Abstract
The cystovirus ϕ6 shares several distinctive features with other double-stranded RNA viruses, including the human pathogen, rotavirus: segmented genomes; non-equivalent packing of 120 subunits in its icosahedral capsid; capsids as compartments for transcription and replication. ϕ6 assembles as a dodecahedral procapsid that undergoes major conformational changes as it matures into the spherical capsid. We determined the crystal structure of the capsid protein, P1, revealing a flattened trapezoid subunit with a novel α-helical fold. We also solved the procapsid by cryo-electron microscopy to comparable resolution. Fitting the crystal structure into the procapsid disclosed substantial conformational differences between the two P1 conformers. Maturation via two intermediate states involves remodeling on a similar scale, besides huge rigid-body rotations. The capsid structure and its stepwise maturation which is coupled to sequential packaging of three RNA segments sets the cystoviruses apart from other dsRNA viruses as a dynamic molecular machine.
Keywords: Bacteriophage ϕ6, Cystoviridae, cryo-electron microscopy, capsid structure, capsid expansion, segmented genome, conformational change, RNA packaging
Introduction
Double-stranded RNA viruses have a wide host range covering animals, plants, fungi and bacteria but nevertheless share a number of distinctive properties (Patton, 2008). Their genomes comprise multiple (up to 12) linear segments and are typically accommodated in multiple nested protein shells (capsids). After cell entry, the genome remains inside the innermost capsid that also houses the viral RNA-dependent RNA polymerase. This particle functions as a replication machine or polymerase complex where mRNAs are transcribed and secreted. Related particles, called procapsids, are formed early in the assembly of next-generation virions. They package ssRNA segments, synthesize second strands, and finally transcribe. The overall architecture of these inner capsids is similar in all dsRNA viruses, consisting of 120 copies of a major capsid protein organized in a T=1 icosahedral shell built from 60 dimers of non-equivalent subunits. Key and still unanswered questions are: how is a single copy of each segment selected and packaged?
High-resolution structures have been determined by X-ray crystallography and cryo-electron microscopy (cryoEM) for the inner capsids of several dsRNA viruses, including several reoviruses (Grimes et al., 1998; Nakagawa et al., 2003; Reinisch et al., 2000), birnaviruses (Coulibaly et al., 2005), picobirnaviruses (Duquerroy et al., 2009), and totiviruses (Naitow et al., 2002). Except for birnaviruses, the shell is formed by 60 dimers of non-equivalent subunits, denoted A and B, an architecture encountered in no other virus family. Twelve pentamers of A subunits are centered on the 5-fold vertices and 20 trimers of B-subunits on the 3-fold axes. Although both subunits have identical sequences and similar folds, they differ somewhat in local features and radically in inter-subunit interactions (Grimes et al., 1998; Jaalinoja et al., 2007; Nakagawa et al., 2003; Reinisch et al., 2000).
Cystoviridae are the only family of dsRNA viruses that infect bacteria. They were also the first dsRNA viruses for which reverse genetics has been developed (Mindich, 1999a, b; Olkkonen et al., 1990), an asset that commended them as a model system for studying assembly and replication of dsRNA viruses. In consequence, much biochemical and genetic data have been collected concerning their replication cycle (Frilander and Bamford, 1995; Mindich, 1999a; Poranen and Bamford, 2012).
Bacteriophage ϕ6, the type member of the Cystoviridae, initially assembles as an RNA-free procapsid with deeply recessed vertices, giving it a dodecahedral morphology (Butcher et al., 1997). The procapsid accommodates the polymerase (P2) and an accessory protein (P7), which has a regulatory function in assembly and RNA packaging (Poranen et al., 2008). P2 is bound to the inner surface of the procapsid (Nemecek et al., 2010; Sen et al., 2008) at sites close to the 3-fold axes that overlap those occupied by P7 (Nemecek et al., 2012; Sun et al., 2012). On the outer surface of the procapsid, hexamers of the packaging NTPase (P4) overlie the 5-fold vertices (de Haas et al., 1999; Pirttimaa et al., 2002).
Packaging of the three ssRNA segments proceeds in order from the shortest segment (s, 2948 nt) to the m-segment (4063 nt) and finally the l-segment (6374 nt) (Mindich, 1999a). Packaging is accompanied by a major structural transformation that yields the spherical mature capsid, with a net volume increase of ~ 250%. It has been proposed (Mindich, 1999a) that the transformation proceeds stepwise, sequentially exposing binding sites for each RNA segment on the outer surface of maturing procapsid. A binding site for the s-segment has been localized to the region between amino acids 98 and 155 of P1 by cross-linking (Qiao et al., 2003b). In support of this scenario, two expansion intermediates have been observed (Nemecek et al., 2011). The shell conformation also controls the activity of the polymerase P2 which begins to synthesize second (minus) RNA strands only after all three segments are packaged (Frilander et al., 1992).
The present study has aimed to gain insight into the mechanisms that control assembly, maturation, and functioning of the polymerase complex. To do so, we set out to crystallize P1 and complemented this approach with cryo-EM and image reconstruction of the procapsid. Both approaches succeeded. Then by fitting the crystal structure into cryo-EM density maps, we were able to characterize the structural alterations undergone by P1 as it adapts to the P1A and P1B conformations. Applying the same approach to earlier reconstructions of the mature capsid (Huiskonen et al., 2006) and two expansion intermediates (Nemecek et al., 2011), we were able to follow the molecular rearrangements undergone during maturation. From these models, we could identify a positively charged cavity that is open on the procapsid and likely to bind the s-segment but closed and therefore inaccessible in the expansion intermediates and mature capsid, observations consistent with the sequential packaging hypothesis.
Results
Crystallization of P1
Our initial crystallization trials with monomeric P1 produced crystals but they diffracted only to ~ 6 Å. Later, a mixture of P1 and P7 yielded crystals diffracting to 3.6 Å. As they could not be separated from a film of denatured protein, we were unable to determine whether they also contained P7. Nevertheless, they sufficed to solve the structure of P1. As the self-rotation function suggested a pentamer in the asymmetric unit, we used the P1 pentamer from an earlier cryo-EM map at 7 Å resolution as a search model for molecular replacement. Phases were extended to 3.6 Å, using non-crystallographic symmetry. In the resulting density map (Figure S1A), the entire P1 subunit was well ordered except for a few C-terminal residues and it was possible to trace the whole Cα chain and almost all side-chains (e.g. Figure S1B), apart from several flexible Arg and Lys residues We did not find any density attributable to P7 but cannot rule out that P7 was present at low occupancy or in a disordered state and somehow promoted P1 assembly into pentamers or stabilized a conformation that allowed the growth into well ordered crystals.
