Abstract
Crimean-Congo hemorrhagic fever virus (CCHFV) belongs to the genus Nairovirus within the family Bunyaviridae and is the causative agent of severe hemorrhagic fever. Despite increasing knowledge about hemorrhagic fever viruses, the factors determining their pathogenicity are still poorly understood. The interferon-induced MxA protein has been shown to have an inhibitory effect on several members of the Bunyaviridae family, but the effect of MxA against CCHFV has not previously been studied. Here, we report that human MxA has antiviral activity against CCHFV. The yield of progeny virus in cells constitutively expressing MxA was reduced up to 1,000-fold compared with control cells, and accumulation of viral genomes was blocked. Confocal microscopy revealed that MxA colocalizes with the nucleocapsid protein (NP) of CCHFV in the perinuclear regions of infected cells. Furthermore, we found that MxA interacted with NP by using a coimmunoprecipitation assay. We also found that an amino acid substitution (E645R) within the C-terminal domain of MxA resulted in a loss of MxA antiviral activity and, concomitantly, in the capacity to interact with CCHFV NP. These results suggest that MxA, by interacting with a component of the nucleocapsid, prevents replication of CCHFV viral RNA and thereby inhibits the production of new infectious virus particles.
The Bunyaviridae family is the largest virus family, comprising over 300 arthropod-borne viruses. It is divided into the five genera Nairovirus, Bunyavirus, Hantavirus, Phlebovirus, and Tospovirus. All viruses within the Bunyaviridae family contain three single-stranded RNA segments of negative sense, designated the large (L), the medium (M), and the small (S) segments. The L segment encodes the RNA-dependent RNA polymerase, while the M segment encodes the glycoproteins (G1 and G2) and the S segment encodes the nucleocapsid protein (NP) (4, 23, 29). In addition, certain members of the Bunyaviridae use an ambisense coding strategy to generate nonstructural proteins (3, 4, 29, 32).
Crimean-Congo hemorrhagic fever virus (CCHFV) is a member of the Nairovirus genus and is the causative agent of Crimean-Congo hemorrhagic fever, a severe disease with a mortality rate of around 30% in humans, with most deaths occurring 5 to 14 days after the onset of illness (23, 34). At present, Crimean-Congo hemorrhagic fever is a public health problem in many regions of the world—e.g., Asia, Eastern Europe, Africa, and Russia (23, 27)—and its potential use as a terrorist agent is of great concern. Transmission to humans occurs through the bites of Ixodid ticks (Hyalomma genus) or by contact with blood or tissue from infected animals. Furthermore, nosocomial CCHFV infections among caregivers have also been reported (6, 33).
The factors determining the pathogenicity of the bunyaviruses remain largely unexplored. A better understanding of virus host-cell interaction is necessary to design efficient strategies for disease control. While earlier studies have demonstrated that human interferons (IFNs) have an antiviral effect against a number of these viruses (25, 35, 36), little is known about the molecular mechanisms of this antiviral action.
Alpha and beta IFNs are important mediators of innate immune responses and are crucial for limiting early replication and spread of viruses (19). They induce a number of proteins involved in antiviral actions, such as the 2′,5′-oligoadenylate synthetase, the double-stranded RNA-activated protein kinase, and the Mx proteins (Mx1 in mice and MxA in humans) (10, 26).
MxA belongs to the dynamin superfamily of large GTPases, which are involved in a variety of intracellular transport processes (13, 16, 31). The human MxA protein is induced exclusively by alpha and beta IFNs and is partly associated with membranes of the smooth endoplasmic reticulum (1, 13). It is a key component in the interferon-induced defense against a number of viruses belonging to several different families, i.e., Bunyaviridae (10, 15, 17), Togaviridae (18), Orthomyxoviridae (11, 24), Paramyxoviridae (30), Rhabdoviridae (24), Picornaviridae (7), and Hepdnaviridae (12). However, the mechanism by which MxA is able to inhibit such diverse viruses is not well understood.
Recently, Kochs and coworkers suggested a mechanism for the action of MxA against La Crosse virus (LACV), another member of the Bunyaviridae (17). They demonstrated that MxA interferes with the replication process of LACV by sequestering an essential virus component, the viral NP, to perinuclear inclusions in the cytoplasm, where this protein is no longer available for generating new viral particles.
Despite increasing knowledge regarding the antiviral action of MxA against bunyaviruses, no data have so far been published for CCHFV. In particular, CCHFV research has been hindered by the biosafety containment procedures required for handling the virus. In the present study, we show that human MxA inhibits the growth of CCHFV. Furthermore we demonstrate that MxA interacts with the nucleocapsid component NP of CCHFV and present evidence that this interaction leads to a block in viral genome replication in infected cells.
MATERIALS AND METHODS
Cells and antibodies.
