Abstract
Human papillomaviruses (HPVs) are present in virtually all cervical cancers. An important step in the development of malignant disease, including cervical cancer, involves a loss of sensitivity to transforming growth factor β (TGF-β). HPV type 16 (HPV16) early gene expression, including that of the E6 and E7 oncoprotein genes, is under the control of the upstream regulatory region (URR), and E6 and E7 expression in HPV16-immortalized human epithelial cells is inhibited at the transcriptional level by TGF-β. While the URR contains a myriad of transcription factor binding sites, including seven binding sites for nuclear factor I (NFI), the specific sequences within the URR or the transcription factors responsible for TGF-β modulation of the URR remain unknown. To identify potential transcription factors and binding sites involved in TGF-β modulation of the URR, we performed DNase I footprint analysis on the HPV16 URR using nuclear extracts from TGF-β-sensitive HPV16-immortalized human keratinocytes (HKc/HPV16) treated with and without TGF-β. Differentially protected regions were found to be located around NFI binding sites. Electrophoretic mobility shift assays, using the NFI binding sites as probes, showed decreased binding upon TGF-β treatment. This decrease in binding was not due to reduced NFI protein or NFI mRNA levels. Mutational analysis of individual and multiple NFI binding sites in the URR defined their role in TGF-β sensitivity of the promoter. Overexpression of the NFI family members in HKc/HPV16 decreased the ability of TGF-β to inhibit the URR. Since the oncoprotein Ski has been shown to interact with and increase the transcriptional activity of NFI and since cellular Ski levels are decreased by TGF-β treatment, we explored the possibility that Ski may provide a link between TGF-β signaling and NFI activity. Anti-NFI antibodies coimmunoprecipitated endogenous Ski in nuclear extracts from HKc/HPV16, confirming that NFI and Ski interact in these cells. Ski levels dramatically decreased upon TGF-β treatment of HKc/HPV16, and overexpression of Ski eliminated the ability of TGF-β to inhibit the URR. Based on these studies, we propose that TGF-β inhibition of HPV16 early gene expression is mediated by a decrease in Ski levels, which in turn dramatically reduces NFI activity.
Cervical cancer is the second most common malignancy in women worldwide, and its etiology has been linked to high-risk human papillomaviruses (HPVs) (reviewed in reference 62). High-risk HPV E6 and E7 oncoproteins, whose expression is controlled by the HPV upstream regulatory region (URR), play a significant role in the malignant conversion of infected cutaneous and mucosal epithelial cells. Transcriptional control, via the URR, of the high-risk HPVs has thus been the focus of numerous investigations. These studies have identified a myriad of transcription factors and their cognate DNA binding elements within the URR and have demonstrated that HPV early gene expression is controlled by a complex interaction of cellular and viral factors that bind to this regulatory region (5, 8, 9, 38, 50, 52).
Transforming growth factor β (TGF-β) signaling pathways play an important role in development, wound healing, immune response, proliferation, differentiation, and apoptosis, and dysregulation of these pathways is a crucial step in the pathogenesis of cancer (reviewed in references 36, 37, 55, and 57). Several studies have explored the cellular pathways leading to enhanced rates of gene transcription in response to TGF-β, and much progress has been recently made in defining the details of these pathways (reviewed in references 31, 32, 37, and 55). However, studies involving the pathways leading to inhibition of gene expression in response to TGF-β have received less attention. A study by Woodworth et al. (58) over a decade ago was the first to report that TGF-β inhibits at the transcriptional level the expression of the HPV type 16 (HPV16) early genes in HPV-immortalized human genital epithelial cells. However, details concerning the mechanism(s) involved in TGF-β modulation of HPV16 URR activity have not been previously reported.
Nuclear factor I (NFI), also known as NF1, NF-1, and CTF (CAAT box transcription factor), is a family of transcription factors that have been shown to control viral and cellular gene expression (reviewed in reference 18). In addition, NFI has been shown to be an important transcription factor regulating the activity of the URR of various HPVs (8, 9, 11, 12, 16, 21, 56). A report by Tarapore et al. (54) described the interaction with and transcriptional activation of NFI by the oncoprotein Ski. This study prompted us to investigate a possible link between the TGF-β signaling pathway and NFI regulation of HPV16 early gene expression by exploring the possibility that NFI-Ski interactions might be involved in TGF-β inhibition of the HPV16 URR.
The goal of the present study was to investigate the nuclear factor(s) and binding site(s) within the HPV16 URR which may be responsible for TGF-β modulation of early gene expression. In this report we provide convincing evidence that NFI-Ski interactions mediate TGF-β inhibition of the HPV16 URR.
MATERIALS AND METHODS
Cell culture and cell lines.
Normal human keratinocytes (HKc) were isolated from neonatal foreskins and immortalized by transfection with a plasmid containing a head-to-tail dimer of HPV16 DNA (HKc/HPV16). Establishment and characteristics of the HKc/HPV16 cell lines used in this study (TGF-β-sensitive HKc/HPV16 and TGF-β-resistant, differentiation-resistant HKc/HPV16 [HKc/DR]) have been described in detail in previous publications (24, 25, 43). HKc/HPV16 were cultured in serum-free MCDB153-Luria-Bertani basal medium supplemented with 5 ng of epidermal growth factor/ml, 35 to 50 μg of bovine pituitary extract protein/ml, 0.2 μM hydrocortisone, 0.1 mM calcium chloride, 10 nM triiodothyronine, 10 μg of transferrin/ml, and 5 μg of insulin/ml (complete medium), while HKc/DR were grown in complete medium containing 1 mM calcium chloride and 5% fetal bovine serum. Cells were split 1:10 when confluent, and medium was replaced every 48 h.
Nuclear extracts.
