Abstract
For various groups of plant viruses, the genomic RNAs end with a tRNA-like structure (TLS) instead of the 3′ poly(A) tail of common mRNAs. The actual function of these TLSs has long been enigmatic. Recently, however, it became clear that for turnip yellow mosaic virus, a tymovirus, the valylated TLSTYMV of the single genomic RNA functions as a bait for host ribosomes and directs them to the internal initiation site of translation (with N-terminal valine) of the second open reading frame for the polyprotein. This discovery prompted us to investigate whether the much larger TLSs of a different genus of viruses have a comparable function in translation. Brome mosaic virus (BMV), a bromovirus, has a tripartite RNA genome with a subgenomic RNA4 for coat protein expression. All four RNAs carry a highly conserved and bulky 3′ TLSBMV (about 200 nucleotides) with determinants for tyrosylation. We discovered TLSBMV-catalyzed self-tyrosylation of the tyrosyl-tRNA synthetase but could not clearly detect tyrosine incorporation into any virus-encoded protein. We established that BMV proteins do not need TLSBMV tyrosylation for their initiation. However, disruption of the TLSs strongly reduced the translation of genomic RNA1, RNA2, and less strongly, RNA3, whereas coat protein expression from RNA4 remained unaffected. This aberrant translation could be partially restored by providing the TLSBMV in trans. Intriguingly, a subdomain of the TLSBMV could even almost fully restore translation to the original pattern. We discuss here a model with a central and dominant role for the TLSBMV during the BMV infection cycle.
Thirty years ago, the exciting discovery was made that the RNAs of certain plant-infecting viruses can be aminoacylated. For the genera Tymovirus, Bromovirus, Tobamovirus, and Furovirus of the family Bromoviridae, the 3′ ends of the viral RNAs can be charged with either valine, tyrosine, or histidine in a way that is comparable to the aminoacylation of the corresponding canonical tRNAs (for reviews, see references 8, 10, and 22). On the basis of chemical and enzymatic probing experiments and functional assays, these 3′ untranslated regions (UTRs) can all be folded into structures more or less resembling those of canonical tRNAs. The full functional meaning of these tRNA-like structures (TLSs) has remained enigmatic.
For the tymovirus turnip yellow mosaic virus (TYMV), it was recently discovered, however, that valylated TLSTYMV entraps ribosomes and directs them to the second open reading frame (ORF) of the single genomic RNA for synthesis of the replicase domain-containing polyprotein (3). Removal of the TLSTYMV from the native TYMV RNA completely abolished polyprotein synthesis, whereas translation of the first ORF into movement protein and that of the subgenomic RNA (sgRNA) into coat protein were unchanged. The 3′-linked valine was found to become incorporated at the N terminus of the polyprotein in a cap-independent and initiator-tRNA-independent fashion. Polyprotein synthesis could also start, however, by the action of a nonaminoacylated or 3′-truncated TLSTYMV. This discovery prompted us to investigate whether the much larger TLSs of a different genus of viruses have a comparable function in translation, and in the present study we focus on the TLSBMV of Brome mosaic virus (BMV), the type species of the genus Bromovirus.
The TLSTYMV-mediated initiation of TYMV polyprotein synthesis is exceptional, since eukaryotic translation normally starts with initiation-factor-dependent recognition of the 5′ m7G(5′)pppN cap and recruitment of the 40S ribosomal subunit. Then, by numerous events that are catalyzed and regulated by at least 12 initiation factors, the 40S ribosomal subunit with initiator tRNA is prepared for scanning towards the first AUG as a start codon. After 60S subunit association, the ribosome can enter the elongation phase of the translation cycle. Efficient translation depends on 5′- to 3′-end communication of the mRNA, established by a 3′ poly(A) stretch that communicates via the poly(A) binding protein (PABP) and eIF4G with eIF4E bound to the capped 5′ end. This circular form is believed to assess the mRNA integrity and to recycle ribosomes for multiple rounds of translation. Indeed, the 5′ cap binding factors and PABP synergistically stimulate translation (33).
As intracellular parasites, viruses need the host translation apparatus for their reproduction, and their RNAs compete with the host mRNAs for ribosomes to start translation. Remarkably, viral RNAs do not always contain the usual 5′ cap and 3′ poly(A) entities, although they are efficiently translated. For TYMV RNA, this circularization appears to be established by 3′-TLSTYMV communication with the start of ORF2 (3). For BMV, with a different genomic organization, the 3′ TLSBMV may also contribute to communication with the 5′ end in a somehow related way.
BMV has a tripartite genome (Fig. 1A): RNA1 codes for a protein with domains homologous to methyltransferases and helicases (Me/He), RNA2 codes for an RNA-dependent RNA polymerase (RdRp), and RNA3 codes for the viral movement protein (MP) and the coat protein (CP). Because of the downstream location of the CP cistron, the latter is exclusively expressed from an sgRNA synthesized from RNA3. All of these RNAs carry a 5′ cap and a highly conserved 3′ TLSBMV, a bulky structure of about 200 nucleotides (Fig. 1B). Though not strongly mimicking a canonical tRNATyr, the TLS is a substrate for tyrosylation both in vivo and in vitro (9, 11, 16, 21), and it functions as a promoter for minus-strand synthesis (25, 31). It also functions as a nucleation site for coat protein assembly and encapsidation of the BMV RNAs into virions (7).
FIG. 1.
Genomic organization of BMV. (A) Schematic view of genomic RNAs 1 to 3 with ORFs for the following proteins (molecular masses are shown in parentheses in the figure): methyltransferase/helicase (Me/He), RdRp, MP, and CP, which is only translated from the sgRNA. All of the RNAs have the same 5′ cap (•) and 3′ TLSBMV. (B) Secondary structure of the TLSBMV with nucleotide numbering in the 3′→5′ direction. The nucleotides in the shaded boxes were hybridized to two DNA oligonucleotides for TLSBMV disruption by RNase H. The various stem-loop structures are indicated (A to E).
