Abstract
The assembly of African swine fever virus (ASFV) at the cytoplasmic virus factories commences with the formation of precursor membranous structures, which are thought to be collapsed cisternal domains recruited from the surrounding endoplasmic reticulum (ER). This report analyzes the role in virus morphogenesis of the structural protein p54, a 25-kDa polypeptide encoded by the E183L gene that contains a putative transmembrane domain and localizes at the ER-derived envelope precursors. We show that protein p54 behaves in vitro and in infected cells as a type I membrane-anchored protein that forms disulfide-linked homodimers through its unique luminal cysteine. Moreover, p54 is targeted to the ER membranes when it is transiently expressed in transfected cells. Using a lethal conditional recombinant, vE183Li, we also demonstrate that the repression of p54 synthesis arrests virus morphogenesis at a very early stage, even prior to the formation of the precursor membranes. Under restrictive conditions, the virus factories appeared as discrete electron-lucent areas essentially free of viral structures. In contrast, outside the assembly sites, large amounts of aberrant zipper-like structures formed by the unprocessed core polyproteins pp220 and pp62 were produced in close association to ER cisternae. Altogether, these results indicate that the transmembrane structural protein p54 is critical for the recruitment and transformation of the ER membranes into the precursors of the viral envelope.
African swine fever virus (ASFV), the only known member of the Asfarviridae family, is a complex enveloped deoxyvirus that infects different species of suids as well as soft ticks of the genus Ornithodoros (22, 23, 48, 58). ASFV is unique among DNA viruses in that it resembles the poxviruses in genome structure and gene expression strategy but morphologically is similar to the iridoviruses (48, 58). The ASFV particles, with an overall icosahedral shape and an average diameter of 200 nm, are composed of several concentric domains: an internal core consisting of a central DNA-containing nucleoid coated by a thick protein layer referred to as core shell, an inner lipid envelope, and an icosahedral protein capsid (8, 9, 17). The extracellular virions usually contain an additional external membrane acquired by budding from the plasma membrane (12). Both intracellular and extracellular mature virions are infectious (10).
ASFV morphogenesis occurs in the cytoplasm at specialized perinuclear sites called virus factories. These assembly sites localize close to the microtubule organizing center and the Golgi complex and are typically surrounded by endoplasmic reticulum (ER) cisternae, recruited mitochondria, and a cage of vimentin filaments (9, 34, 37, 46). Current models propose that ASFV assembly begins with the modification of ER membranes, which are subsequently recruited to the viral factories and transformed into characteristic open curved membrane structures (9, 47). These ER-derived viral membranes represent the precursors of the inner viral envelope, which is thought to be a collapsed cisternal domain consisting of two juxtaposed lipid membranes that are usually indistinguishable in the mature particles (9, 47). During this poorly understood process of membrane acquisition, host cell proteins are generally excluded from the viral membranes as deduced from immunocytochemical evidence (9). Other large enveloped DNA viruses, such as herpesviruses and poxviruses, also become enwrapped by membrane cisternae during virion morphogenesis (29, 36, 53, 54, 56). In the case of ASFV, the envelope precursors develop into icosahedral structures by the progressive assembly of the outer capsid layer (8, 28) through an ATP- and calcium-dependent process (21). At the same time, the core shell is formed underneath the concave face of the viral envelope and the viral DNA and nucleoproteins are packaged and condensed to form the innermost electron-dense nucleoid (8, 13). Finally, the intracellular particles move from the assembly sites to the plasma membrane by a microtubule-dependent mechanism (5, 19) before being released by budding from the infected cell (12).
An open question regarding the formation of the envelope precursors is the nature of the molecular mechanisms underlying the recruitment and modification of the ER membrane cisternae, a process that may involve the targeting of viral proteins to the ER. The best candidates for this role are the structural membrane proteins of the intracellular particles. The ASFV genome encodes 26 putative transmembrane proteins (43, 61), 8 of which (pEP153R [27], pEP402R [44], p12 [16], p17 [52], p22 [15], p54 [j13L] [39], pE199L [j18L] [55], and pH108R [j5R] [14]) being found in purified virus particles. Protein p54, encoded by the late gene E183L in the BA71V strain, contains a potential membrane-spanning domain close to its N terminus (42). The term “p54” is not related to its molecular mass, which is about 25 kDa, but to its relative position in two-dimensional gels (1). Protein p54 has been localized in the envelope precursors as well as in both intracellular and extracellular mature particles (14, 42). Also, it has been shown to be essential for virus viability and to be involved in the early steps of virus infection (42). Thus, purified protein p54 and antibodies against it prevent infection by blocking the attachment of the virus particles to susceptible cells (30, 31). Recently, it has been shown that p54 binds to the LC8 subunit of cytoplasmic dynein, a minus-end-directed microtubule-associated motor protein, which may represent a mechanism for microtubule-mediated virus transport (4).
The present report analyzes the role of protein p54 in ASFV morphogenesis. Our results indicate that p54 is a type I transmembrane protein that forms disulfide-linked homodimers and is targeted to the ER membranes when expressed in transfected cells. We also describe the construction and properties of a conditional lethal ASFV mutant, vE183Li, with an inducible copy of gene E183L. The analysis of recombinant vE183Li indicates that p54 protein is essential for the recruitment and transformation of the ER-derived envelope precursors and, consequently, for the assembly of ASFV particles.
MATERIALS AND METHODS
Cells and viruses.
Vero and COS-7 cells were obtained from the American Type Culture Collection and grown in Dulbecco's modified Eagle's medium (DMEM) containing 5 or 10% fetal calf serum (FCS), respectively. The ASFV strain BA71V, adapted to grow in Vero cells, and vGUSREP, a BA71V-derived recombinant that expresses the Escherichia coli lac repressor, have been already described (25, 28). The recombinant vaccinia virus (VV) vTF7-3 expressing bacteriophage T7 RNA polymerase (26) was kindly provided by Bernard Moss.
Antibodies.
The following antibodies against ASFV structural proteins have been described previously: the pig anti-p54 serum (39), the mouse monoclonal antibody (MAb) 17L.D3 against protein p72 (49), and the rabbit polyclonal sera against proteins p150 and p35 (50, 51). The rat anti-p54 antibody was raised against the synthetic peptide CENLRQRNTYTHKDLENS (residues 166 to 182), which was coupled to keyhole limpet hemocyanin with an Imject-activated immunogen conjugation kit (Pierce, Rockford, Ill.). The MAb specific for protein disulfide isomerase (PDI) was purchased from Stressgen. The rabbit serum against a purified extract of membrane ER glycoproteins (anti-MERG) was a gift from D. I. Meyer (University of California, Los Angeles).
Plasmid construction.
The intermediate transfer vectors pIND1 and pIND2, designed to allow the inducible expression of a target gene after homologous recombination with virus vGUSREP, have been described previously (10). They contain a cassette formed by the viral inducible promoter p72.I, the lacZ gene under the control of the strong late promoter p72 (28), and two multiple-cloning sites to allow the cloning of the target gene and the corresponding upstream- and downstream-flanking sequences. The vectors pIND1 and pIND2 differ in the direction of transcription of the lacZ gene with respect to the inducible promoter.
(i) pIND1.E183L and pIND2.E183L.
A synthetic DNA fragment of 1,359 bp, which contains the nucleotide sequence from −1333 to + 4 relative to the translation initiation codon of the ASFV E183L gene, was obtained by PCR using the oligonucleotides 5′-aaagcggccgcCCATCATTTAAGTCCAAG and 5′-attcaggtaccATTTAAGATACTATT (these oligonucleotides include NotI and KpnI restriction sites [in lowercase type] at their respective 5′ ends). Plasmids pIND1.E183L.Fl and pIND2.E183L.Fl were generated by inserting this PCR fragment, cut with KpnI and NotI, into KpnI/NotI-digested pIND1 and pIND2, respectively. A complete copy of the E183L gene, including 137 nucleotides downstream, flanked by BamHI restriction sites was generated by PCR using the oligonucleotides 5′-GCGCGGATCCATGGATTCTGAATTTTTTCAACC and 5′-GCGCGCGGATCCGTAGCTAATAAGCTCTGAGC. The 712-bp PCR product was cloned into the BamHI site of plasmid pLitmus29 (NEB, Beverly, Mass.), generating the plasmid pL29.E183L. The complete inserted sequence was excised from pL29.E183L using XbaI and PstI restriction enzymes and cloned into XbaI/PstI-digested plasmids pIND1.E183L.Fl and pIND2.E183L.Fl to obtain the final transfer vectors pIND1.E183L and pIND2.E183L.
(ii) pcDNA.E183L.
To generate pcDNA.E183L the complete coding sequence of gene E183L was excised using BamHI from pL29.E183L and cloned into BamHI-digested pcDNA 3.1/mycHisA (Invitrogen).
Generation of recombinant virus vE183Li.
Vero cells were infected with virus vGUSREP and transfected with plasmids pIND1.E183L or pIND2.E183L in the presence of different concentrations of IPTG. At 48 h postinfection (hpi), the cells were harvested and the recombinant viruses were isolated by sequential rounds of plaque purification in the presence of IPTG. The inducer concentration at which virus production was maximal, 0.15 mM, was chosen for the growth and purification of the recombinant virus. Similar results were obtained with the two plasmids used and one virus clone from the pIND2.E183L-transfected cells was selected for further characterization. The genomic structure of this recombinant virus, named vE183Li, was confirmed by DNA hybridization analysis.
Plaque assays.
Preconfluent monolayers of Vero cells seeded in six-well plates were infected with 600 PFU of recombinant vE183Li or parental BA71V. After 1 h, the inoculum was removed and the cells were overlaid with DMEM containing 0.6% Noble agar and 2% FCS in the presence or absence of 0.15 mM IPTG (isopropyl-β-d-thiogalactopyranoside). Five days later, the medium was removed and the monolayers were stained with 1% crystal violet.