TGF-β-sensitive HKc/HPV16 and TGF-β-resistant HKc/DR were grown to 40 to 50% confluency in 100-mm-diameter tissue culture plates and then treated in complete medium without or with TGF-β1 (40 pM, solubilized in 4 mM hydrochloric acid containing 1 mg of bovine serum albumin/ml, from R&D Systems) for two consecutive 24-h treatments (48-h total). The cells were then rinsed twice with ice-cold phosphate-buffered saline and collected on ice in phosphate-buffered saline containing 1 mM EDTA with the use of cell lifters. The cells were collected by centrifugation (200 × g, 5 min, 4°C) and resuspended in ice-cold hypotonic buffer (10 mM HEPES [pH 7.9], 10 mM potassium chloride, 1.5 mM magnesium chloride, 0.5 mM dithiothreitol, 0.2 mM phenylmethylsulfonyl fluoride, 1 μg of leupeptin/ml, 1 μg of pepstatin A/ml). Cells were then immediately repelleted, resuspended in ice-cold hypotonic buffer (7 ml/20 100-mm-diameter tissue culture dishes), and allowed to swell on ice for 10 min. Nuclei were obtained by centrifugation (1,500 × g, 10 min, 4°C) following disruption of the cells with a Dounce-type mortar and pestle and then resuspended in extraction buffer (250 μl/20 100-mm-diameter tissue culture dishes; 20 mM HEPES [pH 7.9], 0.5 M potassium chloride, 1.5 mM magnesium chloride, 0.2 mM EDTA, 25% glycerol, 0.5 mM dithiothreitol, 0.2 mM phenylmethylsulfonyl fluoride, 1 μg of leupeptin/ml, 1 μg of pepstatin A/ml) by gentle pipetting. Nuclear proteins were extracted by gentle rocking (30 min at 4°C) followed by centrifugation (16,000 × g, 30 min, 4°C). The supernatant containing the nuclear extract was dialyzed (Slide-A-Lyzer cassette; Pierce) for 45 min on ice with gentle stirring in 100 ml (400 volumes) of ice-cold dialysis buffer (20 mM HEPES [pH 7.9], 0.1 M potassium chloride, 0.2 mM EDTA, 20% glycerol, 0.5 mM dithiothreitol, 0.2 mM phenylmethylsulfonyl fluoride). Nuclear extracts were aliquoted and stored at −80°C after removal of precipitates by centrifugation (16,000 × g, 20 min, 4°C). Protein concentration in the nuclear extracts was determined by the DC protein assay (Bio-Rad Laboratories).
DNase I footprint analysis.
Double-stranded DNA segments of about 200 bp in length, representing the keratinocyte-dependent enhancer (KDE), were obtained by PCR with two primer sets: set 1, upper, 5′ CGC CAG GCC CAT TTT GTA GC 3′, and lower, 5′ GGC CCA TAG TGC TTA AGT TTA TAT GAC AC 3′, and set 2, upper, 5′ CAT TGT TTT TTA CAC TGA ATT CTG TGC AAC TAC TG 3′, and lower, 5′ CCT TTA CAC ACT TAA GGT ATG AAC TAG G 3′. Each PCR fragment was digested with either EcoRI or Bst981 to enable only the coding or noncoding strand to be 3′ end labeled with [α-32P]dTTP and [α-32P]dATP (Amersham) by filling in with the large fragment of DNA polymerase I. Probes were purified using Wizard PCR Preps (Promega). Primer set 1 yielded a probe corresponding to nucleotides 7455 to 7665 of the URR, while set 2 yielded a probe corresponding to nucleotides 7615 to 7800. DNase I footprinting was performed using the Core Footprinting system (Promega). Nuclear extract (40 μg of protein) from TGF-β-sensitive HKc/HPV16 treated with and without TGF-β (40 pM, 48 h) was incubated with each probe at room temperature for 15 min. Excess specific, nonspecific (NS), and specific mutant oligonucleotides were added to the binding reaction mixtures to demonstrate specificity. After treatment with DNase I, the DNA fragments were precipitated and resuspended in loading buffer. Maxam and Gilbert A + G sequencing reactions for each probe were performed to create sequence markers (53). Probe combined with 6 μg of genomic DNA in a 30-μl reaction mixture was modified using 2 μl of 1 M formic acid (20 min in a 37°C water bath), ethanol precipitated, and cleaved with 150 μl of 0.25 M piperidine (15 min in a 90°C water bath). Butanol (1 ml) was added, and the mix was incubated for 5 min at room temperature. After centrifugation, the pellet was resuspended in 150 μl of water and precipitated again with 1 ml of butanol. Probe markers were dried, resuspended in loading buffer, and resolved on a 6% denaturing polyacrylamide gel alongside each DNA footprinting reaction mixture. Gels were dried and visualized using a Bio-Rad K-screen and phosphorimager.
EMSAs and supershift analysis.
Electrophoretic mobility shift assays (EMSAs) were performed using double-stranded oligonucleotides between 20 and 25 bases in length (Fig. 3A) as probes, 5′ end labeled with [γ-32P]ATP (Amersham) by using T4 polynucleotide kinase (Promega). Nuclear extracts (12 μg of protein) from TGF-β-sensitive HKc/HPV16 treated with and without TGF-β (40 pM, 48 h) were incubated with 10-fold-concentrated binding buffer (100 mM Tris-HCl [pH 7.5], 0.5 M sodium chloride, 10 mM dithiothreitol, 50% glycerol), 1 μg of poly(dI · dC) (Pharmacia), 0.5 μg of sonicated herring sperm DNA, and 125-fold unlabeled specific or NS oligonucleotides (as competitors to determine binding specificity) in a final volume of 10 μl for 15 min on ice. Probe (100 ng) was added to each reaction mixture and allowed to incubate at room temperature for an additional 15 min. The entire reaction mixture was loaded without dye and resolved on a 5% nondenaturing Tris-glycine polyacrylamide gel (2.5 h at 150 V). Gels were dried and visualized using a Bio-Rad K-screen and phosphorimager. Supershift analysis was performed by adding anti-NFI antiserum (2 μl, provided by Naoko Tanese) or control rabbit immunoglobulin G (IgG; 1 μg) to the binding reaction mixtures, with an incubation of 1 h at room temperature.
