Highlights
-
•
Poliovirus infection induces the co-localization of SRp20 and TIA-1 in the cytoplasm of infected cells.
-
•
SRp20 is a host mRNA splicing factor that is recruited by poliovirus as an IRES trans-acting factor (ITAF) to stimulate viral protein synthesis.
-
•
TIA-1 is a host RNA binding protein that is a marker for stress granules.
-
•
SRp20-containing stress granules appear to be distinct from those that form under conditions of arsenite stress.
-
•
Formation of SRp20-containing stress granules/cytoplasmic foci may control the levels of viral translation during mid to late times of poliovirus infection.
Keywords: Picornavirus, Poliovirus, Stress granules, SRp20, TIA-1, Protein co-localization
Abstract
Different types of environmental stress cause mammalian cells to form cytoplasmic foci, termed stress granules, which contain mRNPs that are translationally silenced. These foci are transient and dynamic, and contain components of the cellular translation machinery as well as certain mRNAs and RNA binding proteins. Stress granules are known to be induced by conditions such as hypoxia, nutrient deprivation, and oxidative stress, and a number of cellular factors have been identified that are commonly associated with these foci. More recently it was discovered that poliovirus infection also induces the formation of stress granules, although these cytoplasmic foci appear to be somewhat compositionally unique. Work described here examined the punctate pattern of SRp20 (a host cell mRNA splicing protein) localization in the cytoplasm of poliovirus-infected cells, demonstrating the partial co-localization of SRp20 with the stress granule marker protein TIA-1. We determined that SRp20 does not co-localize with TIA-1, however, under conditions of oxidative stress, indicating that the close association of these two proteins during poliovirus infection is not representative of a general response to cellular stress. We confirmed that the expression of a dominant negative version of TIA-1 (TIA-1-PRD) results in the dissociation of stress granules. Finally, we demonstrated that expression of wild type TIA-1 or dominant negative TIA-1-PRD in cells during poliovirus infection does not dramatically affect viral translation. Taken together, these studies provide a new example of the unique cytoplasmic foci that form during poliovirus infection.
1. Introduction
The presence of certain environmental stress stimuli can cause the accumulation of translationally silenced mRNPs in cytoplasmic foci, termed stress granules, which are thought to represent temporary sites of mRNA sorting and storage (for reviews, see Anderson and Kedersha, 2002, Anderson and Kedersha, 2008, Kedersha and Anderson, 2002, Kedersha et al., 2005). Stress granules are generated when conditions of cellular stress result in the inhibition of mRNA translation initiation, although the mechanism by which they form is still unknown. These foci contain stalled pre-initiation complexes consisting of small, but not large, ribosomal subunits, mRNA, and canonical translation initiation factors such as eIF4E, eIF4G, eIF4A, eIF4B, and eIF3. While stress granule assembly has not been completely defined, it has been shown to involve the motor proteins dynein and kinesin, and movement along microtubules (Chernov et al., 2009, Ivanov et al., 2003, Loschi et al., 2009, Tsai et al., 2009, Zurla et al., 2011). RNA binding proteins have been proposed to be required for the formation of stress granules, including T-cell restricted intracellular antigen 1 (TIA-1), TIA-1-related protein (TIAR), and RasGAP-SH3 domain binding protein (G3BP) (Kedersha et al., 1999, Tourriere et al., 2003). The self-oligomerization properties of these proteins contribute to their assembly of cytoplasmic aggregates under conditions of stress. A truncated form of TIA-1 (TIA-1-PRD) containing only the C-terminal domain of the protein has been shown to be a dominant negative inhibitor of stress granule assembly (Gilks et al., 2004). Expressing only the central domain of G3BP also prevents stress granule formation (Tourriere et al., 2003).
In addition to conditions such as hypoxia, nutrient deprivation, and oxidative stress, some viruses are also known to induce the formation of stress granules (for review, see White and Lloyd, 2012). Respiratory syncytial virus, coronaviruses, and reovirus induce stress granule formation as a way to down-regulate host cell translation (Lindquist et al., 2010, Raaben et al., 2007, Smith et al., 2006, Sola et al., 2011). Semliki Forest virus and hepatitis C virus initially induce the formation of stress granules upon infection, but then subsequently inhibit their formation over the course of infection (Ariumi et al., 2011, McInerney et al., 2005). Rotavirus and cardioviruses induce eIF2α phosphorylation (a canonical pathway for translation inhibition) but additionally inhibit stress granule formation (Borghese and Michiels, 2011, Montero et al., 2008). Infection with poliovirus, a positive-strand RNA virus, also induces the formation of stress granules (Mazroui et al., 2006). One study reported that poliovirus infection initially induced the formation of stress granules, but disrupted their formation as infection progressed (White et al., 2007). G3BP was cleaved by the viral proteinase 3C, and it was proposed that this cleavage led to the dissociation of stress granules during infection. A subsequent report was published suggesting that stress granules form and persist even at later times of poliovirus infection (Piotrowska et al., 2010). This latter study examined the stress granule marker protein TIA-1 and found that TIA-1 positive foci could still be observed over the course of infection, despite lacking G3BP and eIF4G. Sam68, an RNA binding protein that has been shown to interact with the poliovirus RNA-dependent RNA polymerase 3D (Lukong and Richard, 2003, McBride et al., 1996), was also found to be a unique component of poliovirus-induced stress granules. A more recent report suggests that poliovirus infection can induce typical stress granules, which contain TIA-1, translation initiation factors, mRNA, and RNA binding proteins, only early in infection (White and Lloyd, 2011). Later in infection the granules are atypical, and still contain TIA-1 although they lack some of the other constituents usually found in stress granules. Importantly, it is still not understood what role (if any) stress granules play during poliovirus infection.
Poliovirus is an enterovirus and a member of the Picornaviridae family of single-stranded, positive-sense RNA viruses. The genome of ∼7.4 kb can be immediately translated in the cytoplasm of the infected cell via an IRES-mediated, cap-independent mechanism of initiation. Poliovirus translation produces the proteins required for viral RNA synthesis as well as for modulation of the subcellular environment (for review, see Rozovics and Semler, 2010). During infection, viral proteinases 2A and 3C cleave several cellular proteins, including eIF4G and PABP, to down-regulate host cell translation (Etchison et al., 1982, Joachims et al., 1999, Krausslich et al., 1987, Kuyumcu-Martinez et al., 2002, Lloyd et al., 1987). Viral proteinases also cause the inhibition of host cellular transcription (Clark et al., 1991, Clark et al., 1993, Yalamanchili et al., 1997a, Yalamanchili et al., 1997b). In addition, several nuclear pore complex proteins have been shown to be cleaved, which is thought to disrupt specific transport pathways and result in the redistribution of certain host cell nuclear factors (Belov et al., 2000, Belov et al., 2004, Gustin and Sarnow, 2001). Our work has demonstrated that poliovirus infection causes a dramatic nucleo-cytoplasmic re-localization of cellular splicing factor SRp20, an important IRES trans-acting factor, or ITAF, that interacts with PCBP2 and stimulates poliovirus translation (Fitzgerald and Semler, 2011). As recently reported, this re-localization is also observed during coxsackievirus B3 infection or human rhinovirus 16 infection, albeit with decreased efficiency in the latter case (Fitzgerald et al., 2013). Interestingly, both the poliovirus and human rhinovirus 16 2A proteinases, when expressed alone, can direct the cytoplasmic redistribution of SRp20. Taken together, it is clear that picornavirus infections generate an altered cellular environment that contributes to specific steps in virus replication.