One-step virus growth curves.
Preconfluent monolayers of Vero cells seeded in 24-well plates were infected with 5 PFU of recombinant vE183Li or parental BA71V per cell. After a 1-h adsorption, the cells were incubated in DMEM supplemented with 2% FCS. IPTG (0.15 mM) was added immediately after the adsorption period or at 12, 18, or 24 hpi. Infected cells with their culture supernatants were harvested at different times postinfection, sonicated, and titrated by plaque assay in the presence of 0.15 mM IPTG.
Subcellular fractionation.
Subcellular fractionation was performed as previously described (6). Briefly, Vero cells were mock infected or infected with 10 PFU/cell of the BA71V strain. At 18 hpi, the cells were resuspended at 107 cells/ml in homogenization buffer (20 mM Tris-HCl [pH 7.5], 0.25 M sucrose, 1 mM EDTA), supplemented with protease inhibitors (Complete EDTA-free cocktail; Roche), and passed through a 25-gauge syringe 20 times. Cell breakage was monitored by phase-contrast microscopy. The homogenate was centrifuged at 750 × g for 5 min to sediment nuclei and unbroken cells, and the postnuclear supernatant was subsequently centrifuged at 100,000 × g for 15 min to separate the cytosolic and membrane/particulate fractions. Membrane/particulate pellets were gently rinsed in homogenization buffer prior to resuspension in the appropriate buffer. For analysis under nonreducing conditions, cells were incubated on ice for 10 min with phosphate-buffered saline (PBS) containing a 25 mM concentration of the sulfhydryl modifying agent N-ethylmaleimide (NEM; Sigma-Aldrich) prior to the fractionation. NEM (25 mM) was also included in the subsequent steps of the process.
Preparation of highly purified ASFV.
Highly purified virus was prepared as described previously (17). For the analysis under nonreducing conditions, the virus was pretreated with 25 mM NEM for 10 min on ice and dissociated in the presence of 25 mM NEM to avoid the formation of artifactual dimers.
In vitro transcription and translation.
In vitro transcription reactions containing 1 μg of linearized pcDNA.E183L template DNA were carried out as described in the mMessage mMachine T7 kit (Ambion). Translation of the mRNAs was performed for 45 min at 30°C using the Flexi Rabbit Reticulocyte Lysate System (Promega) in the presence or absence of canine pancreatic microsomal membranes (Promega).
For membrane sedimentation analysis, an aliquot of the translation mixture was diluted with physiological salt buffer (38), adjusted to 3 mM tetracaine, and incubated for 5 min on ice. Samples were layered on a 0.5 M sucrose cushion and centrifuged at 100,000 × g for 15 min at room temperature. Membrane sediments were gently rinsed in physiological salt buffer before dissociation. Analysis of equivalent amounts of supernatant and pellet fractions was performed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) followed by autoradiography. For the analysis of co- and posttranslational membrane association, microsomes were added for 45 min at 30°C during or after the translation reaction. In the latter case, translation was stopped with 1.25 mM puromycin prior to the addition of microsomal membranes.
Protease protection assays.
Protease protection assay was performed essentially as described by Ooi and Weiss (38) with minor modifications. Translation mixtures were diluted with 50 mM Tris-HCl (pH 7.5)-100 mM NaCl and incubated on ice for 5 min in the presence of 3 mM tetracaine. Proteinase K was added to a final concentration of 0.5 mg/ml in the absence or presence of 1% Triton X-100, and digestion was carried out at 20°C for 60 min. The reaction was terminated by the addition of 100 mM phenylmethylsulfonyl fluoride to a final concentration of 10 mM. Samples were immunoprecipitated using the pig anti-p54 serum, separated by SDS-PAGE on a 15-to-20% polyacrylamide gradient gel, and detected by autoradiography.
Triton X-114 and sodium carbonate extraction of membranes.
Membrane/particulate cell fractions or microsomal pellets from the in vitro translation reactions were treated with 2% Triton X-114 or sodium carbonate buffer (0.1 M sodium carbonate, pH 11.3) as previously described (6). Equivalent amounts of the resulting fractions were analyzed by Western immunoblotting.
Metabolic labeling and immunoprecipitation.
Preconfluent monolayers of Vero cells were mock infected or infected with vE183Li or BA71V at 5 PFU/cell. vE183Li-infected cells were maintained either in the presence or absence of 0.15 mM IPTG. At 12 hpi, cells were pulse labeled for 6 h with [35S]methionine-[35S]cysteine (Promix in vitro cell labeling mix [500 μCi/ml]; Amersham Pharmacia Biotech). At the end of the pulse period, the cells were washed with cold PBS and lysed at 4°C with immunoprecipitation buffer (0.01 M Tris-HCl [pH 7.5], 0.15 M NaCl, 1% sodium deoxycholate, 1% IGEPAL CA-630, 0.1% SDS supplemented with protease inhibitors [Complete EDTA-free cocktail; Roche]). Cell extracts were analyzed by SDS-PAGE and autoradiography or previously immunoprecipitated with anti-p54 pig antibodies immobilized on protein A-Sepharose (Sigma-Aldrich). Quantitation of protein bands was performed with a Bio-Rad GS710 densitometer and Quantity One software (Bio-Rad).
Indirect immunofluorescence.
For the immunofluorescence of transfected cells, COS-7 cells were transfected for 1 h at 37°C with 250 ng of DNA per 105 cells of the plasmids pcDNA 3.1 and pcDNA.E183L using Lipofectamine Plus Reagent (Life Technologies) and following the manufacturer's indications. The transfected cells were then infected with 5 PFU of the recombinant VV vTF7-3 expressing T7 RNA polymerase per cell. Infected cells were incubated in the presence of cytosine arabinoside (40 μg/ml), an inhibitor of VV DNA replication and late protein synthesis. At 7 hpi, the cells were rinsed twice with PBS and fixed with methanol at −20°C for 5 min. For the immunofluorescence of cells infected with recombinant vE183Li, preconfluent Vero cells grown on coverslips were infected at 1 PFU per cell in the presence or absence of 0.15 mM IPTG. At 12 hpi, vE183Li-infected cells were fixed with methanol at −20°C for 5 min.
Fixed cells were incubated at 37°C with DNase-free RNase (1 mg/ml)-1% bovine serum albumin in PBS. After 45 min, coverslips were washed twice with PBS and blocked for 30 min at 37°C with blocking buffer (1% cold fish skin gelatin, 0.1% Triton X-100, 1× PBS). Cells were then sequentially incubated for 1 h with primary and the corresponding secondary antibodies diluted with blocking buffer. Propidium iodide (10 μg/ml) or DAPI (4′,6-diamidino-2-phenylindole) was used along with the secondary antibodies to stain DNA in nuclei and virus factories. Finally, coverslips were mounted with Mowiol/Dabco on glass slides. Preparations were examined with a Bio-Rad Radiance 2000 confocal laser-scanning microscope. Images were processed using Adobe Photoshop software.
For the immunofluorescence of streptolysin O (SLO)-permeabilized cells, Vero cells grown in coverslips were infected with ASFV at a multiplicity of infection of 0.2 PFU per cell. At 12 h, the cells were incubated on ice for 30 min with SLO (100 U/ml; Sigma-Aldrich) in HEPES buffer (25 mM HEPES [pH 7.4], 115 mM potassium acetate, 2.5 mM MgCl2,1 mM dithiothreitol), washed twice with HEPES buffer, and then incubated for 30 min at 37°C in the same buffer. SLO-treated cells were incubated for 1 h with the rat antibody against the C-terminal region of p54 (1/250 dilution) and the MAb against PDI (1/250 dilution). The cells were fixed for 1 h with 1% paraformaldehyde in HEPES buffer and further fixed and permeabilized with methanol at −20°C for 5 min. Fixed cells were incubated twice for 10 min in PBS containing 50 mM NH4Cl, blocked for 30 min at 37°C, and then treated for 45 min with secondary antibodies. The cells were finally stained with DAPI and mounted with Mowiol/Dabco on glass slides. Two controls were carried out. (i) Cells were subjected to the same treatments, but SLO was omitted. (ii) Cells were subjected to the same treatments as SLO-treated cells, but primary antibodies were omitted; instead, the incubation with the primary and secondary antibodies was performed after the fixation and permeabilization of the cells. Fluorescence images were collected with a Coolsnap color camera (Roper Scientific) on a Zeiss Axioskop microscope. Images were processed using Adobe Photoshop software.
The primary antibodies were used at the following dilutions: swine serum anti-p54 (1/250 dilution), rat serum anti-p54 (1/200 dilution), mouse MAb anti-PDI (1/200 dilution), rabbit serum against pp200/p150 (1/500), rabbit serum anti-MERG (1/150 dilution). The secondary antibodies used were goat anti-swine immunoglobulin G (IgG) coupled to Texas red (1/300 dilution; Jackson Immunoresearch), donkey anti-mouse IgG coupled to Cy5 (1/200 dilution; Jackson Immunoresearch), goat anti-rat, goat anti-rabbit, and goat anti-mouse IgG antibodies coupled to Alexa 488 or Alexa 594 (1/500 dilution; Molecular Probes).
Electron microscopy.
For conventional Epon section analysis, Vero cells were infected with 10 PFU per cell and fixed at the indicated times with 2% glutaraldehyde in 200 mM HEPES (pH 7.4) for 1 h at room temperature. Postfixation was carried out with 1% OsO4 and 1.5% K3Fe(CN)6 in H2O at 4°C for 30 min. Samples were dehydrated with acetone and embedded in Epon according to standard procedures. For immunoelectron microscopy, the cells were fixed for 1 h with 4% formaldehyde and 0.1% glutaraldehyde in 200 mM HEPES (pH 7.4) on ice. Freeze substitution, cryosectioning, and immunogold labeling were performed as previously described (9, 11). Specimens were examined at 80 kV in a JEOL 1010 electron microscope.