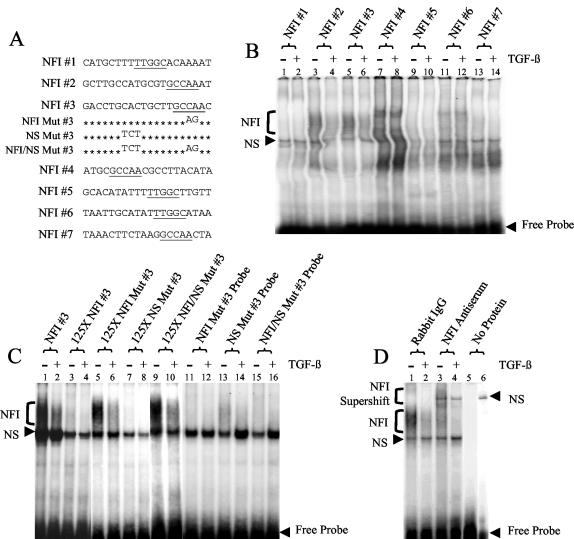
FIG. 3.
Binding to multiple NFI sites in the HPV16 URR decreases upon treatment of HKc/HPV16 with TGF-β. (A) The nucleotide sequence is provided for each of the oligonucleotides representing the seven NFI half-sites in the HPV16 URR. Mutations made to the NFI 3 oligonucleotides are also shown. Putative NFI binding sites are underlined. (B) EMSAs were performed using each NFI site of the HPV16 URR as a probe. Nuclear extract from TGF-β-sensitive HKc/HPV16 treated with (even-numbered lanes) and without (odd-numbered lanes) 40 pM TGF-β for 48 h was incubated with each probe. Protein-probe complexes were separated from the free probe on a 5% nondenaturing polyacrylamide gel. Specific NFI binding (bracket) as well as NS binding is noted. (C) EMSAs were performed using probes made from the differentially protected area found around NFI site 3 of the HPV16 URR. Nuclear extract from TGF-β-sensitive HKc/HPV16 treated with (even-numbered lanes) and without (odd-numbered lanes) 40 pM TGF-β for 48 h was incubated with either a radiolabeled NFI 3 probe (lanes 1 to 10) or a radiolabeled mutant NFI 3 probe (lanes 11 to 16). Excess unlabeled oligonucleotides (125-fold) containing the intact NFI site (lanes 3, 4, 7, and 8) or the mutated NFI site (lanes 5, 6, 9, and 10) were added to the binding reaction mixture to demonstrate specificity. (D) NFI was verified as the transcription factor responsible for the differential binding by performing supershift analysis using NFI 3 oligonucleotide as the probe. Rabbit IgG (lanes 1 and 2) or anti-NFI antiserum (lanes 3 and 4) was added to the binding reaction mixtures (described above). NFI (bracket) and NS binding, as well as the resulting NFI supershift (bracket), is shown. Lane 5 contains labeled probe only (no nuclear extract). Lane 6 contains labeled probe and anti-NFI antiserum (no nuclear extract).
Coimmunoprecipitation and Western immunoblot analysis.
Protein G agarose (50 μl; Pierce) was washed and resuspended in 25 μl of immunoprecipitation buffer (250 mM sodium chloride, 0.1% NP-40, 50 mM HEPES [pH 7.0], 1 mM phenylmethylsulfonyl fluoride, 5 mM EDTA, 5 μg of aprotinin/ml) and then incubated with 5 μg of anti-NFI antibody (rabbit; Santa Cruz Biotechnology) at room temperature for 30 min. Nuclear extracts (850 μg of protein) from TGF-β-sensitive HKc/HPV16 and TGF-β-resistant HKc/DR treated with and without TGF-β (40 pM, 24 h) were incubated (2 h, 25°C) with the protein G-anti-NFI antibody complex. After immunoprecipitation, the protein G agarose beads were washed three times with 500 μl of immunoprecipitation buffer, resuspended in fivefold-concentrated loading dye (Pierce) with 5 μl of 0.5 M dithiothreitol, and boiled for 5 min. Agarose beads were removed by centrifugation prior to Western immunoblot analysis. Either the entire immunoprecipitate or nuclear extracts (30 μg of protein) from TGF-β-sensitive HKc/HPV16 and TGF-β-resistant HKc/DR treated with and without TGF-β (40 pM, 48 h) were separated on a 10% polyacrylamide gel. Proteins were transferred to nitrocellulose and probed with an anti-Ski antibody (mouse; Cascade Bioscience) at 4°C overnight (3 μg/ml in a blocking solution containing 4% nonfat dry milk and 0.05% Tween 20 in phosphate-buffered saline). The blot was then incubated with an appropriate horseradish peroxidase-conjugated secondary antibody (Boehringer Mannheim; diluted 1:5,000 in blocking solution) for 1 h at room temperature. The Super Signal West Pico chemiluminescence detection kit (Pierce) was used for detection. Alternatively, nuclear extracts (30 μg of protein) from TGF-β-sensitive HKc/HPV16 treated with and without TGF-β (40 pM, 48 h) were separated on a sodium dodecyl sulfate (SDS)-12% polyacrylamide gel, transferred, and probed with the anti-NFI antibody at room temperature for 2 h (1 μg/ml in a blocking solution containing 5% nonfat dry milk and 0.05% Tween 20 in phosphate-buffered saline).
Plasmid constructs and mutagenesis.
