Abstract
Replication of positive-strand caliciviruses is mediated by a virus-encoded RNA-dependent RNA polymerase (RdRp). To study the replication of Norovirus (NV), a member of the family Caliciviridae, we used a recombinant baculovirus system to express an enzymatically active RdRp protein from the 3D region of the NV genome and defined conditions for optimum enzymatic activity. Using an RNA template from the NV 3′ genomic region, we observed similar levels of enzymatic activity in assays with and without a poly(A) tail. RdRp activity was not significantly affected by the addition of an RNA primer to the reaction mixture. Thus, the NV RdRp exhibited primer- and poly(A)-independent RNA polymerase activity. While the RdRp inhibitor phosphonoacetic acid inhibited NV RdRp activity, another gliotoxin did not. The active recombinant NV RdRp will be of benefit to studies of NV replication and will facilitate the development of specific inhibitors of NV proliferation.
Norovirus (NV), a member of the family Caliciviridae, is the leading cause of epidemic acute, nonbacterial gastroenteritis. NV infection causes nausea, vomiting, low-grade fever, and diarrhea, which can be severe in infants and young children. For these reasons, it is a major public health concern (7). An effective vaccine or other therapeutic agent would be valuable for preventing the significant morbidity and potential mortality associated with NV infections. However, the lack of an efficient culture system has hampered the biochemical characterization of the NV proteins, and thus far, molecular biological techniques have been the most useful tools for the study of NV.
The NV virion contains a polyadenylated plus-strand RNA genome of ∼7.7 kb (11, 15). The structures of the full-length genome, phylogenetic trees, and genetic recombination among distinct genogroups have been analyzed in detail (13). Based on sequence similarities with other single-stranded RNA viruses, the NV open reading frame 1 (ORF1) is predicted to encode a large polyprotein that is cleaved into several viral proteins, including NTPase, proteinase, and RNA-dependent RNA polymerase (RdRp) (11, 15).
The RdRp encoded by the 3D region has a conserved amino acid motif, glycine-aspartic acid-aspartic acid (GDD), which is found in the active site of many viral RdRps (14), and thus might have an important role in NV replication. As in other positive-strand RNA viruses (2), NV genomic RNA likely acts as a template for the synthesis of minus-strand RNA. The minus-strand RNA then, in turn, serves as a template for the synthesis of progeny genomic plus-strand RNA molecules. Thus, RdRp is central to the synthesis of both plus- and minus-strand RNA molecules. Among the Caliciviridae, Rabbit hemorrhagic disease virus (RHDV) and Feline calicivirus (FCV) express an enzymatically active RdRp protein (27, 28). However, caliciviruses that infect humans have not been examined for RdRp activity.
The aims of this study were to develop a cell-free system that permits the identification of RdRp activity in vitro and to characterize the biochemical properties of RdRp in NV replication. We expressed the RdRp protein from the 3D region of the NV genome with a baculovirus vector and then tested the activity of the purified protein.
MATERIALS AND METHODS
Construction of plasmids.
The primer sequences used for plasmid construction are listed in Table 1.
TABLE 1.
Sequences of DNA primers used for plasmid construction
| Primer | Polarity | Sequencea (5′→3′) |
|---|---|---|
| U201LV1S-BglII | + | GAagatctTCGTGAATGAAGATGGCGTCTAACGAC |
| TX30SXN | − | GACTAGTTCTAGATCGCGAGCGGCCGCCCTTTTTTTTTTTTTTTTTTTTTTTTTTTTTT |
| EagI-3D | + | TTTcggccgATGGGAGGTGACGACAAGGGCACCTAT |
| 3D-EcoRI | − | AAAgaattcTTATTATTCGACGCCATCTTCATTCACAAA |
| 3D-GAA-F | + | TTCTATGGTG CTGCTGAGATTGTA |
| 3D-GAA-R | − | CTCA GCAGCACCATAGAAAGAGTA |
| 3D-GAD-F | + | TTCTATGGTG CTGATGAGATTGTA |
| 3D-GAD-R | − | CTCATCA GCACCATAGAAAGAGTA |
| BamH-3B | + | CGCggatccATGGGAAAGAAAGGCAAGAACAAGTCC |
| 3B-Xba | − | TGCtctagaTTATTATTCAAAATCTATCCTCTCATTGTA |
| KpnT7ORF3 | + | CGGggtaccTAATACGACTCACTATAGGGAAGGATTCAATAATGGCTGGAG |
| 201pAMS | − | TACGGCATGCacgcgtTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTAAAAGATTCTAAATCAAATT |
| T7S608 | + | TTATAATACGACTCACTATAGGACAGCTTTTGTCACTCCACCATC |
| 201ASdelA-Xba | − | GtctagaAAAGATTCTAAATCAAATTTAGG |
Restriction sites used for cloning are represented by lowercase letters. The underlined nucleotides were changed to introduce amino acid substitutions at the conserved GDD motif.