RESULTS
Protein p54 is an integral membrane polypeptide that forms disulfide-linked dimers.
As mentioned in the introduction, p54-specific label appears mainly associated with membranous structures as well as with immature and mature particles within the virus factories (14, 42). Since one of the major structural features of protein p54 is the presence of a putative membrane-anchor domain close to its N terminus (amino acid residues 33 to 53), we decided to characterize the mechanism of membrane association of p54. For this, ASFV-infected cells were disrupted and fractionated at 18 hpi into a membrane/particulate and a cytosolic fraction. Equivalent amounts of both fractions, together with purified virus particles, were then analyzed by Western immunoblotting using a rat anti-p54 antibody raised against the C terminus of p54 (residues 166 to 182). As a control, a similar fractionation was carried out with mock-infected cells. As shown in Fig. 1A, a 25-kDa band corresponding to protein p54 was exclusively present in the membrane fraction of the infected cells and in the virus preparation.
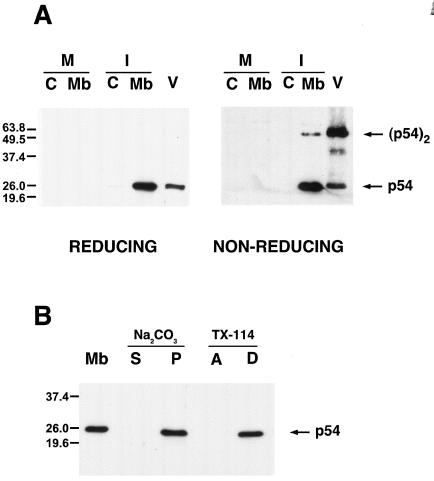
FIG. 1.
p54 is a disulfide-bonded dimeric integral membrane protein. (A) Mock-infected (M) or ASFV-infected (I) cells were disrupted at 18 hpi and fractionated into cytosolic (C) and membrane/particulate (Mb) fractions. Equivalent amounts of both fractions, together with purified ASFV particles (V), were resolved by reducing and nonreducing SDS-PAGE and analyzed by Western blotting using an anti-p54 antibody raised against its C terminus. The positions of the monomeric and dimeric forms of p54 are indicated. (B) Membrane fractions (Mb) from infected cells were treated with sodium carbonate. After centrifugation, the supernatant (S) and sediment (P) were analyzed by Western immunoblotting with anti-p54 antibody. Alternatively, the membrane fraction was extracted with Triton X-114 and subjected to temperature-induced phase separation. After centrifugation, the aqueous (A) and detergent-rich (D) phases were analyzed as described for panel A. Molecular masses are indicated (in kilodaltons).
Next, the possibility that p54 forms disulfide-bonded dimers through its unique cysteine residue at position 17 was studied. For this, cell fractions and purified virions were subjected to nonreducing SDS-PAGE and analyzed as above. As shown in Fig. 1A, the anti-p54 antibody recognized in the membrane fraction a major band of 25-kDa and, in a minor proportion, a protein of 50 kDa, the size expected for a disulfide-bonded p54 homodimer. At variance, the 50-kDa band was the major form of p54 found in highly purified virions, which suggests that covalent dimers are selectively recruited during virion assembly or that they are formed at a very late stage of virus morphogenesis.
We then analyzed whether p54 behaves as an integral membrane protein. Membrane fractions were incubated with 100 mM sodium carbonate, pH 11.3, a treatment that extracts peripherally associated, but not integral proteins from membranes. As shown in Fig. 1B, p54 remained associated to the membrane sediment after such treatment. Membrane fractions were also extracted with Triton X-114 and subjected to a temperature-induced phase separation. As expected for an integral membrane protein, p54 partitioned entirely into the detergent phase (Fig. 1B). Our results together with the previous immunolocalization data (14, 42) strongly support that p54 is a transmembrane protein of the inner viral envelope, the only membranous structure of the intracellular virus.
Protein p54 is a type I transmembrane protein that associates with microsomal membranes in a cotranslational manner.
To study in more detail the insertion of protein p54 into membranes, the protein was translated in vitro in the presence or in the absence of rough microsomal membranes. Membrane proteins were separated from soluble proteins by centrifugation, and subjected to SDS-PAGE. As shown in Fig. 2A, most of p54 was detected in the supernatant fraction in the absence of microsomes, whereas the protein was more abundant in the pellet fraction when microsomes were present. To test whether p54 protein is inserted into the membranes in a posttranslational manner, the translation was performed in the absence of microsomes, then puromycin was added to prevent further protein synthesis and finally the incubation was continued in the presence of microsomes. After centrifugation, most of protein p54 was present into the supernatant (Fig. 2A) suggesting that it associates with membranes in a cotranslational manner.
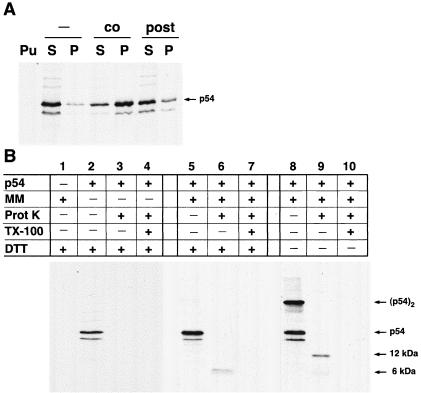
FIG. 2.
Membrane association and topology of in vitro-synthesized protein p54. (A) p54 was translated in vitro in the presence (co) or absence of microsomal membranes (−). Soluble (S) and membrane-associated (P) fractions were separated by centrifugation and analyzed by SDS-PAGE. Note that in the absence of microsomes, most of p54 is detected in the supernatant, whereas in the presence of microsomes, the protein is more abundant in the membrane sediment. When microsomes were added after the translation was blocked with puromycin (post), p54 did not associate significantly with membranes, suggesting that no posttranslational insertion occurs. As a control, no p54 was detected when translation was carried out in the presence of puromycin (Pu). The samples were resolved by SDS-PAGE under reducing conditions. (B) In vitro translations were carried out in the absence (lane 1) or presence (lanes 2 to 10) of mRNA specific for p54 and in the absence (lanes 2 to 4) or presence (lanes 1 and 5 to 10) of microsomal membranes (MM). After translation, reactions were left untreated (lanes 1, 2, 5, and 8) or were incubated with proteinase K (lanes 3, 6, and 9) or with proteinase K in the presence of Triton X-100 (lanes 4, 7, and 10). The samples were immunoprecipitated using the swine anti-p54 antibody against the whole protein and resolved by SDS-PAGE under reducing (lanes 1 to 7) or nonreducing (lanes 8 to 10) conditions. The size of the protected fragments and the positions of the monomeric and dimeric forms of p54 are indicated on the right. The presence of the protected fragments of 6 and 12 kDa is consistent with a type I topology for protein p54.
To analyze the topology of p54 protein, the protein was translated in vitro with or without membranes and then incubated with proteinase K. The signal-anchor sequence of p54 divides the protein in two unequal halves, a small N-terminal portion of about 6 kDa containing the unique cysteine residue and a large C-terminal fragment of about 19 kDa. As shown in Fig. 2B, in the absence of membranes no protected product was detected with a pig anti-p54 serum raised against the whole protein. In contrast, when microsomal membranes were added, a protected fragment of approximately 6 kDa was observed. Under nonreducing conditions, a protected fragment of approximately 12 kDa was detected in addition to the 6-kDa form. Finally, when the microsomal membranes were solubilized with Triton X-100 before proteinase digestion no protected fragments were detected. As can be noticed in the figure, these protected fragments show a weaker intensity than the undigested p54, which can be explained by the loss of two methionine residues due to the proteolytic digestion and by a lower efficiency of immunoprecipitation of the protected fragments compared with the complete molecule (not shown). These results indicate that p54 adopts a type I topology whereby the small N terminus portion of p54 is oriented to the relatively oxidizing luminal space, thus allowing its covalent dimerization through its unique cysteine 17 residue.
To confirm the topology of p54 in infected cells, an immunofluorescence assay on cells permeabilized with SLO was performed (24). The selective permeabilization of the plasma membrane was verified using antibodies against the cytoplasmic protein tubulin, which was readily detected (not shown), and against the ER luminal protein PDI, which was not accessible to the antibodies (Fig. 3). Under these conditions, antibodies directed against the C terminus of protein p54 labeled discrete structures in the perinuclear region of the cell coincidental with the DAPI-stained DNA at the viral factory (Fig. 3), thereby confirming the cytosolic orientation of the large C terminus of protein p54. To assess that the ER integrity was not affected by the SLO treatment, SLO-permeabilized cells were then fully permeabilized with cold methanol and incubated with the anti-PDI antibody. As shown in Fig. 3, the PDI label displays the characteristic pattern of the ER that excludes the viral factory (9).
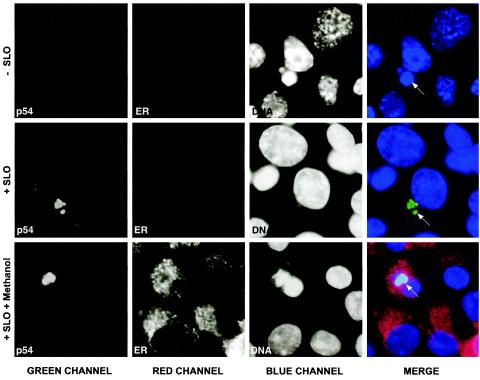
FIG. 3.
