Abstract
Avermectins produced by Streptomyces avermitilis are potent anti-parasitic agents that are useful in animal health care, agriculture, and the treatment of human infections. In a search for novel regulators that affect avermectin biosynthesis, comparative transcriptome analysis was performed between wild-type strain ATCC31267 and avermectin overproducing strain 76-02-e, revealing some differentially expressed genes. SAV576, which is downregulated in 76-02-e and encodes a TetR family transcriptional regulator (TFR), was shown to inhibit avermectin production by indirectly affecting the expression of ave genes. SAV576 directly repressed the transcription of its gene SAV576 and of adjacent genes SAV575 (encodes cytochrome P450/NADPH-ferrihemoprotein reductase) and SAV574. The SAV576-binding sites within the bidirectional SAV575-SAV576 promoter region were determined by DNase I footprinting assays. A consensus 15-bp palindromic sequence CCRTACRVYGTATGS was found in these binding sites and shown to be important for SAV576-binding activity. SAV575, an important target gene of SAV576, was shown to exert a positive effect on avermectin production. The study findings extend our limited knowledge of the complex regulation of avermectin biosynthesis and provide a basis for rational genetic manipulation of S. avermitilis to improve avermectin production through control of SAV576 and its target gene.
Introduction
Streptomyces species are gram-positive filamentous soil bacteria known for their ability to produce a wide range of bioactive secondary metabolites during their complex life cycle. These metabolites include many useful antibiotics that display antibacterial, anticancer, anthelmintic, and/or immunosuppressive activities [1], [2]. The genes responsible for the biosynthesis of these antibiotics are usually clustered, and are co-regulated by pathway-specific regulatory genes and various higher-level pleiotropic regulatory genes [3]. The initiation of the expression of these regulatory genes is affected by a variety of environmental and physiological factors, including growth rate, imbalances in metabolism [4], nutrient levels (carbon, nitrogen, and phosphate) [5]–[10], and small signaling molecules such as γ-butyrolactone [11]–[13] and ppGpp [14]–[16]. The production of the antibiotics is thus a complex process that is tightly regulated at multiple genetic levels.
Avermectins are a series of potent anthelmintic and insecticidal macrolide antibiotics (A1a, A1b, A2a, A2b, B1a, B1b, B2a, and B2b) produced by S. avermitilis. They are used commercially for broad-spectrum parasite control in medical, veterinary, and agricultural fields [17], [18]. The avermectin biosynthetic pathway has been well elucidated [18]–[20], and the complete S. avermitilis genome has been sequenced [21], [22]; however, the complex regulatory mechanisms of avermectin production remain poorly understood. The regulatory genes that are reportedly involved in avermectin biosynthesis include: aveR [19], [23]–[25], aveR1/aveR2 [26], orfX [27], afsK-av [28], aveI [29], [30], SAV3818 [31] and avaR3 [32]. Further studies are required to identify other yet-unknown regulatory genes, which will contribute to better understanding of the regulatory networks of avermectin biosynthesis and to the practical construction of avermectin high-producing strains.
Comparative transcriptome analysis has been applied increasingly during the past decade to identify alterations of gene expression in antibiotic-overproducing Streptomyces strains [33]–[35] and has been shown to be an efficient technique for the discovery of novel regulatory genes. In the present study, we compared the transcriptomes of S. avermitilis wild-type strain ATCC31267 and avermectin high-producer 76-02-e using a S. avermitilis whole-genome microarray chip and thereby revealed some putative regulatory genes that may be related to avermectin biosynthesis. We further characterized a previously unknown TetR family transcriptional regulator (TFR) gene, SAV576, as an important avermectin downregulator, and demonstrated that SAV576 inhibits avermectin production by modulating the transcription level of its target genes and ave genes.
Materials and Methods
Strains, Plasmids, and Growth Conditions
S. avermitilis wild-type strain ATCC31267 was used as a host strain for gene propagation and gene disruption. 76-02-e, an avermectin high-producer, was derived from ATCC31267 by continuous mutagenesis (two rounds of high frequency electronic flow mutagenesis, five rounds of NTG mutagenesis, Co60 mutagenesis, and three rounds of UV mutagenesis) and was collected in our laboratory [36]. S. avermitilis strains were grown at 28°C on solid YMS medium [6] for sporulation or in liquid YEME medium [37] containing 25% sucrose for growth of mycelia. RM14 medium [38] was used for regeneration of protoplasts and for selection of transformants. MM agar [37] was used for observation of S. avermitilis phenotype. Seed medium and fermentation medium FM-I used for avermectin production were as described previously [39]. Because FM-I contains insoluble yeast meal, soluble fermentation medium FM-II [25] was used to cultivate mycelia for growth and ChIP analysis and for RNA isolation. In comparison to ATCC31267, 76-02-e produced amounts of avermectins that were approximately 40-fold higher (∼5000 µg/ml) when grown in FM-I, and 5-fold higher (∼600 µg/ml) when grown in FM-II.
E. coli strains JM109 and BL21 (DE3) (Novagen) were used as the cloning host and the expression host, respectively. E. coli ET12567 (dam dcm hsdS) [38] was used to propagate non-methylated DNA for transformation into S. avermitilis. E. coli strains were grown at 37°C in Luria-Bertani (LB) medium [40]. The antibiotics used were described previously [41]. Multiple-copy vector pKC1139 [42] was used for gene disruption and overexpression in S. avermitilis. pSET152 [42] was used to introduce a single-copy gene into S. avermitilis. pET-28a (+) (Novagen) was used for production of recombinant His6-tagged protein in E. coli.
Microarray Assays
Mycelia of ATCC31267 or 76-02-e grown in FM-II were collected on days 2 and 6, flash-frozen in liquid nitrogen, and ground into a fine powder. RNA was extracted using Trizol reagent (Tiangen, China) according to the manufacturer’s instructions. Agilent microarrays (8×15K) for the analysis of S. avermitilis gene expression were designed and manufactured by Shanghai Biochip Co. Ltd (SBC, China), based on publicly available complete genome sequence information (http://avermitilis.ls.kitasato-u.ac.jp). For each gene, two different 60-mer oligonucleotides were designed. Each slide contained a total of 7670 open reading frames (ORFs). The microarray assays, including labeling, hybridization, washing, and microarray data normalization, were performed by SBC.
Microarray Dataset Accession Number
The raw microarray dataset used in this study has been submitted to NCBI Gene Expression Omnibus under the accession number GSE47223.
Gene Disruption, Complementation, and Overexpression
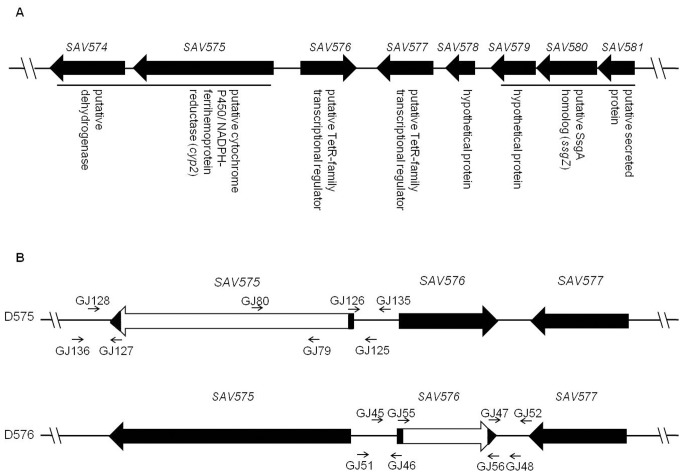
To construct a SAV576 deletion mutant, two fragments flanking SAV576 were prepared by PCR from the genomic DNA of ATCC31267. A 796-bp 5′ flanking region was amplified with primers GJ45 and GJ46, and a 746-bp 3′ flanking region was amplified with primers GJ47 and GJ48. The two PCR fragments were ligated into pKC1139 to generate a SAV576-deletion vector pDGJ576. Transformation of pDGJ576 into ATCC31267 and selection of double-crossover recombinant strains were performed as described previously [41]. The SAV576-deleted mutant D576 was confirmed by PCR using primers GJ51, GJ52, GJ55, and GJ56 (Fig. 1B). When primers GJ51 and GJ52, which flank the exchange regions, were used for PCR analysis of putative SAV576 deletion mutant, a 1.75-kb band appeared, whereas a 2.36-kb band was detected when genomic DNA of ATCC31267 was used as the template. When using primers GJ55 and GJ56 located within the deletion region of SAV576 gene, only ATCC31267 produced a 0.9-kb PCR fragment as predicted (data not shown). For complementation of D576, a 1.57-kb DNA fragment carrying the SAV576 ORF and its putative promoter was amplified by PCR with primers GJ45 and GJ56*. The PCR product was inserted into pSET152 to generate SAV576 complementation vector pSET152-576, which was then introduced into D576 to obtain the complemented strain. The 1.57-kb EcoRI/XbaI fragment containing the SAV576 from pSET152-576 was cloned into pKC1139 to produce pKC1139-576, which was used for overexpession of SAV576 in ATCC31267.