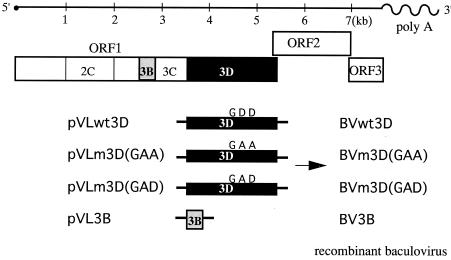
To obtain a cDNA fragment corresponding to the full-length genome of the NV genogroup 2 strain U201, we used primers U201LV1S-BglII and TX30SXN to amplify viral RNA extracted from a stool specimen (U201) (13). RNA extraction, reverse transcription, and PCR were performed as previously described (13). The 3D region was amplified from the full-length cDNA of strain U201 with the primers EagI-3D and 3D-EcoRI, digested with EagI and EcoRI, and cloned into the EagI-EcoRI site of the pVL1392 vector (BD Bioscience, San Jose, Calif.) to obtain pVLwt3D (Fig. 1).
FIG. 1.
Expression constructs for RdRp proteins. The genome structure of NV is shown to scale. The 3D region with the wild-type GDD motif (wt3D) and with amino acid substitutions of GDD to GAA [m3D(GAA)] or GAD [m3D(GAD)] and the 3B region of the NV cDNA were inserted into baculovirus transfer vectors to construct recombinant baculoviruses.
The plasmid pVLwt3D was used as a template for mutagenesis, and DNA oligonucleotide primers 3D-GAA-F, 3D-GAA-R, 3D-GAD-F, and 3D-GAD-R were used to introduce amino acid substitutions at the conserved GDD motif. To obtain a mutant RdRp containing an amino acid change of the GDD motif to GAA, we amplified the 5′ and 3′ halves of the mutant 3D region by using primer set 3D-GAA-R and EagI-3D or 3D-GAA-F and 3D-EcoRI, respectively. Since the resulting two cDNA fragments partially overlapped each other, they were mixed, denatured, annealed, and filled in with Tbr EXT DNA polymerase (Fynzyme, Espoo, Finland) to obtain the full-length construct with a mutant 3D region. The resulting products were digested with EagI and EcoRI and ligated into the pVL1392 vector to form the plasmid pVLm3D(GAA) (Fig. 1).
We amplified two additional cDNA fragments. To obtain another mutant RdRp that changes the GDD motif to GAD, we amplified the 5′ or 3′ half of the mutant 3D region by using primer set 3D-GAD-R and EagI-3D or 3D-GAD-F and 3D-EcoRI with the same strategy as that used for the construction of plasmid pVLm3D(GAA). To obtain the full-length 3D construct with a GAD mutant, the resulting two cDNA fragments were mixed, denatured, annealed, and filled in with Tbr EXT DNA polymerase. The resulting products were digested with EagI and EcoRI and ligated into the pVL1392 vector to form the plasmid pVLm3D(GAD) (Fig. 1). Finally, the 3B region of the NV genome was amplified, using primers BamH-3B and 3B-Xba. The resulting cDNA was digested with BamHI and XbaI and cloned into the BamHI-XbaI site of the pVL1393 vector (BD Bioscience) to obtain pVL3B (Fig. 1).
A series of plasmids (i.e., pMT-ORF3pA, pUC608-pA, and pUC608delA) was used for in vitro transcription to prepare the RNA templates or RNA size markers for the analysis of RdRp products. These were constructed by PCR with the oligonucleotide primers described below. For the plasmid pMT-ORF3pA, primers KpnT7ORF3 and 201pAMS were used with the full-length cDNA template of U201 to amplify a cDNA fragment from ORF3 to the poly(A) region under the T7 promoter sequence. The resulting product was digested with KpnI and MluI and then cloned into the KpnI and MluI sites of the pMT1 vector (kindly provided by M. Tatsumi, National Institute of Infectious Diseases, Tokyo, Japan). Primers T7S608, which contained the T7 promoter, and 201pAMS were used with template plasmid pMT-ORF3pA to amplify a cDNA fragment corresponding to the 3′-terminal 232 nucleotides of the NV genome. The amplified product was ligated to the pT7Blue vector (Takara Bio, Shiga, Japan) to obtain pT7608-pA. The HindIII-EcoRI fragment of the cDNA was inserted into the HindIII-EcoRI site of the pUC119 vector and the plasmid was designated pUC608-pA. Primers T7S608 and 201AsdelA-Xba were used with template plasmid pMT-ORF3pA to amplify a cDNA fragment that corresponded to plasmid pUC608-pA except for a deletion of the poly(A) sequence. The PCR product was inserted into the pT7Blue vector to obtain pT7608delA. By the same strategy, the HindIII-EcoRI fragment of the cDNA was cloned into the pUC119 vector to obtain pUC608delA.
Preparation of RNA.
The oligo(U)15 RNA primer was purchased from Takara Bio. The RNA dimer UpU was purchased from Sigma (Tokyo, Japan). ORF3-pA, 608-pA, and 608-delA RNAs were transcribed in vitro by T7 RNA polymerase (Promega, Tokyo, Japan) from the templates MluI-digested pMT-ORF3pA, MluI-digested pUC608-pA, and XbaI-digested pUC608delA, respectively. The RNAs were purified, and their integrity was verified as described previously (6).