Topology of p54 in infected cells. ASFV-infected Vero cells were treated with SLO to selectively permeabilize the plasma membrane (+ SLO). The cells were then incubated with a MAb against the luminal ER marker PDI (ER) and the antipeptide antibody against the C terminus of p54 (p54) followed by secondary antibodies coupled to Alexa 488 and 594, respectively. Cells were counterstained with DAPI to visualize cellular and viral DNA. As a negative control (−SLO), the cells were treated as described above but SLO was omitted. As a positive control (+SLO + methanol), SLO-treated cells were fully permeabilized with cold methanol prior to the incubation with the antibodies. Arrows indicate the position of the viral factories. Notice that the C terminus of p54 is detected in SLO-treated cells, whereas PDI is only accessible after the full permeabilization of the cells.
p54 is targeted to ER membranes in transfected cells.
To test whether p54 can associate with ER membranes independently of other ASFV proteins, we analyzed its subcellular localization in transfected cells. COS cells were transfected with a plasmid containing the E183L gene under control of the T7 promoter and subsequently infected for 7 h with a recombinant VV expressing the T7 RNA polymerase. To avoid any interference with VV morphogenesis, transient expression of p54 was performed in the presence of cytosine arabinoside, an inhibitor of DNA replication and consequently of VV late gene expression. Transfected cells were analyzed by confocal immunofluorescence after double labeling with the pig anti-p54 antibody and a serum raised against a purified extract of membrane ER glycoproteins. As shown in Fig. 4, the p54 signal essentially colocalized with the ER-specific labeling. Similar results were obtained with a MAb against the luminal ER marker PDI (not shown). These results indicate that p54 is targeted to the ER, the cell compartment from which the viral envelope precursors are originated. However, the possibility that early VV proteins might alter the localization of p54 cannot be completely ruled out.
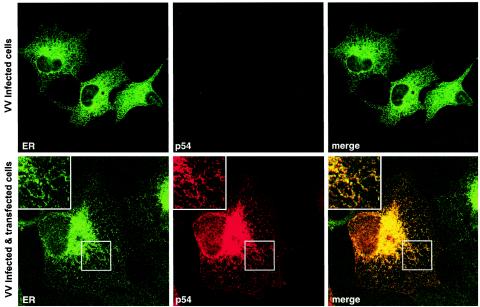
FIG. 4.
Protein p54 is targeted to the ER in transfected cells. Transfected COS cells expressing p54 were fixed and double labeled with a swine anti-p54 antibody (p54) and a rabbit antibody against ER glycoproteins (ER). Labeling was detected with a Texas red-linked anti-swine antibody (p54) and an Alexa 466-linked anti-rabbit antibody (ER). Confocal laser scanning images recorded in green and red channels are presented separately, or as a merged image. The inserts show a higher magnification of the areas delimited by squares. As a control of the antibody specificity, nontransfected COS cells, infected with VV, are also shown (upper panels).
Generation of ASFV recombinant vE183Li.
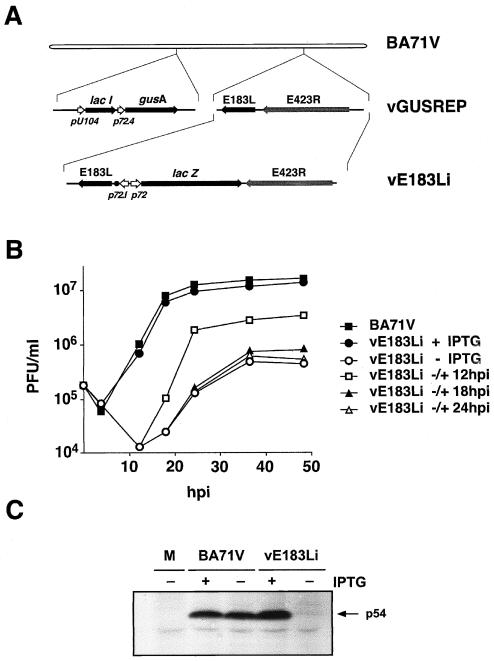
To further study the role of protein p54 in virus replication, we constructed the ASFV recombinant vE183Li in which the expression of gene E183L is controlled by the E. coli lac operator/repressor system (Fig. 5A). vE183Li virus was obtained from vGUSREP, a recombinant virus derived from the BA71V strain that constitutively expresses the E. coli lac repressor (28). To allow the inducible expression of protein p54, vGUSREP was modified by replacing the original promoter of gene E183L by an inducible promoter, p72.I, composed of the strong late viral promoter p72.4 and the operator sequence O1 (28). The genome structure of the resulting vE183Li virus was confirmed by DNA hybridization (data not shown).
FIG. 5.
(A) Genomic structure of the ASFV recombinant virus vE183Li. The recombinant virus vE183Li was obtained from vGUSREP, a BA71V-derived recombinant virus, which contains the lac repressor-encoding gene lacI inserted into the nonessential thymidine kinase locus. In vE183Li virus, the promoter of the p54-encoding gene E183L was replaced by an inducible promoter, p72.I, which is composed by a strong late promoter (p72.4) and the operator sequence O1 (•) from the E. coli lac operon. The reporter genes lacZ and gusA, used for selection and purification of the recombinants, are also represented. (B) One-step growth curves of vE183Li. Vero cells were infected with 5 PFU of vE183Li per cell in the presence or the absence of 0.15 mM IPTG. At the indicated times of infection, the total virus titer of each sample was determined by plaque assay on Vero cells in the presence of the inducer. Parental BA71V infections were also titrated as a control. Recombinant vE183Li was also grown under restrictive conditions for 12, 18, or 24 h and then induced with 0.15 mM IPTG. At different times after induction, the infectious virus was titrated as described above. (C) Inducible expression of protein p54. Vero cells were either mock infected (M) or infected with parental BA71V or recombinant vE183Li viruses in the presence (+) or absence (−) of IPTG. The cells were pulse labeled with [35S]methionine-[35S]cysteine from 12 to 18 hpi, lysed, and immunoprecipitated with a serum against protein p54 and analyzed by SDS-PAGE. The position of protein p54 is indicated.
Inducer dependence of recombinant vE183Li.
To test the inducer dependence of recombinant vE183Li virus, a plaque assay was performed with different concentrations of the inducer ranging from 0 to 1 mM IPTG. Lysis plaque number was maximal at 0.15 mM IPTG. Under these conditions, the plaque numbers and the plaque sizes were similar for both parental and recombinant viruses. Omission of IPTG caused a dramatic decrease in plaque formation by vE183Li virus when compared with control BA71V virus (not shown).
In a further approach, one-step growth curves of vE183Li virus were performed in the presence or in the absence of 0.15 mM IPTG. As shown in Fig. 5B, under permissive conditions the growth curve of recombinant vE183Li virus was similar to that obtained with parental BA71V virus. In contrast, under restrictive conditions, the vE183Li yields were reduced by 2.5 log units from 12 to 18 hpi and by 1.5 log units from 24 h onwards. In the same experiment, we also tested the ability of vE183Li grown for different times under nonpermissive conditions to produce infectious particles upon IPTG addition. As shown in Fig. 5B, the later the time of induction was, the lower the final virus yield was. When IPTG was added at 12 hpi the virus yield increased significantly, being about 80% of that observed for vE183Li grown under permissive conditions throughout the infection. When the inducer was added at 18 or 24 hpi, the maximal titers were only 10 and 6%, respectively, of those obtained in the control. Altogether, plaque assays and one-step growth curves indicate that recombinant vE183L is an IPTG-dependent lethal conditional mutant.
Inducible expression of protein p54.
Next, we verified that the expression of protein p54 can be regulated by the inducer during vE183Li infections. Cells infected with the recombinant under permissive or nonpermissive conditions were labeled with [35S]methionine-[35S]cysteine from 12 to 18 hpi, a period in which late gene expression is in progress. Mock- and BA71V-infected cells, labeled under the same conditions, were used as negative and positive controls, respectively. Similar overall protein profiles were observed in the infections with parental BA71V and recombinant vE183Li, with the exception that the major core protein p150, one of the processing products of polyprotein pp220, was not detected under nonpermissive conditions (not shown). Since protein p54 expression is not easily detectable in total extracts, its expression was analyzed by immunoprecipitation with an anti-p54 pig antibody. As shown in Fig. 5C, p54 synthesis was drastically inhibited under nonpermissive conditions. Densitometric quantification revealed that, under permissive conditions, protein p54 expression was about 110% of that observed in control BA71V infections whereas, under nonpermissive conditions, it was reduced to about 5% of the control. In summary, these results confirm that the conditional lethal phenotype of recombinant vE183Li is related to the inducer-dependent expression of protein p54.
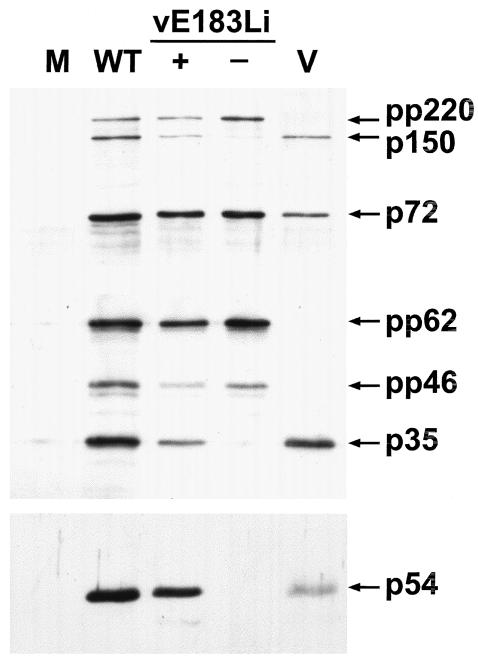
p54 shutoff impairs polyprotein processing.