Figure 1. Organization of SAV576 and its adjacent genes on the chromosome of S. avermitilis (A) and schematic representation of the strategy used for deletion of SAV575 and SAV576 genes (B).
(A) Gene notations are based on the Genome Project of S. avermitilis (http://avermitilis.ls.kitasato-u.ac.jp/). The two transcriptional units are indicated by black bars. (B) Long black arrows indicate genes and their directions. Short arrows indicate the positions of primers used for cloning exchange regions and confirming gene deletions. White blocks represent in-frame deletions in the corresponding genes.
To construct a SAV575 deletion mutant, a 739-bp 5′ flanking region and a 517-bp 3′ flanking region were amplified with primer pairs GJ125/GJ126 and GJ127/GJ128, respectively. The two PCR fragments were cloned into pKC1139 to produce a SAV575-deletion vector pDGJ575, which was then introduced into ATCC31267. The resulting SAV575-deleted mutant D575 was confirmed by PCR using primers GJ135, GJ136, GJ79, and GJ80 (Fig. 1B). When using primers GJ135 and GJ136, which flank the exchange regions, a 1.48-kb band appeared, whereas the theoretical 4.46-kb band was too large to be detected when genomic DNA of ATCC31267 was used as the template. When using primers GJ79 and GJ80 located within the deletion region of SAV575, only ATCC31267 yielded a 369-bp PCR fragment (data not shown). A 4.13-kb DNA fragment carrying the promoter and the coding region of SAV575 was amplified with primers GJ89 and GJ90, and was cloned into pKC1139 to give pKC1139-575 for overexpression of SAV575 in ATCC31267 or in D576.
The construction of SAV575-576 double mutant D575-576 was similar to that of D575, using D576 as host strain. All primers used in this study are listed in Table S1.
Semiquantitative and Real-time RT-PCR Analyses
RNA extractions were carried out with Trizol (Tiangen) from cultures of S. avermitilis at various times. To remove chromosomal DNA contamination, each RNA sample was treated with DNase I and tested by PCR to confirm the absence of chromosomal DNA. The treated RNA sample (2 µg) was reverse transcribed using M-MLV (RNase H-, TaKaRa), random hexamers (25 µM), and a dNTP mixture (10 mM each). Semiquantitative RT-PCR analysis was performed to determine the transcription levels of various genes using the obtained cDNA as template and the primers listed in Table S1. The hrdB gene, which encodes the major sigma factor in Streptomyces, was used as a positive internal control in the RT-PCR assays. The obtained cDNAs were also used as templates for real-time PCR analysis. The experiments were performed using FastStart Universal SYBR Green Master (ROX) (Roche) with analysis by an ABI 7900HI Sequence Detection System using optical-grade 96-well plates. Template cDNA, 10 µl FastStart Universal SYBR Green Master (ROX), and forward and reverse primers (each 300 nM) were mixed in each reaction system (total volume 20 µl). The PCR protocol consisted of 95°C for 10 min, 40 cycles of 95°C for 10 s, and 60°C for 30 s with a single fluorescence measurement.
Overexpression and Purification of Recombinant His6-tagged SAV576
A DNA fragment encoding the predicted 218 amino acids of SAV576 protein was generated by PCR with primers GJ71 and GJ72. The PCR fragment was inserted into the expression vector pET-28a (+) to generate pET28-576, which was then introduced into E. coli BL21 (DE3) for protein overexpression. Following induction by IPTG, the resulting recombinant His6-tagged SAV576 protein was purified on a Ni2+-NTA spin column according to the manufacturer’s instructions (Qiagen). The purified protein was used for antibody induction, EMSA, and DNase I footprinting assays.
Preparation of Antibodies against SAV576 Protein
Polyclonal antibodies against SAV576 were prepared by Beijing Protein Innovation (China). 200 µg purified recombinant protein His6-SAV576 was mixed with Freund’s complete adjuvant and injected into a rabbit. After 2 weeks, the antigen was injected into the same rabbit with Freund’s incomplete adjuvant. Further booster immunizations were given at 11-day intervals. The rabbit was bled 10 days after each boost, and serum was prepared. Each serum was stored at 4°C, and its potency was checked by ELISA. After three booster immunizations, the immune serum attained a high potency and was used as a source of anti-SAV576 antibodies.
Chromatin Immunoprecipitation (ChIP) Assay
The ChIP protocol was as described previously [25]. Briefly, S. avermitilis cultures grown in FM-II for 72 h were fixed in cross-linked buffer [0.4 M sucrose, 10 mM Tris·Cl (pH 8.0), 1 mM EDTA] containing 1% formaldehyde for 20 min at 28°C. Chromatin immunoprecipitation was performed using anti-SAV576 antibody. After DNA extraction, pellets were washed with 70% ethanol and resuspended in 50 µl TE, and 2 µl DNA solution was used for PCR using the primer sets listed in Table S1.
Western Blotting
Western blotting analyses were performed as described previously [25]. Polyclonal antibody against AveR [25] or SAV576 was used at a dilution of 1∶1,000. Western blots were developed with polyclonal antibodies using an ECL detection system (Roche).
Electrophoretic Mobility Shift Assay (EMSA)
EMSAs were performed using a DIG Gel Shift Kit, 2nd Generation (Roche). The probes were amplified by PCR and labeled at the 3′ ends with nonradioactive digoxigenin (DIG). The 20 µl reaction mixture contained the probes, proteins, and 1 µg poly [d(I-C)] in a binding buffer. The mixture was incubated at 25°C for 30 min and then added with 5 µl loading buffer with bromophenol blue. Protein-bound and free DNA were separated by electrophoresis on non-denaturing 5% polyacrylamide gels with 0.5×TBE as running buffer, and were then transferred onto nylon membranes by electroblotting. The membranes were baked for 10 min at 80°C, and the DNA fragments were cross-linked by exposure to UV radiation for 10 min. Chemiluminescence detection was performed according to the manufacturer’s instructions, and the membranes were exposed to X-ray film (Fuji) for 15–30 min.
DNase I Footprinting
A non-radiochemical capillary electrophoresis method was used for DNase I footprinting [43]. To characterize the binding sites of SAV576 protein in the SAV575-SAV576 intergenic region, two fluorescence-labeled DNA fragments were synthesized by PCR using primer pairs FAM-GJ78/GJ77 and FAM-GJ228/GJ227. The resulting 547-bp and 478-bp DNA fragments covered the entire intergenic region. Following purification, labeled DNA fragments (400 ng) and appropriate concentrations of His6-tagged SAV576 protein were added to a final reaction volume of 50 µl and incubated for 30 min at 25°C. Digestion with DNase I (0.016 units) was performed for 40 s at 37°C and stopped by the addition of EDTA at a final concentration of 60 mM. The reaction mixture was heated to 80°C for 10 min to totally inactivate DNase I. The samples were subjected to phenol-chloroform extraction, ethanol precipitation, and capillary electrophoresis by loading into an Applied Biosystems 3730 DNA Genetic Analyser together with the internal-lane size standard ROX-500 (Applied Biosystems). Electrophoregrams were analyzed using the GeneMarker program, v1.8 (Applied Biosystems).