P1 has a novel α-helical fold
The P1 subunit (Figure 1B) has a trapezoid shape with sides of ~ 91 Å by 73 Å. The thickness varies between 14 Å and 38 Å at the edges, reaching 47 Å at the center. The structure is mostly α-helical (46% of residues; Figure S1C,D), in agreement with Raman spectroscopy data (Benevides et al., 2002) and cryo-EM reconstructions at ~ 7 Å resolution that resolved many rod-like densities – putatively α-helices - in both the procapsid (Nemecek et al., 2012) and the nucleocapsid (Huiskonen et al., 2006). The fold appears to be unique among known structures as we found no similar fold – and in particular, no similar capsid protein fold - upon using the DALI server (Holm and Rosenstrom, 2010).
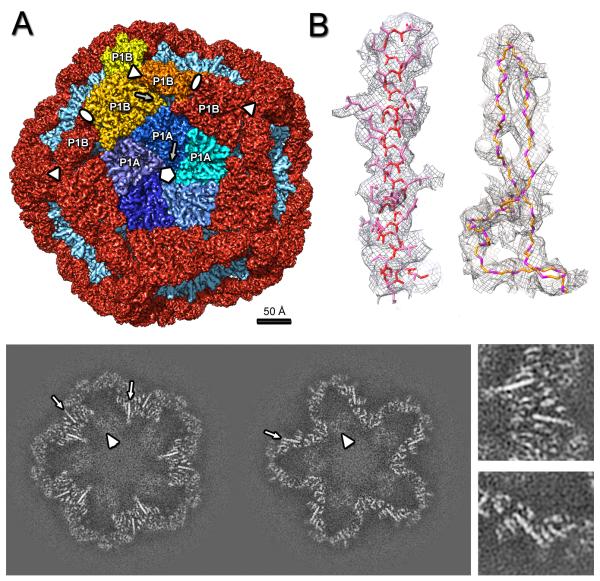
Figure 1.
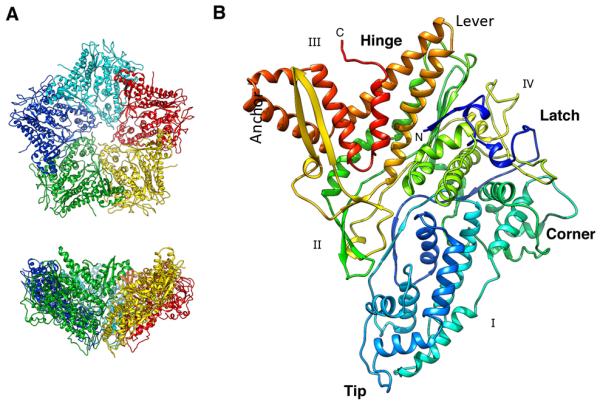
Crystal structure of P1. (A) Top and side views of the funnel-shaped P1 pentamer (the 5 subunits are in different colors). (B) The P1 subunit (rainbow-colored from blue at the N-terminus to red at the C-terminus) has a trapezoid shape with four edges labeled I - IV. The long helix-turn-helix (in gold) forms a “lever” that rotates during maturation of the procapsid. Five copies of the “tip” line the axial channel through the pentamer. The “corner” of one pentamer subunit fits against the “anchor” of a neighboring subunit. See also Figure S1.
The N-terminus forms a “latch” over two helices located in the middle of the structure (blue in Figure 1B). This is followed by a long loop connected to the “tip” region, featuring six α-helices. The next part is a set of four helices forming the “corner” of the trapezoid. The central part of the polypeptide chain is composed of long extended loops interspersed with a few helices. This region largely separates the N- and C-terminal thirds. Finally, the C-terminal part starts with a two-strand β-sheet forming a protruding hairpin (yellow in Figure 1B). This is followed by a long helix-loop-helix (the “lever”), three helices denoted the “anchor”, and the C-terminal helix and loop. These C-terminal helices form an interface that maps to the P1B subunits in the procapsid where a large “hinge” motion contributes to the expansion of the shell (see below). The hinge region also interfaces the P1A to the P1B subunits around the rim of the P1A pentamer (Figure 2A).
Figure 2.
Cryo-EM reconstruction of the ϕ6 procapsid. (A) Segmentation of the outer surface viewed along a 5-fold axis. The 12 inverted 5-fold vertices are occupied by P1A pentamers (the 5 subunits are in shades of blue) set in a dodecahedral frame of sixty P1B subunits (red, except for the three subunits around one 3-fold axis which are in shades of yellow). (B) Density for one α-helix (left) and two β-strands with the corresponding atomic model (side-chains are shown for the α-helix only, for clarity). (C) Slices through the cryoEM reconstruction viewed along the 5-fold axis at 20 Å (left) and 40 Å (right) from the procapsid center. Elongated densities representing α-helices that are approximately in-plane are indicated by arrows and enlarged in the right panels. Scale bar: 100 Å. See also Movie S2.
In order to describe the inter-subunit interfaces in the procapsid (below), we designate the four edges of the trapezoid as I – IV (Figure 1B). Only edges I and II are involved in inter-subunit interactions in the crystallized pentamer (Figure 1A), and they show good complementarity of positive and negative residues The buried surface at this interface is 9248 Å2, suggesting a very stable pentamer.
Expression of quasi-equivalence in the procapsid
We determined the procapsid structure by cryo-EM to a resolution of 4.5 Å (Table 2). The features visualized, such as regular helical grooves on α-helices, separation of loops and β-strands, and densities for bulky side-chains, validate the calculated resolution (Figure S2). With some discrepancies (see below), the crystal P1 pentamer fits into the P1A pentamer, and the P1 monomer could be placed into the P1B-related density without ambiguity. The molecular boundaries (Figure 2A) agree with those inferred from previous cryo-EM maps of the nucleocapsid (Huiskonen et al., 2006) and the procapsid (Nemecek et al., 2012). Each edge of the trapezoid forms two different interfaces with other P1 subunits. In the P1A pentamer, edges I and II are apposed. Edge II in P1B fits across two edges from different P1A subunits (one an edge III and the other an edge IV). The P1B edges I and IV constitute the intra-trimer interfaces around the three-fold axis. At the two-fold axis, the apposing P1B edge III interfaces to form the major hinge involved in expansion (see below).
Table 2.
Single particle analysis: statistics
| Number of micrographs | 154 |
| Number of particle images | 28194 (65% used) |
| Defocus range (^m) | 1.3 – 3.2 |
| FSC0.3 (Å) | 3.9 |
| FSC0.5 (Å) | 4.4 |
We performed flexible fitting of crystal P1 into the P1A density. The quality of the density in the cryo-EM map gave confidence in the reliability of the results. Overall there is good correspondence but with significant differences that reflect adaptation of the protein conformation to the context of the procapsid. The largest difference is in the hinge region, where four helices (H24, H25, H28 and H29) are rotated (by ~18°) to accommodate the adjacent P1B in the procapsid (Figure 3B). The C-terminal helix is also shifted, positioning the C-terminus of P1A into a hydrophobic groove in the neighboring P1B subunit.