It is not self-evident to see an analogy between the TLSBMV and the TLSTYMV with respect to the latter's functioning as an initiator of translation of a second and overlapping ORF, since the genomic organization of BMV is quite different, without overlapping ORFs. Our experimental strategy was as follows. We first studied the effect of TLSBMV disruption on the translation of the BMV RNAs in an in vitro wheat germ system and discovered clear effects on genomic RNAs 1 to 3, but no effect on the sgRNA. We looked for TLSBMV-mediated tyrosine incorporation in BMV translation products but did not get a final answer due to the discovery of an unexpectedly high level of self-tyrosylation of the tyrosyl-tRNA synthetase (TyrRS) induced by the TLSBMV. Finally, we did complementation studies with the TLSBMV added in trans to BMV RNAs with 3′-disrupted TLSs and localized the complementation activity to a specific TLSBMV subdomain. On the basis of our results, we discuss a model with a prime role for the TLSBMV during the BMV infection cycle.
MATERIALS AND METHODS
RNA preparations.
Native BMV RNA was purchased from Promega (Madison, Wis.). Infectious BMV clones were kind gifts from P. Ahlquist (17). For the synthesis of 5′-capped T7 transcripts of BMV RNA1, -2, and -3, a T7 RiboMAX transcription kit (Promega) was used, with the following adaptations to the protocol. The reaction was initiated in the presence of 1 mM cap analogue, m7G(5′)ppp(5′)G (New England Biolabs, Beverly, Mass.), together with a 5 mM concentration (each) of ATP, CTP, and UTP. After a 15-min initial incubation at 37°C, the mixture was supplemented with 1 mM GTP to allow further chain elongation. This was repeated every 15 min until the transcription mixture contained 5 mM GTP. The total incubation time was 210 min. 32P-labeled transcripts were obtained by internal incorporation of [α-32P]CTP (3,000 Ci/mmol). The T7 transcripts of tRNATyr from Saccharomyces cerevisiae and of the different TLSBMV species were synthesized without m7G(5′)ppp(5′)G addition. Prior to runoff transcription, the plasmids for the transcription of BMV RNAs 1 to 3 were linearized with EcoRI, and the plasmids for (mutant) TLSBMV and tRNATyr were linearized with BstNI (11). Small transcripts were purified by 8% (wt/vol) polyacrylamide gel electrophoresis under denaturing conditions (PAGE/DC), followed by electroelution and ethanol precipitation (11), and large transcripts were purified by phenol extraction and ethanol precipitation. The resulting RNA pellets were dissolved in 50 μl of H2O and applied to a Micro Bio-Spin (P-30) column (Bio-Rad, Hercules, Calif.) in order to fully remove unincorporated nucleotides and low-molecular-weight contaminants. This step was repeated once. The integrity of the RNAs was checked by gel electrophoresis in agarose.
TLS disruption.
For the disruption of TLSBMV, two different DNA oligonucleotides (Invitrogen Europe) were designed, with one being complementary to stem-loop structure C and part of structure D and the other being complementary to stem-loop structures B3 and E (Fig. 1B). These regions were chosen because of their 100% conservation between the different BMV RNAs. A 30-fold molar excess of each DNA oligonucleotide compared to BMV RNA was allowed to anneal with the RNA, and the TLSBMV was disrupted by digestion with RNase H (New England Biolabs) (3). Treated samples underwent phenol extraction and further purification in Micro Bio-Spin (P-30) columns. Although the DNA oligonucleotides did not encompass the whole TLSBMV, this procedure allowed the complete removal of the TLSBMV. The latter was checked in a reverse transcription assay with SuperScript II reverse transcriptase (using a protocol from Invitrogen) by using the same DNA oligonucleotides as 5′ 32P-end-labeled primers, followed by PAGE/DC analysis, and no more products were observed. The specificity of the RNase H cleavage procedure was also checked with [α-32P]CTP-labeled T7 transcripts of BMV RNA, and no additional cleavage sites upstream of the TLSBMV were detected (not illustrated). This specific and restricted cleavage also appeared from the almost full complementation of the disturbed BMV RNA (−TLS) translation by the addition in trans of TLSBMV(-A, -B1, -B3, -E), as shown in Fig. 5.
FIG. 5.
Translational complementation in trans with shortened TLSBMV variants and with yeast tRNATyr as a reference molecule. The effect of increasing concentrations of TLSBMV(-B2) and TLSBMV(-C) (left gel) and of TLSBMV(-A, -B1, -B3, -E) and yeast tRNATyr (right gel) on the translation of 3′-truncated BMV RNAs 1 to 3 was monitored. Above each gel, the schematic RNA structures of the TLSBMV variants are shown, with lacking parts indicated in gray. Relative band intensities were calculated from three independent experiments. See the legend for Fig. 2 for further information.
Translation.
All in vitro translation assays were carried out in a wheat germ system (Promega) as described previously (3), with the incorporation of in vitro translation-grade [35S]Met (37 TBq/mmol) (Amersham Pharmacia Biotech) for autoradiographic analysis on a PhosphorImager (Bio-Rad) after sodium dodecyl sulfate-PAGE separation.
For measurements of the half-lives of the different BMV RNAs under these translation conditions, samples programmed with [α-32P]CTP-labeled BMV RNAs were taken at different time points and mixed with 2 volumes of stop solution (98% [vol/vol] formamide, 10 mM EDTA, 0.025% [wt/vol] bromophenol blue) for subsequent PAGE/DC analysis. Band intensities were scanned on a PhosphorImager.