Expression and purification of RdRp protein.
The procedures for generating recombinant baculoviruses and for subsequent large-scale production of the proteins were adapted from the work of Li et al. (16). Briefly, transfer plasmid pVLwt3D, pVLm3D (GAA), pVLm3D (GAD), or pVL3B was cotransfected into insect SF9 cells with linearized BaculoGold DNA (BD Bioscience) to obtain recombinant baculovirus BVwt3D, BVm3D(GAA), BVm3D(GAD), or BV3B, respectively. Insect Tn5 cells were infected with these recombinant baculoviruses for the large-scale expression of recombinant RdRp proteins. After 4 days of incubation at 27°C, cells were harvested, collected by centrifugation, and washed twice with phosphate-buffered saline.
The cell pellet was resuspended in ice-cold cell lysis buffer (20 mM Tris-HCl [pH 7.5], 1.0 mM EDTA, 10 mM dithiothreitol [DTT], 2% Triton X-100, 500 mM NaCl, 10 mM MgCl2, 50% glycerol, and EDTA-free protease inhibitors) (Roche Diagnostics, Tokyo, Japan). The extract was sonicated, cleared of debris by centrifugation at 15,000 × g for 30 min, dialyzed against SP buffer (25 mM morpholineethanesulfonic acid [pH 6.4], 1.0 mM EDTA, and 1% Triton X-100), and applied to a HiTrap SP column (Amersham Biosciences) that was pre-equilibrated with SP buffer. Bound proteins were eluted by a linear gradient of 0 to 1.0 M NaCl in SP buffer and were analyzed by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis and Coomassie blue staining. The RdRp protein was eluted with 320 to 550 mM NaCl, dialyzed against RdRp sample buffer (20 mM Tris-HCl [pH 7.7], 1.0 mM EDTA, 100 mM NaCl, 10 mM DTT, 2% Triton X-100, and 50% glycerol), and tested for RdRp activity. The fractions containing active RdRp protein were combined, dialyzed against Q buffer (20 mM Tris-HCl [pH 7.7], 1.0 mM EDTA, and 1% Triton X-100), and applied to a HiTrap Q column (Amersham Biosciences) that was pre-equilibrated with Q buffer. Bound proteins were eluted with a linear gradient of 0 to 1.0 M NaCl in Q buffer and were analyzed by SDS-polyacrylamide gel electrophoresis and Coomassie blue staining. RdRp fractions eluted at ∼520 mM NaCl. They were combined, dialyzed against RdRp sample buffer, and examined for RdRp activity.
RdRp and terminal nucleotidyl transferase (TNTase) assays.
The RdRp reaction was performed in a 15-μl volume with 375 ng of RdRp protein and 5.0 pmol of in vitro-transcribed ORF3-pA RNA, 608-polyA RNA, or 608-delA RNA in a reaction buffer containing, unless otherwise specified, 20 mM Tris-HCl (pH 6.8), 2.0 mM MnCl2, 100 mM NaCl, 20 mM DTT, 20 U of RNase inhibitor (Promega), 50 μg of actinomycin D/ml, 250 μM GTP, 125 μM ATP, 125 μM CTP, 5.0 μM UTP, and 4.0 μCi of [33P]UTP (>2,500 Ci/mmol) (Amersham Biosciences).
Nuclease digestion was performed as described by Ishii et al. (10). TNTase assays were performed in the same buffer with specified nucleoside triphosphates. RdRp and TNTase reactions were done at 30°C for 90 min and stopped by the addition of 60 μl of a stop solution (10 mM Tris-HCl [pH 7.5], 10 mM EDTA, 100 mM NaCl). The RNA products were extracted with TRISOL LS reagent (Invitrogen, Tokyo, Japan) and precipitated with isopropanol. Products were dissolved with RNA sample buffer containing 80% formamide, 1 mM EDTA, and 0.1% bromophenol blue. After heat denaturation, the RNA products were separated in 6.0 or 10.0% polyacrylamide gels in 8.0 M urea. Radiolabeled RNA products were analyzed with the BAS 1000 system (Fuji Film, Tokyo, Japan).
For the examination of polymerase inhibitors, phosphonoacetic acid (PAA) and gliotoxin were purchased from Sigma. These materials were dissolved in H2O, and then increasing amounts (25, 100, and 250 μM) of the inhibitors were mixed with the RdRp reaction buffer.
RESULTS
Expression and purification of enzymatically active RdRp proteins.
In FCV, a member of the family Caliciviridae, the proteinase-polymerase precursor (3CD) rather than the 3D region itself is highly active, which may be attributed to inefficient proteolytic processing of the linker domain of 3CD (25, 28). Since efficient proteolytic processing of the NV polyprotein into 3D is observed in both Escherichia coli and mammalian cell expression systems (17, 24), we cloned and expressed the 3D region itself to study the biochemical properties of NV RdRp (Fig. 1). A recombinant baculovirus system was used to express NV RdRp. Soluble lysates of infected cells were separated by cation-exchange chromatography (Fig. 2A), and fractions were examined for RdRp activity.
FIG. 2.