Stress granule formation has recently been shown to be among the many subcellular changes induced by poliovirus infection. It is not clear whether poliovirus directs the formation of stress granules, or if these cytoplasmic foci are a type of general cellular stress response to viral infection. It is also not known precisely how stress granules affect poliovirus infection, but expression of an altered version of a key stress granule component (G3BP) that is resistant to cleavage by the poliovirus 3C proteinase resulted in an eight-fold reduction in viral titer (White et al., 2007). Our previous work has demonstrated that SRp20 displays a punctate pattern of localization in the cytoplasm of poliovirus-infected cells, reminiscent of the stress granules observed to develop during infection. In this study we sought to further define the role of stress granule formation during poliovirus infection, with particular focus on the cytoplasmic patterning of SRp20, as well as the effect of stress granules on viral translation. We found that SRp20 partially co-localized with a stress granule marker protein (TIA-1) in the cytoplasm of poliovirus-infected cells at 3 h post-infection. SRp20 was observed to be present in some, but not all, TIA-1 positive cytoplasmic foci that formed. We demonstrated that oxidative stress via arsenite treatment of the cells did not result in SRp20 co-localization with TIA-1. Expression of a dominant negative version of TIA-1 (TIA-1-PRD) was confirmed to disrupt stress granule formation during oxidative stress. However, we observed that with expression of either wild type TIA-1 or dominant negative TIA-1-PRD, levels of poliovirus translation were not dramatically affected. Collectively, these results highlight the unique composition of poliovirus-induced stress granules as part of a myriad of virus-induced host modifications brought about during infection.
2. Methods
2.1. Cell culture and DNA constructs
HeLa cells were grown as monolayers in Dulbecco's Modified Eagle's Media (DMEM) supplemented with 8% newborn calf serum (NCS). SK-N-SH cells (American Type Culture Collection number: HTB-11) were grown in DMEM supplemented with 20% fetal calf serum (FCS). COS cells were grown in DMEM supplemented with 10% FCS. pEGFP and pEGFP-SRp20 plasmids were kindly provided by Dr. Roz Sandri-Goldin. The pMT2, pMT2-HA-TIA-1, and pMT2-HA-TIA-1-PRD plasmids were generously provided by Dr. Paul Anderson (Gilks et al., 2004, Kedersha et al., 2000).
2.2. DNA transfection and virus infection
HeLa cells or SK-N-SH cells were seeded on coverslips and allowed to grow to approximately 70% confluency. Cells were transfected with EGFP-SRp20 (or EGFP) using Fugene transfection reagent (Roche). Twenty-four hours post-transfection, cells were mock-infected or infected with poliovirus at an MOI of 25. At 3 h post-infection, cells were washed once with 1× phosphate-buffered saline (1× PBS) and processed for imaging.
2.3. Arsenite stress experiments
SK-N-SH cells were seeded on coverslips and allowed to grow to approximately 70% confluency. Cells were transfected with either EGFP-SRp20, or with HA-TIA-1 or HA-TIA-1-PRD (or empty vector, not shown). Transfections were carried out using Fugene transfection reagent. Twenty-four hours post-transfection, media was replaced and oxidative stress was induced via the addition of 0.5 mM sodium arsenite (Sigma). Cells were stressed for 30 min or 1 h at 37 °C, washed once with 1× PBS and processed for imaging.
2.4. Fluorescence microscopy and laser scanning confocal microscopy
Following transfection and virus infection or arsenite stress (as described), cells were fixed with 3.7% paraformaldehyde at room temperature for 20 min. Cells were washed with 1× PBS twice, and cell membranes were permeabilized with 0.5% NP-40 in PBS for 5 min. Cells for initial SRp20 cytoplasmic granule examination were washed with 1% NCS in PBS and incubated with DAPI to stain nuclei. For the rest of the imaging experiments, cells were washed with 1% NCS in PBS and incubated with normal donkey serum (Jackson Immunoresearch) for 1 h to block nonspecific interactions. Cells were then incubated with an anti-TIA-1 (kindly provided by Dr. Kurt Gustin) or anti-HA (Sigma) monoclonal antibody for 1 h. Cells were washed and incubated with AlexaFluor594 donkey anti-goat or AlexaFluor594 goat anti-mouse secondary antibody (Molecular Probes), respectively, for 45 min. Cells were washed with 1% NCS-PBS and incubated with DAPI to stain nuclei. Following DAPI incubation, all cells were washed with PBS; coverslips were mounted on microscope slides with mounting media (Biomeda) and allowed to dry overnight at room temperature. Coverslips were sealed with nail polish, and cells were imaged using a Zeiss Axiovert 200 inverted fluorescence microscope or a Zeiss LSM 510 multi-photon laser scanning confocal microscope. Images were processed using the Axiovision software or the LSM 510 software, respectively.
2.5. Poliovirus infection and [35S]-methionine-labeling of viral proteins
COS cells were seeded and allowed to grow to approximately 70% confluency. Cells were transfected with pMT2, HA-TIA-1, or HA-TIA-1-PRD using Lipofectamine transfection reagent (Invitrogen). Twenty-four hours post-transfection, cells were mock-infected or infected with poliovirus at an MOI of 25 in methionine-free media (MP Biomedicals). After 2 h of methionine starvation, 23 μCi of [35S]-Met (PerkinElmer) was added to each well and infection was carried out for an additional 6 h. Cells were harvested every 2 h from the beginning of infection, from 0 to 8 h post-infection. Mock-infected cells were collected at 8 h post-infection. At each time point, cells were washed with 1× PBS and harvested by scraping. Cells were pelleted and lysed in RIPA buffer (50 mM Tris–HCl pH 7.4, 150 mM NaCl, 1 mM EDTA, 5 μg/ml Aprotinin, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS) on ice for 10 min. Debris was pelleted by centrifugation at 16,000 × g for 20 min, and equal volumes of cleared lysate were subjected to SDS-PAGE. Radiolabeled proteins were visualized by autoradiography.