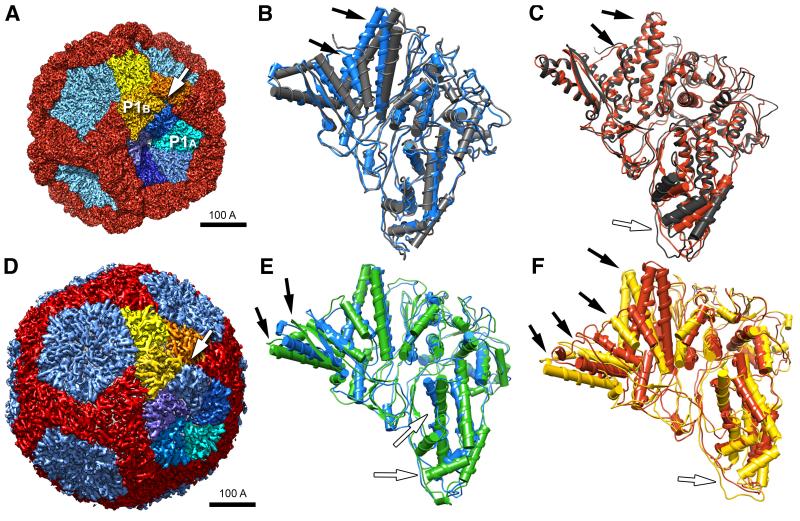
Figure 3.
(A) Cryo-EM reconstruction of the procapsid with subunits color-coded as in Figure 2A but viewed from a different angle. (B) Differences between the P1A subunit in the procapsid (blue) and the P1 crystal structure (dark gray) are localized mainly to the hinge region (black arrows, white arrow in (A)). (C) Comparison of P1A (black) and P1B (red) in the procapsid, showing differences of similar magnitude to those in (B): black arrows point out features most affected and additional differences at the tip (white arrow). (D) Cryo-EM reconstruction of the nucleocapsid (EMD-1206 by Huiskonen et al. (2006) segmented as in (A). (E) Changes in the P1A structure on maturation (procapsid – blue; nucleocapsid – green) localize in the helices next to the hinge region (black arrows, white arrow in (C)). (F) Changes in the P1B structure on maturation (procapsid, red; nucleocapsid, yellow) involve the whole of edge III (black arrows) as well as the tip (white arrow). Scale bar: 100 Å.
In P1B, on the other hand, the hinge region and the C-terminal helix are essentially as in crystal P1, as are the loops and helices at the II-IV interface with the adjacent P1B subunit. The most pronounced differences in P1B affect the “tip” region at the interface with P1A (Figure 3C). Here, two helices are rotated about 27° and 9° respectively to accommodate displacement of loops at the P1A/P1B interface.
Changes in P1 during capsid maturation
Procapsid maturation during RNA packaging involves massive conformational changes and accompanying changes in size and shape (cf. Figure 3A and Figure 3D). To determine the changes in the conformations and interactions of P1A and P1B subunits, we performed flexible fitting with crystal P1 into a nucleocapsid map at 7.5 Å resolution (EMDB ID: emd1206 (Huiskonen et al., 2006)) (Figure 3E, F). This exercise showed that the principal mechanism is large-scale hinging movements about inter-subunit interfaces (Figure 4). The accompanying conformational changes, mainly affecting P1B, are on the same order as those that distinguish procapsid subunits from crystal P1. In P1A, the greatest change is in the long β-hairpin and underlying helices of the anchor that tilt through ~20° (Figure 3E). Interactions with neighboring subunits also change; e.g. the long helix-turn-helix (the lever) is no longer connected to the neighboring P1B subunit by the Arg655 salt bridge.
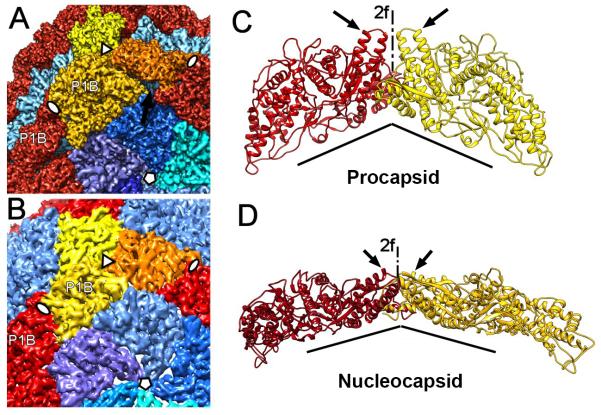
Figure 4.
Transformation at the hinge. (A) P1B subunits (red and yellow) are tightly connected to P1A subunits (blues) of the inverted vertices in the procapsid. This shell contains cavities between the P1B and P1A subunits (arrows). (B) In the nucleocapsid, the P1B subunits are rotated so that the planes of these flat molecules concide with the tangential plane of the shell, leaving no significant cavities between the subunits. (C) Orientation of P1B subunits (red and yellow ribbons) on either side of a 2-fold icosahedral axis in the procapsid. The subunit planes are almost perpendicular to each other, and their helix-turn-helix motifs are aligned with the 2-fold axis (arrows). (D) Corresponding representation of two P1B subunits in the nucleocapsid. The two subunits are now almost coplanar. (The views shown in C and D are rotated around the 2-fold axis so that the dihedral angles appear considerably larger.) See also Movie S2.
During expansion, adjacent P1B subunits rotate around a “pivot” axis connecting the three-fold axes through the two-fold axis (Figure 4A, B). The planes of the two subunits meet at an angle of ~98° in the procapsid, changing to ~148° in the nucleocapsid (Figure 4C,D). This movement results in much larger buried surfaces for both subunits (~50% increase for P1B and ~16% for P1A) and a better match of complementary charged residues. At the intramolecular level, the lever shifts away from the 2-fold icosahedral axis and is further tilted ~25° within the subunit, pointing towards the β-hairpin in the apposing P1B subunit (arrows in Figure 4C,D, Movie S2). Concomitantly, the hydrophobic C-terminus is moved into the uncharged groove in the opposing P1B subunit.
Staging-posts in the expansion reaction
We also had available two expansion intermediates at lower resolution (18 Å) (EMDB: 5355 and EMDB: 5357). With them, we performed rigid-body fitting, using the P1A and P1B conformations of both the procapsid and the mature capsid. In both particles, the best fits were obtained with P1A in its procapsid conformation and P1B in its nucleocapsid conformation. Figure 5 and Movie S3 show the sequence of transitions, starting with the compact procapsid and expanding to the almost spherical mature capsid and a markedly thinner P1 shell. The large change in the hinge region occurs mainly in the first transition from the procapsid to expansion intermediate 1, which has the lowest-energy state of the P1 shell (not the fully mature shell - (Nemecek et al., 2011)). Subsequent changes in the expansion involve smaller rearrangements, mainly around the hinge and tip regions .