Stably transfected African green monkey kidney cells (Vero E-6), constitutively expressing human MxA (clones VA9, VA12, and VA3) (11) or the variant MxA (E645R) [clone VA(E645R)] (17), and control cells expressing only the neomycin resistance gene (clones VN36 and VN41) (11) were grown in Dulbecco's modified Eagle's minimal essential medium supplemented with 10% fetal bovine serum, 200 mM l-glutamine and 2 mg of G-418 (Geneticin; Roche, Basel, Switzerland) per ml. Antibodies used in this study included a rabbit polyclonal anti-CCHFV NP antibody and a mouse monoclonal anti-MxA antibody (M143) (9).
Indirect immunofluorescence.
Subconfluent cells were infected with CCHFV (strain IbAr 10200) at different multiplicities of infection (MOI). At 24 h postinfection (hpi), the cells were fixed with 4% formalin for 48 h at 4°C and permeabilized with ice-cold acetone-methanol (1:1) or Triton X-100. The cells were incubated with primary antibodies for 1 h in phosphate-buffered saline (PBS) containing 0.2% bovine serum albumin and 0.1% Triton X-100 at 37°C. After being rinsed three times with PBS, the cells were incubated with anti-mouse or anti-rabbit fluorescein isothiocyanate-Texas red-conjugated antibodies (Jackson, Baltimore, Md.) for 1 h at 37°C. After three washes, the slides were mounted and analyzed by microscopy.
For double immunofluorescence studies, the cells were incubated for 1 h with mouse anti-MxA and rabbit anti-CCHFV NP antibodies at 37°C. After washing in PBS, the cells were incubated with Texas red-conjugated anti-mouse antibodies and fluorescein isothiocyanate-conjugated anti-rabbit antibodies for 1 h. After three washes, the slides were mounted and analyzed. All handling of virus was performed in a biosafety level 4 laboratory.
Assay for determination of fluorescence focus units.
Vero E6 cells, VA3, VA9, VA12, VN36, VN41, and VA(E645R) cells were cultured in six-well plates and infected with native CCHFV (strain IbAr 10200) at an MOI of 0.01. After absorption for 1 h in 37°C, the virus inoculum was removed and the cells were washed twice with PBS. Infected cells and supernatants were harvested at 24 or 48 hpi. The amount of total infectious virus (extracellular plus intracellular) was determined by infecting Vero E6 cells with serial dilutions (10-fold) of the harvested material. Cells were fixed at 24 hpi and stained by indirect immunofluorescence as described above, followed by enumeration of fluorescent foci in the last well showing positive immunofluorescence. All handling of virus was performed in a biosafety level 4 laboratory.
Confocal microscopy.
Confocal imaging was achieved using a Sarastro 2000 (Molecular Dynamics, Sunnyvale, Calif.) equipped with an air-cooled argon laser (10 mW, 30% efficiency). Activation was done with argon laser wavelengths, including 488 and 514 nm, primary beam splitter 535DRLP, secondary beam splitter 595 DRLP, and barrier filter EFLP600 (red channel; Texas red) and line-pass filter 545DF30 (green channel; fluorescence). The high voltage on photomultiplier defectors was in both case 850 V. To reduce noise in the merges, each section was scanned four to six times. The intensity of the red and green labels was adjusted to approximately equal intensity to avoid channel mixing. Only negligible bleeding between green fluorescence and red fluorescence was observed.
Immunoprecipitation.
Confluent cells were infected with CCHFV (IbAr 10200) at an MOI of 0.1. At 24 hpi the cells were lysed in ice-cold gentle lysis buffer(2% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate [CHAPS]), 150 mM NaCl, 50 mM HEPES plus protease inhibitors (3 μg of antipain [Sigma, St. Louis, Mo.] per ml, 6 μg of leupeptin [Sigma] per ml, and 1 μg of pepstatin [Sigma] per ml). One hundred microliters of the cell lysate was incubated with 10 μl of anti-CCHFV NP antibody and 400 μl of radioimmunoprecipitation assay (RIPA) buffer (150 mM NaCl, 50 mM HEPES, 0.5% CHAPS) at 4°C overnight. Forty microliters of Staphylococcus aureus protein A-Sepharose CL-4B (Amersham Pharmacia, Little Chalfont, Buckinghamshire, United Kingdom) was added, and the mixture was incubated for 1 h under constant shaking. The immune complexes were washed four times with RIPA buffer, resuspended in reducing sample buffer and boiled for 5 min. The samples were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
SDS-PAGE.
Polypeptide separation was performed by reducing SDS-PAGE using a 5% concentration gel and a 10% separation gel. Electrophoresis was carried out at a constant current of 25 A as previously described (21).
Western blotting.