The lack of detection of protein p150 under restrictive conditions led us to explore a possible general defect in the proteolytic processing of the ASFV polyproteins. Polyprotein processing was monitored by Western immunoblotting of extracts of vE183Li-infected cells maintained under permissive or restrictive conditions for 24 h using antibodies specific for polyprotein pp220 and its mature product p150 and polyprotein pp62 and its mature product p35 (Fig. 6). As a control, uninfected cells or cells infected with the parental BA71V were analyzed as above. Also, the expression of protein p54 and the major capsid protein p72 were analyzed for reference. Under permissive conditions, polyprotein processing occurred to a similar extent as in the control BA71V infections. On the contrary, under restrictive conditions, the proteolytic cleavage of both core precursors was severely impaired.
FIG. 6.
Polyprotein processing requires the expression of protein p54. Vero cells were mock infected (lane M) or infected with parental BA71V (lane WT) or recombinant vE183Li virus in the presence (lane +) or absence (lane −) of IPTG. At 24 hpi the cells were lysed and analyzed, together with purified ASFV particles (lane V), by Western immunoblotting with antibodies against the capsid protein p72, protein p54, polyprotein pp220 and its mature product p150, and polyprotein pp62 and its mature product p35. The positions of the detected proteins are indicated. Protein pp46 is an intermediate in the processing of pp62 polyprotein.
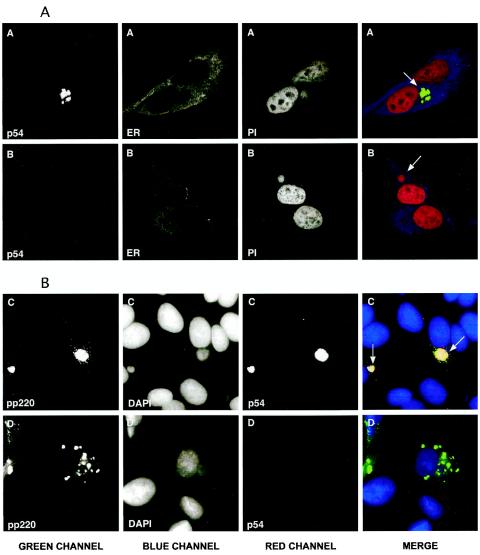
The repression of protein p54 results in an aberrant localization of ASFV polyproteins.
To study in more detail the mutant phenotype of recombinant vE183Li, we performed immunofluorescence analyses on vE183Li-infected cells maintained for 12 h with or without 0.15 mM IPTG. In a first approach, the cells were labeled with a rat anti-p54 antibody and a mouse MAb against the ER marker PDI. The virus factories were visualized by DNA staining with propidium iodide. As shown in Fig. 7A, in the presence of the inducer the p54 signal was essentially confined to the perinuclear assembly sites whereas the PDI labeling was mostly excluded from these areas, staining the surrounding cytoplasm. Similar labeling patterns have been described for p54 and PDI in virus infections performed with parental BA71V virus (9, 14, 39). Under restrictive conditions (Fig. 7B), the fluorescence signal of p54 was almost undetectable, while PDI and DNA labeling were essentially the same as those under permissive conditions.
FIG. 7.
Immunofluorescence microscopy analysis of vE183Li-infected Vero cells. Vero cells were fixed 12 h after infection with vE183Li in the presence (series A and C) or absence (series B and D) of 0.15 mM IPTG. Samples were incubated with a MAb against PDI (ER), the antipeptide antibody against p54 (p54), and a rabbit antibody specific for polyprotein pp220 and its major product p150 (pp220). Antigens were visualized by secondary antibodies coupled to Alexa 488, 594, or Cy5. Cells were counterstained with propidium iodide (PI) or DAPI to visualize cellular and viral DNA. Samples were examined with a Bio-Rad Radiance 2000 confocal laser-scanning microscope (panel A) or a Coolsnap color camera (Roper Scientific) on a Zeiss Axioskop microscope (panel B). Arrows indicate the position of the viral factories.
Polyprotein processing in ASFV is an essential maturational process related with the assembly of the viral core (3, 7, 33). Since the proteolytic processing of the core precursors requires the expression of p54, we investigated their localization in vE183Li-infected cells. Figure 7C shows a immunofluorescence of vE183Li-infected cells maintained under permissive conditions and labeled with rat anti-p54 and rabbit anti-pp220 antibodies. As expected, p54 and pp220 polyprotein colocalized with the viral DNA at the viral factory. In addition, the anti-pp220 antibody labeled virus particles scattered throughout the cytoplasm. In contrast, under restrictive conditions the labeling pattern of pp220 was severely altered (Fig. 7D). The pp220 signal was found predominantly in discrete structures around the virus factories or scattered in the cytoplasm (Fig. 7D). Similar results were obtained with an antibody against polyprotein pp62 (not shown). These findings indicate that p54 expression is essential for the proper localization of the ASFV polyproteins in the virus assembly sites.
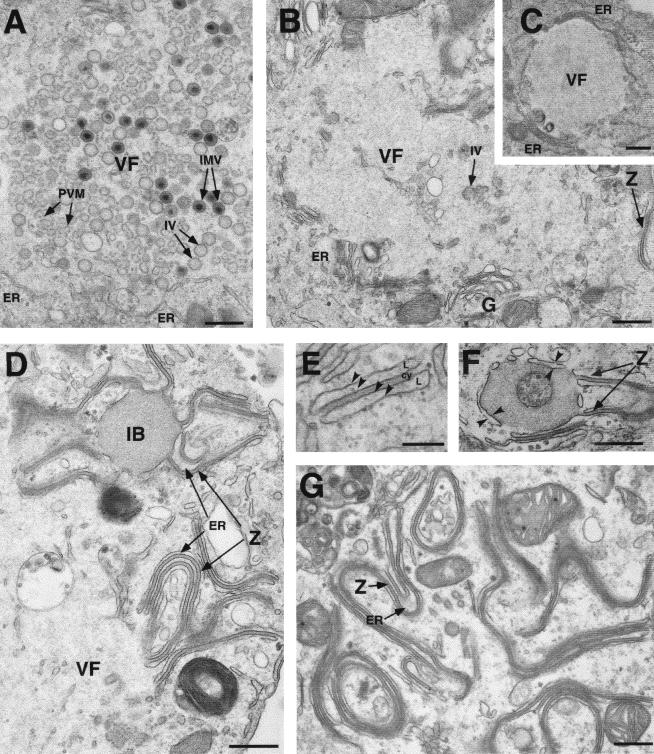
Electron microscopy of vE183Li-infected cells.
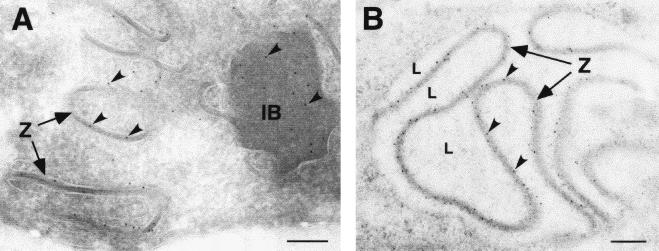
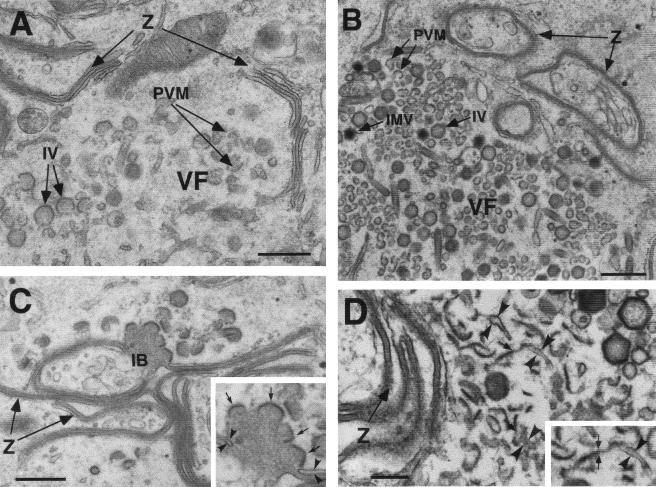
To investigate the stage at which the virus replication was blocked in the absence of p54 expression, cells infected with vE183Li in the presence or absence of IPTG were analyzed by transmission electron microscopy. As shown in Fig. 8A, after 12 h under permissive conditions, the cytoplasmic virus factories contained the expected repertoire of viral structures, namely, envelope precursors and immature and mature icosahedral particles. In contrast, when IPTG was omitted, viral factories were essentially devoid of membrane precursors and other developmental forms of ASFV (Fig. 8B and C). Thus, the assembly sites appeared as discrete electron-lucent areas close to the Golgi complex and surrounded by ER membranes, mitochondria, and other membrane organelles (Fig. 8B and C). Strikingly, outside the virus factories, large amounts of aberrant zipper-like structures were detected in tight association with ER cisternae (Fig. 8D and E). As previously described, the zipper-like structures, which appear occasionally in normal infections, are elongated laminar domains reminiscent of the viral core shell but flanked by adjacent ER cisternae (9). Interestingly, they are composed of the unprocessed core precursors pp220 and pp62 (7). Together with the abundant zipper-like structures, the absence of p54 expression also produced the accumulation of large and electron-dense masses of granular appearance. These inclusion bodies appeared usually at the periphery of the virus factories, being frequently enwrapped by membrane cisternae and connected to zipper-like structures (Fig. 8D and F). When analyzed at 24 hpi, the proportion of the zipper-like structures increased considerably, covering large extensions of the cytoplasm (Fig. 8G).
FIG. 8.