The parent cloning vector (pCH) and hemagglutinin-tagged NFI expression vectors (pCHA1.1, mouse NFI-A1.1; pCHB, mouse NFI-B2; pCHC, mouse NFI-C2; and pCHX, mouse NFI-X2) were kindly provided by Richard Gronostajski (7), and human Ski parent (pRSVpL) and expression (pRSVpLHuSkiEE and pcDNA 3-hSki) vectors were obtained from Ed Stavnezer (39). A luciferase reporter vector under control of the HPV16 URR (pGL3/URR) was constructed by cloning the entire URR (Fig. 1) into the HindIII multiple cloning site of pGL3-basic (Promega) by using the following PCR primers, which contain incorporated HindIII sites and an SP6 site for sequencing: forward primer, 5′ TAA ATA TTA AGT TGT AAG CTT GTT TGT TAT TTA GGT GAC ACT ATA GAG GGC CCA TGT GTT TTT AAA TGC TTG TG 3′; reverse primer, 5′ CTC CTG TGG GTC CTG AAA GCT TGC AGG GCC CTT TTG GTG CAT AAA ATG TC 3′. Deletion constructs were made in a similar manner by using various lower PCR primers yielding HPV16 URR products of decreasing size. Mutant constructs were created using the Quick Change site-directed mutagenesis kit (Stratagene). Point mutations were introduced by PCR with various primers containing mutations in the NFI site(s) (GCCAA changed to GCAGA) or YY1 site(s) (CCAT or ACAT changed to TACG), and the parent plasmid was removed by digestion with DpnI. All constructs and mutations were screened by PCR for the presence and orientation of their respective inserts by using primers from both the plasmid and the insert. The nucleotide sequence of each construct was verified by direct sequencing.
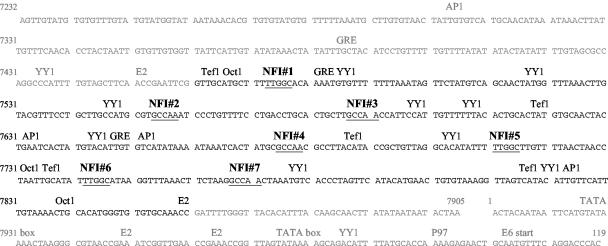
FIG. 1.
Nucleotide sequence of the entire HPV16 URR (nucleotides 7232 to 119). Putative transcription factor binding sites are noted. The KDE is shown in black, and 3′ and 5′ segments of the URR are shown in gray. The seven potential NFI binding sites are numbered in sequential order, and their nucleotide sequences are underlined.
Transfections and luciferase assay.
Plasmid constructs were transfected, in triplicate, into TGF-β-sensitive HKc/HPV16 cultured in 60-mm-diameter dishes at 30 to 40% confluency by using Transfast (Promega). Transfection medium (complete medium containing no antibiotics, 1.5 ml) and 1.5 μg of reporter plasmid and/or 2 μg of expression plasmids were combined with 5 μl of Transfast per dish. After 30 min, the transfection medium was removed and replaced with complete medium containing no antibiotics. To control for variations in transfection efficiencies, the triplicate plates were trypsinized, and the cells were pooled and replated onto six 60-mm-diameter dishes 12 to 15 h posttransfection. At 6 to 8 h postreplating, half of the dishes were treated and half were not treated with 40 pM TGF-β. Dishes were retreated with or without TGF-β after 24 h, and luciferase activity was determined 68 to 72 h posttransfection with the luciferase assay system (Promega). Experiments were performed at least three times, and each construct was tested in triplicate dishes. Relative light units were determined using a luminometer (Berthold Lumat LB9501).
RNA extraction and real-time PCR analysis.
TGF-β-sensitive HKc/HPV16 (60 to 75% confluent in 100-mm-diameter tissue culture dishes) overexpressing either Ski or individual NFI family members were treated with and without TGF-β (40 pM, 46 h), and the cells were harvested 68 to 72 h posttransfection. RNA was collected using RNeasy columns (Qiagen). RNA was treated with twice the suggested DNase I concentration for 25 min to ensure complete digestion of any residual DNA. mRNA expression was determined by real-time PCR analysis. Specific primer sets were designed to detect transcripts for each NFI family member, Ski, or RPLP0 (ribosomal protein, large protein 0): NFIA, upper, 5′ GAC TGC CTG CGC CAG GC 3′; lower, 5′ GTC CTG GAA GCC CAA ATG TCC ATT 3′; upper, 5′ AAA GTC CCA GCC AGC CAA GTG AA 3′; lower, 5′ CCT CCT CAT TGC TCC TGG ACT 3′; NFIB, upper, 5′ GAC TGC CTG CGC CAG GC 3′; lower, 5′ GGC TTG GAC TTC CTG ATT GTC CAG AA 3′; upper, 5′ GAA GTC CAA GCC ACA GTG ATC CT 3′; lower, 5′ CTG CAG GTT CAC ACC AGA GTT; NFIC, upper, 5′ GGT CAT CCT GTT CAA GGG CAT 3′; lower, 5′ ATG GGC TTG CTG TCC TCC TGG TC 3′; upper, 5′ CAG CCC CCG GAC AGG TGT 3′; lower, 5′ GGA GGT GCT GGG TAG AGT CCT TCT 3′; NFIX, upper, 5′ GAC TGC CTG CGC CAG GC 3′; lower, 5′ GGG CAG TGG TTT GAT GTC CGC AT 3′; upper, 5′ CAA TCA GAT AGT TCA AAC CAG CAA 3′; lower, 5′ CCT TCC CAG GGT CAC TTG ATT 3′; Ski, upper, 5′ GCG CCT TCC GAA AAG GAC AA 3′; lower, 5′ GCT CTT TCT CAC TCG CTG ACA CT 3′; upper, 5′ GAG GCG GAG GTG GAA GTT GAAA 3′; lower, 5′ GCA GGA ACT TCT CTT TGG CTT CCT T 3′. Each primer set spanned an intron and yielded products of differing melting temperatures to ensure specificity. Reverse transcription was performed with random hexamers with the GeneAmp RT-PCR kit (Perkin-Elmer). The length of each product was verified by agarose gel electrophoresis. The SYBR Green PCR core reagent kit (PE Biosystems) was used to amplify each reverse transcription-PCR product. Real-time PCR was performed using the iCycler iQ detection system (Bio-Rad Laboratories) with the following parameters: one cycle, 95°C for 8.5 min; 50 cycles, 30 s at 95°C, 30 s at 55°C, and 30 s at 72°C, plus a melting curve of 55°C at 0.5°C intervals for 10 s for 80 cycles; and 58°C for 30 s for one cycle, ending with a hold at 10°C. mRNA expression was calculated assuming 100% primer efficiency for each primer set. For the NFI family members and Ski, expression is given as induction compared to that for the control primer set (RPLP0). For the verification of overexpression of the NFI family members and Ski, expression was determined as induction over endogenous levels upon transfection of the respective empty expression vector. Cycle differences were calculated by subtracting the cycle number for each NFI family member or Ski from the cycle number for RPLP0 or the respective empty expression vector. Induction was then calculated by raising 2 (which assumes 100% primer efficiency) to the power of the cycle difference. At least two experiments were performed on each NFI family member and Ski, and samples were run in duplicate. The cycle differences and induction were calculated by averaging two experiments from only one primer set.