For measurements of the 5′ cap dependencies of translation of the different BMV RNAs, wheat germ translation mixtures with increasing concentrations of the cap analogue m7G(5′)ppp(5′)G (New England Biolabs) were preincubated for 5 min before the addition of the BMV RNAs. The preincubation mixtures were complemented with additional MgCl2 (0.8 mol/mol of cap analogue) in order to compensate for the chelating effect of the cap analogue (2).
Tyrosylation.
All tyrosylation reactions, with 1 μM TyrRS from yeast and 4 μM [3,5-3H]Tyr (1.81 TBq/mmol) (Amersham Pharmacia Biotech), were essentially performed as described by Fechter et al. (11). [3H]Tyr incorporation was analyzed by liquid scintillation counting or by PAGE as described by Barends et al. (3).
RESULTS
TLSBMV regulates BMV RNA translation.
In order to establish whether the TLSBMV might have any function in protein synthesis, we disrupted the TLSBMV of the population of native BMV RNAs isolated from virions by adding an excess of a mixture of two different DNA oligonucleotides complementary to two conserved parts of the TLSBMV (Fig. 1B) and by site-directed cleavage of the heteroduplex with RNase H. As a result (Fig. 2), the translation of RNA1 and RNA2 decreased about 10-fold, whereas the translation of RNA3 into MP decreased about 2- to 5-fold, depending on the other RNA concentrations. The translation of sgRNA into CP remained completely unaffected. This effect on translation was independent of the amount of oligonucleotides added in the range of a 20- to 600-fold excess over BMV RNA during the TLSBMV disruption procedure (not illustrated). Apparently, the TLSBMV does regulate translation of the genomic RNAs. One could argue that the TLSBMV functionally mimics a poly(A) tail and that its removal would thus simply reduce translation by an increased susceptibility of the 3′ end and upstream regions to degradation. However, sgRNA translation into CP (Fig. 2) was completely unaffected by the removal of the TLSBMV, which argues against such an explanation. As shown below, degradation cannot possibly be the explanation, because the addition of the TLSBMV in trans effectively restored translation of the genomic RNAs. Just as for the population of native RNAs, the same pattern of reduced translation activity was observed after the removal of the TLSBMV from the T7 transcripts of RNAs 1 to 3, and Fig. 2 illustrates the effects for the separate transcripts. The removal of the TLSBMV caused the enhanced appearance of a protein band of about 50 kDa (see asterisk in Fig. 2) for the mixture of native RNAs. The product apparently originates from RNA2 (−TLS) and may be due to a change in RNA folding which causes ribosomal pausing.
FIG. 2.
Effects of TLSBMV disruption on BMV RNA translation in a wheat germ system. The separate genomic RNAs 1, 2, and 3 (15 nM for each T7 transcript), their mixture (RNA1-3) (25 nM for each T7 transcript, i.e., 75 nM for the total population), and a native BMV RNA population mixture (8.6 nM for the total population) were subjected to TLSBMV disruption (−) and used for in vitro translation. The translation patterns of [35S]Met-labeled protein bands are compared to those for intact RNAs (+). The positions of molecular weight markers and the BMV proteins are indicated on the left and the right, respectively. The asterisk indicates an incomplete product of enhanced intensity after TLSBMV disruption. The bottom panels show the scanned relative intensities (%) of full-length protein bands together with their incomplete products, taking the intensities produced by the intact (+) RNAs as 100% (average values from at least two independent experiments).
In order to check the translational activities of the 5′-capped genomic BMV T7 transcripts in our wheat germ system in terms of RNA stability and cap dependency of translation, we determined their half-lives during translation and their responses to increasing concentrations of the cap analogue m7GpppG as a competitive inhibitor of translation initiation (the inhibitor concentrations at half-maximum inhibition [IC50 values] are presented). Table 1 shows that both RNA stability and cap dependency are very similar for the three different RNAs. Also, for the native BMV RNAs, we found comparable IC50 values of m7GpppG for RNA1 to -3 and sgRNA, as already reported by Ali et al. (2). The similarity of the two sets of IC50 values, with all at about 20 μM m7GpppG, underscores the efficiency of the in vitro capping procedure for T7 transcripts described above. Because of the similar stabilities and cap dependencies of all the BMV RNAs at rate-limiting concentrations, we think that the different effects on their translational activities caused by TLSBMV removal may be related to a difference in initiation efficiencies, a situation that was previously found for the genomic and subgenomic TYMV RNAs (3).
TABLE 1.
The stabilities of BMV RNAs 1 to 3 in a wheat germ translation system and the dependence of their translational activities on a 5′ cap structurea
| RNA species | Half-lifeb (min) | IC50 of m7GpppGc (μM) |
|---|---|---|
| RNA1 | 73 ± 15 | 20 ± 6 |
| RNA2 | 60 ± 18 | 18 ± 8 |
| RNA3 | 58 ± 9 | 20 ± 1 |
All values were measured for 5′-capped T7 transcripts under standard translation conditions at 25°C (3) and are average values from two independent experiments.
Determined by scanning the band intensities of intact 32P-labeled RNAs after PAGE/DC.
Measured by scanning the [35S]Met-labeled translation products after SDS-PAGE.
Is tyrosylation of TLSBMV important for translation of BMV RNA?
In a way that is analogous to that for the Val-TLSTYMV of TYMV RNA, the tyrosine residue of the Tyr-TLSBMV might become incorporated as the N-terminal amino acid of BMV proteins. For the isolation of tyrosylated BMV RNAs and free TLSBMV, we tried several approaches but only obtained unexpectedly low recoveries of tyrosylated RNAs after a standard acidic phenol extraction (phenol solutions saturated with 0.2 M sodium acetate at pHs 4.5, 5.0, 5.5, and 7.5 were tested). Only <1% [3H]Tyr-TLSBMV could be retrieved from a TLSBMV fraction after phenol extraction (as measured by liquid scintillation counting of trichloroacetic acid (TCA)-precipitated aliquots before and after phenol extraction), whereas typically 60% [3H]Tyr-tRNATyr could be retrieved by the same procedure.