Purification and enzymatic activity of NV RdRp. (A) Eluted proteins from a HiTrap SP (cation exchange) column (left) and a HiTrap Q (anion exchange) column (right) were separated by SDS-polyacrylamide gel electrophoresis and visualized by Coomassie blue staining. The portions of fractions containing RdRp protein eluted by 320 to 550 mM NaCl in SP buffer from the cation-exchange column were used to examine enzymatic activity. The fractions with active RdRp from the cation-exchange column were then subjected to further purification in the anion-exchange column. (B) Genome structure of NV shown to scale. The RNA template used for RdRp assays is represented by a black bar. (C) RdRp reaction with the fractions from a cation-exchange column. At the same time, m3D(GAA) and m3D(GAD), which had substitutions in the GDD motif to GAA and GAD, respectively, and 3B protein as a negative control were separated in cation-exchange columns, and the eluted fractions were subjected to an RdRp assay using the same strategy as that used for the wild-type protein (wt3D). RNA products were separated in a denaturing polyacrylamide gel, analyzed by the BAS1000 system, and presented together with [33P]-incorporated molecular size markers transcribed by T7 RNA polymerase from MluI-digested pMT-ORF3pA (T7-labeled RNA).
Several investigators have shown that viral RdRps have primer-dependent RNA polymerase activities with homopolymeric RNA templates (5, 10, 28). To clarify the molecular mechanism of NV replication, it is more important to test the activity with the viral RNA template than with the homopolymeric RNA template. In this study, we examined the NV RdRp activity with synthetic RNA templates that corresponded to the 3′-terminal region of the NV genome. We believe that our strategy for measuring NV RdRp activity better replicates the production of minus-strand RNA from plus-strand genomic RNA.
The template ORF3-pA RNA (Fig. 2B) was incubated with RdRp fractions in the presence of [33P]UTP and cold ribonucleotide triphosphates. Radiolabeled reaction products of RdRp assays were analyzed in denaturing polyacrylamide gels. [33P]UTP was incorporated in fractions from BVwt3D-infected cell lysates that eluted at 320 to 550 mM NaCl (Fig. 2C).
We next examined whether the nucleotide length of the RNA product obtained by the RdRp assay was the same as that obtained with the template ORF3-pA RNA. As a size marker, we prepared T7 RNA polymerase-labeled RNA transcribed from MluI-digested plasmid pMT-ORF3pA (Fig. 2C, lane 1). The template RNA for the RdRp assays was also transcribed from the same starting material by T7 RNA polymerase (see Materials and Methods). The lengths of the RNA products from the RdRp assays were identical to that of the template, indicating that the RNA products of the RdRp reaction were transcripts of the template RNA. On the other hand, incorporation was at background levels in the lysates of negative control cells infected with BVm3D(GAA) and BV3Dm(GAD), which carried amino acid substitutions at a GDD motif (Fig. 2C).
To analyze the activity of the wild-type RdRp protein in detail, we collected the active RdRp fractions from cation-exchange chromatography and subsequent anion-exchange chromatography steps (Fig. 2A). Purified RdRp protein was used for further analyses.
Characterization of RdRp activity.
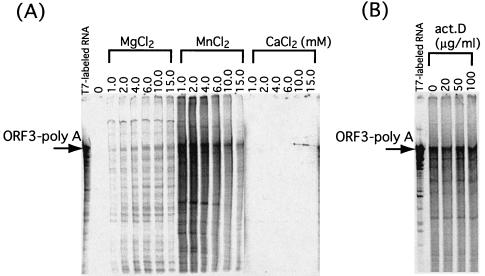
We next examined the effects of pH and divalent cation concentrations, including MgCl2, MnCl2, and CaCl2, on NV RdRp activity. ORF3-pA RNA synthesized in vitro was used as a template. Since the requirements of divalent cations are different for primer-dependent and primer-independent synthesis (21), the oligonucleotide primer was not used in the reaction. RdRp activity was markedly increased in the presence of 2.0 mM Mn2+. A slight increase was detected with increased concentrations of Mg2+, although to a much lesser extent than with Mn2+. No increase was seen with Ca2+ (Fig. 3A). The RdRp activity was found to be optimal at pHs 6.8 to 7.5 (data not shown). Based on these results, we used pH 6.8 and 2.0 mM Mn2+ for further studies of NV RdRp activity.
FIG. 3.
Optimization of RdRp assay conditions and effects of actinomycin D on enzymatic activity. The RNA template used for RdRp assays is shown in Fig. 2B. (A) Divalent cation optimization. (B) Effects of increasing amounts of actinomycin D on enzymatic activity. RNA products were analyzed using methods similar to those described for Fig. 2.
We also examined the possibility that the synthesis was due to a DNA-dependent RNA polymerase in the extracts of insect Tn5 cells. To do this, we added actinomycin D to the reaction mixtures. Actinomycin D forms a complex with DNA and interferes with RNA synthesis. The production of labeled RNA with RdRp assays was not inhibited in the presence of various amounts of the inhibitor (Fig. 3B), indicating that the labeled RNA had not originated from the action of a DNA-dependent RNA polymerase.
Characterization of RdRp products.