3. Results
3.1. SRp20 partially co-localizes with a stress granule marker protein during poliovirus infection
We have previously demonstrated the dramatic nucleo-cytoplasmic re-localization of SRp20 during poliovirus infection, in both neuroblastoma cells (SK-N-SH) and HeLa cells (Fitzgerald and Semler, 2011). Beginning around 3 h post-infection, we also observed that SRp20 displayed a readily identifiable punctate pattern of localization in the cytoplasm of infected cells, reminiscent of stress granules that form under certain conditions of cellular stress. We observed this cytoplasmic punctate pattern in SK-N-SH cells (Fig. 1A) as well as in HeLa cells (Fig. 1B) visualized by expression of GFP-tagged SRp20. We have previously demonstrated that overexpression of GFP-SRp20 does not affect the kinetics of poliovirus infection (Fitzgerald and Semler, 2011). We have also carried out experiments using a monoclonal antibody to label endogenous SRp20, and we found that the endogenous protein re-localizes from the nucleus to the cytoplasm during poliovirus infection (data not shown). Transfection of the GFP-tagged version of the protein provided the advantage of a consistent signal from the GFP fluorescence, in contrast to labeling SRp20 with the monoclonal antibody, which resulted in a higher variability of the signal obtained. GFP-SRp20 has been previously shown to localize and function similarly to endogenous SRp20 (Escudero-Paunetto et al., 2010). While SRp20 was localized throughout the cytoplasm, a portion of the protein could be seen forming granule-like clusters. We investigated whether these cytoplasmic puncta also contained the RNA binding protein TIA-1, a protein that appears to be critical for stress granule formation and is known to localize to stress granules during poliovirus infection (Piotrowska et al., 2010, White and Lloyd, 2011). We transfected SK-N-SH cells with a GFP-SRp20 construct (or GFP empty vector), and subsequently infected the cells with poliovirus for 3 h (the time point during infection at which SRp20 cytoplasmic granules could first be detected). We used SK-N-SH cells for these experiments because they have a more distinct cytoplasmic space compared to other cell types (e.g., HeLa cells). Cells were labeled with an anti-TIA-1 antibody to identify stress granules and then analyzed for co-localization of SRp20 and TIA-1. The results of these experiments are shown in Fig. 2 , which displays the localization of SRp20 and TIA-1 in cells that were mock-infected (top panels) or poliovirus-infected (bottom panels). As we have shown previously, SRp20 was predominantly nuclear in mock-infected cells but re-localized to the cytoplasm during poliovirus infection, where a punctate pattern of localization could also be observed (Fitzgerald and Semler, 2011). TIA-1 was found in both the nucleus and the cytoplasm of mock- and poliovirus-infected cells, but during infection we observed TIA-1 positive puncta in the cytoplasm, consistent with stress granule formation. These TIA-1 containing stress granules were observed with or without the expression of GFP-SRp20 (data not shown; also refer to Fig. 2B). Interestingly, we observed co-localization of SRp20 and TIA-1 in the cytoplasm of cells during poliovirus infection at 3 h post-infection (see the merged panels in Fig. 2A; co-localization denoted with white arrows). Additional experiments confirmed the co-localization of SRp20 and TIA-1 in some, but not all, TIA-1 positive cytoplasmic foci (data not shown). As a control, in Fig. 2B we demonstrated that GFP alone did not co-localize with TIA-1 in either mock- or poliovirus-infected cells (although stress granules were still observed during infection; bottom panels), indicating that the GFP tag alone did not contribute to the localization of SRp20. Therefore, SRp20 and TIA-1 can be visualized in very close proximity to each other in some of the TIA-1-containing cytoplasmic stress granules that form during poliovirus-infection.
Fig. 1.
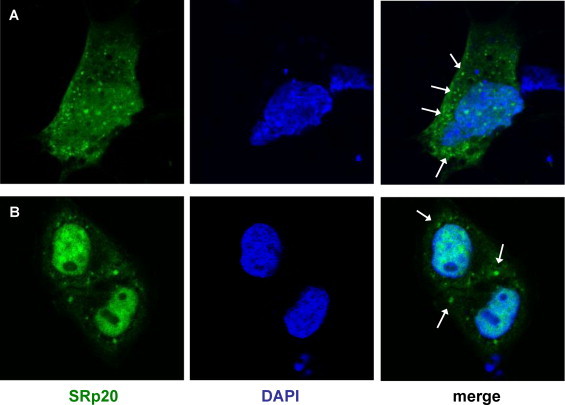
SRp20 punctate pattern of localization during poliovirus infection. SK-N-SH cells (A) or HeLa cells (B) were transfected with GFP-SRp20 and infected with poliovirus at an MOI of 25. Cells were fixed at 3 h post-infection and imaged. SRp20 localization was determined using confocal microscopy; nuclei were identified by DAPI staining. The cytoplasmic granules observed are indicated with white arrows.
Fig. 2.
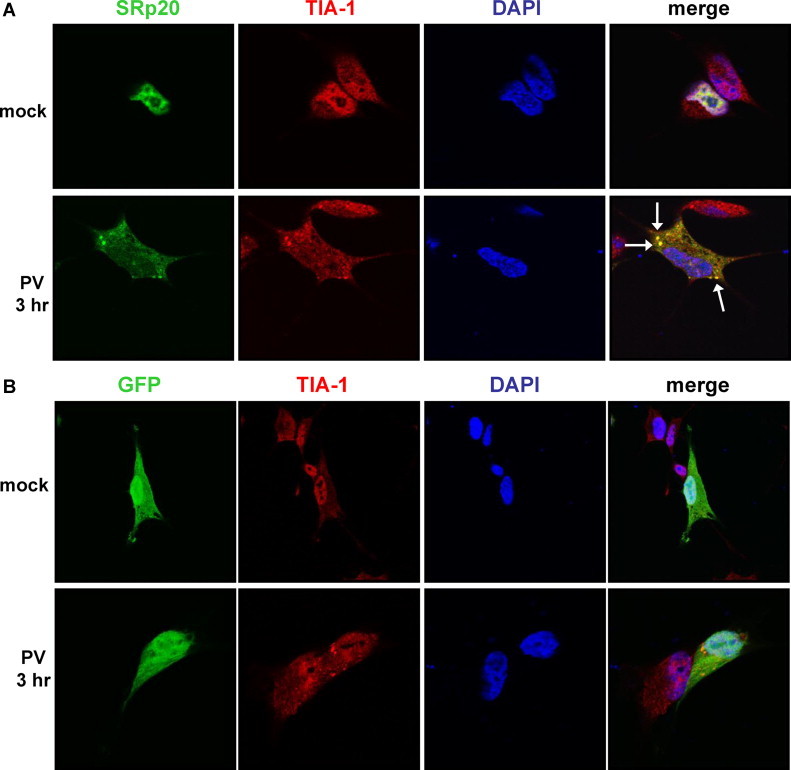
SRp20 partial co-localization with a stress granule marker protein during poliovirus infection. SK-N-SH cells were transfected with GFP-SRp20 (A) or GFP empty vector (B) and either mock-infected (top panels in A and B) or infected with poliovirus (bottom panels in A and B) at an MOI of 25. At 3 h post-infection cells were fixed and incubated with an anti-TIA-1 monoclonal antibody, and subsequently AlexaFluor594 donkey anti-goat secondary antibody. Cells were examined for co-localization of TIA-1 with SRp20 (shown in A, the merged image in yellow and highlighted by white arrows) or with GFP (B) using confocal microscopy; nuclei were identified by DAPI staining. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of the article.)