Identification of the Transcriptional Start Point Using 5′-RACE
To determine the transcriptional start point of SAV575 and SAV576, total RNA was extracted from an 84-h culture of ATCC31267 grown on YMS medium. 2 µg total RNA was used for reverse transcription with 20 pmol of gene-specific primer 575SP1 or 576SP1 using a 5′/3′ RACE Kit (2nd Generation, Roche). The sample was purified using a PCR Product Purification Kit (Beijing HT-Biotech Co. Ltd, China). An oligo-dA tail was added to the 3′ end of the cDNA using terminal deoxynucleotidyl transferase, followed by direct amplification of the tailed cDNA using the oligo dT-anchor primer (GACCACGCGTATCGATGTCGACTTTTTTTTTTTTTTTTV) and a second inner gene-specific primer 575SP2 or 576SP2. An additional round of PCR was performed with a 1,000-fold dilution of the original PCR product as template, using an anchor primer (GACCACGCGTATCGATGTCGAC) and a nested 575SP3 or 576SP3 primer, to obtain a single specific band. The final PCR products were cloned into pMD18-T vector (TaKaRa) for sequencing. The first nucleotide following the oligo-dA sequence was considered to be the transcriptional start point.
Fermentation and HPLC Analysis of Avermectin Production
Fermentation of S. avermitilis ATCC31267 and its mutants was performed, and avermectins in the fermentation culture were identified by HPLC analysis as described previously [39].
Results
Transcriptome Comparison between Wild-type and Avermectin Overproducing Strains
Total RNA was isolated from mycelia of S. avermitilis wild-type strain ATCC31267 and avermectin overproducing strain 76-02-e grown in FM-II at day 2 (early exponential phase) and day 6 (stationary phase), and was used for microarray assays. Comparative transcriptome analysis revealed that 162 genes were upregulated and 150 were downregulated 2-fold or more in 76-02-e relative to ATCC31267 at both of the studied time points. Most of the differentially expressed genes encoded unknown or unclassified proteins (upregulated 73, downregulated 85). Smaller numbers of genes were involved in regulatory functions (upregulated 16, downregulated 16), transport and binding proteins (upregulated 11, downregulated 7), mobile and extrachromosomal element functions (upregulated 14, downregulated 6), or secondary metabolism (upregulated 20, downregulated 18). The remaining genes were associated with sig-regulons, amino acid metabolism, fatty acid and phospholipid metabolism, carbohydrate metabolism, purine/pyrimidine metabolism, biosynthesis of cofactors and prosthetic groups, protein synthesis, folding and modification, RNA recombination, cell envelope, cell division/morphological differentiation, or gas vesicle cluster (File S1). As expected, most of the genes (17 of 19) in the avermectin biosynthetic gene cluster, including aveR (encoding a pathway-specific activator) and aveA1 (encoding polyketide synthase AVES1), were significantly overexpressed in 76-02-e (File S1), which was consistent with the increased avermectin production.
Real-time RT-PCR was used to test five upregulated genes (aveR, aveA1, SAV292, SAV880, SAV4189) and two downregulated genes (SAV576, SAV151). The PCR results were mainly consistent with the microarray data (Fig. S1), demonstrating the general reliability of the microarray data. Among the tested genes, SAV292, SAV880, SAV576 and SAV151 encode TFRs, and SAV4189 encodes a MarR-family transcriptional regulator. The functions of these regulatory genes were not determined. The SAV576 gene was of particular interest. The microarray data indicated that SAV576 in 76-02-e was downregulated 37-fold at day 2 and 93-fold at day 6 (File S1), suggesting that this gene is involved in the regulation of avermectin biosynthesis. We therefore performed functional analysis of SAV576.
Characterization of SAV576 and Its Adjacent Genes
The SAV576 gene, located in the left arm of the chromosome, contains 657 nucleotides and encodes a TFR of 218 amino acids, including a common N-terminal helix-turn-helix DNA-binding domain and a C-terminal all-alpha domain homologous to TetR. The divergently transcribed genes SAV575 and SAV574 are located upstream of SAV576. SAV575 (cyp2) encodes a putative cytochrome P450/NADPH-ferrihemoprotein reductase, and SAV574 encodes a putative dehydrogenase (Fig. 1A). SAV577 is another TFR gene located downstream of SAV576. SAV578 and SAV579 both encode hypothetical proteins. SAV580 (ssgZ) is an ssgA homologous gene, and SAV581 encodes a putative secreted protein. RT-PCR analysis using primers that amplify intergenic regions revealed that SAV574 and SAV575 are co-transcribed, and that SAV579-SAV580-SAV581 forms another transcriptional unit (Fig. S2). The SAV576, SAV577, SAV578, SAV579, and SAV581 genes are specific to S. avermitilis and have no orthologs in other sequenced Streptomyces genomes.
SAV576 Plays an Important Role in Avermectin Production
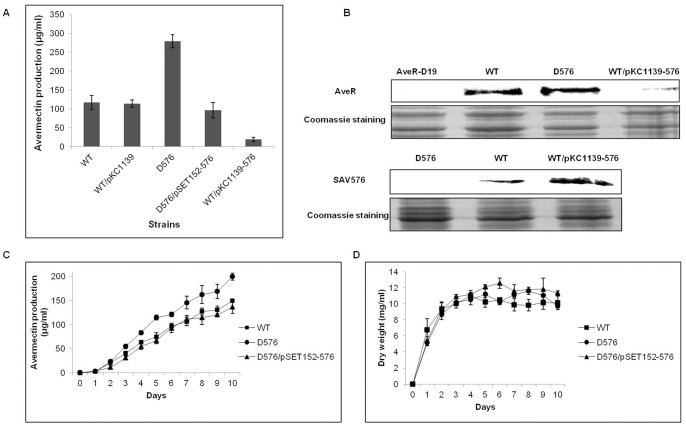
To assess the role of SAV576 in S. avermitilis, the SAV576 deletion mutant D576 was constructed (Fig. 1B). D576 displayed no obvious differences from the wild-type strain when grown on YMS, RM14, or MM solid medium (data not shown). HPLC analysis of the fermentation products following cultivation in FM-I for 10 days showed that avermectin production in D576 was ∼2.3-fold compared to that in ATCC31267 (Fig. 2A). Avermectin production was restored in the SAV576 gene complementation strain (D576/pSET152-576), confirming that the increased avermectin production in D576 was due solely to the deletion of SAV576. The enhancement of SAV576 expression in wild-type strain (WT/pKC1139-576) led to a clear reduction in avermectin yield (Fig. 2A). The deletion and overexpression of SAV576 in wild-type strain were confirmed by Western blotting (Fig. 2B). Western blotting analysis of the avermectin pathway-specific activator AveR showed that, in agreement with the avermectin yield, AveR was overexpressed in D576 but underexpressed in WT/pKC1139-576 (Fig. 2B). These findings suggest that SAV576 inhibits avermectin production by directly or indirectly affecting the expression of aveR.
Figure 2. Avermectin production and growth of wild-type ATCC31267 and SAV576 mutant strains.
(A) Comparison of avermectin production in various S. avermitilis strains grown in FM-I medium for 10 days. WT, wild-type strain ATCC31267; WT/pKC1139, ATCC31267 carrying control plasmid pKC1139; D576, SAV576 deletion mutant; D576/pSET152-576, complementation strain of D576; WT/pKC1139-576, SAV576 overexpression strain. (B) Western blotting analysis of SAV576 and AveR protein in cells grown in FM-I for 6 days. Approximately 100 µg total protein of each sample was subjected to Western blot and Coomassie Blue staining for the loading control. AveR-D19, aveR mutant. (C and D) Effect of SAV576 deletion on avermectin production (C) and growth (D) of S. avermitilis grown in FM-II. Solid squares, ATCC31267; Solid circles, D576; Solid triangles, complementation strain of D576.
To investigate whether the avermectin overproduction in D576 was due to increased cell growth, we analyzed the growth and avermectin production of ATCC31267, D576, and D576/pSET152-576 cultured in FM-II. The deletion of SAV576 resulted in increased avermectin production (Fig. 2C), but did not affect cell growth (Fig. 2D). The growth and avermectin production of D576/pSET152-576 were both similar to those of ATCC31267. These findings indicate that SAV576 acts to inhibit avermectin biosynthesis, but has no effect on cell growth.