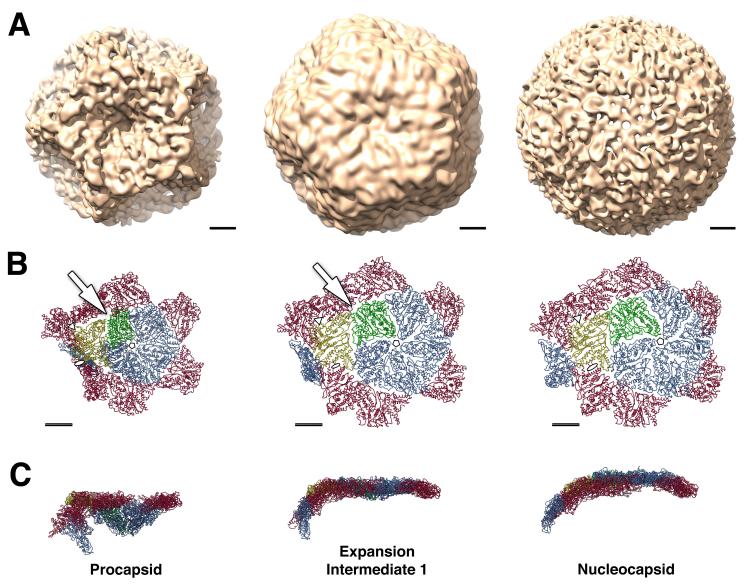
Figure 5.
Procapsid expansion. (A) CryoEM reconstructions of three conformational states of the ϕ6 capsid at 16 Å resolution. (B, C) Models of a portion of capsid comprising a pentamer of P1A subunits (blue, green) and surrounding P1B subunits (red, yellow) subunits, viewed from above (B) and from the side (C). Expansion to intermediate 1 is the major transition of the maturing capsid, achieved by rotation of P1B subunits around an axis connecting the 3-fold icosahedral axes (bar in panel A). This rotation appears to stabilize the P1B/P1B interface at the 2-fold axis (Figure 6) and seals gaps between P1A and P1B subunits near the 3-fold axis (arrow). Further expansion to intermediate 2 is achieved by outward movement of the P1A subunits. The final step to the nucleocapsid state is accompanied by local conformational changes in the P1A subunits that correlate with increased outward curvature at the 5-fold axis. Scale bar: 100 Å. See also Movie S3.
Discussion
Cystoviruses are of interest as double-stranded RNA viruses that provide tractable experimental systems. The molecular composition and assembly of the procapsid have been studied extensively, and high resolution structures have been determined for three of its four proteins: the P2 RNA-dependent RNA polymerase of ϕ6 (Butcher et al., 2001); the P4 packaging motor of ϕ12 (Mancini et al., 2004); and the P7 packaging facilitator of ϕ12 (Eryilmaz et al., 2008). Here we complete the ensemble with the structure of P1 from ϕ6 and show how the procapsid serves as a framework to which the other three proteins bind, and how it transforms during maturation.
P1 appears to have a novel fold: in particular, we detected no similarity with the known folds of the capsid proteins of other dsRNA viruses. To the extent that capsid protein fold may be viewed as a hallmark of common ancestry (Bamford et al., 2005), this distinction would suggest that cystoviruses originated in a different lineage than other dsRNA viruses. P1 does share with them the distinctive, nonequivalent, 120-subunit capsid geometry but the in-plane shape of the subunit and of the pentamer of A-subunits, which is quite pentagonal in the case of ϕ6, are quite different between the respective systems.
Another property that sets cystoviruses apart from other dsRNA viruses is the large-scale conformational changes undergone during maturation. In this respect, they are more akin to the capsids of tailed bacteriophages (Conway et al., 2001; Gertsman et al., 2009; Lata et al., 2000) than to other dsRNA viruses, although the respective folds (the HK97 archetype - (Wikoff et al., 2000) - in the case of tailed phages) and architecture differ. (Tailed phages assume quasi-equivalent geometries, and a variety of T-numbers – (Dearborn et al., 2012) (Duda et al., 2006)). In the same vein, there is no evidence that the inner capsids of other dsRNA viruses undergo comparable structural changes. Insofar as these stepwise transitions of the maturing ϕ6 procapsid may afford a mechanism for selecting one copy each of the three RNA segments - a proposition for which there is supporting evidence but not yet a conclusive proof - this would also suggest that the other dsRNA viruses select their complements of segments for packaging by a different mechanism.
Mapping functional sites on the P1 lattice
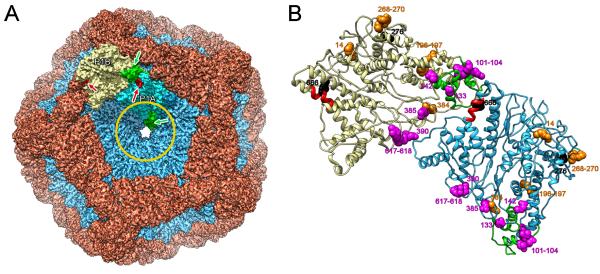
Previous studies have identified the residues involved in the scissile sites on the procapsid for two proteases and in the epitope for a monoclonal antibody (Qiao et al., 2003b). The procapsid structure demonstrates the locations of these sites on its outer surface (Figure 6 & Figure S3) - where indeed they should be, for accessibility. As such, these observations help to validate the determined structure.
Figure 6.
(A) Regions of P1 interacting with the s-segment (green) and a monoclonal antibody (red) are shown respectively for the P1A and P1B subunits in the context of the procapsid shell. The circle represents the area covered by the P4 hexamer. (B) Locations of mutations in P1 affecting s- and m-segment binding (magenta) and packaging (orange) are shown for the P1A (cyan) and P1B (yellow) subunits. The proteolytically susceptible sites for factor Xa and trypsin are also shown (black). The subunits are oriented as in the procapsid (panel A) and viewed from the outside. See also Figure S3.
The three other proteins interact only with P1A: P2 and P7 bind on the inside of the procapsid (Katz et al., 2012; Nemecek et al., 2012; Sen et al., 2008) and P4 on the outside (de Haas et al., 1999; Pirttimaa et al., 2002). The overlapping footprints of P2 and P7 cover the “tip” region of P1A. Crosslinking of the s-segment and mutants that affect s-segment packaging (Qiao et al., 2003a) also map to the tip (Figure 6), indicating this region as the likely s-segment binding site. This region is exposed to the exterior in P1B whereas it is covered by P4 hexamers in P1A (Figure 6A). Consequently, the s-segment most likely binds to P1B on the procapsid exterior, while the positively charged tips of P1A interact with RNA that has translocated through the axial channel. The overlap with sites of mutants affected in m-segment packaging (Qiao et al., 2003a) suggests some commonality in the binding sites of the first two segments to be packaged.