3.2. Oxidative stress via arsenite treatment of cells causes TIA-1 positive foci formation but does not result in co-localization of SRp20 and TIA-1
We next investigated if SRp20 could be observed to co-localize, at least in part, with TIA-1 during other conditions of cellular stress in addition to virus infection. Our goal was to determine whether the localization of SRp20 to stress granules was potentially a general response to stress within the cells. For these experiments, we transfected SK-N-SH cells with GFP-SRp20 and subsequently treated the cells with sodium arsenite, which induces oxidative stress. We then labeled the cells with anti-TIA-1 antibody to detect arsenite-induced stress granules and determined the localization of SRp20 in mock- and arsenite-treated cells. Oxidative stress was carried out for 30 min (Fig. 3A) or for 1 h (Fig. 3B) prior to cell fixation, immuno-labeling, and fluorescence imaging. In the mock-treated cells (top panels in Fig. 3A and B), TIA-1 was localized in both the nucleus and the cytoplasm without any distinct subcellular pattern of localization. However, after 30 min of stress treatment, TIA-1-containing stress granules were apparent in the cytoplasm of the cells (bottom panels in Fig. 3A), and were even more pronounced after 1 h of stress (bottom panels in Fig. 3B). Despite the readily detectable cytoplasmic stress granules, SRp20 remained localized in the nucleus of the arsenite-stressed cells and did not co-localize with TIA-1 (see merged panels in Fig. 3A and B). Thus, SRp20 does not localize to TIA-1-containing stress granules formed during conditions of oxidative stress. Taken together, we conclude that the partial co-localization of SRp20 and TIA-1 observed during poliovirus infection does not represent a general response to cellular stress.
Fig. 3.
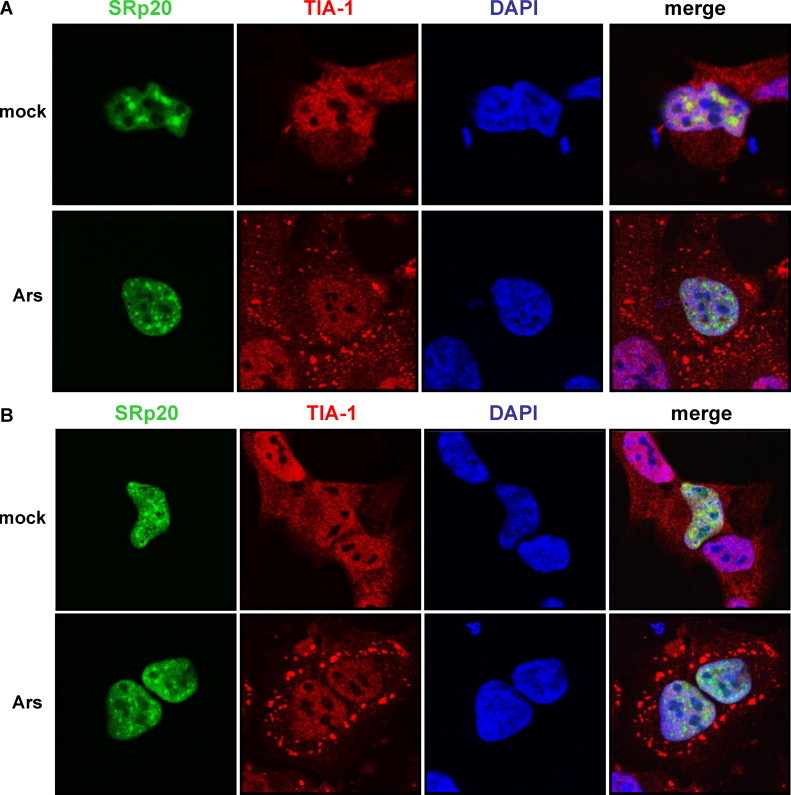
Effect of oxidative stress on the co-localization of SRp20 and TIA-1. SK-N-SH cells were transfected with GFP-SRp20 and either mock-stressed (top panels in A and B) or stressed with sodium arsenite (final concentration 0.5 mM; bottom panels in A and B) for 30 min (A) or 1 h (B) at 37 °C. Cells were washed and fixed, incubated with an anti-TIA-1 monoclonal antibody, and subsequently AlexaFluor594 donkey anti-goat secondary antibody. Cells were examined for stress granule formation and TIA-1/SRp20 localization using confocal microscopy; nuclei were identified by DAPI staining.
3.3. Expression of wild type or dominant negative TIA-1 does not dramatically stimulate or inhibit poliovirus translation
Because poliovirus infection induces the formation of stress granules, we investigated the effect of disrupting these cytoplasmic foci specifically on viral translation. If a portion of the cytoplasmic SRp20 is aggregated in stress granules during infection, it may be unavailable for viral translation. We hypothesized that disruption of stress granules during infection may boost levels of poliovirus translation, as more SRp20 is made available to the virus. To disrupt stress granule formation, we utilized a construct encoding a dominant negative version of TIA-1, termed TIA-1-PRD (Gilks et al., 2004, Kedersha et al., 2000). This protein contains only the C-terminal domain of TIA-1 and its over-expression has been previously shown to disrupt stress granule formation during arsenite stress (Gilks et al., 2004) as well as during poliovirus infection (Piotrowska et al., 2010). We first validated the effect of expressing either wild type TIA-1 or dominant negative TIA-1-PRD on stress granule formation, utilizing arsenite stress to induce stress granules in the cells. SK-N-SH cells were transfected with HA-tagged wild type TIA-1 or TIA-1-PRD (or vector only, data not shown) and subsequently mock- or arsenite-treated for 30 min. Cells were fixed and incubated with an HA antibody to label the wild type or dominant negative TIA-1 proteins, and then processed for imaging. The results are shown in Fig. 4 . Wild type HA-tagged TIA-1 was localized in both the nucleus and the cytoplasm in mock-treated cells (Fig. 4A, top panels). Upon arsenite treatment, the HA-TIA-1 formed cytoplasmic foci as expected (Fig. 4A, bottom panels). Dominant negative HA-tagged TIA-1-PRD was localized predominantly in the cytoplasm of mock-treated cells and formed nonspecific aggregates, consistent with published data for this protein (Fig. 4B, top panels). After arsenite treatment, HA-TIA-1-PRD was dispersed throughout the cytoplasm, and no authentic stress granule formation was observed (Fig. 4B, bottom panels).
Fig. 4.
