancement and phorbol ester responsiveness of primate cytomegalovirus major immediate-early promoters in monocyte and T-lymphocyte cell types. J. Virol. 70:8590-8605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang, P.-J., D. Shedd, L. Gradoville, M.-S. Cho, L.-W. Chen, J. Chang, and G. Miller. 2002. Open reading frame 50 protein of Kaposi's sarcoma-associated herpesvirus directly activates the viral PAN and K12 genes by binding to related response elements. J. Virol. 76:3168-3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang, Y., E. Cesarman, M. S. Pessin, F. Lee, J. Culpepper, D. M. Knowles, and P. S. Moore. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 266:1865-1869. [DOI] [PubMed] [Google Scholar]
- 9.Chang, Y.-N., D. L. Dong, G. S. Hayward, and S. D. Hayward. 1990. The Epstein-Barr virus Zta transactivator: a member of the bZIP family with unique DNA-binding specificity and a dimerization domain that lacks the characteristic heptad leucine zipper motif. J. Virol. 64:3358-3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, H.-L., J. Lee, Y. Wang, D. Huang, R. M. Ambinder, and S. D. Hayward. 1999. The Epstein-Barr virus latency BamHI-Q promoter is positively regulated by STATs and ZTA interference with JAK/STAT activation leads to loss of BamHI-Q promoter activity. Proc. Natl. Acad. Sci. USA 96:9339-9344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10a.Chen, J., K. Ueda, S. Sakakibara, T. Okuno, C. Parravicini, M. Corbellino, and K. Yamanishi. 2001. Activation of latent Kaposi's sarcoma-associated herpesvirus by demethylation of the promoter of the lytic transactivator. Proc. Natl. Acad. Sci. USA 98:4119-4124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chiou, C.-J., L. J. Poole, P. Kim, D. M. Ciufo, J. S. Cannon, C. M. ap Rhys, D. J. Alcendor, J. C. Zong, R. F. Ambinder, and G. S. Hayward. 2002. Patterns of gene expression and a transactivation function exhibited by the vGCR (ORF74) chemokine receptor protein of Kaposi's sarcoma-associated herpesvirus. J. Virol. 76:3421-3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ciufo, D. M., J. S. Cannon, L. J. Poole, F. Y. Wu, P. Murray, R. F. Ambinder, and G. S. Hayward. 2001. Spindle cell conversion by Kaposi's sarcoma-associated herpesvirus: formation of colonies and plaques with mixed lytic and latent gene expression in infected primary dermal microvascular endothelial cell cultures. J. Virol. 75:5614-5626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Curran, T., and B. R. Franza, Jr. 1988. Fos and Jun: the AP-1 connection. Cell 55:395-397. [DOI] [PubMed] [Google Scholar]
- 14.Curreli, F., F. Cerimele, S. Muralidhar, L. J. Rosenthal, E. Cesarman, A. E. Friedman-Kien, and O. Flore. 2002. Transcriptional downregulation of ORF50/Rta by methotrexate inhibits the switch of Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8 from latency to lytic replication. J. Virol. 76:5208-5219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deng, H., A. Young, and R. Sun. 2000. Auto-activation of the RTA gene of human herpesvirus-8/Kaposi's sarcoma-associated herpesvirus. J. Gen. Virol. 81:3043-3048. [DOI] [PubMed] [Google Scholar]
- 16.Deng, H., M. J. Song, J. T. Chu, and R. Sun. 2002. Transcriptional regulation of the interleukin-6 gene of human herpesvirus 8 (Kaposi's sarcoma-associated herpesvirus). J. Virol. 76:8252-8264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flemington, E., and S. H. Speck. 1990. Identification of phorbol ester response elements in the promoter of Epstein-Barr virus putative lytic switch gene BZLF1. J. Virol. 64:1217-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Francis, A. L., L. Gradoville, and G. Miller. 1997. Alteration of a single serine in the basic domain of the Epstein-Barr virus ZEBRA protein separates its functions of transcriptional activation and disruption of latency. J. Virol. 71:3054-3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gradoville, L., J. Gerlach, E. Grogan, D. Shedd, S. Nikiforow, C. Metroka, and G. Miller. 2000. Kaposi's sarcoma-associated herpesvirus open reading frame 50/Rta protein activates the entire viral lytic cycle in the HH-B2 primary effusion lymphoma cell line. J. Virol. 74:6207-6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gu, W., H. Huang, and G. S. Hayward. 1995. Multiple tandemly repeated binding sites for the YY1 repressor and transcription factors AP-1 and SP-1 are clustered within intron-1 of the gene encoding the IE110 transactivator of herpes simplex virus type 1. J. Biomed. Sci. 2:203-226. [DOI] [PubMed] [Google Scholar]