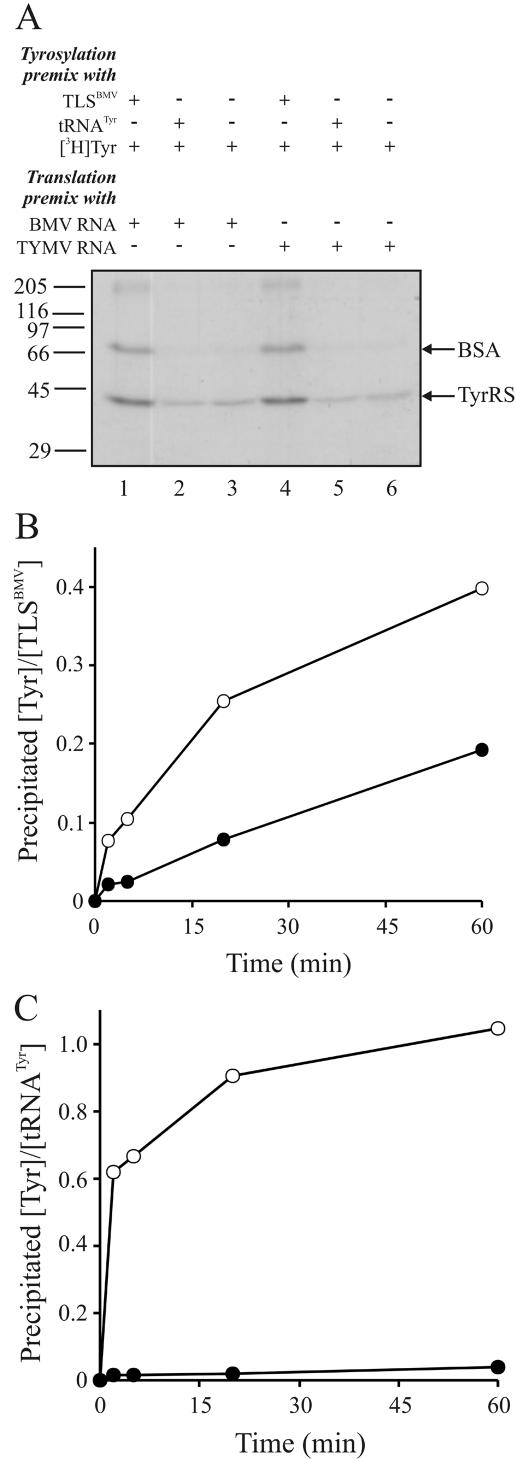
To study Tyr-TLSBMV behavior during BMV RNA translation, we then tried a coupled reaction system by preincubating a TLSBMV [3H]tyrosylation mixture (with 1 μM TLSBMV, with or without tRNATyr as controls) for 60 min and adding it to a standard translation reaction mixture with a 1,500-fold excess of unlabeled Tyr and with ribosomes preprogrammed with 8.6 nM native BMV RNA mixture or with 100 nM native TYMV RNA (as a control). The subsequent incubation step of the coupled tyrosylation-translation reaction took 90 min. No BMV-encoded proteins with TLSBMV-derived [3H]Tyr were detected by PAGE and autoradiography. However, two protein bands, of about 45 and 70 kDa, did appear in either case (Fig. 3A, lanes 1 and 4). The same band pattern was also observed when the preincubated tyrosylation mixtures were analyzed without the addition of the wheat germ system (not illustrated). Surprisingly, these [3H]Tyr-containing proteins appeared to correspond to the yeast TyrRS and bovine serum albumin (BSA), which were present at 1 and 5 μM, respectively, in our standard tyrosylation reactions. During the 60-min preincubation step in the absence of translation, the TLSBMV must have strongly induced Tyr-AMP formation in an idling reaction of the TyrRS, resulting in the subsequent tyrosylation of TyrRS itself and of BSA. In contrast, complex formation between TyrRS and yeast tRNATyr only led to background levels of idling Tyr-AMP formation and protein modification, comparable to the levels without tRNATyr added (Fig. 3A, lanes 2 and 5 and lanes 3 and 6, respectively). In a kinetic tyrosylation assay with the TLSBMV, one set of time points was stopped in the usual way by TCA precipitation to reveal the accumulated amounts of [3H]tyrosylated RNAs and proteins, and another set was stopped by the addition of a 150-fold molar excess of RNase A prior to TCA precipitation in order to identify exclusively the amount of [3H]tyrosylated proteins (Fig. 3B). After 60 min, up to 50% of the total counts of TCA-precipitable [3H]Tyr appeared to originate from the side reaction with TyrRS and BSA. In a parallel experiment with the tRNATyr aminoacylation reaction, only background levels of protein-linked [3H]Tyr were retained after RNase A treatment (Fig. 3C). For unknown reasons, the remaining 50% of the [3H]Tyr linked to the TLSBMV (Fig. 3B) could not be recovered in significant amounts by a standard acidic phenol extraction (see above), whereas for Tyr-tRNATyr a significant level of recovery was obtained. Due to these complications and to the high background levels of [3H]Tyr modification of TyrRS and BSA in coupled BMV RNA [3H]tyrosylation and translation experiments, we could not answer the question about a putative role of the Tyr-TLSBMV as a ribosomal substrate for the N-terminal donation of Tyr to BMV proteins.
FIG. 3.