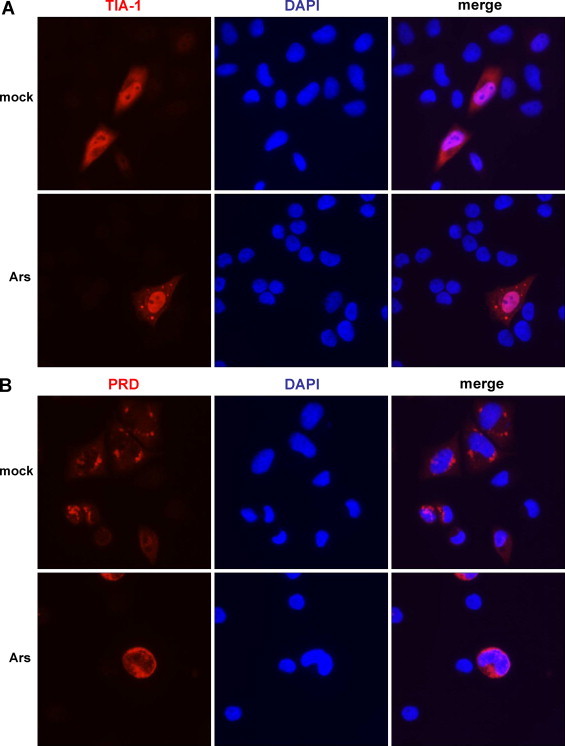
Stress granule formation following expression of a dominant negative version of TIA-1. SK-N-SH cells were transfected with HA-tagged TIA-1 or TIA-1-PRD (or empty vector, not shown) and either mock-stressed (top panels in A and B) or stressed with sodium arsenite (final concentration 0.5 mM; bottom panels in A and B) for 30 min at 37 °C. Cells were washed and fixed, incubated with an anti-HA monoclonal antibody, and subsequently AlexaFluor594 goat anti-mouse secondary antibody. Cells were examined for the formation of stress granules via fluorescence microscopy; nuclei were identified by DAPI staining.
We next determined the effect of expressing wild type or dominant negative TIA-1 (or vector alone) on poliovirus translation via [35S]-methionine-labeling of viral proteins during infection. These experiments were carried out in COS cells, a cell line permissive for poliovirus infection with virus growth kinetics similar to HeLa cells, often utilized due to high efficiency uptake and replication of plasmid DNA following transfection (Gluzman, 1981, Mellon et al., 1981). In addition, previously published work has demonstrated the induction of stress granules in COS cells during poliovirus infection, as well as the disruption of stress granules during infection with expression of TIA-1-PRD in this cell type (Piotrowska et al., 2010). We transfected COS cells with HA-TIA-1 or HA-TIA-1-PRD (or empty vector), and then mock-infected cells or infected cells with poliovirus at an MOI of 25 in methionine-free media. After 2 h of methionine starvation, [35S]-methionine was added to radiolabel the viral proteins produced. Infections were carried out for 8 h total, and every 2 h cells were harvested and lysed. Equal volumes of lysates were subjected to SDS-PAGE, and labeled proteins were analyzed via autoradiography. The results are shown in Fig. 5 . Of note, the expression vectors utilized in these experiments allowed for continual production of their encoded products (indicated by the asterisks) during the course of poliovirus infection. Compared to empty vector (Fig. 5A, lanes 2–7), expression of wild type TIA-1 appeared to just slightly decrease the levels of viral proteins produced (lanes 8–13), although it did not dramatically inhibit viral translation. Expression of the dominant negative TIA-1-PRD appeared to have only a slight stimulatory effect on viral translation (Fig. 5B, lanes 8–13) compared to empty vector (lanes 2–7). Thus, in the transfection/infection assays described here, neither wild type TIA-1 nor dominant negative TIA-1-PRD expression resulted in a dramatic difference in poliovirus translation.
Fig. 5.
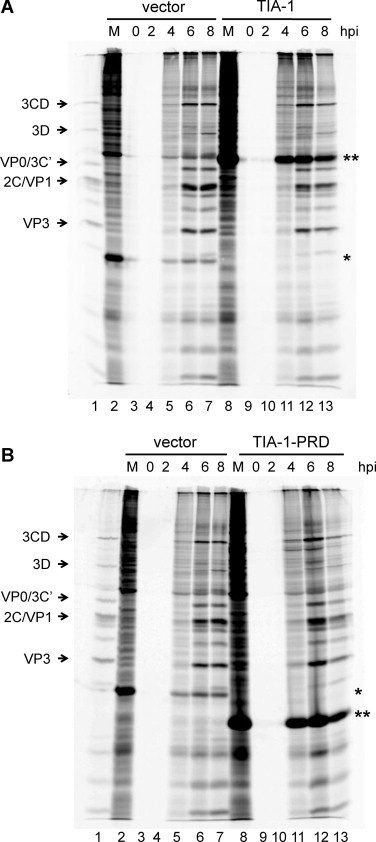
Effect of TIA-1 or TIA-1-PRD expression on poliovirus translation. COS cells were transfected with HA-TIA-1 (A) or HA-TIA-1-PRD (B), or the pMT2 empty vector (A and B) as a control, and either mock-infected or infected with poliovirus at an MOI of 25. After 2 h of methionine starvation, 23 μCi of [35S]-methionine (PerkinElmer) was added and infection was carried out an additional 6 h to radiolabel viral proteins. Cells were harvested every 2 h from the beginning of infection, from 0 to 8 h post-infection (hpi). Lysates were generated and subjected to SDS-PAGE; radiolabeled proteins were visualized by autoradiography. The migration of different viral proteins is indicated to the left of the autoradiograph. (A) Lane 1, radiolabeled poliovirus proteins from an infection of HeLa cells as a marker; lane 2, cells transfected with pMT2 (vector alone) and mock-infected (M); lanes 3–7, cells transfected with pMT2 (vector alone) and poliovirus-infected for 0, 2, 4, 6, and 8 h; lane 8, cells transfected with TIA-1 expression vector and mock-infected (M); lanes 9–13, cells transfected with TIA-1 expression vector and poliovirus-infected for 0, 2, 4, 6, and 8 h. (*) indicates dihydrofolate reductase, encoded in the empty vector; (**) indicates HA-TIA-1. (B) Lane 1, poliovirus radiolabeled in vivo marker; lane 2, cells transfected with pMT2 (vector alone) and mock-infected (M); lanes 3–7, cells transfected with pMT2 (vector alone) and poliovirus-infected for 0, 2, 4, 6, and 8 h; lane 8, cells transfected with TIA-1-PRD expression vector and mock-infected (M); lanes 9–13, cells transfected with TIA-1-PRD expression vector and poliovirus-infected for 0, 2, 4, 6, and 8 h. (*) indicates dihydrofolate reductase, encoded in the empty vector; (**) indicates HA-TIA-1-PRD. It should be noted that lanes 3,4 and 9,10 in the gel images are blank because cells were initially infected with poliovirus in methionine-free media. Methionine starvation was carried out for 2 h prior to the addition of 35S-methionine to the media. Since viral proteins are radiolabeled with 35S-methionine for visualization, cells harvested during the methionine starvation period (up to 2 h post-infection) will not contain any radiolabeled proteins.