Overexpression of NFI and Ski.
Both pGL3/URR (1.5 μg/60-mm-diameter tissue culture dish) and parent or expression constructs (2 μg/60-mm-diameter tissue culture dish) were transfected and analyzed essentially as described above. Overexpression of NFI family members was confirmed by Western analysis with an antihemagglutinin antibody (mouse; Boehringer Mannheim) or an anti-NFI antibody (rabbit; Santa Cruz Biotechnology). Ski overexpression was confirmed by Western analysis with an anti-Ski antibody (mouse; Cascade Bioscience). Upregulation of mRNA was confirmed by real-time PCR analysis.
RESULTS
DNase I footprinting demonstrates differential binding of nuclear proteins around multiple NFI sites in the HPV16 URR following TGF-β treatment.
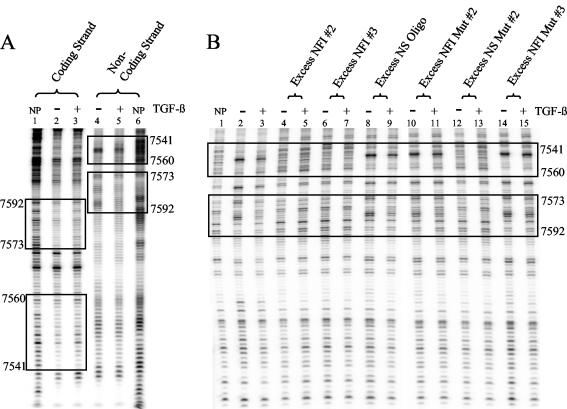
TGF-β, a powerful growth inhibitor of epithelial cells, has been shown to inhibit HPV16 early gene expression at the transcriptional level (58). The HPV16 URR, which is responsible for regulating the transcription of all the viral early genes, contains numerous binding sites for many transcription factors, including seven half-sites (labeled NFI#1 to NFI#7) for NFI (Fig. 1). In order to determine which transcription factor binding sites may be responsible for TGF-β modulation of early gene expression, we performed DNase I footprinting on the entire KDE (Fig. 1, KDE sequence shown in black type). Using nuclear extracts collected from TGF-β-sensitive HKc/HPV16 treated with and without TGF-β, we observed areas of differential binding around several of the NFI sites (Fig. 2A, boxed areas, and data not shown). A difference in the banding pattern intensity represents a change in protection and/or binding of nuclear protein(s) (Fig. 2A, boxed areas). Upon TGF-β treatment, protection by nuclear protein(s) was altered around NFI sites 2 (nucleotides 7541 to 7560) and 3 (nucleotides 7573 to 7592) on the coding (Fig. 2A, lanes 1 to 3) and noncoding (Fig. 2A, lanes 4 to 6) strands. Similar results were observed around additional NFI sites present in the KDE (data not shown).
FIG. 2.
DNase I footprint analysis of the HPV16 URR. (A) A double-stranded segment of the KDE (nucleotides 7502 to 7677) was end labeled with 32P on either the coding (lanes 1 to 3) or the noncoding (lanes 4 to 6) strand. Nuclear extract from TGF-β-sensitive HKc/HPV16 treated for 48 h with (lanes 3 and 5) and without (lanes 2 and 4) 40 pM TGF-β was incubated with the labeled probes. Each probe was also incubated without protein (NP, lanes 1 and 6) to form a DNase I ladder. After DNase I digestion, unprotected fragments were resolved on a 6% denaturing polyacrylamide gel. Maxam and Gilbert A + G sequencing was performed on each probe for nucleotide identification (data not shown). Two areas of differential protection are located around NFI binding sites and are shown in boxes on each strand. The URR nucleotide number is given on the left and right sides of the footprint. NFI 2 is located between nucleotides 7541 and 7560, while NFI 3 is located between nucleotides 7573 and 7592. (B) Specificity of the differential binding was confirmed using the labeled noncoding strand probe described for panel A. Excess unlabeled oligonucleotides containing either intact NFI binding sites (lanes 4 to 7, 12, and 13) or without intact NFI binding sites (lanes 8 to 11, 14, and 15) were added to the binding reaction mixtures (described above). See Fig. 3A for the complete nucleotide sequence of the unlabeled competitor oligonucleotides, NFI 2, NFI 3, and NFI mutant (Mut) 3. The nucleotide sequence of the NS oligonucleotide (Oligo) was GCT TGT ACG GCG TGC AGA AT, the sequence of the NFI mutant (Mut) 2 was GCT TGC CAT GCG TGC AGA AT, and the sequence of NS mutant (Mut) 2 was GCT TGT ACG GCG TGC CAA AT. Competition and destruction of the differential binding (boxed areas, lanes 2 and 3) can be observed only in lanes containing oligonucleotides with intact NFI binding sites (lanes 4 to 7, 12, and 13).