The binding site for the s-segment has been mapped to the polypeptide segment between Cys98 and Cys155 of P1 (Qiao et al., 2003b). This segment is located near the “tip” (Figure 6). In P1A, it lines the RNA-packaging channel at the 5-fold vertex. In P1B, its residues are exposed on the procapsid surface near a cavity between tilted P1B subunits at the periphery of the P1A pentamer. The cavity, which is ~ 60Å from the 5-fold axis (Figure 5B), is positively charged with contributions from Arg104 and Lys105 on P1B and Arg523 on the neighboring P1B. Presumably, the negatively charged sugar-phosphate backbone of the s-segment pac site (about 200 nucleotides near the 5′ end of the ssRNA) binds in this cavity. Subsequently, the cavity closes as the procapsid converts to the first expansion intermediate (Figure 5).
It is likely that the segment-binding sites involve more than one molecule of P1 and are located some distance away from the P4 hexamers since deletion of 22 nucleotides in the s-sequence 5′ to the pac sequence was able to prevent packaging but allowed binding and competition with normal binding and packaging (Qiao et al., 1997).
Mutations that alter RNA packaging also affect the procapsid conformation
Even small changes in the pac sequence have drastic effects on RNA packaging. Point mutations in P1 can suppress pac sequence mutations or, conversely, may prevent the wild-type RNA sequences from binding (Qiao et al., 2003b). The amino acid replacements that eliminate s-segment binding (WR103VA, R385A, RR617AA) are located near edge II of P1 on the procapsid exterior (Figure S3A). These residues are not clustered and the mutations probably act independently. Residue R196 is located inside the P1 fold in the tip (Figure S3B) and the RR617AA mutant probably disrupts interactions in the adjacent s-segment binding site. Suppressors of this mutation (T316I, A402T) are not close to this site, implying that long-range allosteric effects are involved. Similarly, the suppressors Y486C or Y486S map in different regions of P1 (Figure S3D). On the other hand, suppressors of the Spac and Mpac mutants all map close to the s-segment binding site, suggesting direct interactions with it (Figure S3E,F). Moreover, they suggest that the binding sites for the s- and m-segments are close together.
Conformations of P1 in the procapsid: implications for assembly
Attachment of P1B subunits around the rims of the P1A pentamers results in conformational changes at the P1A/P1B interface. The lever in P1A is bent through ~ 18° and interacts with two adjacent P1B subunits. The adjacent C-terminal helix in P1A is also affected, being bent towards the P1A/P1B interface, with the hydrophobic C-terminus inserted into a hydrophobic pocket in P1B.
The conformation of P1B subunits in the procapsid is also altered at the interface with P1A subunits at the tip. Loops and helices are bent in this region to accommodate attachment to helix bundles in P1A. However, the conformation of the P1B/P1B interface at the 2-fold icosahedral axis corresponds to the conformation in the crystal structure (Figure 3C). Although there are complementary electrostatic interaction across this interface, the negatively charged tips of the levers come into close proximity. The P1B subunits are relatively tilted against each other by ~ 90°, resulting in an interface with the neighboring subunits that is ~ 20% smaller compared with P1A in the procapsid (Figure 4). Similarly, the interfaces between P1B subunits around the 3-fold icosahedral axis are relatively small and there are appreciable cavities between the P1B subunits and the P1A vertices (Figure 5B). These considerations suggest that P1B subunits may attach to preassembled P1A pentamers and connect them into a dodecahedral shell in an overall conformation that is suboptimal but is later optimized during maturation.
Conformations of P1A and P1B: implications for maturation
In maturation, the conformation of P1A subunits changes mainly in the hinge region, where the large β-hairpin and the underlying helices bend through ~20°. Also, loops at their interface with P1B alter to accommodate new inter-subunit interactions (Figure 3E). As for P1B conformation, the entire subunit is affected, particularly in the hinge region. The lever is bent about 26° from the icosahedral 2-fold axis and its position is replaced by the C-terminal helix (Figures 3F, 4D). This change accompanies a large rigid-body rotation of P1B (Movie S2) that interlocks P1B subunits around the 3-fold icosahedral axes and closes pre-existing cavities between subunits. In this way, the buried surface area per P1B subunit increases by ~50% in the nucleocapsid and becomes comparable with that of P1A.
Knowledge of the procapsid and nucleocapsid structures illuminates numerous aspects of assembly, maturation and packaging. However, weighty issues remain. How does the s-segment engage with P4, the packaging NTPase after attaching to its binding site? Do the P2 polymerase molecules remain in place during maturation? and how does their location affect transcription and replication? In particular, why is packaging of s-segment confined to a single copy when 60 binding sites are distributed over the procapsid surface? Here, one possibility is kinetics; i.e. binding of a single s-segment may be sufficient to induce rapid conversion of the procapsid to the expansion intermediate 1 conformation, precluding further s-segment packaging (Nemecek et al., 2011).
Experimental Procedures
Expression and purification of P1 and P7 subunits
Genes for expression of the full-length P1 and truncated P7 (residues 1-150) subunits were extracted from plasmids pLM3572 and pLM3623 respectively and recloned into the first cassette of the pRSFD vector. The plasmids were transformed into E. coli BL21(DE3) Star cells and over-expressed overnight at 20°C after induction with 0.5 mM isopropyl thiogalactoside (IPTG) at OD = 0.8. The proteins contained a C-terminal His6-tag and were purified from the cell extract by affinity chromatography, using Ni-NTA resin (QIAGEN). P1 was further purified by size-exclusion chromatography using a Superdex 200 column (GE Healthcare) equilibrated in 20 mM Tris, pH = 7.4, 75 mM NaCl, 3 mM DTT buffer. The protein eluted in two peaks corresponding to monomer and dimer. Fractions from the monomer peak were pooled and concentrated to ~8 mg/ml, flash-frozen in liquid nitrogen and stored at −80 °C. P7 was further purified using a Superdex 75 column (GE Healthcare) equilibrated in 10 mM Tris, pH = 7.4, 30 mM NaCl, 4 mM DTT, 1 mM EDTA buffer. P7-containing fractions were pooled, concentrated to 2.5 mg/ml, and stored at −80 °C.