[3H]Tyr-labeled proteins and RNAs in a coupled tyrosylation-translation system. (A) Autoradiogram of a protein gel after sodium dodecyl sulfate-PAGE of coupled tyrosylation-translation reaction mixtures. The premix compositions of the various samples are indicated above the gel, molecular weight markers are shown on the left, and reference positions of BSA and TyrRS are shown on the right. For further details, see the text. (B and C) Tyrosylation kinetics were monitored for the TLSBMV (B) and for yeast tRNATyr (C). During the 60-min incubation of the [3H]tyrosylation mixture, samples were either directly quenched in TCA (○) or incubated in the presence of a 150-fold molar excess of RNase A for 2 min at 37°C prior to TCA precipitation, filtration, and liquid scintillation counting (•). All data are representative of three independent experiments.
TLSBMV complementation in trans.
To examine whether the TLSBMV could also function in trans during translation, just like the TLSTYMV, we analyzed translation in mixtures of the 3′-truncated RNAs 1 to 3 in the presence of increasing concentrations of free TLSBMV, and indeed, the TLSBMV partially restored the translation of the 3′-truncated RNAs (Fig. 4). An optimum for this complementation mechanism was observed with about an eightfold excess of TLSBMV over BMV RNA (Fig. 4). A further increase of added TLSBMV reduced translation. Importantly, the addition of excess amounts of yeast tRNATyr did not have significant effects on the translation efficiency of the 3′-truncated BMV templates (Fig. 5).
FIG. 4.
Translational complementation in trans by the free and complete TLSBMV. The defective translation of T7 transcripts of BMV RNAs 1 to 3 with a disrupted 3′ TLSBMV (−) was complemented with increasing concentrations of free TLSBMV (indicated in molar excess over the 3′-truncated RNAs) and was compared to the translation pattern of the intact RNAs (+). Optimal complementation was observed at about an eightfold molar excess of free TLSBMV. Relative band intensities were calculated from three independent experiments. See the legend for Fig. 2 for further information.
In order to further localize the complementation activity, we tested TLSBMV variants that lacked certain substructures and had been studied previously for their substrate activities with TyrRS (11). The TLSBMV variant without hairpin structure B2, TLSBMV(-B2), restored translation even better than the complete TLSBMV (Fig. 5). Again, this complementation showed an optimum, at about a 2.5-fold molar excess of TLSBMV over the 3′-truncated BMV RNAs. In contrast, TLSBMV(-C), which lacks hairpin C, could not complement much for translation in trans, and already at a 2.5-fold molar excess of TLSBMV(-C) or higher, the translation of the 3′-truncated BMV RNAs was inhibited even further, until there was no translation with a 12.5-fold excess. When increasing concentrations of TLSBMV(-C) were added to a translation mixture programmed with an intact and native population of BMV RNAs, all translation, including that of sgRNA, was inhibited, with half-maximum inhibition at a 50-fold excess of TLSBMV(-C) (not illustrated). TLSBMV(-C) possibly inhibits translation by binding and inactivating an essential factor. Intriguingly, TLSBMV(-A, -B1, -B3, -E), which actually only harbors stem-loop structures B2, C, and D, was surprisingly efficient at complementation (Fig. 5) and could restore translation almost completely (70 to 80%) to the original pattern (a result also showing the absence of significant upstream RNA cleavage after 3′-TLSBMV removal). Apparently, a combination of stem-loop structures B2, C, and D is not only sufficient for the initiation of translation from the 5′ ends of the genomic RNAs, but can initiate even more efficiently than the complete free TLSBMV.
DISCUSSION
This study was aimed at investigating to what extent the TLS of BMV RNA is involved in the translation of viral protein, as recently reported for TYMV (3). In contrast to the single genomic TYMV RNA with its overlapping ORFs 1 and 2, the genome of BMV is located on three different genomic RNAs (Fig. 1) without any overlapping ORFs. Although the TLSBMV does have functional properties resembling those of a canonical tRNATyr, its structural tRNA mimicry is not as obvious as that of the TLSTYMV for tRNAVal.
Effects of TLSBMV removal.
In our wheat germ system, disruption of the 3′ TLSBMV part of the BMV RNAs strongly reduced the translation of RNA1 and -2 into Me/He and RdRp (needed early in infection), whereas the translation of RNA3 into movement protein was less dependent on an intact TLSBMV. Coat protein expression from sgRNA was not significantly affected by TLSBMV disruption (MP and CP both function later in infection). Interestingly, this resembles the effect observed when the TLSTYMV of TYMV RNA is disrupted: the expression of polyprotein (with domains for Me/He and RdRp) is blocked, whereas that of MP and CP is not affected (3).
The 3′ UTR of the BMV RNAs, with three pseudoknot structures just upstream of the TLSBMV, is known to stimulate translation (as a 3′ fusion construct with reporter genes in carrot protoplasts) by functionally substituting for a poly(A) tail, as was also observed for a comparable 3′ UTR of tobacco mosaic virus (TMV) (13). Work by others (19) indicated the importance of the RNA context of 3′ UTRs; results from experiments in vivo (barley protoplasts) and in vitro (wheat germ extract) with a series of partial UTR deletions confined to only BMV RNA3 and sgRNA showed no UTR stimulation of translation but an essential stimulation of RNA replication for RNA3. For the single genomic TMV RNA, the 3′-UTR pseudoknot domain (also called UPD) just upstream of the 3′ TLSTMV appeared to specifically bind eIF1A (34). The single genomic TYMV RNA does not contain such a prominent pseudoknot domain, and its polyprotein translation in a wheat germ system is fully dependent on the presence of the TLSTYMV (note that its translation of MP from the genomic RNA and of CP from the sgRNA is TLSTYMV independent [3]). Whereas this TLSTYMV-mediated polyprotein translation of TYMV RNA was found to be cap independent (but not that of MP and CP [3]), translation of the three BMV RNA transcripts was fully cap dependent in a wheat germ system (Table 1). The same is true for the subgenomic BMV RNA4 and has also been reported for translation of the native BMV RNAs in a rabbit reticulocyte lysate (2). Apparently, both the pseudoknot domain and the TLSBMV regulate BMV translation. Since our BMV RNAs with a deleted TLSBMV still have an intact 3′ pseudoknot domain, the TLSBMV may play a dominant role for the differential stimulation of translation.