The Transcription Levels of SAV576 Vary at Different Life Stages
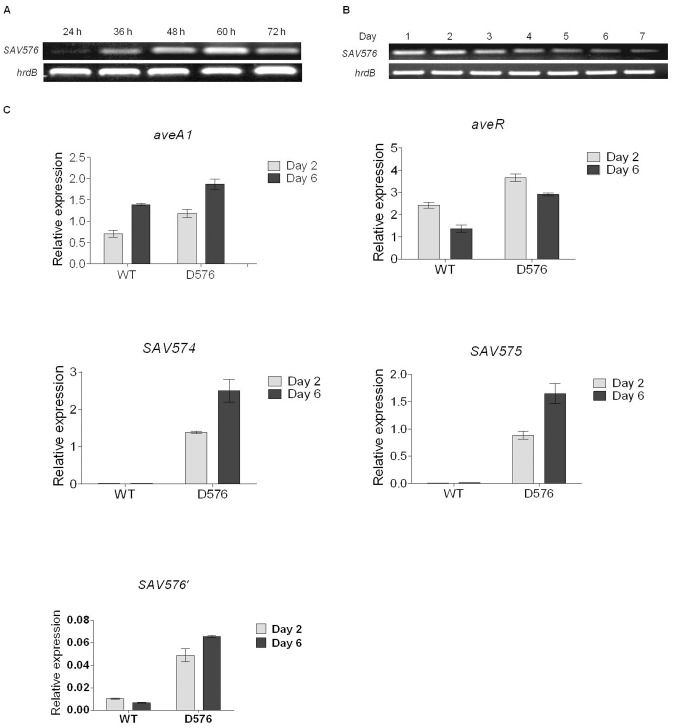
To determine the transcription levels of SAV576 at various life stages, semiquantitative RT-PCR was performed using RNA isolated from ATCC31267 grown on solid sporulation medium YMS, or in liquid fermentation medium FM-II for various durations. Various stages of S. avermitilis development were clearly observable on YMS: vegetative (substrate mycelia) growth (represented by the 24 h RNA sample) was followed by aerial growth (the 36-h, 48-h and 60-h RNA samples) and then by sporulation (>60 h, represented by the 72-h RNA sample). SAV576 transcription was detectable throughout the life cycle, but its level varied depending on the time point. The transcription level was low at 24 h when the strain was growing as substrate mycelia, increased during aerial hyphae growth with a peak at 60 h, and then decreased at 72 h, when mature spores were evident (Fig. 3A). These data suggest that the transcription of SAV576 is controlled by some factor(s) related to morphological development.
Figure 3. Transcriptional analysis of SAV576 and related genes.
(A and B) Semiquantitative RT-PCR analysis of transcription levels of SAV576 in ATCC31267 grown on solid medium YMS (A) and in liquid medium FM-II (B) for various durations. The 214-bp SAV576 transcript was amplified from the internal coding region of SAV576 with primers GJ83 and GJ84. hrdB was used as a positive internal control. (C) Real-time RT-PCR analysis of aveR, aveA1, SAV574, SAV575 and SAV576 transcription levels from ATCC31267 (WT) and D576 grown in FM-II on days 2 and 6. Relative values were obtained using hrdB as a reference. SAV576′, 142-bp transcript amplified from the SAV576 promoter region and the remainder ORF of D576 with primers GJ46* and GJ55*.
In the liquid medium FM-II, S. avermitilis was unable to sporulate but produced larger amount of avermectins than it did on solid medium. Avermectin production was not observable by HPLC on day 1, but then increased gradually from day 2 onward (Fig. 2C). RT-PCR analysis showed that SAV576 transcription reached its maximal level on day 2 and then gradually declined (Fig. 3B). This finding is consistent with the inhibitory role of SAV576 in avermectin biosynthesis.
Transcription of Certain Genes is Affected by SAV576 Deletion
To find potential targets regulated by SAV576, RT-PCR was performed using RNAs isolated from ATCC31267 and from D576 grown for 2 or 6 days in FM-II, corresponding to the stages of the microarray data. Real-time RT-PCR analysis showed that both aveR and aveA1 were upregulated in D576 (Fig. 3C), indicating that SAV576 affects avermectin production by downregulating the expression of ave genes.
The transcription levels of SAV576 and its adjacent divergently transcribed genes SAV574 and SAV575 were tested using the same RNA preparations. Each of these genes was significantly upregulated in D576 on both days (Fig. 3C), indicating that the SAV576 gene product acts either directly or indirectly to downregulate the transcription of its own gene and of the adjacent genes.
Binding of SAV576 to the Intergenic Region between SAV575 and SAV576
TFRs are a common class of transcriptional factors that regulate target genes by binding to their promoters. To assess whether the regulation of the genes listed in the preceding section by SAV576 was direct, we performed ChIP assays and EMSAs.
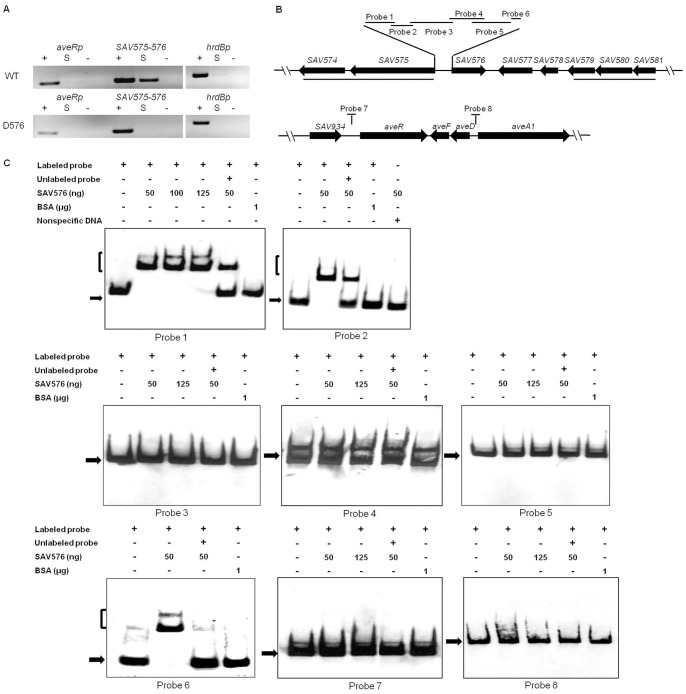
ChIP assays are widely used to determine where DNA-binding proteins bind to the genome in vivo. Following 48 h growth in FM-II, avermectin production in D576 was higher than that in ATCC31267 (Fig. 2C), suggesting that the SAV576 protein binds to its DNA targets and thereby downregulates avermectin biosynthesis prior to this time point. ChIP assays were performed using S. avermitilis strains treated with formaldehyde at 72 h to cross-link the SAV576 protein to its DNA targets. The cross-linked DNA was extracted and fragmented by sonication, and immunoprecipitation was then performed using anti-SAV576 antibodies to screen the DNA fragments that were attached to the SAV576 protein. Two putative promoter regions were chosen on the basis of sequence analysis: a 200-bp region upstream of aveR and a 207-bp region at the SAV575-SAV576 intergenic region containing two divergent promoters. The primer pairs used for PCR detection of the two promoter regions are listed in Table S1. All of the PCR bands of correct size were obtained from the positive control DNA of ATCC31267 or D576, whereas no such bands were detected when the negative control DNA without antibody was used as template (Fig. 4A). In comparison with the control hrdB promoter, it was clear that only the PCR product of the SAV575-SAV576 intergenic region was selectively enriched when the immunoprecipitated DNA of ATCC31267 was used as template. In contrast, no correct PCR bands were amplified from the immunoprecipitated DNA of D576 (Fig. 4A). These results indicate that the SAV576 protein binds specifically to the bidirectional SAV575-SAV576 promoter region, but not to the aveR promoter region in vivo.
Figure 4. Analysis of SAV576 protein binding to target promoter regions.