Proteins were transferred to nitrocellulose membranes using a transfer buffer containing 25 mM Tris, 192 mM glycine, and 20% (vol/vol) methanol at 100 V for 1 h, and membranes were blocked in 5% nonfat dry milk overnight at 4°C. After being washed in PBS containing 0.01% Tween (PBST), the membranes were incubated with a rabbit polyclonal anti-CCHFV NP antibody or a mouse monoclonal anti-MxA antibody in dilution buffer (5% nonfat dry milk in PBST) for 1 h at room temperature. The membranes were then washed three times with PBST before the addition of goat anti-rabbit or rabbit anti-mouse immunoglobulin G horseradish peroxidase-conjugated antibody (Bio-Rad, Hercules, Calif.) diluted in dilution buffer. The membranes were incubated for 1 h at room temperature. After careful washing, detection was carried out using ECL Plus Western blotting detection reagents (Amersham Pharmacia) according to the manufacturer's instructions.
RNA preparation.
Mock- or CCHFV-infected cells were washed with PBS and lysed in TRIZOL reagent (Gibco/Life Technologies/Invitrogen, Groningen, The Netherlands). Fifty microliters of chloroform was added to 200 μl of the TRIZOL-treated samples before centrifugation at 12,000 × g for 15 min at 4°C. RNA was extracted from the aqueous phase using the QIAamp Viral RNA Mini Kit (Qiagen, Valencia, Calif.) according to the manufacturer's instructions.
Oligonucleotide design and reversed transcription.
Primers and the corresponding 5′-nuclease probes were designed with the Primer Express software (Applied Biosystems). All probes contained a 5′ reporter, 6-carboxy-fluorescein (FAM), and a 3′ quencher, 6-carboxy-tetramethylrhodamine (TAMRA). The probe for CCHFV-NP was 5′-FAM ACC TTC CCG ACG GTG TCA CAG TTC C TAMRA-3′, forward primer 5′-GGC TGG CGT GAC TCC TGA-3′ and reverse 5′-TGG CTG TTT CCC TAG CTC AAA. The probe for CCHFV-GP was 5′-FAM CAG AAT GTC AAA CCA CCC CAA AAC AAC C TAMRA-3′, forward primer 5′-TTC CAT TAT TGG GCA AAA TGG-3′ and reverse primer 5′-CAA AGC TGA ACC AGA AGA GGA AA-3′.
The reversed transcription was performed on GeneAmp PCR System 2700 (Applied Biosystems). The 20-μl assay mixture contained 1 μl of RNA, 1 μM forward primer, a 1 mM concentration of each deoxynucleoside triphosphate (Invitrogen, Groningen, Netherlands), RNase OUT RNase Inhibitor (2 U/μl; Invitrogen), 4 μl of 5× First Strand Buffer (Invitrogen), 1 μl of dithiothreitol (Invitrogen), and Superscript II RNase H− reverse transcriptase (5 U/μl; Invitrogen). The cycling profiles used for reverse transcription were 25°C for 10 min, 42°C for 60 min, 70°C for 15 min, and cooling at 4°C.
Quantitative real-time PCR.
Real-time PCR was performed on an ABI Prism 7900 HT sequence detector system (PE Applied Biosystems). One microliter of the cDNA was analyzed with the PCR mixture in a total volume of 25 μl. The PCR mixture consisted of 1× TaqMan Universal PCR Master Mix, 3 mM MgCl2, 200 nM fluorogenic probe, a 300 nM concentration of forward and reverse primers for CCHFV-NP and for CCHFV-GP, and 600 and 400 nM concentrations of forward and reverse primers, respectively. A plasmid (pCR-script) with a segment of the NP or GP gene of CCHFV was used as a standard in dilutions between 5 and 5 × 104 copies. An EBV standard (8) also was used as external standard. The real-time PCR conditions were as follows: 2 min at 50°C, 10 min at 95°C, 45 repeats of 15 s at 95°C, and 1 min at 60°C. All samples were analyzed in triplicate.
RESULTS
Human MxA protein inhibits CCHFV.
To study the antiviral potential of human MxA protein against CCHFV, we used permanently transfected Vero cells expressing human MxA protein (VA9, VA3, and VA12), transfected Vero cells expressing only the neomycin resistance gene (VN36 and VN41), and Vero cells expressing a mutant form of MxA [VA(E645R)]. The mutant MxA has a single amino acid substitution (glutamic acid to arginine) in the C-terminal domain and has specifically lost its antiviral activity against LACV and vesicular stomatitis virus, but not against influenza A virus and Thogoto virus (17, 40).
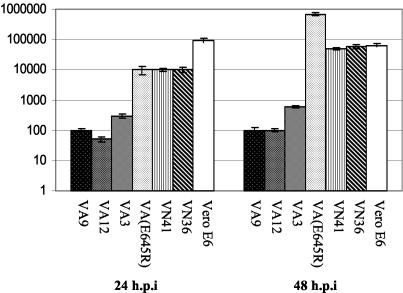
Monolayer cultures were infected with CCHFV at an MOI of 0.01. At 1 hpi, the monolayers were washed twice and incubated with fresh media. Infected cells were harvested at different time points postinfection and the virus yields were assayed as described in Materials and Methods. Figure 1 shows that the virus titers from cells expressing wild-type MxA protein infected with CCHFV were 10- to 1,000-fold lower than the titers from VN36 and VN41 cells, nontransfected Vero E6 cells, or Vero cells expressing the mutant form of MxA (VA(E645R).