- 21.Gwack, Y., H. Byun, S. Hwang, C. Lim, and J. Choe. 2001. CREB-binding protein and histone deacetylase regulate the transcriptional activity of Kaposi's sarcoma-associated herpesvirus open reading frame 50. J. Virol. 75:1909-1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gwack, Y., H. J. Baek, H. Nakamura, S. H. Lee, M. Meisterernst, R. G. Roeder, and J. U. Jung. 2003. Principal role of TRAP/mediator and SWI/SNF complexes in Kaposi's sarcoma-associated herpesvirus RTA-mediated lytic reactivation. Mol. Cell. Biol. 23:2055-2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hardwick, J. M., P. Lieberman, and S. D. Hayward. 1988. A new Epstein-Barr virus transactivator, R, induces expression of a cytoplasmic early antigen. J. Virol. 62:2274-2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hardwick, J. M., L. Tse, N. Applegren, J. Nicholas, and M. A. Veliuona. 1992. The Epstein-Barr virus R transactivator (Rta) contains a complex, potent activation domain with properties different from those of VP16. J. Virol. 66:5500-5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hunninghake, G. W., M. M. Monick, B. Liu, and M. F. Stinski. 1989. The promoter-regulatory region of the major immediate-early gene of human cytomegalovirus responds to T-lymphocyte stimulation and contains functional cyclic AMP response elements. J. Virol. 63:3026-3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Izumiya, Y., S.-F. Lin, T. Ellison, L.-Y. Chen, C. Izumiya, P. Luciw, and H.-J. Kung. 2003. Kaposi's sarcoma-associated herpesvirus K-bZIP is a coregulator of K-Rta: physical association and promoter-dependent transcriptional repression. J. Virol. 77:1441-1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Landschulz, W. H., P. F. Johnson, and S. L. McKnight. 1988. The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science 240:1759-1764. [DOI] [PubMed] [Google Scholar]
- 28.Lee, B.-S., M. Paulose-Murphy, Y.-H. Chung, M. Connlole, S. Zeichner, and J. U. Jung. 2002. Suppression of tetradecanoyl phorbol acetate-induced lytic reactivation of Kaposi's sarcoma-associated herpesvirus by K1 signal transduction. J. Virol. 76:12185-12199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liang, Y., J. Chang, S. J. Lynch, D. M. Lukac, and D. Ganem. 2002. The lytic switch protein of KSHV activates gene expression via functional interaction with RBP-Jk (CSL), the target of the Notch signaling pathway. Genes Dev. 16:1977-1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liang, Y., and D. Ganem. 2003. Lytic but not latent infection by Kaposi's sarcoma-associated herpesvirus requires host CSL protein, the mediator of Notch signaling. Proc. Natl. Acad. Sci. USA 100:8490-8495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liao, W., Y. Tang, Y. L. Kuo, B. Y. Liu, C. J. Xu, and C. Z. Giam. 2003. Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8 transcriptional activator Rta is an oligomeric DNA-binding protein that interacts with tandem arrays of phased A/T-trinucleotide motifs. J. Virol. 77:9399-9411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lieberman, P. M., J. M. Hardwick, J. Sample, G. S. Hayward, and S. D. Hayward. 1990. The Zta transactivator involved in induction of lytic cycle gene expression in Epstein-Barr virus-infected lymphocytes binds to both AP-1 and ZRE sites in target promoter and enhancer regions. J. Virol. 64:1143-1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin, S. F., D. R. Robinson, G. Miller, and H. J. Kung. 1999. Kaposi's sarcoma-associated herpesvirus encodes a bZIP protein with homology to BZLF1 of Epstein-Barr virus. J. Virol. 73:1909-1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lukac, D. M., L. Garibyan, J. R. Kirshner, D. Palmeri, and D. Ganem. 2001. DNA binding by Kaposi's sarcoma-associated herpesvirus lytic switch protein is necessary for transcriptional activation of two viral delayed early promoters. J. Virol. 75:6786-6799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lukac, D. M., J. R. Kirshner, and D. Ganem. 1999. Transcriptional activation by the product of open reading frame 50 of Kaposi's sarcoma-associated herpesvirus is required for lytic viral reactivation in B cells. J. Virol. 73:9348-9361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lukac, D. M., R. Renne, J. R. Kirshner, and D. Ganem. 1998. Reactivation of Kaposi's sarcoma-associated herpesvirus infection from latency by expression of the ORF 50 transactivator, a homolog of the EBV R protein. Virology 252:304-312. [DOI] [PubMed] [Google Scholar]