Novel TyrRS reaction induced by TLSBMV.
During our attempts to purify substantial amounts of [3H]Tyr-TLSBMV, the TLSBMV-triggered protein tyrosylation by TyrRS was an unexpected discovery. Self-aminoacylation was already known for AspRS from yeast, and this activity has been shown to be abolished by the presence of tRNAAsp (18). Therefore, the specific stimulation by the TLSBMV of TyrRS-catalyzed protein tyrosylation and the contrasting absence of such a stimulation by the cognate tRNATyr are now interesting. The protein tyrosylation rate of the enzyme was much slower, however, than the initial TLSBMV aminoacylation rate and did not significantly affect the kcat and Km values reported earlier for the TLSBMV (11; data not shown). We did not succeed in detecting a signal of TLSBMV-donated N-terminal Tyr to specific viral translation products in the high background noise of TyrRS-mediated tyrosylation of other proteins. On account of our complementation studies with TLSBMV subfragments, however, we concluded that the presence of a 3′-linked Tyr is not strictly essential for the start of RNA1 and -2 translation and that their translation products are not necessarily initiated with Tyr. This finding is parallel to observations for TYMV RNA (3). It is worthwhile to mention that in the latter study, no side reactions of ValRS-mediated [3H]valylation of proteins were found and the N-terminal [3H]Val incorporation took place with purified [3H]Val-TLSTYMV.
Complementation in trans by TLSBMV fragments.
In our wheat germ system, the defective translation of BMV RNAs 1 to 3 caused by the removal of their 3′ TLSs can be partially complemented by the addition in trans of a small RNA fragment corresponding to a free TLSBMV. Strikingly, even a subfragment with only the hairpin conformation of the structures B2, C, and D and without an acceptor-arm-like structure is sufficient for restoring this translation and complements even better than the complete TLSBMV fragment. Apparently, just as for the TLSTYMV, a 3′-terminal structure upstream of the final 3′ acceptor arm is crucial for translation. This also implies that the complete TLSBMV may represent a compromise for efficient functioning in both translation and replication (6), whereas the folding of the shortened fragment may be more favorable for translation. For the distantly related Alfalfa mosaic virus (AMV), an Alfamovirus also belonging to the family Bromoviridae, a structural switch of the 3′ UTR from a hairpin array into a sort of TLS regulates the functional transition from translation to replication (24, 27). For this TLSAMV, only recognition by a CCA-adding enzyme was detected, but no aminoacylation occurred; the TLS conformation was found to be required for the initiation of replication. The translationally active hairpin conformation that is required during the first steps of the AMV infection and that no longer contains an acceptor-arm-like structure, is essentially induced by coat protein binding to this region, which prevents the initiation of replication. Accordingly, for efficient translation at the early stage of virus infection, a few molecules of coat protein are required in the inoculum. Either the coat protein or the exposed loop sequences of the hairpin structures bind translation factors and establish 5′- to 3′-end communication of the viral RNA (5). For BMV, a similar structural transition has been suggested (1, 29), but the presence of coat protein in the inoculum is not required for an infection. Interestingly, a mutant AMV with a mutant coat protein with a lost affinity for the 3′ UTR, hence displaying a lost infectivity for protoplasts, can be rescued by replacing the 3′ UTR of AMV with that of BMV (24a).
Stem-loop structure C has been extensively investigated and is known to be an essential docking site for the replicase complex in order to start minus-strand synthesis (31). Especially its terminal triloop is important for the association of the replicase, either directly or via an as yet unidentified host factor. Hairpin structure B2 has been suggested to structurally mimic the anticodon stem-loop structure of canonical tRNATyr. Although B2 interacts with the TyrRS enzyme, extensive mutational analysis revealed that the sequence of the loop structure can be changed without any loss of tyrosylation efficiency (11). In the current three-dimensional model of the TLSBMV, hairpin D is positioned such that it may mimic the TΨC hairpin of canonical tRNAs, and indeed the base of this hairpin structure makes strong contacts with the TyrRS enzyme. However, no functional role for this hairpin has been reported thus far (11, 12), and it is absent from the related Tyr-accepting TLSBBMV of broad bean mottle virus (6).
Model for a role of TLSBMV in RNA 5′- to 3′-end communication.
Assuming a 5′- to 3′-end communication in the case of BMV RNAs, one can envisage an intramolecular RNA-RNA interaction such as that found for barley yellow dwarf virus. In the latter case, a stem-loop structure in the 3′ UTR specifically base pairs via a kissing-loop interaction with the loop sequence of a hairpin structure in the 5′ UTR (15). Could BMV RNA1 and -2 interact through a hypothetical kissing-loop interaction between 3′ TLSBMV structures B2, C, and/or D and a structure in the 5′ UTR, whereas the 5′ UTRs of RNA3 and sgRNA interact less or not at all on account of a totally different sequence and folding? If so, this may explain why TLSBMV disruption does not have such pronounced effects on the translation of RNA3 and sgRNA as it does for RNA1 and RNA2. Otherwise, a 5′- to 3′-end communication for efficient eukaryotic translation can also be established by protein-protein interactions other than those between initiation factors and PABP, such as those shown for AMV RNA and for various cellular mRNAs (14, 20, 23, 24, 30, 33). Recently, the translation of BMV RNAs 1 to 3 (but not that of sgRNA or cellular mRNAs) was shown to depend on host protein factors that are normally involved in deadenylation-dependent mRNA decapping (26).