Electron microscopy of vE183Li-infected cells. Cells infected with vE183Li virus in the presence (A) or in the absence (B to G) of IPTG were fixed at 12 hpi (A to F) and 24 hpi (G) and processed for electron microscopy. Under permissive conditions (A), the viral factories (VF) contain large amounts of precursor viral membranes (PVM), immature virions (IV), and intracellular mature virions (IMV). Under restrictive conditions (B and C), defective factories appear as electron-lucent areas close to the Golgi complex (G) and enclosed by ER membranes (ER). Whereas minor signs of virus assembly (IV in B) are detected within the factories, large amounts of aberrant zipper-like structures (Z) are bound to ER cisternae at the periphery of (D and F) and beyond (G) the assembly sites. As shown in panel E, the zipper-like structures are cytosolic (cy) laminar arrangements flanked by cisternal (L) membrane profiles (arrowheads). Also, large dense inclusion bodies (IB) of granular appearance appear usually near the assembly sites (D and F). Note that these granular masses of core-like material are enwrapped by ER cisternae (arrowheads in F) and frequently connected to zipper-like structures (Z in D and F). Bars, 500 nm (A to D, F, and G) and 200 nm (E).
To analyze the presence of the polyprotein precursors at the abnormal structures produced when p54 synthesis is inhibited, immunoelectron microscopy was performed on cells infected under nonpermissive conditions with vE183Li for 12 h. As expected, antibodies to both polyproteins strongly labeled the cytoplasmic ER-associated zipper-like structures (Fig. 9A and B). Interestingly, the granular inclusion bodies were also found to contain the pp220 (Fig. 9A) and pp62 (not shown) precursors.
FIG. 9.
Polyproteins pp220 and pp62 localize at the aberrant zipper-like structures and granular bodies. Vero cells were infected with recombinant vE183Li in the absence of IPTG. At 12 hpi, the cells were fixed and processed for cryosectioning (A) or freeze-substitution and Lowicryl-embedding (B). Ultrathin sections were incubated with anti-pp220 (A) or anti-pp62 (B) antibodies followed by protein A-gold (10-nm diameter). Arrowheads indicate labeling on the zipper-like structures (Z) and on granular inclusion bodies (IB). In some cases, luminal spaces (L) flanking zipper-like structures are indicated. Bars, 100 nm.
In summary, electron microscopy of vE183Li-infected cells indicates that p54 repression prevents the appearance of the envelope precursor membranes at the virus factories while promoting the aberrant assembly of the core precursors pp220 and pp62 outside the assembly sites. This strongly supports that p54 is essential for the recruitment of the ER membranes, which are then targets for the core precursors and the capsid proteins.
Effect of p54 postinduction on virus assembly.
Next, we analyzed the reversibility of the mutant vE183Li phenotype after IPTG addition to infected cells previously maintained under restrictive conditions. Figure 10 shows electron micrographs of vE183Li-infected cells maintained during 12 h in the absence of inducer and then incubated with IPTG for additional periods of 4 and 12 h. After a 4-h induction (Fig. 10A), evident signals of normal virus assembly were observed within the viral factories. Although intracellular mature particles (IMV) were barely found, significant amounts of envelope precursors as well as icosahedral immature particles were detected. When IPTG was added for 12 h (Fig. 10B to D), larger amounts of precursor membranes as well as immature and mature icosahedral virions were observed. Outside the factories, the previously formed zipper-like structures did not evolve apparently to normal assembling intermediates (Fig. 10A, B, and D). In contrast, numerous icosahedral assembling capsids were found at the border of the granular bodies (Fig. 10C), suggesting that these dense masses could represent an aberrant accumulation of core material. Whether these developing structures could give rise to normal virus particles is unknown at present. The presence of polyhedral coats, on the other hand, suggests a collapse of the membrane cisternae that usually encompassed the core-like material or, alternatively, the attachment of previously formed envelope precursors. Regarding this, a close inspection of the virus factories revealed evident continuities between cisternal profiles and developing precursor virus membranes at the assembly sites (Fig. 10D). Altogether, these observations indicate that the significant restoration of the infectivity observed after the addition of IPTG at 12 hpi (Fig. 5B) is a consequence of the generation of new envelope precursors from membrane cisternae.
FIG. 10.
Effect of p54 induction on ASFV assembly. Electron microscopy of vE183Li-infected cells maintained for 12 h in the absence of IPTG and then incubated for 4 h (A) or 12 h (B to D) in the presence of inducer. The samples were processed by conventional Epon embedding. (A) After a 4-h induction, the presence of precursor viral membranes (PVM) as well as immature particles (IV) is already evident at the virus factories (VF). (B) After a 12-h induction, larger amounts of precursor membranes (PVM) as well as icosahedral immature (IV) and intracellular mature virions (IMV) are detected at the assembly sites. (C) Following p54 induction, the zipper-like structures (Z) do not apparently evolve to normal virus intermediates whereas icosahedral capsids assemble on the border of the granular inclusion bodies (IB). Note the polyhedral coats (small arrows, insert in C) enwrapping the underlying core material. Note also the cisternal profiles (arrowheads, insert in C) that enclose the zipper-like structures. (D) When examined in detail, obvious continuities between cisternal profiles (arrowheads) and envelope precursors (arrows) can be detected at the viral factories. A detail is shown in the insert. Bars, 500 nm (A to C) and 200 nm (D).
DISCUSSION
The first morphological stage of ASFV morphogenesis is the formation of open curved membranous structures within the assembly sites. Viral membranes develop subsequently into the inner viral envelope, the lipoprotein cisternal domain that encloses the internal core structure and supports the outer capsid layer (8, 9, 11, 28). Based on biochemical, morphological and immunocytochemical approaches, we and others have previously proposed that the envelope precursors are derived from collapsed ER cisternae recruited by the virus factory (9, 20, 47). Our present data further support this model and indicate an essential role of the structural protein p54 in the generation of the envelope precursors.
Biochemical and immunocytochemical approaches showed that protein p54 is a transmembrane protein that is cotranslationally inserted into microsomal membranes and is targeted to the ER membranes in transfected cells. These findings are consistent with the abundant presence of p54 at the ER-derived membrane precursors (14, 42) and sustain its localization at the inner viral envelope of the mature virions. Protein p54 has been proposed both as an attachment protein and a neutralizing target protein (30, 31). Such a role could be explained by the fact that the surrounding capsid is arranged in a hexagonal lattice that leaves an intercapsomer space. Also, the capsomers have a central hole (18) that would allow protein p54 to be exposed at the surface of the intracellular particles, which have been found to be infectious (10).
The targeting of p54 to the ER membranes is also indirectly supported by an early report showing that, when expressed by a VV recombinant, the protein blocks the virus assembly prior to the formation of the IMV (35). By using immunoelectron microscopy, the protein p54 was found to be present in the aberrant VV membranes and partially assembled virions produced under these conditions. Because the VV IMV membranes are also originated from the ER (53, 54), it seems probable that p54 interferes with the normal trafficking of VV proteins or prevents protein-protein interactions needed for IMV assembly. It should be noticed, however, that protein p54 does not obviously colocalize with the bona fide ER that surrounds the ASFV factories. The same behavior has been reported for the membrane-anchored virus protein trans-prenyltransferase, which also localizes at the membrane precursors within the virus factories of the ASFV-infected cells and at the ER when it is independently expressed (2). A possible explanation is that nascent p54 polypeptides (as well as other ASFV membrane proteins) are cotranslationally inserted into the rough ER membranes contiguous to the assembly sites. Subsequently, these virus-modified cisternal membranes would be rapidly recruited (see below) and converted into the envelope precursors inside the assembly sites, a process that seems to exclude cellular ER proteins (9). A transient colocalization of the virus proteins and the ER markers could be undetectable by immunofluorescence microscopy.
Our results support the argument that p54 adopts in vitro and in vivo a type I topology, with the small N terminus of the protein inside the luminal space. This topology agrees with the topogenic signals present in the sequence of protein p54 (the positive-inside rule [59]) and determines that the large C terminus of the protein, which contains the recently described binding site to the light-chain subunit (LC8) of the cytoplasmic dynein (4), will face the cytosol, thus allowing this interaction to occur in vivo. It is also interesting to notice that, as put forward by Sodeik and Krijnse-Locker (53) for the IMV membrane proteins of VV, the limited space resulting from the tightly apposed membranes forming the inner viral envelope appears to favor topologies in which the smaller part of the protein resides inside the luminal space.
Dimerization of p54 must occur through the unique cysteine present in the N terminus of the protein that resides in the relatively oxidant ER lumen. Approximately one-half of the protein appears as a dimer when the protein is translated in vitro in the presence of microsomes, and a significant proportion of p54 dimers is also detected when the protein is transiently expressed in transfected COS cells (our unpublished results). Surprisingly, the majority of p54 is found as a monomer in infected cells, while it is mainly present as a covalent dimer in purified virions. These data suggest that, in infected cells, the covalent dimerization of p54 might be a relatively inefficient process, which would be required, however, for the later formation of mature virions at the assembly sites. The selective recruitment of the p54 dimers by the developing particles might account for these findings.