Crystallization, data collection and structure determination
To obtain crystals that diffracted beyond ~6 Å resolution, we used a P1:P7 mixture in 2:1 molar ratio at a total concentration of 2 mg/ml. These crystals were grown in hanging drops consisting of a 1:1 mixture of the protein and well solution (100 HEPES, pH = 7.5, 180 mM calcium acetate, 10 mM EDTA, 39% PEG 400) at 293 K. Datasets were collected from three crystals using a MAR CCD detector at beamline 22-ID, Advanced Photon Source (Chicago, IL) and merged in the program HKL2000 (Otwinowski and Minor, 1997), yielding a complete dataset at 3.6 Å. The self-rotation function suggested a pentamer in the asymmetric unit and therefore we used a 7Å EM map of the P1A pentamer extracted from the wildtype procapsid (EMDB code EMD-2341 (Nemecek et al., 2012) as a search model for molecular replacement. We found a good solution with LLG = 470 and Rfactor = 50.4%. Thereafter, the phases were extended to 3.6Å resolution using non-crystallographic symmetry. The initial model was built by the program Buccaneer of the CCP4 package (Cowtan et al., 2011), (Cowtan, 2012) and manually refined using Phenix (Adams et al., 2010) and Coot (Emsley and Cowtan, 2004). The model was refined to Rwork = 21.7% and Rfree = 27.4% (Table 1). The crystal structure has been deposited in the PDB (code 4K7H).
Table 1.
Crystallographic statistics
| Space group | C 2221 |
| Cell dimension | a = 182.59Å |
| b = 278.85Å | |
| c = 246.47Å | |
| X-ray source | SER-CAT 22 ID |
| Wavelength, Å | 0.97899 |
| Resolution (å) | 3.60 (3.66 – 3.60) |
| Total reflections | 1043858 |
| Unique reflections | 66561 |
| IσI) | 11.15 (2.04) |
| Data Completeness (%) | 91.07 (41.67) |
| Multiplicity | 15.7 (2.3) |
| Rwork, % | 21.7 |
| Rfree, % | 27.4 |
| Rmerge, % | 19.2 |
| rms bond angle deviation, ° | 1.41 |
| rms bond length deviation, Å | 0.008 |
| Ramachandran favored (%) | 95.0 |
| Ramachandran outliers (%) | 0.03 |
| Number of non-hydrogen atoms | 29435 |
Preparation and purification of ϕ6 procapsids
Procapsids were produced in E. coli strain JM109 using the plasmid pLM687 (Mindich et al., 1994) to co-express wild-type P1, P2, P4 and P7 proteins (as P1247 procapsids), then extracted and purified as described previously (Nemecek et al., 2011). Prior to experiments, the samples were transferred into buffer P (10 mM potassium phosphate, 5 mM MgCl2, pH 8.0) using Zeba-midi buffer exchange columns with a 7-kDa cutoff (Thermo Scientific, Rockford, IL).
Cryo-electron microscopy
Drops of ϕ6 procapsids at ~10 mg/ml protein concentration were applied to glow-discharged C-flat grids (Protochips Inc., Raleigh, NC), blotted, and plunge-frozen in liquid ethane using a Vitrobot (FEI, Hillsboro, OR) that had been equilibrated at 25°C and 90% humidity. The vitrified specimens were imaged at 47,000x nominal magnification and 1.0-2.0 μm underfocus with the FEI Titan Krios microscope at UCLA (courtesy of Dr Z.H. Zhou and FEI). The microscope was operated at 300 kV and micrographs were recorded on SO163 Kodak film at ~15 e−/Å2 per exposure.
Image processing
Micrographs were digitized using a Nikon Super Coolscan 9000ED at 4000 dpi. Image processing was done with the EMAN1 package (Baker et al., 2010; Ludtke, 2010; Ludtke et al., 1999) as described in (Wu et al., 2013) and using Bsoft (Heymann and Belnap 2007) as described in (Nemecek et al. 2012). The initial model was derived from the previously determined procapsid map at ~ 8 Å resolution (Nemecek et al., 2012). An angular step size of 0.6° was used for global grid searches over the asymmetric unit. Icosahedral reconstructions were calculated using e2proc3d.py in EMAN2 and their resolution estimated by Fourier shell correlation between reconstructions from two half datasets at the 0.5 cut-off. The structure has been deposited in the EMDB (code 2364).
Flexible fitting of the P1 structure into the procapsid
Initially, the P1 crystal structure was rigid body-fitted into the procapsid map filtered to 7 Å resolution, using Chimera. (Goddard et al., 2005; Pettersen et al., 2004). P1A and P1B were fitted independently, while the EM map was rescaled from from 1.351 Å/pixel to 1.397 Å/pixel to maximize cross-correlation of the fits. Regions where the crystal structure deviated significantly from the EM density were roughly adjusted in Coot (Emsley and Cowtan, 2004) and then the P1A and P1B structures were flexibly fitted using the MDFF package (Trabuco et al., 2009). The fitted structures were further fitted into the EM map of the procapsid filtered to 4 Å resolution together with all neighboring subunits to avoid clashes at the P1A and P1B interfaces. Finally, both structures were visually inspected and refined in Coot.
Flexible fitting of P1 into the nucleocapsid
Here, the cryo-EM map of the ϕ6 nucleocapsid (EMDB ID: emd-1206 (Huiskonen et al., 2006)) was used. This map was rescaled and sharpened to match the current cryoEM map of the procapsid and bandpass-filtered to 7 Å resolution. As in the flexible fitting of P1 into the procapsid map, P1 was initially rigid body-fitted into the map using Chimera and then manually adjusted in Coot. The rod-like densities in the map corresponded well to the α-helices of the fitted P1 structure. We further refined the adjusted structures by flexible fitting with MDFF. All subunits surrounding a given subunit (P1A or P1B) were included in the fit.
Rigid body-fitting of P1 into the expansion intermediates
The P1A and P1B structures were transformed into EM density and bandpass-filtered to 16 Å resolution using bsf in Bsoft (Heymann and Belnap, 2007) and fitted into cryo-EM maps of the two expansion intermediates, using the symmetry-fitting algorithm in Chimera. The fit was done in two steps: initially, P1A and P1B subunits were fitted into the asymmetric unit. Then, their positions were refined taking into account all symmetry-related P1A-P1B dimers in the icosahedral shell. Several starting positions were tested and most solutions converged to the same P1A and P1B orientations.