4. Discussion
Certain conditions within cells can generate dramatic alterations in the intracellular environment. Beyond the changes that occur during development and cell cycling, conditions of cellular stress can affect the translation of mRNAs. Many RNAs are translationally silenced and/or degraded during stress conditions, while others are up-regulated in response to stress. Some of these conditions include hypoxia, oxidative stress, heat shock, as well as virus infection. Cellular stressors can induce the formation of cytoplasmic stress granules, which are foci considered to be a temporary holding place for mRNAs that are not being actively translated, but are not targeted for degradation. These foci contain the specific proteins required for their formation, stalled translation complexes, and mRNPs. Recent studies have identified unique stress granules that form during poliovirus infection (Piotrowska et al., 2010, White et al., 2007, White and Lloyd, 2011). It has been shown that stress granules formed early in infection contain some proteins and mRNAs that are common to stress granules that form during other conditions of stress. While the foci persist throughout infection, the stress granules found later in infection appear compositionally distinct, although they consistently contain the RNA binding protein TIA-1 (Piotrowska et al., 2010, White and Lloyd, 2011). For this reason, TIA-1 is an appropriate marker protein to identify the stress granules that form under many conditions of stress, including poliovirus infection.
During our initial studies of the re-localization of SRp20 during poliovirus infection, we observed a punctate pattern of localization for this protein in the cytoplasm of cells beginning around 3 h post-infection. We considered the possibility that this signal represented the presence of SRp20 in poliovirus-induced stress granules and investigated whether SRp20 co-localized with TIA-1, a protein that appears to be required for stress granule assembly. We determined that SRp20 co-localized with TIA-1 in some, but not all, TIA-1 positive foci at 3 h post-infection. The partial co-localization with TIA-1 suggests that SRp20 is found in some of the unique stress granules that form during poliovirus infection. It is not clear why SRp20 is only localized to a subset of TIA-1-containing stress granules, and this would suggest that even the unique virally induced granules that form are not all compositionally uniform. Indeed, stress granules in general are considered to be transient, dynamic accumulations of different proteins and mRNAs. The SRp20-containing granules may represent accumulations of proteins that function in a manner that is distinct from the foci that do not contain SRp20. Additional studies are necessary to investigate other proteins that co-localize with SRp20 and TIA-1 within these cytoplasmic foci during infection and to examine whether viral proteins and viral RNA are present in these foci.
If SRp20 localizes to stress granules during poliovirus infection, we hypothesized that this localization may be a general response to cellular stress. We investigated whether SRp20 co-localized with TIA-1 during other conditions of stress, by examining the stress granules that form in cells during oxidative stress (i.e., following arsenite treatment). We found that SRp20 did not co-localize with TIA-1 in any of the stress granules induced upon arsenite treatment. We interpret these results to suggest that stress alone is not sufficient to cause the co-localization of TIA-1 and SRp20. It is becoming increasingly apparent that different stress conditions direct the assembly of foci that are inherently different and partially dependent on the specific stress that is applied. Indeed, poliovirus infection provides another example for differences that exist in stress granule composition. G3BP (Ras-GAP SH3 domain binding protein) is cleaved by the viral proteinase 3C during infection, and no longer localizes to stress granules; however, TIA-1-containing stress granules still persist during poliovirus infection (Piotrowska et al., 2010, White et al., 2007, White and Lloyd, 2011). G3BP is thought to be required for stress granule assembly and is consistently localized to stress granules formed under other conditions of stress. We also take into account that poliovirus infection directs the nucleo-cytoplasmic redistribution of some nuclear proteins, including SRp20 (Belov et al., 2004, Fitzgerald et al., 2013, Fitzgerald and Semler, 2011, Gustin and Sarnow, 2001). Arsenite stress alone did not induce any significant re-localization of SRp20 to the cytoplasm of stressed cells, and this may contribute to the lack of SRp20 stress granule localization. Although some portion of cellular SRp20 shuttles continuously between the nucleus and the cytoplasm (Caceres et al., 1998), stress granule association (or potentially the visualization of this association) may require a greater abundance of cytoplasmic SRp20. It is not clear whether cytoplasmic accumulation of SRp20 is required for its partial co-localization with TIA-1. However, our confocal images do not show extensive co-localization of SRp20 and TIA-1 in the nuclei of uninfected or arsenite-treated cells (refer to Fig. 3).
A key unanswered question is whether stress granule formation is actively directed by poliovirus infection, or if the foci formation is a cellular response to the virus infection. For example, cells may respond to poliovirus infection by directing stress granule formation in an attempt to globally down-regulate translation. It is also not clear if stress granules aid or impede the viral infection itself. Published studies have not specifically determined the effect of disrupting stress granule formation on poliovirus RNA replication, translation, or overall growth kinetics. One study examined the effect of maintaining an uncleavable version of G3BP in virus-induced stress granules and found an eight-fold decrease in virus yield (White et al., 2007). The basis for this decrease remains to be determined, because G3BP is not maintained in stress granules over the course of poliovirus infection. To this end, we investigated whether disrupting stress granules had any effect on poliovirus translation during infection. A recent study demonstrated that expression of the C-terminal domain of TIA-1 (TIA-1-PRD) interferes with the formation of stress granules during poliovirus infection (Piotrowska et al., 2010), but no functional studies were presented. Our results suggested that expression of wild type TIA-1 or the dominant negative TIA-1-PRD had no significant effect on poliovirus IRES-mediated translation. While our transfection efficiencies were estimated at around 70% in COS cells (data not shown), it is possible that some deleterious effects on translation were masked by the population of untransfected cells. This potential shortcoming could be addressed by electroporation of cells as an alternative to transfection or by FACS sorting of transfected cells using the HA tag prior to poliovirus infection.
Overall, it remains to be determined what role, if any, stress granule formation plays during poliovirus infection or why SRp20, an important ITAF for poliovirus IRES-mediated translation, is partially localized to these cytoplasmic foci during infection. Considering that stress granules typically contain stalled translation complexes, 40S ribosomal subunits, and other canonical translation initiation factors and RNA binding proteins, it is tempting to speculate that stress granule formation during poliovirus infection may actually contribute to a temporal shift from high levels of viral translation to increasing levels of RNA replication. Such a shift has been previously ascribed, in part, to viral-mediated cleavage of ITAFs during the course of infection (Back et al., 2002, Perera et al., 2007).
Acknowledgements
We are grateful to Andrea Cathcart for critical comments on the manuscript and to Kurt Gustin for insightful discussions. We thank Rozanne Sandri-Goldin for providing the GFP-SRp20 expression plasmid, Paul Anderson for providing TIA-1 expression plasmids, and Kurt Gustin for the gift of antibodies to TIA-1. Confocal images were generated with assistance from Adeela Syed at the UCI Optical Biology Core, which is supported by Comprehensive Cancer Center award P30CA062203 from the National Cancer Institute, and at the UCI Stem Cell Research Center, with assistance from Marek Mandau. K.D.F. was a predoctoral trainee of Public Health Service training grant AI 07319. BLS was a Senior Investigator of the American Asthma Foundation. This work was supported by Public Health Service Grant AI 26765 from the National Institutes of Health.