The role of protein p54 in virus morphogenesis was investigated using the lethal conditional recombinant vE183Li. The repression of p54 expression arrested virus assembly before the formation of the envelope precursors. The viral factories appeared usually devoid of virus particles and membrane elements, whereas evident signals of abnormal assembly were detected at the periphery and outside these areas. Thus, large amounts of aberrant zipper-like structures were found in close association with ER cisternae. To a lesser extent, dense granular masses of core material frequently enclosed by cisternal membrane profiles were also detected. We have recently shown that the zipper-like structures are membrane-bound laminar arrangements composed by the unprocessed core polyproteins pp220 and pp62 (7, 9). Also, the membrane attachment has been shown to be dependent on the N-myristoylation of polyprotein pp220 (11). It is noteworthy that the zipper-like structures, which are unusual in normal infections (9), are abundantly produced when the expression of the major capsid protein p72 is blocked by means of the inducible recombinant vA72 (28). In the absence of p72 expression, the proteolytic processing of both polyproteins is drastically reduced, as also occurs in nonpermissive vE183Li infections (7). Nevertheless, two striking differences distinguish the mutant phenotypes of both ASFV recombinants. First, whereas the zipper-like structures formed in vE183Li-infected cells appear outside the viral factories, those formed in the absence of capsid assembly accumulate massively within the assembly sites (28). Second, the vE183Li zippers associate with genuine ER cisternae while most of the vA72 zippers are bound to the derived viral envelope precursors, which do not exhibit obvious luminal spaces. Collectively, these data strongly suggest that the absence of protein p54 interrupts the trafficking of the ER cisternae to the interior of the assembly sites, thus hampering their conversion into envelope precursors and the subsequent formation of virus particles.
An intriguing question is how protein p54 could be essential for the membrane recruitment if, as mentioned above, it resides at the assembly sites rather than at the surrounding ER membranes. A possible explanation is that, soon after its insertion at the adjacent ER membranes, protein p54 itself promotes the rapid transport of the virus-targeted cisternae to the virus factories. In relation to this, a suggestive hypothesis is that the membrane recruitment involves the minus-end-directed microtubular transport by means of the recently described binding of p54 to dynein light chain LC8. Since both proteins have also been shown to colocalize in transfected cells as well as in ASFV-infected cells at late times of infection (4), it seems conceivable that ASFV uses the centripetal microtubular transport to recruit the virus-targeted membranes to the perinuclear virus factories. Regarding the interaction of the polyproteins pp220 and pp62 with the ER membranes observed during nonpermissive vE183Li infections, the available data point out that p54 is not involved in the membrane targeting of the core precursors. Rather, this could be a consequence of their specific interaction with other ER-targeted virus proteins that normally reside at the derived envelope precursors of the virus factories. The interruption of the membrane trafficking would cause such interactions to occur outside the assembly areas. In support of this view is the finding that the transient coexpression of the pp220 and pp62 genes produces zipper-like structures that associate mainly with the plasma membrane and with endosomal/lysosomal membranes instead of ER membranes (7). Finally, as a result of these deleterious effects on virus assembly, the polyprotein processing, which constitutes a late maturational event related to the developing of the full icosahedral virions (3, 7), would be also impaired.
Together with the zipper elements, the formation of inclusion bodies represents a second sign of abnormal assembly that also involves the core polyproteins pp220 and pp62. However, the different organization of these masses of core-like material suggests that, at variance with the zippers, they could also contain other core proteins present in the assembly sites. It is noteworthy that similar structures, either with or without enclosing membranes, are produced when VV morphogenesis is arrested prior to the formation of the characteristic crescent membranes. This is the case of a number of VV conditional lethal mutants affected in the targeting and recruitment of the ER membranes to the factories, the attachment of the core material to the viral membranes, or the formation of the crescent precursors (32, 40, 41, 45, 57, 60, 62).
An important aspect of the vE183Li recombinant is its partially reversible phenotype. A significant restoration of the infectious-virus yield was achieved when p54 expression was induced at 12 hpi, but not at 18 or 24 hpi. We interpret this result as a consequence of the progressive hijacking of the ER membranes by the zipper-like structures, which would decrease the availability of “free” cisternal membranes to be recruited to the virus factories. The possibility to arrest and then reestablish the formation of the envelope precursors at early times makes recombinant vE183Li a powerful tool to further analyze key morphogenetic events such as membrane recruitment, the trafficking of the virus membrane proteins, and the interactions of virus membrane proteins with the core and capsid components.
Acknowledgments
We thank J. Salas for helpful discussions and critical reading of the manuscript. We also thank M. Guerra and M. Rejas for technical assistance.
This study was supported by grants from the Ministerio de Ciencia y Tecnología (BMC 2000-1485 and AGL2002-10220-E) and the European Community (QLRT-2000-02216) and by an institutional grant from Fundación Ramón Areces.
REFERENCES
- 1.Alcaraz, C., A. Brun, F. Ruiz-Gonzalvo, and J. M. Escribano. 1992. Cell culture propagation modifies the African swine fever virus replication phenotype in macrophages and generates viral subpopulations differing in protein p54. Virus Res. 23:173-182. [DOI] [PubMed] [Google Scholar]
- 2.Alejo, A., G. Andrés, and M. L. Salas. 1999. The African swine fever virus prenyltransferase is an integral membrane trans-geranylgeranyl-diphosphate synthase. J. Biol. Chem. 276:18033-18039. [DOI] [PubMed] [Google Scholar]
- 3.Alejo, A., G. Andrés, and M. L. Salas. 2003. African swine fever virus proteinase is essential for core maturation and infectivity. J. Virol. 77:5571-5577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alonso, C., J. Miskin, B. Hernáez, P. Fernández-Zapatero, L. Soto, C. Cantó, I. Rodríguez-Crespo, L. Dixon, and J. M. Escribano. 2001. African swine fever virus protein p54 interacts with the microtubular motor complex through direct binding to light-chain dynein. J. Virol. 75:9819-9827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alves de Matos, A. P., and Z. G. Carvalho. 1993. African swine fever virus interaction with microtubules. Biol. Cell. 78:229-234. [DOI] [PubMed] [Google Scholar]
- 6.Andrés, G., A. Alejo, C. Simón-Mateo, and M. L. Salas. 2001. African swine fever virus protease: a new viral member of the SUMO-1-specific protease family. J. Biol. Chem. 276:780-787. [DOI] [PubMed] [Google Scholar]
- 7.Andrés, G., A. Alejo, J. Salas, and M. L. Salas. 2002. African swine fever virus polyproteins pp220 and pp62 assemble into the core shell. J. Virol. 76:12473-12482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andrés, G., C. Simón-Mateo, and E. Viñuela. 1997. Assembly of African swine fever virus: role of polyprotein pp220. J. Virol. 71:2331-2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Andrés, G., R. García-Escudero, C. Simón-Mateo, and E. Viñuela. 1998. African swine fever virus is enveloped by a two-membraned collapsed cisterna derived from the endoplasmic reticulum. J. Virol. 72:8988-9001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Andrés, G., R. García-Escudero, E. Viñuela, M. L. Salas, and J. M. Rodríguez. 2001. African swine fever virus structural protein pE120R is essential for virus transport from the assembly sites to the plasma membrane but not for infectivity. J. Virol. 75:6758-6768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Andrés, G., R. García-Escudero, M. L. Salas, and J. M. Rodríguez. 2002. Repression of African swine fever virus polyprotein pp220-encoding gene leads to the assembly of icosahedral core-less particles. J. Virol. 76:2654-2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Breese, S. S. Jr., and C. J. DeBoer. 1966. Electron microscope observation of African swine fever virus in tissue culture cells. Virology 28:420-428. [DOI] [PubMed] [Google Scholar]
- 13.Brookes, S. M., A. D. Hyatt, T. Wise, and R. M. E. Parkhouse. 1998. Intracellular virus DNA distribution and the acquisition of the nucleoprotein core during African swine fever virus particle assembly: ultrastructural in situ hybridisation and DNase-gold labelling. Virology 249:175-188. [DOI] [PubMed] [Google Scholar]
- 14.Brookes, S. M., H. Sun, L. K. Dixon, and R. M. E. Parkhouse. 1998. Characterization of African swine fever virion proteins j5R and j13L: immuno-localization in virus particles and assembly sites. J. Gen. Virol. 79:1179-1188. [DOI] [PubMed] [Google Scholar]
- 15.Camacho, A., and E. Viñuela. 1991. Protein p22 of African swine fever virus: an early structural protein that is incorporated into the membrane of infected cells. Virology 181:251-257. [DOI] [PubMed] [Google Scholar]
- 16.Carrascosa, A. L., I. Sastre, and E. Viñuela. 1991. African swine fever virus attachment protein. J. Virol. 65:2283-2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carrascosa, A. L., M. del Val, J. F. Santarén, and E. Viñuela. 1985. Purification and properties of African swine fever virus. J. Virol. 54:337-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carrascosa, J. L., J. M. Carazo, A. L. Carrascosa, N. García, A. Santisteban, and E. Viñuela. 1984. General morphology and capsid fine structure of African swine fever virus. Virology 132:160-172. [DOI] [PubMed] [Google Scholar]
- 19.Carvalho, Z. G., A. P. Alves de Matos, and C. Rodrigues-Pousada. 1988. Association of African swine fever virus with the cytoskeleton. Virus Res. 11:175-192. [DOI] [PubMed] [Google Scholar]
- 20.Cobbold, C., J. T. Whittle, and T. Wileman. 1996. Involvement of the endoplasmic reticulum in the assembly and envelopment of African swine fever virus. J. Virol. 70:8382-8389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cobbold, C., S. M. Brookes, and T. Wileman. 2000. Biochemical requirements of virus wrapping by the endoplasmic reticulum: involvement of ATP and endoplasmic reticulum calcium store during envelopment of African swine fever virus. J. Virol. 74:2151-2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Costa, J. V. 1990. African swine fever virus, p. 247-270. In G. Darai (ed.), Molecular biology of iridoviruses. Kluwer Academic Publishers, Boston, Mass.
- 23.Dixon, L. K., J. V. Costa, J. M. Escribano, D. L. Rock, E. Viñuela, and P. J. Wilkinson. 2000. The Asfarviridae, p. 159-165. In M. H. V. Van Regenmortel, C. M. Fauquet, D. H. L. Bishop, E. B. Carsten, M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and R. B. Wickner (ed.), Virus taxonomy. Seventh report of the International Committee for the Taxonomy of Viruses. Academic Press, New York, N.Y.