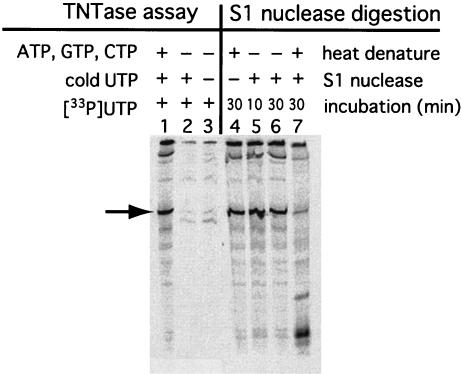
Hepatitis C virus (HCV) RdRp expressed in recombinant baculovirus systems shows TNTase activity (20). TNTase activity might confound the interpretation of the actual properties of RdRp. Since TNTase labels the 3′ ends of RNA, the RNA product in our RdRp assays could result from the addition of [33P]UTP to the 3′ end of the template RNA by TNTase. To examine this possibility, we performed TNTase assays without cold ATP, CTP, and GTP. The template 608-pA RNA contained the NV 3′-terminal region and 30 nucleotides of poly(A) sequence (Fig. 4). [33P]-labeled RNA products were not detected in the TNTase assays in the presence of [33P]UTP only or [33P]UTP plus cold UTP, without any other NTPs (Fig. 4, lanes 2 and 3). The RdRp protein did not display any activity that added UTP to the 3′ end of the template RNA. Therefore, the labeled RNA product was not the result of TNTase activity.
FIG. 4.
TNTase assay and S1 nuclease digestion. An in vitro-transcribed RNA, 606-polyA, corresponding to the 3′-terminal 232 nucleotides of the NV genome, was used as a template for the RdRp reaction. For TNTase assays, reactions were performed without ATP, GTP, and CTP (lane 2) or without ATP, GTP, CTP, and cold UTP (lane 3). An RNA product from a standard reaction mixture is shown (lane 1) as a control. For the S1 nuclease digestion analyses, RdRp products were heat denatured at 95°C for 2 min (lanes 4 and 7) before treatment without (lane 4) or with (lanes 5, 6, and 7) S1 nuclease for the indicated times. The 232-nucleotide band, identified by an arrow, is missing from lanes 2, 3, and 7. RNA products were analyzed by methods similar to those described for Fig. 2.
We next determined whether the products synthesized from the NV RNA template were single or double stranded. Double-stranded RNA would indicate that cRNA is synthesized from the template RNA by RdRp. Since S1 nuclease catalyzes the specific degradation of single-stranded RNA to mononucleotides, the RdRp product was incubated with S1 nuclease. The 608-pA RNA containing the 3′-terminal region of the NV genome and the poly(A) sequence was used as a template. The RNA product was resistant to S1 nuclease digestion (Fig. 4, lanes 5 and 6). However, S1 nuclease treatment after heat denaturation almost completely degraded the RNA (lane 7). This result indicated that the product formed by the RdRp reaction was double-stranded RNA.
Poly(A)- and primer-independent RdRp activity.
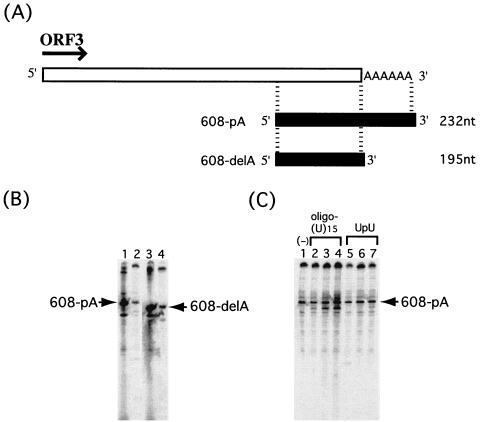
The results described above indicated that NV RdRp was able to synthesize cRNA without the primer at the 3′ region of the NV genome. Since the NV genome contains a poly(A) sequence at the 3′ end (11, 15), we investigated whether the poly(A) sequence is required for the initiation of RNA synthesis by NV RdRp. RdRp activity was measured by using NV 3′ genomic RNA with or without a poly(A) tail. The 608-pA template RNA contained the 3′-terminal region of the NV genome and 30 nucleotides of poly(A) sequence; 608-delA RNA contained the same NV sequence but not the poly(A) sequence (Fig. 5A). [33P]UTP incorporation was similar in both reactions. Furthermore, the labeled RNA size markers driven by T7 RNA polymerase confirmed that the RdRp product was the same size as the template RNA, irrespective of the presence of the poly(A) tail on the template (Fig. 5B). The results strongly indicated that the 3′ poly(A) sequence is not necessary for the initiation of synthesis or for synthesis to the exact length of the cRNA.
FIG. 5.