To determine the specificity of this differential binding, excess unlabeled oligonucleotides (see Fig. 3A for nucleotide sequences) were added to the footprint binding reaction mixtures as competitors (Fig. 2B). When unlabeled oligonucleotides containing intact NFI sites (Fig. 2B, lanes 4 to 7, 12, and 13) were added, differential binding was abolished. Conversely, differential binding was retained when oligonucleotides without intact NFI binding sites were added (Fig. 2B, lanes 8 to 11, 14, and 15). The results of this DNase I footprint screen prompted us to explore further the role of NFI sites as mediators of TGF-β modulation of HPV16 URR activity.
Oligonucleotides representing all seven NFI half-sites were constructed (Fig. 3A) and used as probes in EMSAs (Fig. 3B to D) to verify the differential binding demonstrated by our DNase I footprint analysis. We compared binding of nuclear proteins to each NFI half-site using TGF-β-sensitive HKc/HPV16 treated with and without TGF-β. Binding to multiple NFI half-sites in the HPV16 URR decreased upon TGF-β treatment, but binding to all NFI sites was not equal (Fig. 3B). For example, very little binding was observed for NFI sites 1, 5, and 7 (Fig. 3B, lanes 1, 2, 9, 10, 13, and 14). NFI sites 4 and 6 showed a different pattern of binding (Fig. 3B, lanes 7, 8, 11, and 12), which is likely due to the fact that these sites are adjacent to Tef-1 binding sites (Fig. 1). NFI sites 2 and 3 showed the most dramatic loss of binding following TGF-β treatment (Fig. 3B, lanes 3 to 6) and were used to test binding specificity (Fig. 3C and data not shown). A smear of binding, which decreased upon TGF-β treatment (Fig. 3C, lanes 1 and 2), was competed using excess unlabeled oligonucleotides containing intact NFI binding sites (Fig. 3C, lanes 3, 4, 7, and 8). Unlabeled oligonucleotides containing mutant NFI sites, however, did not compete the shift (Fig. 3C, lanes 5, 6, 9, and 10), and mutant NFI probes (Fig. 3A) were not able to bind the same complex (Fig. 3C, lanes 11, 12, 15, and 16). These data confirmed that binding of the complex was specific for the target sequence of NFI and that binding to a subset of the consensus NFI sites within the HPV16 URR decreased upon TGF-β treatment.
NFI sites 2 (data not shown) and 3 (Fig. 3D) were also used to perform supershift analysis. An NFI supershift smear resulted upon addition of NFI antiserum (Fig. 3D, lanes 3 and 4) but not of rabbit IgG (Fig. 3D, lanes 1 and 2) to the binding reaction mixtures. The presence of an NS supershifted band can also be observed when there is antiserum but no protein (nuclear extract) present in the binding reaction mixture (Fig. 3D, lane 6), demonstrating some cross-reactivity of the NFI antiserum with the probe. Similar results were observed using NFI site 2 as the probe (data not shown), verifying the presence of NFI in the bound complex. These results demonstrate that there is a decrease in NFI binding at multiple sites in the HPV16 URR upon TGF-β treatment, which suggests that NFI is involved in TGF-β modulation of HPV16 early gene expression.
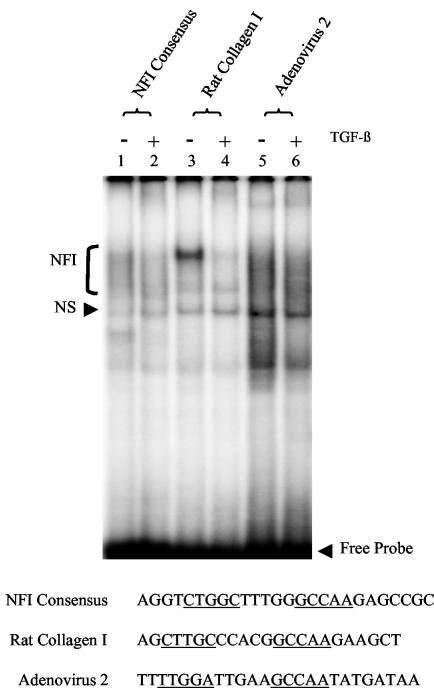
Using EMSAs and nuclear extracts from HKc/HPV16 treated with and without TGF-β, we tested two known TGF-β-responsive promoters containing NFI binding sites to determine whether TGF-β treatment also reduced NFI binding to these promoters (Fig. 4). An NFI consensus oligonucleotide was used as a control (27). Upon TGF-β treatment, we observed a slight reduction in NFI binding to labeled probes containing the NFI consensus sequence (Fig. 4, lanes 1 and 2) and NFI binding sequences present in the adenovirus type 2 promoter (61) (Fig. 4, lanes 5 and 6). A dramatic loss of NFI binding occurred to the rat α1 (I) collagen promoter (48) (Fig. 4, lanes 3 and 4) upon TGF-β treatment. These results demonstrate that TGF-β can modulate NFI binding to TGF-β-responsive promoters in addition to the URR.
FIG. 4.
TGF-β modulation of NFI binding to TGF-β-responsive promoters. EMSAs were performed using an NFI consensus binding site (lanes 1 and 2) and sequences containing NFI binding sites from the rat collagen I (lanes 3 and 4) and adenovirus 2 (lanes 5 and 6) promoters as probes. The sequence of each probe is listed, and the NFI binding sites are underlined. Nuclear extract from TGF-β-sensitive HKc/HPV16 treated with (even-numbered lanes) and without (odd-numbered lanes) 40 pM TGF-β for 48 h was incubated with each probe. Protein-probe complexes were separated from the free probe on a 5% nondenaturing polyacrylamide gel. Specific NFI binding (bracket) as well as NS binding is noted.