(A) ChIP assays in vivo. Anti-SAV576 antibodies were used to immunoprecipitate SAV576-DNA complexes from ATCC31267 and D576 cells treated with formaldehyde. The DNAs used for PCR were total DNA prior to immunoprecipitation (positive control: lanes “+”), immunoprecipitated DNA (experimental sample: lanes “S”), and negative control DNA without antibody (lanes “−”). The hrdB promoter region was used as a control. (B) Schematic representation of the relative positions of probes used for EMSAs in vitro. Probe 1, 98-bp DNA fragment from +2 to −96 relative to the translational start codon of SAV575; probe 2, 43-bp fragment from −98 to −140; probe 3, 276-bp fragment from −118 to −393; probe 4, 257-bp fragment from −306 to −562; probe 5, 333-bp DNA fragment from −533 to −865; probe 6, 43-bp DNA fragment from −866 to −908. Probe 7, 200-bp DNA fragment from −116 to −315 relative to the start codon of aveR. Probe 8, 328-bp DNA fragment from −14 to −341 relative to the start codon of aveA1. Probes 7 and 8 cover the putative transcriptional start points of aveR and aveA1, respectively. (C) EMSAs of the interaction of the probes with purified His6-SAV576 protein. Each lane contained 0.3 nM labeled probe. The labeled probe and an approximately 100-fold excess of the unlabeled probe were used in competitive assays. BSA was used as a negative control for SAV576 protein. Labeled non-specific DNA was used to eliminate non-specific binding of SAV576 protein. The free probes are indicated by solid arrows, and the retarded DNA fragments are indicated by parentheses.
To confirm that SAV576 protein binds directly to the above target promoters, we performed in vitro EMSAs using a full-length recombinant His6-SAV576 protein expressed in E. coli. The entire intergenic region between SAV575 and SAV576 was 900-bp in length, and six probes (designated probes 1, 2, 3, 4, 5, and 6) labeled with DIG were designed to cover this region. The 200-bp aveR promoter region used in the ChIP assays and the 327-bp aveD-aveA1 intergenic region were labeled as probes 7 and 8, respectively (Fig. 4B). Results showed that the His6-SAV576 protein clearly retarded probes 1, 2, and 6, but not probes 3, 4, 5, 7, or 8 (Fig. 4C). A labeled nonspecific DNA probe and BSA were used as negative controls. These findings indicate that the SAV576 protein binds to the bidirectional SAV575-SAV576 promoter region directly by binding DNA sites located within the sequences of probes 1, 2, and 6. These in vitro EMSA results are in agreement with those from the in vivo ChIP assays, indicating that the SAV576 protein indirectly regulates avermectin biosynthesis, but directly controls the transcription of its own gene and adjacent gene SAV575 through interaction with their promoter regions. SAV574 is co-transcribed with SAV575 (Fig. S2), indicating that it is also a target gene of SAV576.
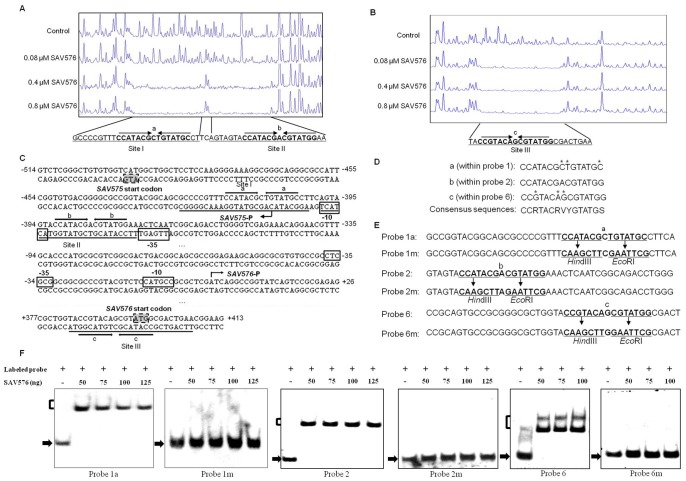
Determination of the Binding Sites of the SAV576 Protein
The results of the transcription experiments and the EMSAs suggest that SAV576 regulates its own gene and SAV575 by direct binding to three regions: probes 1, 2, and 6. To clarify the regulation mechanism of SAV576, we determined the promoter structures of SAV575 and SAV576 and the specific binding sites of SAV576 on the SAV575-SAV576 intergenic region. The transcriptional start point (tsp) of SAV575 was localized by 5′ RACE to C at position 87 nt upstream of the translational start codon of SAV575 (Fig. 5C and S3A). The SAV576 tsp was localized to A at position 405 nt upstream of the translational start codon of SAV576 (Fig. 5C and S3B).
Figure 5. Determination of the binding sites of the SAV576 protein.
(A and B) DNase I footprinting assay of SAV576 on the SAV575 (A) and SAV576 (B) promoter regions, respectively. The fluorograms correspond to the control DNA (10 µM BSA) and to the protection reactions with increasing concentrations of His6-SAV576 protein, respectively. (C) Nucleotide sequence of the SAV575-SAV576 promoter region and SAV576-binding sites. The numbers indicate the distance (nt) from the transcriptional start point of SAV576. Solid lines, SAV576-binding sites; arrows, inverted repeats; bent arrows, transcriptional start points and transcription orientation; boxed areas, putative −10 and −35 regions; shaded areas, translational start codon. (D) Three 15-bp palindromic sequences “a”, “b”, and “c”. The mismatched nucleotides in comparison with sequence b are indicated by asterisks. (E) Mutations introduced into the 15-bp palindromic sequences. Each of the probes used was 43-bp. Probes 1a, 2, and 6 contained sequences a, b, and c, respectively. HindIII and EcoRI sites were generated at sequences a, b, and c to produce mutated probes 1m, 2m, and 6m, respectively. The nucleotides changed are indicated by underlining. (F) EMSAs using the mutated DNA probes. The free probes are indicated by solid arrows, and the retarded DNA fragments are indicated by parentheses.
DNase I footprinting assays of the bidirectional SAV575-SAV576 promoter region were performed in the presence and absence of His6-SAV576 protein, using a capillary sequencer to analyze the protected regions. As expected, three protected regions (sites I, II, and III) were identified on the coding strand of SAV576 (Fig. 5A and B). These three sites were located respectively within the sequences of probes 1, 2, and 6, confirming the direct interaction of SAV576 with these probes. Site I extends from positions +19 to −7, and site II extends from positions −10 to −33, relative to the SAV575 tsp. Sites I and II are separated by 2 nt, and site II overlaps the potential −10 region of the SAV575 promoter (Fig. 5C), indicating that SAV576 negatively regulates SAV575 transcription by blocking the access of RNA polymerase to its promoter region. Site III is located far downstream of the SAV576 tsp (from positions +383 to +407) and overlaps the start codon of SAV576 (Fig. 5C). This finding is analogous to a previous report that the binding site of JadR1 on the cmlJ promoter is far downstream of the cmlJ tsp, and that JadR1 negatively regulates cmlJ expression [44]. SAV576 may negatively regulate the transcription of its own gene by directly interfering with transcription elongation or by recruiting other repressors.
A palindromic sequence is a typical feature of TFR-binding targets. Analysis of the SAV576-binding sites using the DNAMAN program revealed that site II contains a perfect 15-bp palindromic sequence (termed “b”), and that sites I and III contain similar 15-bp sequences (termed “a” and “c”, respectively) (Fig. 5A and B). Sequences “a” and “c” have three mismatched nucleotides in comparison with “b”. The comparison of sequences “a”, “b” and “c” provided a consensus sequence, CCRTACRVYGTATGS (R: A or G; V: A, G, or C; Y: T or C; S: G or C) (Fig. 5D).
To estimate the relative contributions of the three 15-bp sequences to SAV576 protein binding, EMSAs were performed using a probe that contained either the intact 15-bp sequence or a mutated sequence (Fig. 5E). The affinity of SAV576 for the mutated probes (termed 1 m, 2 m, and 6 m), which lacked inverted repeats, was abolished completely in comparison with the corresponding wild-type 43-bp probes 1a, 2, and 6 (Fig. 5F). These results indicate that each of the three 15-bp sequences is important for SAV576-binding activity.