FIG. 1.
Antiviral activity of human MxA protein against CCHFV. Vero cells constitutively expressing wild-type MxA (VA9, VA12, and VA3), nontransfected Vero cells, cells expressing only the neomycin resistance gene (VN36 and VN41) and cells expressing a mutant form of MxA [VA(E645R)] were infected with CCHFV at an MOI of 0.01. At 24 and 48 hpi, the cells and supernatants were harvested and the amount of infectious virus was determined as described in Materials and Methods. The mean log titers given are the average of three independent experiments. Error bars indicate standard deviations.
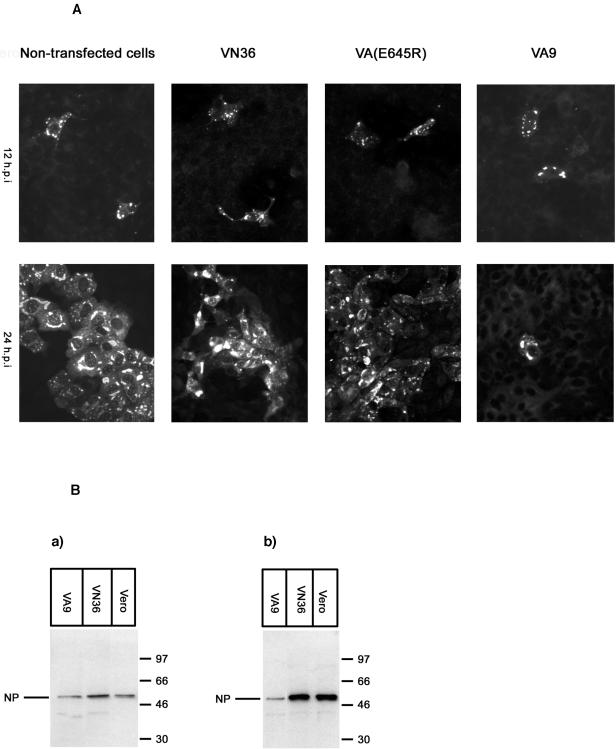
To study whether wild-type MxA could prevent formation of viral foci, cells were infected with CCHFV at an MOI of 0.01. At 12 or 24 hpi the cells were fixed and analyzed for NP expression by indirect immunofluorescence. These experiments revealed that CCHFV was not able to form foci in cell cultures expressing MxA. In contrast, large foci formed in VN36 cells, Vero E6 cells, and Vero cells expressing the mutant form of MxA at 24 hpi (Fig. 2A), demonstrating that wild-type MxA specifically interfered with the multiplication of CCHFV.
FIG. 2.
(A) Formation of foci was inhibited in cells expressing MxA protein. Nontransfected Vero cells, Vero cells expressing only the neomycin resistance gene (VN36), Vero cells expressing MxA(E645R) [VA(E645R)] or cells expressing wild-type MxA (VA9) were grown in chamber slides and infected with CCHFV at MOI of 0.01. At 12 or 24 hpi, the cells were fixed and analyzed for CCHFV NP expression by indirect immunofluorescence. (B) Monolayers of control cells (Vero E6 cells and VN36) and cells constitutively expressing MxA (VA9) were infected with CCHFV at MOI of 0.1. At 12 (a) or 24 (b) hpi, the cells were harvested and analyzed by Western blot using an anti-CCHFV NP antibody. Molecular markers are shown at the right side of the panel.
Next, we studied the NP synthesis in MxA-expressing cells. Monolayer cultures were infected with CCHFV at an MOI of 0.1, lysed at 12 and 24 hpi, and analyzed for expression of CCHFV NP by Western blotting. The expression levels of NP were comparable in VA9 and control cell lines at 12 hpi, indicating that transcription of the S segment was not affected and NP synthesis was almost normal (Fig. 2B, subpanel a). At later time points, NP expression increased in control cells but not in MxA-expressing cells (Fig. 2B, subpanel b). Double immunofluorescence staining clearly demonstrated that most of the infected cells indeed expressed MxA (data not shown). These results suggest that replication rather than transcription of CCHFV was blocked by MxA, as also shown previously for LACV (17).
Replication of CCHFV is blocked in MxA-expressing cells.
To study the role of MxA in the replication process, VA9, VN36, and Vero E6 cells were infected with CCHFV at an MOI of 0.1. Mock- and CCHFV-infected cells were lysed at different time periods after infection, and total RNA was purified and analyzed by real-time PCR as described in Materials and Methods. At 12 h after infection, the amounts of S segment viral RNA (vRNA) were significantly reduced in VA9 cells compared to those in the control cells (Fig. 3a). The same findings were obtained for the M segment (Fig. 3b). The amounts of S and M vRNAs remained unchanged in MxA-expressing cells during prolonged periods of time postinfection (Fig. 3). These results suggest that MxA blocks the replication step of CCHFV and thereby prevents the generation of infectious virus particles.