Supplementary Material
Acknowledgements
This work was supported by the intramural research programs of NIAMS and NIDDK and by IATAP grants to J.H. for E.B. and to A.C.S. and by ASCR (RVO: 61388963) to E.B.. Crystallographic data were collected at Southeast Regional Collaborative Access Team 22-ID beamline at the Advanced Photon Source, Argonne National Laboratory. Use of the Advanced Photon Source was supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract W-31-109-Eng-38.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta crystallographica Section D, Biological crystallography. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker ML, Zhang J, Ludtke SJ, Chiu W. Cryo-EM of macromolecular assemblies at near-atomic resolution. Nature protocols. 2010;5:1697–1708. doi: 10.1038/nprot.2010.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamford DH, Grimes JM, Stuart DI. What does structure tell us about virus evolution? Curr Opin Struct Biol. 2005;15:655–663. doi: 10.1016/j.sbi.2005.10.012. [DOI] [PubMed] [Google Scholar]
- Benevides JM, Juuti JT, Tuma R, Bamford DH, Thomas GJ., Jr. Characterization of subunit-specific interactions in a double-stranded RNA virus: Raman difference spectroscopy of the phi6 procapsid. Biochemistry. 2002;41:11946–11953. doi: 10.1021/bi0201623. [DOI] [PubMed] [Google Scholar]
- Butcher SJ, Dokland T, Ojala PM, Bamford DH, Fuller SD. Intermediates in the assembly pathway of the double-stranded RNA virus phi6. The EMBO journal. 1997;16:4477–4487. doi: 10.1093/emboj/16.14.4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butcher SJ, Grimes JM, Makeyev EV, Bamford DH, Stuart DI. A mechanism for initiating RNA-dependent RNA polymerization. Nature. 2001;410:235–240. doi: 10.1038/35065653. [DOI] [PubMed] [Google Scholar]
- Conway JF, Wikoff WR, Cheng N, Duda RL, Hendrix RW, Johnson JE, Steven AC. Virus maturation involving large subunit rotations and local refolding. Science. 2001;292:744–748. doi: 10.1126/science.1058069. [DOI] [PubMed] [Google Scholar]
- Coulibaly F, Chevalier C, Gutsche I, Pous J, Navaza J, Bressanelli S, Delmas B, Rey FA. The birnavirus crystal structure reveals structural relationships among icosahedral viruses. Cell. 2005;120:761–772. doi: 10.1016/j.cell.2005.01.009. [DOI] [PubMed] [Google Scholar]
- Cowtan K. Completion of autobuilt protein models using a database of protein fragments. Acta crystallographica Section D, Biological crystallography. 2012;68:328–335. doi: 10.1107/S0907444911039655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowtan K, Emsley P, Wilson KS. From crystal to structure with CCP4. Acta crystallographica Section D, Biological crystallography. 2011;67:233–234. doi: 10.1107/S0907444911007578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haas F, Paatero AO, Mindich L, Bamford DH, Fuller SD. A symmetry mismatch at the site of RNA packaging in the polymerase complex of dsRNA bacteriophage phi6. J Mol Biol. 1999;294:357–372. doi: 10.1006/jmbi.1999.3260. [DOI] [PubMed] [Google Scholar]
- Dearborn AD, Laurinmaki P, Chandramouli P, Rodenburg CM, Wang S, Butcher SJ, Dokland T. Structure and size determination of bacteriophage P2 and P4 procapsids: function of size responsiveness mutations. J Struct Biol. 2012;178:215–224. doi: 10.1016/j.jsb.2012.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duda RL, Hendrix RW, Huang WM, Conway JF. Shared architecture of bacteriophage SPO1 and herpesvirus capsids. Current biology : CB. 2006;16:R11–13. doi: 10.1016/j.cub.2005.12.023. [DOI] [PubMed] [Google Scholar]
- Duquerroy S, Da Costa B, Henry C, Vigouroux A, Libersou S, Lepault J, Navaza J, Delmas B, Rey FA. The picobirnavirus crystal structure provides functional insights into virion assembly and cell entry. The EMBO journal. 2009;28:1655–1665. doi: 10.1038/emboj.2009.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta crystallographica Section D, Biological crystallography. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Eryilmaz E, Benach J, Su M, Seetharaman J, Dutta K, Wei H, Gottlieb P, Hunt JF, Ghose R. Structure and dynamics of the P7 protein from the bacteriophage phi 12. J Mol Biol. 2008;382:402–422. doi: 10.1016/j.jmb.2008.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frilander M, Bamford DH. In vitro packaging of the single-stranded RNA genomic precursors of the segmented double-stranded RNA bacteriophage phi 6: the three segments modulate each other’s packaging efficiency. J Mol Biol. 1995;246:418–428. doi: 10.1006/jmbi.1994.0096. [DOI] [PubMed] [Google Scholar]
- Frilander M, Gottlieb P, Strassman J, Bamford DH, Mindich L. Dependence of minus-strand synthesis on complete genomic packaging in the double-stranded RNA bacteriophage phi 6. J Virol. 1992;66:5013–5017. doi: 10.1128/jvi.66.8.5013-5017.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gertsman I, Gan L, Guttman M, Lee K, Speir JA, Duda RL, Hendrix RW, Komives EA, Johnson JE. An unexpected twist in viral capsid maturation. Nature. 2009;458:646–650. doi: 10.1038/nature07686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard TD, Huang CC, Ferrin TE. Software extensions to UCSF chimera for interactive visualization of large molecular assemblies. Structure. 2005;13:473–482. doi: 10.1016/j.str.2005.01.006. [DOI] [PubMed] [Google Scholar]
- Grimes JM, Burroughs JN, Gouet P, Diprose JM, Malby R, Zientara S, Mertens PP, Stuart DI. The atomic structure of the bluetongue virus core. Nature. 1998;395:470–478. doi: 10.1038/26694. [DOI] [PubMed] [Google Scholar]
- Heymann JB, Belnap DM. Bsoft: image processing and molecular modeling for electron microscopy. J Struct Biol. 2007;157:3–18. doi: 10.1016/j.jsb.2006.06.006. [DOI] [PubMed] [Google Scholar]
- Holm L, Rosenstrom P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huiskonen JT, de Haas F, Bubeck D, Bamford DH, Fuller SD, Butcher SJ. Structure of the bacteriophage phi6 nucleocapsid suggests a mechanism for sequential RNA packaging. Structure. 2006;14:1039–1048. doi: 10.1016/j.str.2006.03.018. [DOI] [PubMed] [Google Scholar]
- Jaalinoja HT, Huiskonen JT, Butcher SJ. Electron cryomicroscopy comparison of the architectures of the enveloped bacteriophages phi6 and phi8. Structure. 2007;15:157–167. doi: 10.1016/j.str.2006.12.004. [DOI] [PubMed] [Google Scholar]