RdRp activity was not dependent on RNA primer and poly(A) sequence. (A) Genome structure downstream of NV ORF3 with poly(A). The RNA templates used for RdRp assays are represented by black bars. (B) The RdRp products from reactions with template RNA 608-pA (lane 2) and 608-delA (lane 4), together with labeled RNA from a T7 polymerase reaction of 608-pA (lane 1) and 608-delA (lane 3), are represented. (C) Effect of RNA primer on RdRp activity. RdRp reactions were performed with 608-pA RNA as a template, with 1.0 pmol (lane 2), 5.0 pmol (lane 3), or 20.0 pmol (lane 4) of oligo(U)15; 10.0 pmol (lane 5), 1.0 nmol (lane 6), or 10.0 nmol (lane 7) of UpU dinucleotide primer; or no primer (lane 1). RNA products were analyzed by methods similar to those described for Fig. 2.
We next examined the influence on RdRp activity of the addition of an oligo(U)15 primer or UpU dimer primer to the template RNA containing the poly(A) tail (Fig. 5C). When the oligo(U)15 primer was added to the reaction, the amounts of RdRp product increased slightly with increasing amounts of primer. Also, a distinct RNA product was observed which migrated faster than the templates. The reason for the presence of this smaller RNA is not known. The presence of the oligo(U)15 primer made little difference on the synthesis of the major product, which corresponded to cRNA of the template RNA (Fig. 5C, lanes 2 to 4). Interestingly, there was no effect on RNA production with the reaction containing the dinucleotide primer UpU (Fig. 5C, lanes 5 to 7). From these results, we concluded that NV RdRp could synthesize RNA complementary to the NV genomic RNA in a primer- and poly(A)-independent manner.
Effect of RdRp inhibitors.
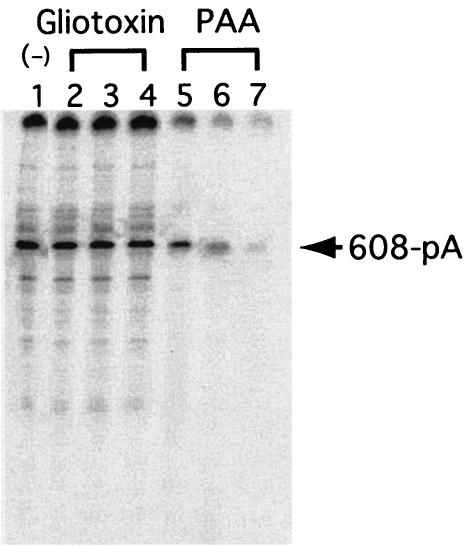
The assay system described above allowed us to screen for potential NV RdRp inhibitors. Various concentrations of gliotoxin or PAA, which inhibits the activities of other viral RdRp proteins, were added to the NV RdRp reaction. PAA inhibited the RdRp activity (Fig. 6). The estimated 50% inhibitory concentration of PAA for NV RdRp activity was <20 μM. No inhibitory effect was observed with gliotoxin, even at concentrations as high as 250 μM.
FIG. 6.
Effects of RdRp inhibitors. RdRp reactions were performed with 608-pA RNA as a template, with 25 μM (lane 2), 100 μM (lane 3), or 250 μM (lane 4) gliotoxin; 25 μM (lane 5), 100 μM (lane 6), or 250 μM (lane 7) PAA; or no inhibitors. The 232-nucleotide band, identified by an arrow, disappeared gradually upon PAA treatment. RNA products were analyzed by methods similar to those described for Fig. 2.
DISCUSSION
Previous studies of viral homology have indicated that the C terminus of NV ORF1 contains a motif in the RdRp that is conserved among various plus-strand RNA viruses. In this study, we report that the 3D region of the NV genome contains an active RdRp. We expressed a soluble recombinant RdRp encoded by the NV 3D genomic region in insect cells without a tagged sequence but with a single additional methionine residue at the N terminus. We purified the protein and established a system for measuring its RNA polymerase activity. That activity was not due to contamination by an endogenous insect polymerase, and actinomycin D did not inhibit the RdRp reaction, also indicating a lack of endogenous RdRp.
We focused on the conserved GDD motif in the 3D region. Found in most RdRps of positive-strand RNA viruses, GDD is important for metal binding and is considered to be the catalytic site of the enzyme (14). When we replaced the GDD motif in RdRp, the RNA polymerase activity was lost, and the expressed 3B protein of NV also lacked activity. The loss of RdRp activity in these mutant RdRp proteins indicated that labeled RNA products obtained by a reaction with the wild-type RdRp protein were driven by an RNA polymerase-related activity and maybe by an intrinsic activity of the expressed NV RdRp protein. The enzymatic activity of NV RdRp depended on Mn2+: the optimal Mn2+ concentration was 2.0 mM. This requirement for divalent cations is similar to that of the primer-independent de novo RNA polymerase activity of poliovirus (1), HCV (18, 21), and human rhinovirus 16 (9). In the presence of Mn2+ ions, RdRp from RHDV is thought to form an active structure (19). Our observations suggest that Mn2+ ions promote conformational changes in NV RdRp that are similar to those of other positive-strand RNA viruses, such as picornaviruses, HCV, and RHDV. We believe that the enzymatic activity in the presence of Mn2+ may reflect a primary biological function of RdRp, and we used Mn2+ for all of our RdRp characterizations.