SAV575 Exerts a Positive Effect on Avermectin Production
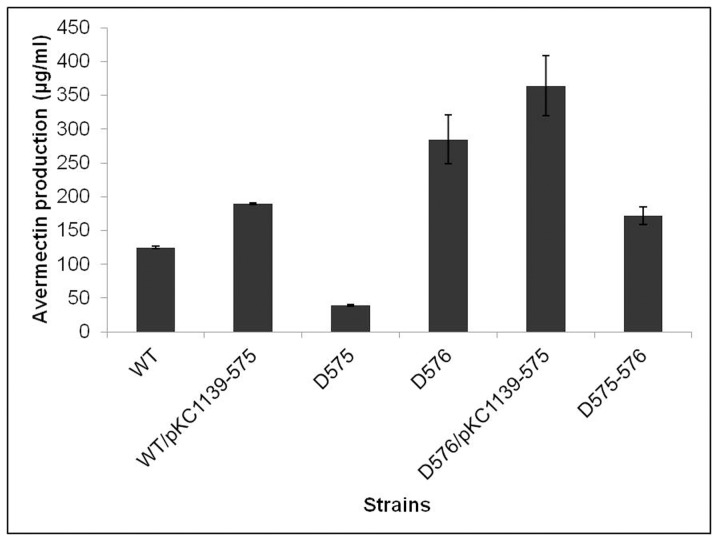
Because SAV575 was found to be a target gene of SAV576, we further investigated its relationship with avermectin production. A comparison of avermectin production among various S. avermitilis strains revealed that the overexpression of SAV575 in wild-type strain (WT/pKC1139-575) and D576 (D576/pKC1139-575) increased avermectin production, whereas the deletion of SAV575 in wild-type strain (D575) and D576 (D575-576) led to decreased avermectin production (Fig. 6). These results indicate that SAV575 has a positive effect on avermectin production.
Figure 6. Comparison of avermectin production in SAV575 mutant strains.
The fermentation products were analyzed by HPLC following cultivation in FM-I for 10 days. WT/pKC1139-575, ATCC31267 containing SAV575 overexpression vector; D575, SAV575 mutant; D576, SAV576 mutant; D576/pKC1139-575, D576 containing SAV575 overexpression vector; D575-576, SAV575-SAV576 double mutant.
Because SAV575 is a cytochrome P450 family gene, it may have no effect on gene expression. To test this hypothesis, we used semiquantitative RT-PCR to measure the transcription of SAV576 and ave genes in ATCC31267 and D575. No marked difference in the transcription levels of these genes was observed (Fig. S4). These results imply that SAV575 exerts its positive effect on avermectin production through some means other than the control of gene expression.
Discussion
TFRs are the third most common transcriptional regulator family in bacteria and regulate a wide range of cellular activities, including antibiotic production, multidrug resistance, amino acid metabolism, osmotic stress, pathogenicity, and development [45]. However, many of the functions of TFRs remain poorly known or unknown. Some Streptomyces species, including S. coelicolor and S. avermitilis, contain over 100 TFR genes. This large number presumably reflects the complex morphological differentiation and secondary metabolism in these species. Of the 115 predicted TFRs in the S. avermitilis genome, only two, SAV3818 [31] and SAV3703 (AvaR3, a γ-butyrolactone-autoregulator receptor) [32] have been characterized as positive regulators of avermectin production. The findings of the present study demonstrate an important role of SAV576, a novel TFR, in the negative control of avermectin production in S. avermitilis. We are currently engaged in characterization of other differentially expressed regulatory genes revealed by transcriptome analysis.
The expression of SAV576 was found to be autoregulated. However, the differing transcription levels of SAV576 at various life stages indicate that the regulation of SAV576 expression is complex and is controlled in part by other yet-unknown upstream factor(s). Studies using a bacterial one-hybrid system will help identify the transcriptional regulators that interact with the SAV576 promoter region. Transcription and binding experiments have shown that SAV576 indirectly downregulates the expression of aveR, which encodes the pathway-specific activator for avermectin biosynthesis [24], [25]. It appears likely that SAV576 controls yet-unknown gene(s) that directly regulate aveR expression. The aveR promoter was shown to be directly recognized and activated by housekeeping EσhrdB in vitro [46]. However, it remains unclear what types of transcriptional regulators directly control aveR expression in S. avermitilis. A transcriptional activator AtrA (SCO4118) was shown to directly regulate the transcription of actII-ORF4, the pathway-specific activator of the actinorhodin biosynthetic gene cluster in S. coelicolor [47]. However, AtrA and its homolog AveI (SAV4110) in S. avermitilis functioned as negative regulators for avermectin biosynthesis, and no direct binding of AtrA or AveI to the promoter region of aveR was observed [29]. Although the γ-butyrolactone-autoregulator receptor AvaR3 positively regulates aveR expression, there is no ARE sequence in the promoter region of aveR, suggesting that such regulation is indirect [32]. Deletion of avaR3 did not affect the transcription level of the adpA homolog SAV5261 [32]. We demonstrated previously that the adpA homolog in S. avermitilis is not involved in the regulation of avermectin production [41]. Taken together, these findings indicate that S. avermitilis has a novel, adpA-independent pathway for the regulation of avermectin biosynthesis. The identification of direct regulators of aveR is essential for better understanding of the regulatory network of avermectin biosynthesis.
Among the SAV576 target genes, we investigated SAV575 in view of its predicted function. SAV575 (CYP102D1) is a member of the cytochrome P450 (CYP) family that catalyzes the monooxygenation of a variety of hydrophobic substrates and plays a key role in primary and secondary metabolic pathways and drug detoxification [48]. S. avermitilis contains 33 CYPs, whereas S. coelicolor contains only 18. Of the CYP genes in S. avermitilis, 11 are located within the secondary metabolite gene cluster and are predicted to be involved in secondary metabolite production [49]. Lamb et al. predicted that some CYP genes that are not linked to a specific gene cluster contribute to secondary metabolite production [49]. SAV575 is not cluster-situated, and its product CYP102D1 is a unique self-sufficient P450 that has no homolog in S. coelicolor. SAV575 or its orthologs in other Actinobacteria (SCAB5931 in S. scabies, SCLAV2750 in S. clavuligerus, SACE4205 in Saccharopolyspora erythraea) had no previously assigned function in the context of antibiotic biosynthesis; however, our findings indicate that SAV575 is involved in avermectin production. The ability of CYP102D1 to catalyze the oxidation of saturated and unsaturated fatty acids [50] suggests that the function of SAV575 is to provide precursors, such as acetate and propionate extender units, for avermectin biosynthesis by oxidizing fatty acids or other compounds. The SAV575 transcription level was very low in wild-type strain ATCC31267. Deletion of SAV576 increased the SAV575 transcription level, presumably increasing the availability of precursors for avermectin biosynthesis and thereby increasing avermectin production. Another possibility is that SAV575 produces molecules that bind to an activator of avermectin biosynthesis to stimulate its DNA-binding affinity or bind to a repressor to eliminate or reduce its DNA-binding affinity. We did not investigate the role of SAV574, another SAV576 target gene, in avermectin biosynthesis. SAV574 encodes a dehydrogenase, and may indirectly provide energy or precursors for avermectin biosynthesis by catalyzing the degradation of certain substrates. Further studies are necessary to evaluate these possibilities and to clarify the function of SAV574.
EMSAs and footprinting assays revealed a 15-bp consensus sequence CCRTACRVYGTATGS that is important for SAV576-binding activity. Similar sequences were also found in many other promoter regions (Table S2). Further experiments will establish which of these genes are SAV576 targets, and will clarify the relationships of the newly discovered SAV576 target genes with aveR expression and avermectin biosynthesis. Improved knowledge of the SAV576 regulatory mechanism will lead to more effective strategies for increasing avermectin production.
Supporting Information
Transcription levels of aveR, aveA1, SAV151, SAV576, SAV292, SAV880, and SAV4189 in avermectin-overproducing strain 76-02-e relative to those in wild-type strain ATCC31267. Samples were collected from each strain grown in FM-II medium after days 2 and 6 of growth. hrdB was used as an internal control. Standard deviations are indicated by error bars (n = 3). Each gene was examined by relative quantification real-time RT-PCR with gene-specific primers.