AP1 is an important positive transcription factor in the regulation of HPV16 early gene expression (6, 8, 9, 40) and has been shown to be involved in TGF-β modulation of various genes (2, 10, 23, 28, 33, 49, 51). However, EMSAs with oligonucleotides representing three AP1 sites within the HPV16 URR showed no reduction in binding upon TGF-β treatment (data not shown). YY1, also an important regulator of the HPV16 URR (13, 34, 41), showed no change in binding upon TGF-β treatment (data not shown). These data further support NFI as the transcription factor that contributes to TGF-β control of HPV16 early gene expression.
NFI binding sites are required for TGF-β modulation of the HPV16 URR.
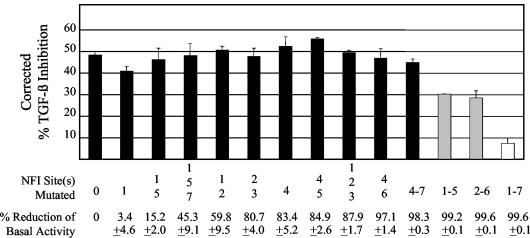
To obtain further evidence that NFI was necessary for TGF-β modulation of HPV16 early gene expression, we utilized reporter constructs containing the entire HPV16 URR. Point mutations of single and multiple NFI sites were made within the context of the HPV16 URR, which was cloned upstream of a luciferase reporter gene (pGL3). Luciferase activity was determined following transfection of these constructs into HKc/HPV16 and treatment of the cells with or without TGF-β. In the absence of TGF-β, pGL3/URR (the reporter construct containing the full-length HPV16 URR) yielded relative light unit numbers of up to 930,000, which were reduced by about 80% following TGF-β treatment. TGF-β also caused about a 30% decrease in luciferase activity from the promoterless pGL3-basic. Since pGL3-basic does not contain a promoter, we considered the decrease in luciferase activity caused by TGF-β on this construct as NS and likely due to causes other than regulation of transcription. Therefore, we subtracted these values from luciferase activities measured when luciferase was expressed under the control of the HPV16 URR, to obtain corrected percent TGF-β inhibition values of each reporter construct (Fig. 5). Corrected TGF-β inhibition of the HPV16 URR was reduced from about 50 to about 30% when five of the seven NFI sites were mutated (gray bars) and further reduced to less than 10% when all seven sites were mutated (white bar) (Fig. 5). Although greater than 99% of the URR basal activity was lost upon mutation of all seven NFI sites (Fig. 5), the luciferase activity of this mutant was still three- to fivefold greater than that of the promoterless pGL3. Furthermore, mutant 2-6, which also had over 99% reduction in basal activity, still retained about 30% inhibition by TGF-β (Fig. 5). These data support the conclusion that NFI binding sites contribute to TGF-β modulation of the HPV16 URR.
FIG. 5.
Effects of single and multiple NFI mutations on TGF-β modulation of the HPV16 URR. The entire HPV16 URR (Fig. 1) was cloned into the luciferase reporter vector pGL3 (Promega) (pGL3/URR) where various NFI sites were mutated from GCCAA to GCAGA, which is unable to bind NFI. These constructs were transfected into TGF-β-sensitive HKc/HPV16 and treated with and without 40 pM TGF-β for 42 h. Luciferase activity was determined 68 h posttransfection. Corrected percent TGF-β inhibition for each construct was determined by subtracting the percent TGF-β inhibition obtained by transfection of a promoterless pGL3 plasmid from the total TGF-β inhibition obtained for each reporter construct. The specific NFI site(s) that was mutated and the percent reduction of basal URR activity are shown for each mutant construct at the bottom of the figure.
YY1 has putative binding sites near five of the seven NFI half-sites in the HPV16 URR (Fig. 1). To investigate any role that the YY1 sites may play in TGF-β modulation of HPV16 URR activity, three YY1 sites were mutated alone or in conjunction with the NFI sites. None of the YY1 mutant reporter constructs or the YY1-NF1 combination mutant reporter constructs exhibited a loss of TGF-β inhibition, suggesting that the YY1 sites do not play a significant role in TGF-β modulation of the HPV16 URR (data not shown).
Reduction of NFI binding is not due to decreased NFI protein or mRNA levels.
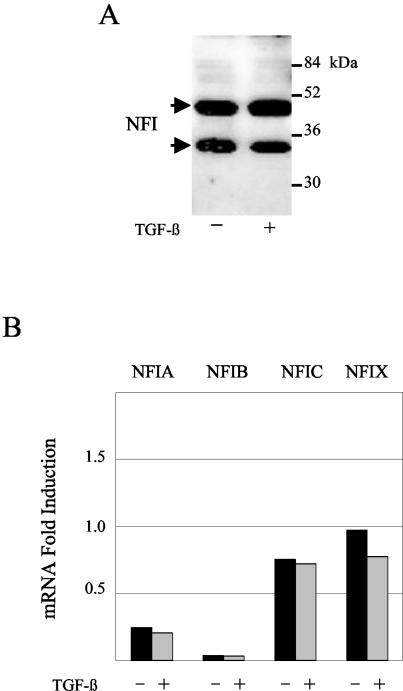
In order to determine if the loss of NFI binding to various sites in the HPV16 URR upon TGF-β treatment was due to a reduction in NFI protein levels, we performed immunoblot analysis of NFI from HKc/HPV16 treated with and without TGF-β (Fig. 6A). NFI is a large, diverse family of transcription factors containing four distinct members that can yield alternatively spliced transcripts (reviewed in reference 18). Immunoblot analysis, using an anti-NFI antibody that recognizes the N-terminal end of the protein and detects all family members, yielded two major NFI bands (likely degradation products) that did not vary upon TGF-β treatment (Fig. 6A). These results demonstrate that overall NFI protein levels do not change with TGF-β treatment.
FIG. 6.