Several investigators have shown that in vitro viral RdRp reactions generate cRNA by a “copy-back” mechanism (3, 26). These reactions produce dimer-sized RNAs, presumably as a result of a self-priming event at the extreme 3′ terminus of the template. In our study, dimer-sized RNAs were not seen when the RdRp products were separated in denatured gels. We speculate that NV RdRp does not have a copy-back mechanism for cRNA synthesis. This notion is supported by the absence of self-priming by an additional oligo(U) at the 3′ end of the template RNA mediated by TNTase activity.
Interestingly, our results indicate that NV RdRp may not recognize poly(A). RNA production was as efficient without a poly(A) tail at the 3′-terminal region of the template RNA as it was with the tail. Furthermore, priming by synthetic oligo(U) or UpU had little effect on the RdRp activity. RdRp was able to synthesize cRNA without a poly(A) tail. Therefore, as reported for HCV RdRp (12), there must be a specific sequence requirement for cis-acting signals for RNA synthesis when NV genomic RNA is used. Additional studies are needed to determine the specific sequence.
Our results revealed that NV RdRp has the activity of de novo RNA synthesis with the NV genomic RNA as a template. Like those of other RNA viruses (1, 12, 18), NV RdRp directs minus-strand RNA synthesis in a primer-independent manner. We also showed an interesting feature of NV, namely that the activity of minus-strand synthesis is independent of a poly(A) tail. The NV genome encodes a VPg-like protein on the genome (11, 15). In picornaviruses, the VPg protein is thought to link to the 5′ end of the genome and serve as a primer for RdRp (23). In the Caliciviridae, a genome-linked VPg protein has also been reported for FCV (8). Therefore, it is important to determine whether the VPg protein is linked to the NV genome and has a role in cRNA synthesis.
Finally, our in vitro RdRp assay may be useful in further studies to develop drugs to treat NV, and the NV RdRp may itself be a useful target for antiviral drugs. It was strongly inhibited by PAA, which interferes with the replication of DNA viruses (4). The 50% inhibitory concentration (<20 μM) for NV RdRp was significantly lower than that reported for HCV RdRp (10), indicating a specific inhibition of NV RdRp activity by PAA. In contrast, gliotoxin, a known inhibitor of poliovirus and HCV RdRps (5, 22), had little effect on NV RdRp activity, despite the high concentrations tested. The differences in sensitivities may be attributed to the specific nature of NV RdRp, which synthesizes minus-strand RNA in a primer- and poly(A)-independent manner.
Acknowledgments
This work was supported by Research on Health Sciences Focusing on Drug Innovation grant KH51048 from the Japan Health Sciences Foundation.
REFERENCES
- 1.Arnold, J. J., S. K. Ghosh, and C. E. Cameron. 1999. Poliovirus RNA-dependent RNA polymerase (3D(pol)). Divalent cation modulation of primer, template, and nucleotide selection. J. Biol. Chem. 274:37060-37069. [DOI] [PubMed] [Google Scholar]
- 2.Bartenschlager, R., and V. Lohmann. 2000. Replication of hepatitis C virus. J. Gen. Virol. 81:1631-1648. [DOI] [PubMed] [Google Scholar]
- 3.Behrens, S. E., L. Tomei, and R. De Francesco. 1996. Identification and properties of the RNA-dependent RNA polymerase of hepatitis C virus. EMBO J. 15:12-22. [PMC free article] [PubMed] [Google Scholar]
- 4.Binne, U. K., W. Amon, and P. J. Farrell. 2002. Promoter sequences required for reactivation of Epstein-Barr virus from latency. J. Virol. 76:10282-10289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ferrari, E., J. Wright-Minogue, J. W. Fang, B. M. Baroudy, J. Y. Lau, and Z. Hong. 1999. Characterization of soluble hepatitis C virus RNA-dependent RNA polymerase expressed in Escherichia coli. J. Virol. 73:1649-1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fukushi, S., M. Okada, T. Kageyama, F. B. Hoshino, K. Nagai, and K. Katayama. 2001. Interaction of poly(rC)-binding protein 2 with the 5′-terminal stem loop of the hepatitis C-virus genome. Virus Res. 73:67-79. [DOI] [PubMed] [Google Scholar]
- 7.Green, K. Y., A. Z. Kapikian, and R. M. Chanock. 2001. Human caliciviruses, p. 841-874. In D. M. Knipe, P. M. Howley, D. E. Griffin, et al. (ed.), Fields virology, 4th ed. Lippincott-Raven, Philadelphia, Pa.