Apparently the TLS of BMV plays a remarkable and central role in the life cycle of the virus. Upon its entry into the host cell, the first step during infection is to start the synthesis of viral enzymes for RNA replication. Here we have shown that (parts of) the TLSBMV plays a crucial role in the expression of those enzymes. When the replicase has initiated minus-strand RNA synthesis and is moving on the plus strand from the 3′ TLSBMV in the 5′ direction, it could clash with ribosomes that are translating this plus strand from 5′ to 3′. However, the dual functioning of the TLSBMV, both as a trigger for translation initiation and as a promoter for minus-strand RNA synthesis, may prevent the simultaneous start of protein and RNA synthesis on the same RNA molecule. TLSBMV binding to the ribosome would prevent TLSBMV binding by the replicase and vice versa. This dual regulation is absent for the sgRNA; its translation into coat protein is not regulated by the TLSBMV and it does not serve as a template for minus-strand synthesis. Perhaps additional host factors function in this dual regulation and serve as a docking site for either the ribosome or the replicase. Quadt et al. (28) reported that a subunit of translation initiation factor 3 from wheat germ as well as from rabbit reticulocytes specifically binds to the RdRp of BMV. At the end of the infection cycle, the virus needs to encapsidate its genomic RNAs for the production of mature virions. Again, the TLS of BMV is a target for regulation, as a nucleation site for coat protein assembly and RNA encapsidation (7). Coat protein binding to the TLSBMV would then prevent alternative interactions with ribosome or replicase. For AMV, there is a different situation, in which deletion of the 3′ UTRs of RNAs 1 and 2 does not interfere with their encapsidation by the coat protein originating from RNA3 (32). For TYMV, the initiation signal for RNA encapsidation appears to be localized on a hairpin structure with characteristic C-C and C-A mismatches located in the 5′ UTR close to the polyprotein start codon, the contact area for the 3′ TLSTYMV (4).
The central role of the TLSBMV in the temporal regulation of the consecutive processes of RNA translation, RNA transcription, and RNA encapsidation in BMV may be of comparable importance for the infection cycles of other TLS-harboring viruses.
Acknowledgments
We are grateful to P. Ahlquist and M. Prins for their kind gifts of infectious BMV clones. We thank J. F. Bol for valuable discussions and Agnès Clénet for technical assistance.
This work was supported by a short-term FEBS fellowship to S.B.
REFERENCES
- 1.Ahlquist, P., R. Dasgupta, and P. Kaesberg. 1981. Near identity of 3′ RNA secondary structure in bromoviruses and cucumber mosaic virus. Cell 23:183-189. [DOI] [PubMed] [Google Scholar]
- 2.Ali, I. K., L. McKendrick, S. J. Morley, and R. J. Jackson. 2001. Truncated initiation factor eIF4G lacking an eIF4E binding site can support capped mRNA translation. EMBO J. 20:4233-4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barends, S., H. H. J. Bink, S. H. E. van den Worm, C. W. A. Pleij, and B. Kraal. 2003. Entrapping ribosomes for viral translation: tRNA mimicry as a molecular Trojan horse. Cell 112:123-129. [DOI] [PubMed] [Google Scholar]
- 4.Bink, H. H. J., K. Hellendoorn, J. van der Meulen, and C. W. A. Pleij. 2002. Protonation of non-Watson-Crick base pairs and encapsidation of turnip yellow mosaic virus RNA. Proc. Natl. Acad. Sci. USA 99:13465-13470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bol, J. F. 2003. Alfalfa mosaic virus: coat protein-dependent initiation of infection. Mol. Plant Pathol. 4:1-8. [DOI] [PubMed] [Google Scholar]
- 6.Bujarski, J. J., T. W. Dreher, and T. C. Hall. 1985. Deletions in the 3′-terminal tRNA-like structure of brome mosaic virus RNA differentially affect aminoacylation and replication in vitro. Proc. Natl. Acad. Sci. USA 82:5636-5640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choi, Y. G., T. W. Dreher, and A. L. Rao. 2002. tRNA elements mediate the assembly of an icosahedral RNA virus. Proc. Natl. Acad. Sci. USA 99:655-660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dreher, T. W. 1999. Functions of the 3′-untranslated regions of positive strand RNA viral genomes. Annu. Rev. Phytopathol. 37:151-174. [DOI] [PubMed] [Google Scholar]
- 9.Dreher, T. W., and T. C. Hall. 1988. Mutational analysis of the tRNA mimicry of brome mosaic virus RNA. Sequence and structural requirements for aminoacylation and 3′-adenylation. J. Mol. Biol. 210:41-55. [DOI] [PubMed] [Google Scholar]
- 10.Fechter, P., J. Rudinger-Thirion, C. Florentz, and R. Giegé. 2001. Novel features in the tRNA-like world of plant viral RNAs. Cell Mol. Life Sci. 58:1547-1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fechter, P., R. Giegé, and J. Rudinger-Thirion. 2001. Specific tyrosylation of the bulky tRNA-like structure of brome mosaic virus RNA relies solely on identity nucleotides present in its amino acid-accepting domain. J. Mol. Biol. 309:387-399. [DOI] [PubMed] [Google Scholar]
- 12.Felden, B., C. Florentz, R. Giegé, and E. Westhof. 1994. Solution structure of the 3′-end of brome mosaic virus genomic RNAs. Conformational mimicry with canonical tRNAs. J. Mol. Biol. 235:508-531. [DOI] [PubMed] [Google Scholar]