FIG. 3.
Accumulation of the CCHFV genome is blocked in cells expressing wild-type MxA. VA9 and VN36 monolayers were infected with CCHFV at an MOI of 0.01. After 12 and 24 hpi, the cells were lysed in TRIZOL reagent. Total RNA was extracted and the amounts of vRNA for the S (a) and M (b) segments were determined by a quantitative real-time PCR as described in Materials and Methods. Values given are the average of three independent experiments. Error bars indicate standard deviations.
MxA colocalizes and interacts with CCHFV NP.
We next investigated whether MxA colocalized with CCHFV NP in infected cells. Monolayers were infected with CCHFV at an MOI of 0.01, fixed at 24 hpi, and analyzed by double immunofluorescence and confocal microscopy. Figure 4 color panels show that wild-type MxA colocalized with CCHFV NP in the perinuclear region of infected cells. This redistribution of MxA with NP resulted in a loss of the typical granular appearance of MxA. In contrast to wild-type MxA, the mutant form MxA(E645R) still remained in a dot-like granular pattern scattered over the cytoplasm of infected cells and did not colocalize with CCHFV NP.
FIG. 4.
(Color panels) Wild-type MxA colocalizes with NP of CCHFV in perinuclear regions of infected cells. VN36, VA(E645R), and VA9, seeded in chamber slides, were infected with CCHFV at an MOI of 0.01. At 24 hpi, the cells were fixed and analyzed for CCHFV NP (green) and MxA (red) under the confocal microscope. Panels at the right show the superimposition of the two images. (Black-and-white panels) MxA coimmunoprecipitated with CCHFV NP. Wild-type MxA-expressing Vero cells (VA9), a mutant form of MxA [VA(E645R)], and cells expressing only the neomycin gene were infected with CCHFV at an MOI of 0.1. (a) At 24 hpi, the cell monolayers were lysed and immunoprecipitated with an anti-CCHFV NP-specific antibody. The immunoprecipitate was analyzed for the presence of MxA by Western blot analysis using specific MxA antibodies. Molecular markers are shown on the left side of the panel. (b) Cell lysates from panel a were analyzed for the expression of MxA and CCHFV NP by Western blot analysis.
To investigate whether MxA interacted directly with NP of CCHFV, we performed coimmunoprecipitation assays. Infected monolayers were lysed at 24 hpi and the lysates were subjected to immunoprecipitation, using an NP-specific polyclonal antibody. The immunoprecipitated protein complexes were subsequently analyzed by Western blotting for the presence of MxA, using an MxA-specific monoclonal antibody. Clearly, wild-type MxA coprecipitated with NP in these assays (Fig. 4 black-and-white panels, subpanel a). In contrast, no coimmunoprecipitation was observed with MxA(E645R), indicating that the mutant form of MxA was unable to recognize CCHFV NP.
The protein expression of MxA and NP in VA9, VA(E645R), and VN36 was assessed by immunoblot analysis of the crude cell lysates. These results demonstrated that both VA9 and VA(E645R) expressed MxA and NP.
DISCUSSION
We demonstrate here that human MxA protein has antiviral activity against CCHFV, an important human pathogen belonging to the genus Nairovirus within the Bunyaviridae family. Nairoviruses differ from other members of the family in having, among other features, generally larger genomes and a more complex processing pattern of their glycoproteins (28). Interestingly, our results suggest that MxA inhibits CCHFV by a similar if not the same mechanism that is also used against LACV, a member of the genus Phlebovirus (17). This notion is supported by several findings. First, yields of CCHFV progeny virus were up to 1,000-fold lower in MxA-expressing cells than in control cells, in agreement with previous findings with LACV and other bunyaviruses (10). Second, a single amino acid substitution (E645R) in the carboxy-terminal effector domain led to a complete loss of antiviral activity against CCHFV, as also reported previously for LACV (17). It should be noted here that MxA(E645R) has normal GTPase activity and is antiviral against other viruses, such as influenza A virus or Thogoto virus (11). Its failure to recognize CCHFV demonstrates the high specificity of the antiviral activity of wild-type MxA. Third, the MxA block seemed to occur at the level of genome replication. Thus, the accumulation of viral proteins, notably NP, was normal up to 12 h of infection but not at later time points, indicating that primary transcription of the incoming genome and translation of viral mRNAs were not affected, as in the case of LACV (17). In contrast, genomic vRNA accumulation was suppressed at early and late time points, as demonstrated by real-time PCR analysis. Fourth, NP representing the major protein component of CCHFV nucleocapsids was suggested as the target of MxA, and MxA was found to interact with NP in coimmunoprecipitation assays. However, we cannot rule out the possibility that MxA interacted with the RNA-containing nucleocapsid structure of CCHFV or, indirectly, by binding to hitherto-unknown cellular factors associated with nucleocapsids.