(TIF)
Confirmation of the transcriptional units SAV574-SAV575 and SAV579-SAV580-SAV581 by RT-PCR. Lanes: RT, RT-PCR; –, negative control with reverse transcriptase omitted; +, positive control with genomic DNA of ATCC31267 as the template. The primers used for amplifying the SAV574-SAV575 intergenic region were GJ109 (GCGCTACCAGCAGGACGT) and GJ110 (CTCCTCCACGGCGAACTT); those used for amplifying the SAV579-SAV581 intergenic region were GJ159 (ACCGCACCCATCAGGAAG) and GJ160 (GGTAAGAAACGAGGGCGTA).
(TIF)
Determination of the transcriptional start points of SAV575 (A) and SAV576 (B) by 5′-RACE PCR. Boxed area: 5′-RACE oligo dT-anchor primer.
(TIF)
Effect of SAV575 deletion on the expression of related genes. Semiquantitative RT-PCR analysis of transcription levels of genes from ATCC31267 (WT) and D575. The strains were grown in FM-II for 6 days. hrdB was used as a positive internal control.
(TIF)
Primers used in this study.
(DOCX)
Putative targets of SAV576.
(DOCX)
List of genes having an expression level at least two-fold higher or lower in avermectin overproducer 76-02-e in comparison with wild-type strain ATCC31267.
(XLS)
Acknowledgments
We thank Prof. G. Liu (Institute of Microbiology, Chinese Academy of Science, China) for critical reading of the draft manuscript and Dr. S. Anderson for English editing.
Funding Statement
This work was supported by grants from the National Natural Science Foundation of China (Grant No. 31170045) and the National Basic Research Program of China (Grant No. 2009CB118905). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Demain AL (2002) Prescription for an ailing pharmaceutical industry. Nat Biotechnol 20: 331. [DOI] [PubMed] [Google Scholar]
- 2. Challis GL, Hopwood DA (2003) Synergy and contingency as driving forces for the evolution of multiple secondary metabolite production by Streptomyces species. Proc Natl Acad Sci USA 100: 14555–14561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bibb MJ (2005) Regulation of secondary metabolism in streptomycetes. Curr Opin Microbiol 8: 208–215. [DOI] [PubMed] [Google Scholar]
- 4. Hood DW, Heidstra R, Swoboda UK, Hodgson DA (1992) Molecular genetic analysis of proline and tryptophan biosynthesis in Streptomyces coelicolor A3(2): interaction between primary and secondary metabolism–a review. Gene 115: 5–12. [DOI] [PubMed] [Google Scholar]
- 5. Aharonowitz Y (1980) Nitrogen metabolite regulation of antibiotic biosynthesis. Annu Rev Microbiol 34: 209–233. [DOI] [PubMed] [Google Scholar]
- 6. Ikeda H, Kotaki H, Tanaka H, Omura S (1988) Involvement of glucose catabolism in avermectin production by Streptomyces avermitilis . Antimicrob Agents Chemother 32: 282–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Martin JF (2004) Phosphate control of the biosynthesis of antibiotics and other secondary metabolites is mediated by the PhoR-PhoP system: an unfinished story. J Bacteriol 186: 5197–5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sanchez S, Chavez A, Forero A, Garcia-Huante Y, Romero A, et al. (2010) Carbon source regulation of antibiotic production. J Antibiot 63: 442–459. [DOI] [PubMed] [Google Scholar]
- 9. van Wezel GP, McDowall KJ (2011) The regulation of the secondary metabolism of Streptomyces: new links and experimental advances. Nat Prod Rep 28: 1311–1333. [DOI] [PubMed] [Google Scholar]
- 10. Martin JF, Santos-Beneit F, Rodriguez-Garcia A, Sola-Landa A, Smith MC, et al. (2012) Transcriptomic studies of phosphate control of primary and secondary metabolism in Streptomyces coelicolor . Appl Microbiol Biotechnol 95: 61–75. [DOI] [PubMed] [Google Scholar]
- 11. Horinouchi S, Beppu T (1994) A-factor as a microbial hormone that controls cellular-differentiation and secondary metabolism in Streptomyces griseus . Mol Microbiol 12: 859–864. [DOI] [PubMed] [Google Scholar]
- 12. Takano E (2006) Gamma-butyrolactones: Streptomyces signalling molecules regulating antibiotic production and differentiation. Curr Opin Microbiol 9: 287–294. [DOI] [PubMed] [Google Scholar]
- 13. Kato JY, Funa N, Watanabe H, Ohnishi Y, Horinouchi S (2007) Biosynthesis of gamma-butyrolactone autoregulators that switch on secondary metabolism and morphological development in Streptomyces . Proc Natl Acad Sci USA 104: 2378–2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Strauch E, Takano E, Baylis HA, Bibb MJ (1991) The stringent response in Streptomyces coelicolor A3(2). Mol Microbiol 5: 289–298. [DOI] [PubMed] [Google Scholar]
- 15. Hesketh A, Chen WJ, Ryding J, Chang S, Bibb M (2007) The global role of ppGpp synthesis in morphological differentiation and antibiotic production in Streptomyces coelicolor A3(2). Genome Biol 8: R161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gomez-Escribano JP, Martin JF, Hesketh A, Bibb MJ, Liras P (2008) Streptomyces clavuligerus relA-null mutants overproduce clavulanic acid and cephamycin C: negative regulation of secondary metabolism by (p)ppGpp. Microbiology 154: 744–755. [DOI] [PubMed] [Google Scholar]
- 17. Burg RW, Miller BM, Baker EE, Birnbaum J, Currie SA, et al. (1979) Avermectins, new family of potent anthelmintic agents: producing organism and fermentation. Antimicrob Agents Chemother 15: 361–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ikeda H, Omura S (1997) Avermectin biosynthesis. Chem Rev 97: 2591–2609. [DOI] [PubMed] [Google Scholar]
- 19. Ikeda H, Nonomiya T, Usami M, Ohta T, Omura S (1999) Organization of the biosynthetic gene cluster for the polyketide anthelmintic macrolide avermectin in Streptomyces avermitilis . Proc Natl Acad Sci USA 96: 9509–9514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ikeda H, Nonomiya T, Omura S (2001) Organization of biosynthetic gene cluster for avermectin in Streptomyces avermitilis: analysis of enzymatic domains in four polyketide synthases. J Ind Microbiol Biotechnol 27: 170–176. [DOI] [PubMed] [Google Scholar]
- 21. Omura S, Ikeda H, Ishikawa J, Hanamoto A, Takahashi C, et al. (2001) Genome sequence of an industrial microorganism Streptomyces avermitilis: Deducing the ability of producing secondary metabolites. Proc Natl Acad Sci USA 98: 12215–12220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ikeda H, Ishikawa J, Hanamoto A, Shinose M, Kikuchi H, et al. (2003) Complete genome sequence and comparative analysis of the industrial microorganism Streptomyces avermitilis . Nat Biotechnol 21: 526–531. [DOI] [PubMed] [Google Scholar]
- 23. Ikeda H, Takada Y, Pang CH, Tanaka H, Omura S (1993) Transposon mutagenesis by Tn4560 and applications with avermectin-producing Streptomyces avermitilis . J Bacteriol 175: 2077–2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kitani S, Ikeda H, Sakamoto T, Noguchi S, Nihira T (2009) Characterization of a regulatory gene, aveR, for the biosynthesis of avermectin in Streptomyces avermitilis . Appl Microbiol Biotechnol 82: 1089–1096. [DOI] [PubMed] [Google Scholar]
- 25. Guo J, Zhao JL, Li LL, Chen Z, Wen Y, et al. (2010) The pathway-specific regulator AveR from Streptomyces avermitilis positively regulates avermectin production while it negatively affects oligomycin biosynthesis. Mol Genet Genomics 283: 123–133. [DOI] [PubMed] [Google Scholar]
- 26.Stutzman-Engwall KJ, Price BS (2001) USA patent no. US 6197591. In., Pfizer Inc.