- 13.Gallie, D. R., and M. Kobayashi. 1994. The role of the 3′-untranslated region of non-polyadenylated plant viral mRNAs in regulating translational efficiency. Gene 142:159-165. [DOI] [PubMed] [Google Scholar]
- 14.Gallie, D. R. 2002. Protein-protein interactions required during translation. Plant Mol. Biol. 50:949-970. [DOI] [PubMed] [Google Scholar]
- 15.Guo, L., E. M. Allen, and W. A. Miller. 2001. Base-pairing between untranslated regions facilitates translation of uncapped, nonpolyadenylated viral RNA. Mol. Cell 7:1103-1109. [DOI] [PubMed] [Google Scholar]
- 16.Hall, T. C., D. S. Shih, and P. Kaesberg. 1972. Enzyme mediated binding of tyrosine to brome mosaic virus ribonucleic acid. Biochem. J. 129:969-976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Janda, M., R. French, and P. Ahlquist. 1987. High efficiency T7 polymerase synthesis of infectious RNA from cloned brome mosaic virus cDNA and effects of 5′ extensions on transcript infectivity. Virology 158:259-262. [DOI] [PubMed] [Google Scholar]
- 18.Kern, D., B. Lorber, Y. Boulanger, and R. Giegé. 1985. A peculiar property of aspartyl-tRNA synthetase from bakers' yeast: chemical modification of the protein by the enzymatically synthesized aminoacyl adenylate. Biochemistry 24:1321-1332. [Google Scholar]
- 19.Lahser, F. C., L. E. Marsh, and T. C. Hall. 1993. Contributions of the brome mosaic virus RNA-3 3′-nontranslated region to replication and translation. J. Virol. 67:3295-3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ling, J., S. J. Morley, V. M. Pain, W. F. Marzluff, and D. R. Gallie. 2002. The histone 3′-terminal stem-loop-binding protein enhances translation through a functional and physical interaction with eukaryotic initiation factor 4G (eIF4G) and eIF3. Mol. Cell. Biol. 22:7853-7867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loesch-Fries, L. S., and T. C. Hall. 1982. In vivo aminoacylation of brome mosaic and barley stripe mosaic virus RNAs. Nature 298:771-773. [Google Scholar]
- 22.Mans, R. M. W., C. W. A. Pleij, and L. Bosch. 1991. tRNA-like structures: structure, function and evolutionary significance. Eur. J. Biochem. 201:303-324. [DOI] [PubMed] [Google Scholar]
- 23.Mazumder, B., V. Seshadri, and P. L. Fox. 2003. Translational control by the 3′-UTR: the ends specify the means. Trends Biochem. Sci. 28:91-98. [DOI] [PubMed] [Google Scholar]
- 24.Neeleman, L., R. C. L. Olsthoorn, H. J. M. Linthorst, and J. F. Bol. 2001. Translation of a nonpolyadenylated viral RNA is enhanced by binding of viral coat protein or polyadenylation of the RNA. Proc. Natl. Acad. Sci. USA 98:14286-14291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24a.Neeleman, L., H. J. M. Linthorst, and J. F. Bol. 2004. Efficient translation of alfamovirus RNAs requires the binding of coat protein dimers to the 3′ termini of the viral RNAs. J. Gen.Virol. 85:231-240. [DOI] [PubMed] [Google Scholar]
- 25.Noueiry, A. O., and P. Ahlquist. 2003. Brome mosaic virus RNA replication: revealing the role of the host in RNA virus replication. Annu. Rev. Phytopathol. 41:77-98. [DOI] [PubMed] [Google Scholar]
- 26.Noueiry, A. O., J. Diez, S. P. Falk, J. Chen, and P. Ahlquist. 2003. Yeast Lsm1p-7p/Pat1p deadenylation-dependent mRNA-decapping factors are required for brome mosaic virus genomic RNA translation. Mol. Cell. Biol. 23:4094-4106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Olsthoorn, R. C. L., S. Mertens, F. T. Brederode, and J. F. Bol. 1999. A conformational switch at the 3′ end of a plant virus RNA regulates viral replication. EMBO J. 18:4856-4864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Quadt, R., C. C. Kao, K. S. Browning, R. P. Hershberger, and P. Ahlquist. 1993. Characterization of a host protein associated with brome mosaic virus RNA-dependent RNA polymerase. Proc. Natl. Acad. Sci. USA 90:1498-1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rietveld, K., C. W. A. Pleij, and L. Bosch. 1983. Three-dimensional models of the tRNA-like 3′ termini of some plant viral RNAs. EMBO J. 2:1079-1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sanchez, R., and W. F. Marzluff. 2002. The stem-loop binding protein is required for efficient translation of histone mRNA in vivo and in vitro. Mol. Cell. Biol. 22:7093-7104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sivakumaran, K., K. Hema, and C. Cheng Kao. 2003. Brome mosaic virus RNA syntheses in vitro and in barley protoplasts. J. Virol. 77:5703-5711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vlot, A. C., L. Neeleman, H. J. M. Linthorst, and J. F. Bol. 2001. Role of the 3′-untranslated regions of alfalfa mosaic virus RNAs in the formation of a transiently expressed replicase in plants and in the assembly of virions. J. Virol. 75:6440-6449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wilkie, G. S., K. S. Dickson, and N. K. Gray. 2003. Regulation of mRNA translation by 5′- and 3′-UTR-binding factors. Trends Biochem. Sci. 28:182-188. [DOI] [PubMed] [Google Scholar]
- 34.Zeenko, V. V., L. A. Ryabova, A. S. Spirin, H. M. Rothnie, D. Hess, K. S. Browning, and T. Hohn. 2002. Eukaryotic elongation factor 1A interacts with the upstream pseudoknot domain in the 3′ untranslated region of tobacco mosaic virus RNA. J. Virol. 76:5678-5691. [DOI] [PMC free article] [PubMed] [Google Scholar]