References
- Anderson P., Kedersha N. Visibly stressed: the role of eIF2, TIA-1, and stress granules in protein translation. Cell Stress and Chaperones. 2002;7:213–221. doi: 10.1379/1466-1268(2002)007<0213:vstroe>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson P., Kedersha N. Stress granules: the Tao of RNA triage. Trends in Biochemical Sciences. 2008;33:141–150. doi: 10.1016/j.tibs.2007.12.003. [DOI] [PubMed] [Google Scholar]
- Ariumi Y., Kuroki M., Kushima Y., Osugi K., Hijikata M., Maki M., Ikeda M., Kato N. Hepatitis C virus hijacks P-body and stress granule components around lipid droplets. Journal of Virology. 2011;85:6882–6892. doi: 10.1128/JVI.02418-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Back S.H., Kim Y.K., Kim W.J., Cho S., Oh H.R., Kim J.E., Jang S.K. Translation of polioviral mRNA is inhibited by cleavage of polypyrimidine tract-binding proteins executed by polioviral 3C(pro) Journal of Virology. 2002;76:2529–2542. doi: 10.1128/jvi.76.5.2529-2542.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belov G.A., Evstafieva A.G., Rubtsov Y.P., Mikitas O.V., Vartapetian A.B., Agol V.I. Early alteration of nucleocytoplasmic traffic induced by some RNA viruses. Virology. 2000;275:244–248. doi: 10.1006/viro.2000.0427. [DOI] [PubMed] [Google Scholar]
- Belov G.A., Lidsky P.V., Mikitas O.V., Egger D., Lukyanov K.A., Bienz K., Agol V.I. Bidirectional increase in permeability of nuclear envelope upon poliovirus infection and accompanying alterations of nuclear pores. Journal of Virology. 2004;78:10166–10177. doi: 10.1128/JVI.78.18.10166-10177.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borghese F., Michiels T. The leader protein of cardioviruses inhibits stress granule assembly. Journal of Virology. 2011;85:9614–9622. doi: 10.1128/JVI.00480-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caceres J.F., Screaton G.R., Krainer A.R. A specific subset of SR proteins shuttles continuously between the nucleus and the cytoplasm. Genes and Development. 1998;12:55–66. doi: 10.1101/gad.12.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernov K.G., Barbet A., Hamon L., Ovchinnikov L.P., Curmi P.A., Pastre D. Role of microtubules in stress granule assembly: microtubule dynamical instability favors the formation of micrometric stress granules in cells. Journal of Biological Chemistry. 2009;284:36569–36580. doi: 10.1074/jbc.M109.042879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark M.E., Hammerle T., Wimmer E., Dasgupta A. Poliovirus proteinase 3C converts an active form of transcription factor IIIC to an inactive form: a mechanism for inhibition of host cell polymerase III transcription by poliovirus. EMBO Journal. 1991;10:2941–2947. doi: 10.1002/j.1460-2075.1991.tb07844.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark M.E., Lieberman P.M., Berk A.J., Dasgupta A. Direct cleavage of human TATA-binding protein by poliovirus protease 3C in vivo and in vitro. Molecular and Cellular Biology. 1993;13:1232–1237. doi: 10.1128/mcb.13.2.1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escudero-Paunetto L., Li L., Hernandez F.P., Sandri-Goldin R.M. SR proteins SRp20 and 9G8 contribute to efficient export of herpes simplex virus 1 mRNAs. Virology. 2010;401:155–164. doi: 10.1016/j.virol.2010.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etchison D., Milburn S.C., Edery I., Sonenberg N., Hershey J.W. Inhibition of HeLa cell protein synthesis following poliovirus infection correlates with the proteolysis of a 220,000-dalton polypeptide associated with eucaryotic initiation factor 3 and a cap binding protein complex. Journal of Biological Chemistry. 1982;257:14806–14810. [PubMed] [Google Scholar]
- Fitzgerald K.D., Chase A.J., Cathcart A.L., Tran G.P., Semler B.L. Viral proteinase requirements for the nucleocytoplasmic relocalization of cellular splicing factor SRp20 during picornavirus infections. Journal of Virology. 2013;87:2390–2400. doi: 10.1128/JVI.02396-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald K.D., Semler B.L. Re-localization of cellular protein SRp20 during poliovirus infection: bridging a viral IRES to the host cell translation apparatus. PLoS Pathogens. 2011;7:e1002127. doi: 10.1371/journal.ppat.1002127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilks N., Kedersha N., Ayodele M., Shen L., Stoecklin G., Dember L.M., Anderson P. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Molecular Biology of the Cell. 2004;15:5383–5398. doi: 10.1091/mbc.E04-08-0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gluzman Y. SV40-transformed simian cells support the replication of early SV40 mutants. Cell. 1981;23:175–182. doi: 10.1016/0092-8674(81)90282-8. [DOI] [PubMed] [Google Scholar]
- Gustin K.E., Sarnow P. Effects of poliovirus infection on nucleo-cytoplasmic trafficking and nuclear pore complex composition. EMBO Journal. 2001;20:240–249. doi: 10.1093/emboj/20.1.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov P.A., Chudinova E.M., Nadezhdina E.S. Disruption of microtubules inhibits cytoplasmic ribonucleoprotein stress granule formation. Experimental Cell Research. 2003;290:227–233. doi: 10.1016/s0014-4827(03)00290-8. [DOI] [PubMed] [Google Scholar]
- Joachims M., Van Breugel P.C., Lloyd R.E. Cleavage of poly(A)-binding protein by enterovirus proteases concurrent with inhibition of translation in vitro. Journal of Virology. 1999;73:718–727. doi: 10.1128/jvi.73.1.718-727.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N., Anderson P. Stress granules: sites of mRNA triage that regulate mRNA stability and translatability. Biochemical Society Transactions. 2002;30:963–969. doi: 10.1042/bst0300963. [DOI] [PubMed] [Google Scholar]