- 37.Miller, G., L. Heston, E. Grogan, L. Gradoville, M. Rigsby, R. Sun, D. Shedd, V. M. Kushnaryov, S. Grossberg, and Y. Chang. 1997. Selective switch between latency and lytic replication of Kaposi's sarcoma herpesvirus and Epstein-Barr virus in dually infected body cavity lymphoma cells. J. Virol. 71:314-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakamura, H., M. Lu, Y. Gwack, J. Souvlis, S. L. Zeichner, and J. U. Jung. 2003. Global changes in Kaposi's sarcoma-associated virus gene expression patterns following expression of a tetracycline-inducible Rta transactivator. J. Virol. 77:4205-4220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nicholas, J., J.-C. Zong, D. J. Alcendor, D. M. Ciufo, L. J. Poole, R. T. Sarisky, C.-J. Chiou, X. Zhang, X. Wan, H. G. Guo, M. S. Reitz, and G. S. Hayward. 1998. Novel organizational features, captured cellular genes, and strain variability within the genome of KSHV/HHV8. J. Natl. Cancer Inst. Monogr. 23:79-88. [DOI] [PubMed] [Google Scholar]
- 40.Ragoczy, T., L. Heston, and G. Miller. 1998. The Epstein-Barr virus Rta protein activates lytic cycle genes and can disrupt latency in B lymphocytes. J. Virol. 72:7978-7984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Renne, R., W. Zhong, B. Herndier, M. McGrath, N. Abbey, D. Kedes, and D. Ganem. 1996. Lytic growth of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) in culture. Nat. Med. 2:342-346. [DOI] [PubMed] [Google Scholar]
- 42.Ruf, I. K., and D. R. Rawlins. 1995. Identification and characterization of ZIIBC, a complex formed by cellular factors and the ZII site of the Epstein-Barr virus BZLF1 promoter. J. Virol. 69:7648-7657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Russo, J. J., R. A. Bohenzky, M. C. Chien, J. Chen, M. Yan, D. Maddalena, J. P. Parry, D. Peruzzi, I. S. Edelman, Y. Chang, and P. S. Moore. 1996. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc. Natl. Acad. Sci. USA 93:14862-14867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sakakibara, S., K. Ueda, J. Chen, T. Okuno, and K. Yamanishi. 2001. Octamer-binding sequence is a key element for the autoregulation of Kaposi's sarcoma-associated herpesvirus ORF50/Lyta gene expression. J. Virol. 75:6894-6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saveliev, A. K., F. X. Zhu, and Y. Yuan. 2002. Transcription mapping and expression patterns of genes in the major immediate-early region of Kaposi's sarcoma-associated herpesvirus. Virology 299:301-304. [DOI] [PubMed] [Google Scholar]
- 46.Seaman, W. T., D. Ye, R. X. Wang, E. E. Hale, M. Weisse, and E. B. Quinlivan. 1999. Gene expression from the ORF50/K8 region of Kaposi's sarcoma-associated herpesvirus. Virology 263:436-449. [DOI] [PubMed] [Google Scholar]
- 47.Shaulian, E., and M. Karin. 2002. AP-1 as a regulator of cell life and death. Nat. Cell Biol. 4:E131-E136. [DOI] [PubMed] [Google Scholar]
- 48.Song, M. J., H. Deng, and R. Sun. 2003. Comparative study of regulation of RTA-responsive genes in Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8. J. Virol. 77:9451-9462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Song, M. J., X. Li, H. J. Brown, and R. Sun. 2002. Characterization of interactions between RTA and the promoter of polyadenylated nuclear RNA in Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8. J. Virol. 76:5000-5013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Soulier, J., L. Grollet, E. Oksenhendler, P. Cacoub, D. Cazals-Hatem, P. Babinet, M. F. d'Agay, J. P. Clauvel, M. Raphael, L. Degos, et al. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman's disease. Blood 86:1276-1280. [PubMed] [Google Scholar]