Moreover, in infected cells, NP was sequestered to a perinuclear location where it colocalized with MxA. This redistribution of MxA in large perinuclear complexes (mostly around the NP inclusion bodies) seemed to be an active process linked to interaction with NP, because the antivirally inactive MxA(E645R) failed to bind to and to form complexes with NP.
In the case of LACV, it has been proposed that targeting of NP into perinuclear complexes would make NP unavailable for the processes of genome replication or packaging which occur near the Golgi compartment. Alternatively, MxA might recognize and bind to NP in the context of viral ribonucleoprotein complexes or nucleocapsid-like structures and prevent their functions (17). We have previously shown that CCHFV NPs localize in the perinuclear region in infected cells (2). It is therefore conceivable that, upon redistribution to the perinuclear area, MxA inhibits the replication process of CCHFV at this location in infected cells. Ultrastructural studies have revealed that MxA forms tight fibrillary bundles with NP in LACV-infected cells (13, 17). Recent data have shown that these MxA/NP filamentous complexes are associated with membranous structures of a still poorly defined intracellular membrane compartment most likely originating from the smooth endoplasmic reticulum (M. Reichelt, S. Stertz, G. Kochs, and O. Haller, Abstr. 12th Int. Conf. Negative Strand RNA Viruses, abstr. 173, 2003). It will be interesting to see whether the interaction of MxA with NP of CCHFV described here involves similar subcellular structures.
The present report adds CCHFV to the list of MxA-sensitive bunyaviruses, demonstrating that members of the Nairovirus genus are also susceptible to the antiviral action of the IFN-induced MxA GTPase. Recently it has been reported that this seems likewise to apply to Dugbe virus, another member of this genus (5). It has previously been shown that members of the genera Bunyavirus and Hantavirus are MxA sensitive, in addition to Phlebovirus (10, 15). Together, these studies allow the conclusion that all animal bunyaviruses are restricted in their intracellular growth by MxA. In all instances analyzed, the NP or nucleocapsid was the target, suggesting a common antiviral mechanism (13).
Work with MxA-transgenic animals demonstrated that the antiviral activity of MxA, as demonstrated here, is not just a cell culture phenomenon. On the contrary, human MxA inhibited LACV growth in MxA-transgenic mice, leading to increased survival even in animals that were IFN nonresponsive for genetic reasons (14). These experiments clearly demonstrate the in vivo antiviral power of the human MxA pathway. Surprisingly, MxA is able to mediate resistance against LACV also in transfected mosquito cells (22), demonstrating that MxA does not require the ingredients of the mammalian host cell for its action but is a powerful antiviral agent on its own. It can be anticipated that MxA is likewise active in human tissues, provided it is properly induced by IFNs. Most likely, it is capable of inhibiting CCHFV production in human blood cells, but this remains to be demonstrated.
Why then does CCHF run its devastating course? It has been estimated that CCHFV causes fatal hemorrhagic disease in almost 30% of infected patients (23, 34). The most likely explanation is that IFN is induced in insufficient amounts or too late in the course of infection to put MxA in place at the time when it is urgently needed. Furthermore, it is conceivable that CCHFV has evolved mechanism to subvert the IFN system as found in other negative strand RNA viruses (37). The antiviral effect against CCHFV is not 100%, and different CCHFV strains or isolates may differ in their individual sensitivity to the MxA action. In recent years, CCHFV outbreaks have occurred in increasingly diverse regions, including Mauritania, Pakistan, Iran, and Russia (20, 27, 38, 39), and novel approaches to combat the disease are urgently needed. Elucidating the mechanisms of MxA action and virus sensitivity may eventually provide new means to prevent and control this dreadful human pathogen.
Acknowledgments
This work was in part supported by a grant from the Deutsche Forschungsgemeinschaft to O.H. (HA 1582/3-1).
REFERENCES
- 1.Accola, M. A., B. Huang, A. Al Masri, and M. A. McNiven. 2002. The antiviral dynamin family member, MxA, tubulates lipids and localizes to the smooth endoplasmic reticulum. J. Biol. Chem. 277:21829-21835. [DOI] [PubMed] [Google Scholar]
- 2.Andersson, I., M. Simon, A. Lundkvist, M. Nilsson, A. Holmstrom, F. Elgh, and A. Mirazimi. 2004. Role of actin filaments in targeting of Crimean Congo hemorrhagic fever virus nucleocapsid protein to perinuclear regions of mammalian cells. J. Med. Virol. 72:83-93. [DOI] [PubMed] [Google Scholar]
- 3.Billecocq, A., M. Vazeille-Falcoz, F. Rodhain, and M. Bouloy. 2000. Pathogen-specific resistance to Rift Valley fever virus infection is induced in mosquito cells by expression of the recombinant nucleoprotein but not NSs non-structural protein sequences. J. Gen. Virol. 81:2161-2166. [DOI] [PubMed] [Google Scholar]
- 4.Bishop, D. H. L. 1996. Biology and molecular biology of bunyaviruses. Plenum Press, New York, N.Y.