- 24.Dupree, P., R. G. Parton, G. Raposo, T. V. Kurzchalia, and K. Simons. 1993. Caveolae and sorting in the trans-Golgi network of epithelial cells. EMBO J. 12:1597-1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Enjuanes, L., A. L. Carrascosa, M. A. Moreno, and E. Viñuela. 1976. Titration of African swine fever virus. J. Gen. Virol. 32:471-477. [DOI] [PubMed] [Google Scholar]
- 26.Fuerst, T. R., E. G. Niles, F. W. Studier, and B. Moss. 1986. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 83:8122-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Galindo, I., F. Almazán, M. J. Bustos, E. Viñuela, and A. L. Carrascosa. 2000. African swine fever virus EP153R open reading frame encodes a glycoprotein involved in the hemadsorption of infected cells. Virology 266:340-351. [DOI] [PubMed] [Google Scholar]
- 28.García-Escudero, R., G. Andrés, F. Almazán, and E. Viñuela. 1998. Inducible gene expression from African swine fever virus recombinants: analysis of the major capsid protein p72. J. Virol. 72:3185-3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gershon, A. A., D. L. Sherman, Z. L. Zhu, C. A. Gabel, R. T. Ambron, and M. D. Gershon. 1994. Intracellular transport of newly synthesized varicella zoster virus: final envelopment in the trans-Golgi network. J. Virol. 68:6372-6390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gómez-Puertas, P., F. Rodríguez, J. M. Oviedo, A. Brun, C. Alonso, and J. M. Escribano. 1998. The African swine fever virus proteins p54 and p30 are involved in two distinct steps of virus attachment and both contribute to the antibody-mediated protective immune response. Virology 243:461-471. [DOI] [PubMed] [Google Scholar]
- 31.Gómez-Puertas, P., F. Rodríguez, J. M. Oviedo, F. Ramiro-Ibáñez, F. Ruiz-Gonzalvo, C. Alonso, and J. M. Escribano. 1996. Neutralizing antibodies to African swine fever virus inhibit through different proteins both virus attachment and internalization. Influence of passage history of the virus. J. Virol. 70:5689-5694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grimley, P. M., E. N. Rosemblum, S. J. Mims, and B. Moss. 1970. Interruption by rifampicin of an early stage in vaccinia virus morphogenesis: accumulation of membranes which are precursors of virus envelopes. J. Virol. 6:519-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heath, C. M., M. Windsor, and T. Wileman. 2003. Membrane association facilitates the correct processing of pp220 during production of the major matrix proteins of African swine fever virus. J. Virol. 77:1682-1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heath, C. M., M. Windsor, and T. Wileman. 2001. Aggresomes resemble sites specialized for virus assembly. J. Cell Biol. 153:449-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jacobs, S. C., L. K. Dixon, S. M. Brookes, and G. L. Smith. 1998. Expression of African swine fever virus envelope protein j13L inhibits vaccinia virus morphogenesis. J. Gen. Virol. 79:1169-1178. [DOI] [PubMed] [Google Scholar]
- 36.Mettenleiter, T. C. 2002. Herpesvirus assembly and egress. J. Virol. 76:1537-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moura Nunes, J. F., J. D. Vigario, and A. M. Terrinha. 1975. Ultrastructural study of African swine fever virus replication in cultures of swine bone marrow cells. Arch. Virol. 49:59-66. [DOI] [PubMed] [Google Scholar]
- 38.Ooi, C. E., and J. Weiss. 1992. Bidirectional movement of a nascent polypeptide across microsomal membranes reveals requirements for vectorial translocation of proteins. Cell 71:87-96. [DOI] [PubMed] [Google Scholar]
- 39.Rodríguez, F., C. Alcaraz, A. Eiras, R. J. Yáñez, J. M. Rodríguez, C. Alonso, J. F. Rodríguez, and J. M. Escribano. 1994. Characterization and molecular basis of heterogeneity of the African swine fever virus envelope protein p54. J. Virol. 68:7244-7252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rodríguez, D., C. Risco, J. R. Rodríguez, J. L. Carrascosa, and M. Esteban. 1996. Inducible expression of the vaccinia virus A17L gene provides a synchronized system to monitor sorting of viral proteins during morphogenesis. J. Virol. 70:7641-7653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rodríguez, D., M. Esteban, and J. R. Rodríguez. 1995. Vaccinia virus A17L gene product is essential for an early step in virion morphogenesis. J. Virol. 69:4640-4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rodríguez, F., V. Ley, P. Gómez-Puertas, R. García, J. F. Rodríguez, and J. M. Escribano. 1996. The structural protein p54 is essential for African swine fever virus viability. Virus Res. 40:161-167. [DOI] [PubMed] [Google Scholar]
- 43.Rodríguez, J. M., R. J. Yáñez, F. Almazán, R. García-Escudero, E. Viñuela, and J. F. Rodríguez. 1995. African swine fever virus membrane-associated proteins, p. 187-200. In G. McFadden (ed.), Viroreceptors, virokines and related inmunomodulators encoded by DNA virus. R. G. Landes Company, Austin, Tex.
- 44.Rodríguez, J. M., R. J. Yáñez, F. Almazán, E. Viñuela, and J. F. Rodríguez. 1993. African swine fever virus encodes a CD2 homolog responsible for the adhesion of erythrocytes to infected cells. J. Virol. 67:5312-5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rodríguez, J. R., C. Risco, J. L. Carrascosa, M. Esteban, and D. Rodríguez. 1998. Vaccinia virus 15-kilodalton (A14L) protein is essential for assembly and attachment of viral crescents to virosomes. J. Virol. 72:1287-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rojo, G., M. Chamorro, M. L. Salas, E. Viñuela, J. M. Cuezva, and J. Salas. 1998. Migration of mitochondria to viral assembly sites in African swine fever virus-infected cells. J. Virol. 72:7583-7588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roullier, I., S. M. Brookes, A. D. Hyatt, M. Windsor, and T. Wileman. 1998. African swine fever virus is wrapped by the endoplasmic reticulum. J. Virol. 72:2373-2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Salas, J., M. L. Salas, and E. Viñuela. 1999. African swine fever virus: a missing link between poxviruses and iridoviruses?, p. 467-480. In E. Domingo, R. G. Webster, and J. J. Holland (ed.), Origin and evolution of viruses. Academic Press, London, United Kingdom.
- 49.Sanz, A., B. García-Barreno, M. L. Nogal, E. Viñuela, and L. Enjuanes. 1985. Monoclonal antibodies specific for African swine fever virus proteins. J. Virol. 54:199-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Simón-Mateo, C., G. Andrés, and E. Viñuela. 1993. Polyprotein processing in African swine fever virus: a novel gene expression strategy for a DNA virus. EMBO J. 12:2977-2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Simón-Mateo, C., G. Andrés, F. Almazán, and E. Viñuela. 1997. Proteolytic processing in African swine fever virus: evidence for a new structural polyprotein, pp62. J. Virol. 71:5799-5804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Simón-Mateo, C., J. M. Freije, G. Andrés, C. Lopez-Otín, and E. Viñuela. 1995. Mapping and sequence of the gene encoding protein p17, a major African swine fever virus structural protein. Virology 206:1140-1144. [DOI] [PubMed] [Google Scholar]
- 53.Sodeik, B., and J. Krijnse-Locker. 2002. Assembly of vaccinia virus revisited: de novo membrane synthesis or acquisition from the host? Trends Microbiol. 10:15-24. [DOI] [PubMed] [Google Scholar]
- 54.Sodeik, B., R. W. Doms, M. Ericsson, G. Hiller, C. E. Machamer, W. van't Hof, G. van Meer, B. Moss, and G. Griffiths. 1993. Assembly of vaccinia virus: role of the intermediate compartment between the endoplasmic reticulum and the Golgi stacks. J. Cell Biol. 121:521-541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sun, H., J. Jenson, L. K. Dixon, and M. E. Parkhouse. 1996. Characterization of the African swine fever virion protein j18L. J. Gen. Virol. 77:941-946. [DOI] [PubMed] [Google Scholar]
- 56.Tooze, J., M. Hollinshead, B. Reis, K. Radsak, and H. Kern. 1991. Progeny vaccinia and cytomegalovirus particles utilize early endosomal cisternae for their envelopes. Eur. J. Cell Biol. 60:163-178. [PubMed] [Google Scholar]
- 57.Traktman, P., K. Liu, J. DeMasi, R. Rollins, S. Jesty, and B. Unger. 2000. Elucidating the essential role of the A14 phosphoprotein in vaccinia virus morphogenesis: construction and characterization of a tetracycline-inducible recombinant. J. Virol. 74:3682-3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Viñuela, E. 1987. Molecular biology of African swine fever virus, p. 31-49. In Y. Becker (ed.), African swine fever. Nijhoff, Boston, Mass.
- 59.von Heijne, G. 1996. Principles of membrane protein assembly and structure. Prog. Biophys. Mol. Biol. 66:113-139. [DOI] [PubMed] [Google Scholar]
- 60.Wolffe, E. J., D. M. Moore, P. J. Peters, and B. Moss. 1996. Vaccinia virus A17L open reading frame encodes an essential component of nascent viral membranes that is required to initiate morphogenesis. J. Virol. 70:2797-2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yáñez, R. J., J. M. Rodríguez, M. L. Nogal, L. Yuste, C. Enriquez, J. F. Rodríguez, and E. Viñuela. 1995. Analysis of the complete nucleotide sequence of African swine fever virus. Virology 208:249-278. [DOI] [PubMed] [Google Scholar]
- 62.Zhang, Y., and B. Moss. 1992. Immature viral envelope formation is interrupted at the same stage by lac operator-mediated repression of the vaccinia virus D13L gene and by the drug rifampicin. Virology 187:643-653. [DOI] [PubMed] [Google Scholar]