- 51.Sun, R., S. F. Lin, K. Staskus, L. Gradoville, E. Grogan, A. Haase, and G. Miller. 1999. Kinetics of Kaposi's sarcoma-associated herpesvirus gene expression. J. Virol. 73:2232-2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang, S., S. Liu, M. Wu, Y. Geng, and C. Wood. 2001. Kaposi's sarcoma-associated herpesvirus/human herpesvirus-8 ORF50 gene product contains a potent C-terminal activation domain which activates gene expression via a specific target sequence. Arch. Virol. 146:1415-1426. [DOI] [PubMed] [Google Scholar]
- 53.Wang, S., S. Liu, M. H. Wu, Y. Geng, and C. Wood. 2001. Identification of a cellular protein that interacts and synergizes with the RTA (ORF50) protein of Kaposi's sarcoma-associated herpesvirus in transcriptional activation. J. Virol. 75:11961-11973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang, S. E., F. Y. Wu, M. Fujimuro, J.-C. Zong, S. D. Hayward, and G. S. Hayward. 2003. Role of the CCAAT/enhancer-binding protein alpha (C/EBPα) in activation of the Kaposi's sarcoma-associated herpesvirus (KSHV) lytic-cycle replication-associated protein (RAP) promoter in cooperation with the KSHV replication and transcription activator (RTA) and RAP. J. Virol. 77:600-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang, S. E., F. Y. Wu, Y. Yu, and G. S. Hayward. 2003. CCAAT/enhancer-binding protein-α is induced during the early stages of Kaposi's sarcoma-associated herpesvirus (KSHV) lytic cycle reactivation and together with the KSHV replication and transcription activator (RTA) cooperatively stimulates the viral RTA, MTA, and PAN promoters. J. Virol. 77:9590-9612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu, F. Y., J.-H. Ahn, D. J. Alcendor, W. J. Jang, J.-S. Xiao, S. D. Hayward, and G. S. Hayward. 2001. Origin-independent assembly of Kaposi's sarcoma-associated herpesvirus DNA replication compartments in transient cotransfection assays and association with the ORF-K8 protein and cellular PML. J. Virol. 75:1487-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu, F. Y., H. Chen, S. E. Wang, C. M. ApRhys, G. Liao, M. Fujimuro, C. J. Farrell, J. Huang, S. D. Hayward, and G. S. Hayward. 2003. CCAAT/enhancer binding protein alpha interacts with ZTA and mediates ZTA-induced p21(CIP-1) accumulation and G1 cell cycle arrest during the Epstein-Barr virus lytic cycle. J. Virol. 77:1481-1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu, F. Y., Q.-Q. Tang, H.-L. Chen, C. M. ApRhys, C. Farrell, J.-M. Chen, M. Fujimuro, M. D. Lane, and G. S. Hayward. 2002. Lytic replication-associated protein (RAP) encoded by Kaposi's sarcoma-associated herpesvirus causes p21CIP-1-mediated G1 cell cycle arrest through CCAAT/enhancer-binding protein-alpha. Proc. Natl. Acad. Sci. USA 99:10683-10688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wu, F. Y., S. E. Wang, Q.-Q. Tang, M. Fujimuro, C.-J. Chiou, Q. Cheng, H.-L. Chen, S. D. Hayward, M. D. Lane, and G. S. Hayward. 2003. Cell cycle arrest by Kaposi's sarcoma-associated herpesvirus replication-associated protein (RAP) is mediated at both the transcriptional and posttranslational levels by binding to CCAAT/enhancer-binding protein alpha and p21CIP-1. J. Virol. 77:8893-8914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zalani, S., E. Holley-Guthrie, and S. Kenney. 1996. Epstein-Barr viral latency is disrupted by the immediate-early BRLF1 protein through a cell-specific mechanism. Proc. Natl. Acad. Sci. USA 93:9194-9199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang, L., R. Wang, A. Sweat, R. Goldstein, R. Horvat, and B. Chandran. 1999. Activation of human herpesvirus 8 (KSHV) thymidine kinase (TK) TATAA-less promoter by KSHV ORF50 gene product is SP1 dependent. DNA Cell Biol. 17:735-742. [DOI] [PubMed] [Google Scholar]
- 62.Zhou, H., A. Lin, Z. Gu, S. Chen, N.-H. Park, and R. Chiu. 2000. 12-O-tetradecanoylphorbol-13-acetate (TPA)-induced cJUN N-terminal kinase (JNK) phosphatase renders immortalized or transformed epithelial cells refractory to TPA-inducible JNK activity. J. Biol. Chem. 275:22868-22875. [DOI] [PubMed] [Google Scholar]
- 63.Zhu, F. X., T. Cusano, and Y. Yuan. 1999. Identification of the immediate-early transcripts of Kaposi's sarcoma-associated herpesvirus. J. Virol. 73:5556-5567. [DOI] [PMC free article] [PubMed] [Google Scholar]