- 5.Bridgen, A., D. A. Dalrymple, F. Weber, and R. M. Elliott. 2004. Inhibition of Dugbe nairovirus replication by human MxA protein. Virus Res. 99:47-50. [DOI] [PubMed] [Google Scholar]
- 6.Burney, M. I., A. Ghafoor, M. Saleen, P. A. Webb, and J. Casals. 1980. Nosocomial outbreak of viral hemorrhagic fever caused by Crimean Hemorrhagic fever-Congo virus in Pakistan, January 1976. Am. J. Trop. Med. Hyg. 29:941-947. [DOI] [PubMed] [Google Scholar]
- 7.Chieux, V., W. Chehadeh, J. Harvey, O. Haller, P. Wattre, and D. Hober. 2001. Inhibition of coxsackievirus B4 replication in stably transfected cells expressing human MxA protein. Virology 283:84-92. [DOI] [PubMed] [Google Scholar]
- 8.Enbom, M., A. Strand, K. I. Falk, and A. Linde. 2001. Detection of Epstein-Barr virus, but not human herpesvirus 8, DNA in cervical secretions from Swedish women by real-time polymerase chain reaction. Sex Transm. Dis. 28:300-306. [DOI] [PubMed] [Google Scholar]
- 9.Flohr, F., S. Schneider-Schaulies, O. Haller, and G. Kochs. 1999. The central interactive region of human MxA GTPase is involved in GTPase activation and interaction with viral target structures. FEBS Lett. 463:24-28. [DOI] [PubMed] [Google Scholar]
- 10.Frese, M., G. Kochs, H. Feldmann, C. Hertkorn, and O. Haller. 1996. Inhibition of bunyaviruses, phleboviruses, and hantaviruses by human MxA protein. J. Virol. 70:915-923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frese, M., G. Kochs, U. Meier-Dieter, J. Siebler, and O. Haller. 1995. Human MxA protein inhibits tick-borne Thogoto virus but not Dhori virus. J. Virol. 69:3904-3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gordien, E., O. Rosmorduc, C. Peltekian, F. Garreau, C. Brechot, and D. Kremsdorf. 2001. Inhibition of hepatitis B virus replication by the interferon-inducible MxA protein. J. Virol. 75:2684-2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haller, O., and G. Kochs. 2002. Interferon-induced Mx proteins: dynamin-like GTPases with antiviral activity. Traffic 3:710-717. [DOI] [PubMed] [Google Scholar]
- 14.Hefti, H. P., M. Frese, H. Landis, C. Di Paolo, A. Aguzzi, O. Haller, and J. Pavlovic. 1999. Human MxA protein protects mice lacking a functional alpha/beta interferon system against La Crosse virus and other lethal viral infections. J. Virol. 73:6984-6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kanerva, M., K. Melen, A. Vaheri, and I. Julkunen. 1996. Inhibition of puumala and tula hantaviruses in Vero cells by MxA protein. Virology 224:55-62. [DOI] [PubMed] [Google Scholar]
- 16.Kochs, G., M. Haener, U. Aebi, and O. Haller. 2002. Self-assembly of human MxA GTPase into highly ordered dynamin-like oligomers. J. Biol. Chem. 277:14172-14176. [DOI] [PubMed] [Google Scholar]
- 17.Kochs, G., C. Janzen, H. Hohenberg, and O. Haller. 2002. Antivirally active MxA protein sequesters La Crosse virus nucleocapsid protein into perinuclear complexes. Proc. Natl. Acad. Sci. USA 99:3153-3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Landis, H., A. Simon-Jodicke, A. Kloti, C. Di Paolo, J. J. Schnorr, S. Schneider-Schaulies, H. P. Hefti, and J. Pavlovic. 1998. Human MxA protein confers resistance to Semliki Forest virus and inhibits the amplification of a Semliki Forest virus-based replicon in the absence of viral structural proteins. J. Virol. 72:1516-1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Le Bon, A., and D. F. Tough. 2002. Links between innate and adaptive immunity via type I interferon. Curr. Opin. Immunol. 14:432-436. [DOI] [PubMed] [Google Scholar]
- 20.Mardani, M., M. K. Jahromi, K. H. Naieni, and M. Zeinali. 2003. The efficacy of oral ribavirin in the treatment of Crimean-Congo hemorrhagic fever in Iran. Clin. Infect. Dis. 36:1613-1618. [DOI] [PubMed] [Google Scholar]
- 21.Mirazimi, A., C. H. von Bonsdorff, and L. Svensson. 1996. Effect of brefeldin A on rotavirus assembly and oligosaccharide processing. Virology 217:554-563. [DOI] [PubMed] [Google Scholar]
- 22.Miura, T. A., J. O. Carlson, B. J. Beaty, R. A. Bowen, and K. E. Olson. 2001. Expression of human MxA protein in mosquito cells interferes with LaCrosse virus replication. J. Virol. 75:3001-3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nichol, S. 2001. Virology, vol. 2. Bunyaviruses. Lippincott, Williams and Wilkins, Philadelphia, Pa.