- Kedersha N., Cho M.R., Li W., Yacono P.W., Chen S., Gilks N., Golan D.E., Anderson P. Dynamic shuttling of TIA-1 accompanies the recruitment of mRNA to mammalian stress granules. Journal of Cell Biology. 2000;151:1257–1268. doi: 10.1083/jcb.151.6.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N., Stoecklin G., Ayodele M., Yacono P., Lykke-Andersen J., Fritzler M.J., Scheuner D., Kaufman R.J., Golan D.E., Anderson P. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. Journal of Cell Biology. 2005;169:871–884. doi: 10.1083/jcb.200502088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N.L., Gupta M., Li W., Miller I., Anderson P. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. Journal of Cell Biology. 1999;147:1431–1442. doi: 10.1083/jcb.147.7.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krausslich H.G., Nicklin M.J., Toyoda H., Etchison D., Wimmer E. Poliovirus proteinase 2A induces cleavage of eucaryotic initiation factor 4F polypeptide p220. Journal of Virology. 1987;61:2711–2718. doi: 10.1128/jvi.61.9.2711-2718.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuyumcu-Martinez N.M., Joachims M., Lloyd R.E. Efficient cleavage of ribosome-associated poly(A)-binding protein by enterovirus 3C protease. Journal of Virology. 2002;76:2062–2074. doi: 10.1128/jvi.76.5.2062-2074.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindquist M.E., Lifland A.W., Utley T.J., Santangelo P.J., Crowe J.E., Jr. Respiratory syncytial virus induces host RNA stress granules to facilitate viral replication. Journal of Virology. 2010;84:12274–12284. doi: 10.1128/JVI.00260-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd R.E., Jense H.G., Ehrenfeld E. Restriction of translation of capped mRNA in vitro as a model for poliovirus-induced inhibition of host cell protein synthesis: relationship to p220 cleavage. Journal of Virology. 1987;61:2480–2488. doi: 10.1128/jvi.61.8.2480-2488.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loschi M., Leishman C.C., Berardone N., Boccaccio G.L. Dynein and kinesin regulate stress-granule and P-body dynamics. Journal of Cell Science. 2009;122:3973–3982. doi: 10.1242/jcs.051383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukong K.E., Richard S. Sam68, the KH domain-containing superSTAR. Biochimica et Biophysica Acta. 2003;1653:73–86. doi: 10.1016/j.bbcan.2003.09.001. [DOI] [PubMed] [Google Scholar]
- Mazroui R., Sukarieh R., Bordeleau M.E., Kaufman R.J., Northcote P., Tanaka J., Gallouzi I., Pelletier J. Inhibition of ribosome recruitment induces stress granule formation independently of eukaryotic initiation factor 2alpha phosphorylation. Molecular Biology of the Cell. 2006;17:4212–4219. doi: 10.1091/mbc.E06-04-0318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride A.E., Schlegel A., Kirkegaard K. Human protein Sam68 relocalization and interaction with poliovirus RNA polymerase in infected cells. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:2296–2301. doi: 10.1073/pnas.93.6.2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInerney G.M., Kedersha N.L., Kaufman R.J., Anderson P., Liljestrom P. Importance of eIF2alpha phosphorylation and stress granule assembly in alphavirus translation regulation. Molecular Biology of the Cell. 2005;16:3753–3763. doi: 10.1091/mbc.E05-02-0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellon P., Parker V., Gluzman Y., Maniatis T. Identification of DNA sequences required for transcription of the human alpha 1-globin gene in a new SV40 host-vector system. Cell. 1981;27:279–288. doi: 10.1016/0092-8674(81)90411-6. [DOI] [PubMed] [Google Scholar]
- Montero H., Rojas M., Arias C.F., Lopez S. Rotavirus infection induces the phosphorylation of eIF2alpha but prevents the formation of stress granules. Journal of Virology. 2008;82:1496–1504. doi: 10.1128/JVI.01779-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera R., Daijogo S., Walter B.L., Nguyen J.H., Semler B.L. Cellular protein modification by poliovirus: the two faces of poly(rC)-binding protein. Journal of Virology. 2007;81:8919–8932. doi: 10.1128/JVI.01013-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piotrowska J., Hansen S.J., Park N., Jamka K., Sarnow P., Gustin K.E. Stable formation of compositionally unique stress granules in virus-infected cells. Journal of Virology. 2010;84:3654–3665. doi: 10.1128/JVI.01320-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raaben M., Groot Koerkamp M.J., Rottier P.J., de Haan C.A. Mouse hepatitis coronavirus replication induces host translational shutoff and mRNA decay, with concomitant formation of stress granules and processing bodies. Cellular Microbiology. 2007;9:2218–2229. doi: 10.1111/j.1462-5822.2007.00951.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozovics J.M., Semler B.L. Genome replication I: the players. In: Ehrenfeld E., Domingo E., Roos R.P., editors. The Picornaviruses. ASM Press; Washington, DC: 2010. pp. 107–125. [Google Scholar]
- Smith J.A., Schmechel S.C., Raghavan A., Abelson M., Reilly C., Katze M.G., Kaufman R.J., Bohjanen P.R., Schiff L.A. Reovirus induces and benefits from an integrated cellular stress response. Journal of Virology. 2006;80:2019–2033. doi: 10.1128/JVI.80.4.2019-2033.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sola I., Galan C., Mateos-Gomez P.A., Palacio L., Zuniga S., Cruz J.L., Almazan F., Enjuanes L. The polypyrimidine tract-binding protein affects coronavirus RNA accumulation levels and relocalizes viral RNAs to novel cytoplasmic domains different from replication-transcription sites. Journal of Virology. 2011;85:5136–5149. doi: 10.1128/JVI.00195-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tourriere H., Chebli K., Zekri L., Courselaud B., Blanchard J.M., Bertrand E., Tazi J. The RasGAP-associated endoribonuclease G3BP assembles stress granules. Journal of Cell Biology. 2003;160:823–831. doi: 10.1083/jcb.200212128. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Tsai N.P., Tsui Y.C., Wei L.N. Dynein motor contributes to stress granule dynamics in primary neurons. Neuroscience. 2009;159:647–656. doi: 10.1016/j.neuroscience.2008.12.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J.P., Cardenas A.M., Marissen W.E., Lloyd R.E. Inhibition of cytoplasmic mRNA stress granule formation by a viral proteinase. Cell Host & Microbe. 2007;2:295–305. doi: 10.1016/j.chom.2007.08.006. [DOI] [PubMed] [Google Scholar]
- White J.P., Lloyd R.E. Poliovirus unlinks TIA1 aggregation and mRNA stress granule formation. Journal of Virology. 2011;85:12442–12454. doi: 10.1128/JVI.05888-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J.P., Lloyd R.E. Regulation of stress granules in virus systems. Trends in Microbiology. 2012;20:175–183. doi: 10.1016/j.tim.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalamanchili P., Banerjee R., Dasgupta A. Poliovirus-encoded protease 2APro cleaves the TATA-binding protein but does not inhibit host cell RNA polymerase II transcription in vitro. Journal of Virology. 1997;71:6881–6886. doi: 10.1128/jvi.71.9.6881-6886.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalamanchili P., Datta U., Dasgupta A. Inhibition of host cell transcription by poliovirus: cleavage of transcription factor CREB by poliovirus-encoded protease 3Cpro. Journal of Virology. 1997;71:1220–1226. doi: 10.1128/jvi.71.2.1220-1226.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurla C., Lifland A.W., Santangelo P.J. Characterizing mRNA interactions with RNA granules during translation initiation inhibition. PLoS ONE. 2011;6:e19727. doi: 10.1371/journal.pone.0019727. [DOI] [PMC free article] [PubMed] [Google Scholar]