- Katz G, Wei H, Alimova A, Katz A, Morgan DG, Gottlieb P. Protein P7 of the cystovirus phi6 is located at the three-fold axis of the unexpanded procapsid. PloS one. 2012;7:e47489. doi: 10.1371/journal.pone.0047489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lata R, Conway JF, Cheng N, Duda RL, Hendrix RW, Wikoff WR, Johnson JE, Tsuruta H, Steven AC. Maturation dynamics of a viral capsid: visualization of transitional intermediate states. Cell. 2000;100:253–263. doi: 10.1016/s0092-8674(00)81563-9. [DOI] [PubMed] [Google Scholar]
- Ludtke SJ. 3-D structures of macromolecules using single-particle analysis in EMAN. Methods Mol Biol. 2010;673:157–173. doi: 10.1007/978-1-60761-842-3_9. [DOI] [PubMed] [Google Scholar]
- Ludtke SJ, Baldwin PR, Chiu W. EMAN: Semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- Mancini EJ, Kainov DE, Grimes JM, Tuma R, Bamford DH, Stuart DI. Atomic snapshots of an RNA packaging motor reveal conformational changes linking ATP hydrolysis to RNA translocation. Cell. 2004;118:743–755. doi: 10.1016/j.cell.2004.09.007. [DOI] [PubMed] [Google Scholar]
- Mindich L. Precise packaging of the three genomic segments of the double-stranded-RNA bacteriophage phi6. Microbiol Mol Biol Rev. 1999a;63:149–160. doi: 10.1128/mmbr.63.1.149-160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mindich L. Reverse genetics of dsRNA bacteriophage phi 6. Adv Virus Res. 1999b;53:341–353. doi: 10.1016/s0065-3527(08)60355-3. [DOI] [PubMed] [Google Scholar]
- Mindich L, Qiao X, Onodera S, Gottlieb P, Frilander M. RNA structural requirements for stability and minus-strand synthesis in the dsRNA bacteriophage phi 6. Virology. 1994;202:258–263. doi: 10.1006/viro.1994.1341. [DOI] [PubMed] [Google Scholar]
- Naitow H, Tang J, Canady M, Wickner RB, Johnson JE. L-A virus at 3.4 A resolution reveals particle architecture and mRNA decapping mechanism. Nat Struct Biol. 2002;9:725–728. doi: 10.1038/nsb844. [DOI] [PubMed] [Google Scholar]
- Nakagawa A, Miyazaki N, Taka J, Naitow H, Ogawa A, Fujimoto Z, Mizuno H, Higashi T, Watanabe Y, Omura T, et al. The atomic structure of rice dwarf virus reveals the self-assembly mechanism of component proteins. Structure. 2003;11:1227–1238. doi: 10.1016/j.str.2003.08.012. [DOI] [PubMed] [Google Scholar]
- Nemecek D, Cheng N, Qiao J, Mindich L, Steven AC, Heymann JB. Stepwise expansion of the bacteriophage varphi6 procapsid: possible packaging intermediates. J Mol Biol. 2011;414:260–271. doi: 10.1016/j.jmb.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemecek D, Heymann JB, Qiao J, Mindich L, Steven AC. Cryo-electron tomography of bacteriophage phi6 procapsids shows random occupancy of the binding sites for RNA polymerase and packaging NTPase. J Struct Biol. 2010;171:389–396. doi: 10.1016/j.jsb.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemecek D, Qiao J, Mindich L, Steven AC, Heymann JB. Packaging accessory protein P7 and polymerase P2 have mutually occluding binding sites inside the bacteriophage φ6 procapsid. J Virol. 2012;86:11616–11624. doi: 10.1128/JVI.01347-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olkkonen VM, Gottlieb P, Strassman J, Qiao XY, Bamford DH, Mindich L. In vitro assembly of infectious nucleocapsids of bacteriophage phi 6: formation of a recombinant double-stranded RNA virus. Proc Natl Acad Sci U S A. 1990;87:9173–9177. doi: 10.1073/pnas.87.23.9173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods Enzymol. 1997:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Patton JT. Segmented Double-Stranded RNA Viruses: Structure and Molecular Biology. Caister Academic Press; Norfolk, UK: 2008. [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Pirttimaa MJ, Paatero AO, Frilander MJ, Bamford DH. Nonspecific nucleoside triphosphatase P4 of double-stranded RNA bacteriophage phi6 is required for single-stranded RNA packaging and transcription. J Virol. 2002;76:10122–10127. doi: 10.1128/JVI.76.20.10122-10127.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poranen MM, Bamford DH. Assembly of large icosahedral double-stranded RNA viruses. Adv Exp Med Biol. 2012;726:379–402. doi: 10.1007/978-1-4614-0980-9_17. [DOI] [PubMed] [Google Scholar]
- Poranen MM, Butcher SJ, Simonov VM, Laurinmaki P, Bamford DH. Roles of the minor capsid protein P7 in the assembly and replication of double-stranded RNA bacteriophage phi6. J Mol Biol. 2008;383:529–538. doi: 10.1016/j.jmb.2008.08.082. [DOI] [PubMed] [Google Scholar]
- Qiao J, Qiao X, Sun Y, Mindich L. Isolation and analysis of mutants of double-stranded-RNA bacteriophage phi6 with altered packaging specificity. J Bacteriol. 2003a;185:4572–4577. doi: 10.1128/JB.185.15.4572-4577.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao X, Qiao J, Mindich L. Stoichiometric packaging of the three genomic segments of double-stranded RNA bacteriophage phi6. Proc Natl Acad Sci U S A. 1997;94:4074–4079. doi: 10.1073/pnas.94.8.4074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao X, Qiao J, Mindich L. Analysis of specific binding involved in genomic packaging of the double-stranded-RNA bacteriophage phi6. J Bacteriol. 2003b;185:6409–6414. doi: 10.1128/JB.185.21.6409-6414.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinisch KM, Nibert ML, Harrison SC. Structure of the reovirus core at 3.6 A resolution. Nature. 2000;404:960–967. doi: 10.1038/35010041. [DOI] [PubMed] [Google Scholar]
- Sen A, Heymann JB, Cheng N, Qiao J, Mindich L, Steven AC. Initial location of the RNA-dependent RNA polymerase in the bacteriophage Phi6 procapsid determined by cryo-electron microscopy. J Biol Chem. 2008;283:12227–12231. doi: 10.1074/jbc.M710508200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Bamford DH, Poranen MM. Probing, by self-assembly, the number of potential binding sites for minor protein subunits in the procapsid of double-stranded RNA bacteriophage Phi6. J Virol. 2012;86:12208–12216. doi: 10.1128/JVI.01505-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trabuco LG, Villa E, Schreiner E, Harrison CB, Schulten K. Molecular dynamics flexible fitting: a practical guide to combine cryo-electron microscopy and X-ray crystallography. Methods. 2009;49:174–180. doi: 10.1016/j.ymeth.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikoff WR, Liljas L, Duda RL, Tsuruta H, Hendrix RW, Johnson JE. Topologically linked protein rings in the bacteriophage HK97 capsid. Science. 2000;289:2129–2133. doi: 10.1126/science.289.5487.2129. [DOI] [PubMed] [Google Scholar]
- Wu W, Chen Z, Cheng N, Watts NR, Stahl SJ, Farci P, Purcell RH, Wingfield PT, Steven AC. Specificity of an anti-capsid antibody associated with Hepatitis B Virus-related acute liver failure. J Struct Biol. 2013;181:53–60. doi: 10.1016/j.jsb.2012.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.