- 24.Pavlovic, J., T. Zurcher, O. Haller, and P. Staeheli. 1990. Resistance to influenza virus and vesicular stomatitis virus conferred by expression of human MxA protein. J. Virol. 64:3370-3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peters, C. J., J. A. Reynolds, T. W. Slone, D. E. Jones, and E. L. Stephen. 1986. Prophylaxis of Rift Valley fever with antiviral drugs, immune serum, an interferon inducer, and a macrophage activator. Antivir. Res. 6:285-297. [DOI] [PubMed] [Google Scholar]
- 26.Samuel, C. E. 2001. Antiviral actions of interferons. Clin. Microbiol. Rev. 14:778-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sanchez, A. J., M. J. Vincent, and S. T. Nichol. 2002. Characterization of the glycoproteins of Crimean-Congo hemorrhagic fever virus. J. Virol. 76:7263-7275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmaljohn, C. 1996. Bunyaviridae: the viruses and their replication, p. 1447-1471. In B. N. Fields, D. M. Knipe, et al. (ed.), Fields virology, 3rd ed. Raven Press, New York, N.Y.
- 29.Schmaljohn, C. S., and J. W. Hooper. 2001. Bunyaviridae: the viruses and their replication, p. 1447-1472. In B. N. Fields, D. M. Knipe, et al. (ed.), Fields virology, 4th ed., vol. 2. Lippincott, Williams and Wilkins, Philadelphia, Pa.
- 30.Schneider-Schaulies, S., J. Schneider-Schaulies, A. Schuster, M. Bayer, J. Pavlovic, and V. ter Meulen. 1994. Cell type-specific MxA-mediated inhibition of measles virus transcription in human brain cells. J. Virol. 68:6910-6917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sever, S., H. Damke, and S. L. Schmid. 2000. Garrotes, springs, ratchets, and whips: putting dynamin models to the test. Traffic 1:385-392. [DOI] [PubMed] [Google Scholar]
- 32.Struthers, J. K., and R. Swanepoel. 1982. Identification of a major non-structural protein in the nuclei of Rift Valley fever virus-infected cells. J. Gen. Virol. 60:381-384. [DOI] [PubMed] [Google Scholar]
- 33.Suleiman, M. N., J. M. Muscat-Baron, J. R. Harries, A. G. Satti, G. S. Platt, E. T. Bowen, and D. I. Simpson. 1980. Congo/Crimean haemorrhagic fever in Dubai. An outbreak at the Rashid Hospital. Lancet ii:939-941. [PubMed] [Google Scholar]
- 34.Swanepoel, R. 1994. Crimean-Congo haemorrhagic fever, p. 723-729. Oxford University Press, Cape Town, South Africa.
- 35.Tamura, M., H. Asada, K. Kondo, M. Takahashi, and K. Yamanishi. 1987. Effects of human and murine interferons against hemorrhagic fever with renal syndrome (HFRS) virus (Hantaan virus). Antivir. Res. 8:171-178. [DOI] [PubMed] [Google Scholar]
- 36.Temonen, M., H. Lankinen, O. Vapalahti, T. Ronni, I. Julkunen, and A. Vaheri. 1995. Effect of interferon-alpha and cell differentiation on Puumala virus infection in human monocyte/macrophages. Virology 206:8-15. [DOI] [PubMed] [Google Scholar]
- 37.Weber, F., G. Kochs, O. Haller, and P. Staeheli. 2003. Viral evasion of the interferon system: old viruses, new tricks. J. Interferon Cytokine Res. 23:209-213. [DOI] [PubMed] [Google Scholar]
- 38.Yashina, L., I. Petrova, S. Seregin, O. Vyshemirskii, D. Lvov, V. Aristova, J. Kuhn, S. Morzunov, V. Gutorov, I. Kuzina, G. Tyunnikov, S. Netesov, and V. Petrov. 2003. Genetic variability of Crimean-Congo haemorrhagic fever virus in Russia and Central Asia. J. Gen. Virol. 84:1199-1206. [DOI] [PubMed] [Google Scholar]
- 39.Yashina, L., O. Vyshemirskii, S. Seregin, I. Petrova, E. Samokhvalov, D. Lvov, V. Gutorov, I. Kuzina, G. Tyunnikov, Y. W. Tang, S. Netesov, and V. Petrov. 2003. Genetic analysis of Crimean-Congo hemorrhagic fever virus in Russia. J. Clin. Microbiol. 41:860-862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zurcher, T., J. Pavlovic, and P. Staeheli. 1992. Mechanism of human MxA protein action: variants with changed antiviral properties. EMBO J. 11:1657-1661. [DOI] [PMC free article] [PubMed] [Google Scholar]