- 8.Herbert, T. P., I. Brierley, and T. D. Brown. 1997. Identification of a protein linked to the genomic and subgenomic mRNAs of feline calicivirus and its role in translation. J. Gen. Virol. 78:1033-1040. [DOI] [PubMed] [Google Scholar]
- 9.Hung, M., C. S. Gibbs, and M. Tsiang. 2002. Biochemical characterization of rhinovirus RNA-dependent RNA polymerase. Antivir. Res. 56:99-114. [DOI] [PubMed] [Google Scholar]
- 10.Ishii, K., Y. Tanaka, C. C. Yap, H. Aizaki, Y. Matsuura, and T. Miyamura. 1999. Expression of hepatitis C virus NS5B protein: characterization of its RNA polymerase activity and RNA binding. Hepatology 29:1227-1235. [DOI] [PubMed] [Google Scholar]
- 11.Jiang, X., M. Wang, K. Wang, and M. K. Estes. 1993. Sequence and genomic organization of Norwalk virus. Virology 195:51-61. [DOI] [PubMed] [Google Scholar]
- 12.Kao, C. C., X. Yang, A. Kline, Q. M. Wang, D. Barket, and B. A. Heinz. 2000. Template requirements for RNA synthesis by a recombinant hepatitis C virus RNA-dependent RNA polymerase. J. Virol. 74:11121-11128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Katayama, K., H. Shirato-Horikoshi, S. Kojima, T. Kageyama, T. Oka, F. Hoshino, S. Fukushi, M. Shinohara, K. Uchida, Y. Suzuki, T. Gojobori, and N. Takeda. 2002. Phylogenetic analysis of the complete genome of 18 Norwalk-like viruses. Virology 299:225-239. [DOI] [PubMed] [Google Scholar]
- 14.Koonin, E. V. 1991. The phylogeny of RNA-dependent RNA polymerases of positive-strand RNA viruses. J. Gen. Virol. 72:2197-2206. [DOI] [PubMed] [Google Scholar]
- 15.Lambden, P. R., E. O. Caul, C. R. Ashley, and I. N. Clarke. 1993. Sequence and genome organization of a human small round-structured (Norwalk-like) virus. Science 259:516-519. [DOI] [PubMed] [Google Scholar]
- 16.Li, T. C., Y. Yamakawa, K. Suzuki, M. Tatsumi, M. A. Razak, T. Uchida, N. Takeda, and T. Miyamura. 1997. Expression and self-assembly of empty virus-like particles of hepatitis E virus. J. Virol. 71:7207-7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu, B. L., G. J. Viljoen, I. N. Clarke, and P. R. Lambden. 1999. Identification of further proteolytic cleavage sites in the Southampton calicivirus polyprotein by expression of the viral protease in E. coli. J. Gen. Virol. 80:291-296. [DOI] [PubMed] [Google Scholar]
- 18.Luo, G., R. K. Hamatake, D. M. Mathis, J. Racela, K. L. Rigat, J. Lemm, and R. J. Colonno. 2000. De novo initiation of RNA synthesis by the RNA-dependent RNA polymerase (NS5B) of hepatitis C virus. J. Virol. 74:851-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ng, K. K., M. M. Cherney, A. L. Vazquez, A. Machin, J. M. Alonso, F. Parra, and M. N. James. 2002. Crystal structures of active and inactive conformations of a caliciviral RNA-dependent RNA polymerase. J. Biol. Chem. 277:1381-1387. [DOI] [PubMed] [Google Scholar]
- 20.Ranjith-Kumar, C. T., J. Gajewski, L. Gutshall, D. Maley, R. T. Sarisky, and C. C. Kao. 2001. Terminal nucleotidyl transferase activity of recombinant Flaviviridae RNA-dependent RNA polymerases: implication for viral RNA synthesis. J. Virol. 75:8615-8623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ranjith-Kumar, C. T., Y. C. Kim, L. Gutshall, C. Silverman, S. Khandekar, R. T. Sarisky, and C. C. Kao. 2002. Mechanism of de novo initiation by the hepatitis C virus RNA-dependent RNA polymerase: role of divalent metals. J. Virol. 76:12513-12525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rodriguez, P. L., and L. Carrasco. 1992. Gliotoxin: inhibitor of poliovirus RNA synthesis that blocks the viral RNA polymerase 3Dpol. J. Virol. 66:1971-1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rueckert, R. R. 1996. Picornaviridae: the viruses and their replication, p. 609-654. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven, Philadelphia, Pa.
- 24.Seah, E. L., J. A. Marshall, and P. J. Wright. 1999. Open reading frame 1 of the Norwalk-like virus Camberwell: completion of sequence and expression in mammalian cells. J. Virol. 73:10531-10535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sosnovtsev, S. V., M. Garfield, and K. Y. Green. 2002. Processing map and essential cleavage sites of the nonstructural polyprotein encoded by ORF1 of the feline calicivirus genome. J. Virol. 76:7060-7072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Steffens, S., H. J. Thiel, and S. E. Behrens. 1999. The RNA-dependent RNA polymerases of different members of the family Flaviviridae exhibit similar properties in vitro. J. Gen. Virol. 80:2583-2590. [DOI] [PubMed] [Google Scholar]
- 27.Vazquez, A. L., J. M. Martin Alonso, R. Casais, J. A. Boga, and F. Parra. 1998. Expression of enzymatically active rabbit hemorrhagic disease virus RNA-dependent RNA polymerase in Escherichia coli. J. Virol. 72:2999-3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wei, L., J. S. Huhn, A. Mory, H. B. Pathak, S. V. Sosnovtsev, K. Y. Green, and C. E. Cameron. 2001. Proteinase-polymerase precursor as the active form of feline calicivirus RNA-dependent RNA polymerase. J. Virol. 75:1211-1219. [DOI] [PMC free article] [PubMed] [Google Scholar]