- 27. Hwang YS, Kim ES, Biro S, Choi CY (2003) Cloning and analysis of a DNA fragment stimulating avermectin production in various Streptomyces avermitilis strains. Appl Environ Microbiol 69: 1263–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rajkarnikar A, Kwon HJ, Ryu YW, Suh JW (2006) Catalytic domain of AfsKav modulates both secondary metabolism and morphologic differentiation in Streptomyces avermitilis ATCC 31272. Curr Microbiol 53: 204–208. [DOI] [PubMed] [Google Scholar]
- 29. Chen L, Lu YH, Chen J, Zhang WW, Shu D, et al. (2008) Characterization of a negative regulator AveI for avermectin biosynthesis in Streptomyces avermitilis NRRL8165. Appl Microbiol Biotechnol 80: 277–286. [DOI] [PubMed] [Google Scholar]
- 30. Chen L, Chen J, Jiang YQ, Zhang WW, Jiang WH, et al. (2009) Transcriptomics analyses reveal global roles of the regulator AveI in Streptomyces avermitilis . FEMS Microbiol Lett 298: 199–207. [DOI] [PubMed] [Google Scholar]
- 31. Duong CT, Lee HN, Choi SS, Lee SY, Kim ES (2009) Functional expression of SAV3818, a putative TetR-family transcriptional regulatory gene from Streptomyces avermitilis, stimulates antibiotic production in Streptomyces species. J Microbiol Biotechnol 19: 136–139. [DOI] [PubMed] [Google Scholar]
- 32. Miyamoto KT, Kitani S, Komatsu M, Ikeda H, Nihira T (2011) The autoregulator receptor homologue AvaR3 plays a regulatory role in antibiotic production, mycelial aggregation and colony development of Streptomyces avermitilis . Microbiology 157: 2266–2275. [DOI] [PubMed] [Google Scholar]
- 33. Lum AM, Huang JQ, Hutchinson CR, Kao CM (2004) Reverse engineering of industrial pharmaceutical-producing actinomycete strains using DNA microarrays. Metab Eng 6: 186–196. [DOI] [PubMed] [Google Scholar]
- 34. Im JH, Kim MG, Kim ES (2007) Comparative transcriptome analysis for avermectin overproduction via Streptomyces avermitilis microarray system. J Microbiol Biotechnol 17: 534–538. [PubMed] [Google Scholar]
- 35. Kang SH, Huang JQ, Lee HN, Hur YA, Cohen SN, et al. (2007) Interspecies DNA microarray analysis identifies WblA as a pleiotropic down-regulator of antibiotic biosynthesis in Streptomyces . J Bacteriol 189: 4315–4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li LL, Guo J, Wen Y, Chen Z, Song Y, et al. (2010) Overexpression of ribosome recycling factor causes increased production of avermectin in Streptomyces avermitilis strains. J Ind Microbiol Biotechnol 37: 673–679. [DOI] [PubMed] [Google Scholar]
- 37.Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA (2000) Practical Streptomyces genetics: a laboratory manual. John Innes Foundation, Norwich, UK.
- 38. Macneil DJ, Klapko LM (1987) Transformation of Streptomyces avermitilis by plasmid DNA. J Indust Microbiol 2: 209–218. [Google Scholar]
- 39. Chen Z, Wen J, Song Y, Wen Y, Li JL (2007) Enhancement and selective production of avermectin B by recombinants of Streptomyces avermitilis via intraspecific protoplast fusion. Chin Sci Bull 52: 616–622. [Google Scholar]
- 40.Sambrook J, MacCallum P, Russell D (2001) Molecular Cloning: A Laboratory Manual, third ed. Cold Spring Harbor Laboratory Press.
- 41. Zhao JL, Wen Y, Chen Z, Song Y, Li JL (2007) An adpA homologue in Streptomyces avermitilis is involved in regulation of morphogenesis and melanogenesis. Chin Sci Bull 52: 623–630. [Google Scholar]
- 42. Bierman M, Logan R, Obrien K, Seno ET, Rao RN, et al. (1992) Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 116: 43–49. [DOI] [PubMed] [Google Scholar]
- 43. Zianni M, Tessanne K, Merighi M, Laguna R, Tabita FR (2006) Identification of the DNA bases of a DNase I footprint by the use of dye primer sequencing on an automated capillary DNA analysis instrument. J Biomol Tech 17: 103–113. [PMC free article] [PubMed] [Google Scholar]
- 44.Xu G, Wang J, Wang L, Tian X, Yang H, et al.. (2010) “Pseudo” gamma-butyrolactone receptors respond to antibiotic signals to coordinate antibiotic biosynthesis. J Biol Chem 285, 27440–27448. [DOI] [PMC free article] [PubMed]
- 45. Yu Z, Reichheld SE, Savchenko A, Parkinson J, Davidson AR (2010) A comprehensive analysis of structural and sequence conservation in the TetR family transcriptional regulators. J Mol Biol 400: 847–864. [DOI] [PubMed] [Google Scholar]
- 46. Zhuo Y, Zhang W, Chen D, Gao H, Tao J, et al. (2010) Reverse biological engineering of hrdB to enhance the production of avermectins in an industrial strain of Streptomyces avermitilis . Proc Natl Acad Sci USA 107: 11250–11254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Uguru GC, Stephens KE, Stead JA, Towle JE, Baumberg S, et al. (2005) Transcriptional activation of the pathway-specific regulator of the actinorhodin biosynthetic genes in Streptomyces coelicolor . Mol Microbiol 58: 131–150. [DOI] [PubMed] [Google Scholar]
- 48. Axarli I, Prigipaki A, Labrou NE (2010) Cytochrome P450 102A2 catalyzes efficient oxidation of sodium dodecyl sulphate: A molecular tool for remediation. Enzyme Res 2010: 125429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lamb DC, Ikeda H, Nelson DR, Ishikawa J, Skaug T, et al. (2003) Cytochrome p450 complement (CYPome) of the avermectin-producer Streptomyces avermitilis and comparison to that of Streptomyces coelicolor A3(2). Biochem Biophys Res Commun 307: 610–619. [DOI] [PubMed] [Google Scholar]
- 50. Choi KY, Jung E, Jung DH, Pandey BP, Yun H, et al. (2012) Cloning, expression, and characterization of CYP102D1, a self-sufficient P450 monooxygenases from Streptomyces avermitilis . FEBS J 279: 1650–1662. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Transcription levels of aveR, aveA1, SAV151, SAV576, SAV292, SAV880, and SAV4189 in avermectin-overproducing strain 76-02-e relative to those in wild-type strain ATCC31267. Samples were collected from each strain grown in FM-II medium after days 2 and 6 of growth. hrdB was used as an internal control. Standard deviations are indicated by error bars (n = 3). Each gene was examined by relative quantification real-time RT-PCR with gene-specific primers.
(TIF)
Confirmation of the transcriptional units SAV574-SAV575 and SAV579-SAV580-SAV581 by RT-PCR. Lanes: RT, RT-PCR; –, negative control with reverse transcriptase omitted; +, positive control with genomic DNA of ATCC31267 as the template. The primers used for amplifying the SAV574-SAV575 intergenic region were GJ109 (GCGCTACCAGCAGGACGT) and GJ110 (CTCCTCCACGGCGAACTT); those used for amplifying the SAV579-SAV581 intergenic region were GJ159 (ACCGCACCCATCAGGAAG) and GJ160 (GGTAAGAAACGAGGGCGTA).
(TIF)
Determination of the transcriptional start points of SAV575 (A) and SAV576 (B) by 5′-RACE PCR. Boxed area: 5′-RACE oligo dT-anchor primer.
(TIF)
Effect of SAV575 deletion on the expression of related genes. Semiquantitative RT-PCR analysis of transcription levels of genes from ATCC31267 (WT) and D575. The strains were grown in FM-II for 6 days. hrdB was used as a positive internal control.
(TIF)
Primers used in this study.
(DOCX)
Putative targets of SAV576.
(DOCX)
List of genes having an expression level at least two-fold higher or lower in avermectin overproducer 76-02-e in comparison with wild-type strain ATCC31267.
(XLS)