NFI protein and mRNA levels do not change following TGF-β treatment of HKc/HPV16. (A) Total NFI protein levels were determined by Western analysis. Nuclear extract (40 μg of protein) from TGF-β-sensitive HKc/HPV16 treated with and without 40 pM TGF-β for 48 h was separated on an SDS-12% polyacrylamide gel, transferred to nitrocellulose, and probed with an anti-NFI antibody (Santa Cruz). Molecular mass markers are shown on the right; arrows pointing to NFI bands are on the left. (B) mRNA expression of each of the four NFI family members (NFIA, NFIB, NFIC, and NFIX) was determined by real-time PCR. RNA was collected using RNeasy columns (Qiagen) from TGF-β-sensitive HKc/HPV16 treated with (gray) and without (black) 40 pM TGF-β for 46 h. Expression was determined using primers specific for each NFI family member, compared with a control set of primers, and expressed as fold induction. The average of two experiments for each family member is shown.
Previous studies have demonstrated that differential expression of one or more NFI family members can influence NFI transcriptional activity (22, 29, 35, 44, 59). To this end, we investigated the expression of each family member in HKc/HPV16 treated with and without TGF-β (Fig. 6B). Real-time PCR analysis was performed using primer pairs specific for each family member. Upon comparing expression of each family member to a control set of primers, we determined that every family member was expressed in HKc/HPV16 and that expression of each family member did not change upon TGF-β treatment (Fig. 6B). Since TGF-β treatment does not reduce overall NFI protein levels or significantly change the expression of any NFI family member, regulation of NFI binding must lie elsewhere.
Ski interacts with NFI and decreases upon TGF-β treatment.
Since our studies indicated that NFI is required for TGF-β modulation of the HPV16 URR, we subsequently wanted to explore links between NFI and the TGF-β signaling pathway. NFI has long been established as an important transcription factor in the upregulation of TGF-β-responsive gene expression (19, 20, 26, 42, 46, 47, 60). Downregulation of TGF-β-responsive genes, however, has been studied to a much lesser extent. We have demonstrated that NFI is an essential positive transcription factor for HPV16, by the dramatic reduction in basal activity upon mutation of the NFI binding sites within the URR (Fig. 5), and that inhibition of HPV16 early gene expression by TGF-β is the result of a reduction of NFI binding. Tarapore et al. (54) established that the Ski oncoprotein interacts with NFI and enhances its transcriptional activation. Ski is known to be directly regulated by the TGF-β signaling pathway via the SMAD proteins that transduce TGF-β signaling (reviewed in references 14 and 32). Based on these observations, we decided to investigate Ski as the possible mediator linking NFI to the TGF-β signaling pathway.
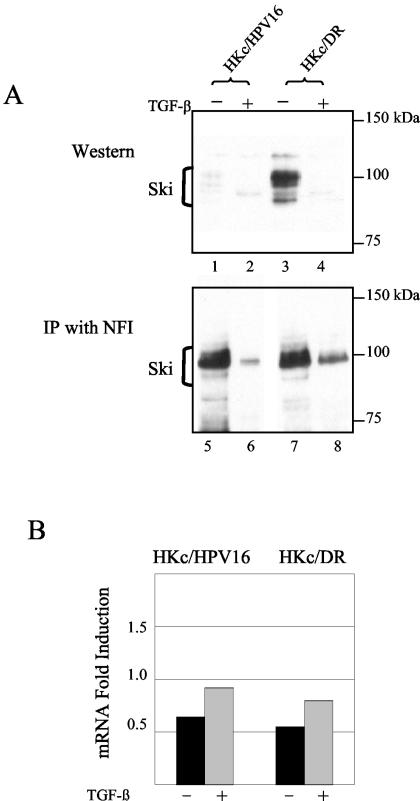
Immunoblot analysis revealed that Ski was expressed at very low levels in nuclear extracts from TGF-β-sensitive HKc/HPV16 (Fig. 7A, lane 1). Interestingly, however, Ski levels were undetectable upon TGF-β treatment (Fig. 7A, lane 2). Ski levels were dramatically increased in nuclear extracts from TGF-β-resistant HKc/HPV16 (HKc/DR) (Fig. 7A, lane 3) but also were reduced upon TGF-β treatment (Fig. 7A, lane 4). A time course experiment showed that Ski falls to undetectable levels between 5 and 10 h of TGF-β treatment of HKc/DR (data not shown). mRNA levels for Ski, however, remain virtually unchanged with TGF-β treatment in both HKc/HPV16 and HKc/DR (Fig. 7B).
FIG. 7.
TGF-β treatment of HKc/HPV16 and HKc/DR decreases nuclear Ski levels, and Ski interacts with NFI. (A) Ski levels were demonstrated by Western analysis (lanes 1 to 4). Nuclear extract (30 μg of protein) from TGF-β-sensitive HKc/HPV16 and TGF-β-resistant HKc/DR treated with and without 40 pM TGF-β for 48 h was separated on an SDS-10% polyacrylamide gel. Proteins were transferred to nitrocellulose and probed with an anti-Ski antibody (Cascade Bioscience), which detects Ski products (brackets) ranging from 95 to 115 kDa. Endogenous Ski coimmunoprecipitated with NFI (lanes 5 to 8). Nuclear extract (850 μg of protein) from TGF-β-sensitive HKc/HPV16 and TGF-β-resistant HKc/DR treated with and without 40 pM TGF-β for 24 h was incubated (2 h, 25°C) with 5 μg of anti-NFI antibody preincubated with protein G agarose. After washing, the immunoprecipitates were resolved by SDS-polyacrylamide gel electrophoresis and probed for Ski as described above. Molecular mass markers are noted on the right. (B) mRNA expression of Ski was determined by real-time PCR. RNA was collected using RNeasy columns (Qiagen) from TGF-β-sensitive HKc/HPV16 treated with (gray) and without (black) 40 pM TGF-β for 46 h. Expression was determined using primers specific for Ski, compared with a control set of primers, and expressed as fold induction. The average of two experiments for each family member is shown.
An interaction between endogenous Ski and NFI was established by coimmunoprecipitating Ski by using anti-NFI antibodies and nuclear extracts from HKc/HPV16 (Fig. 7A, lane 5) or HKc/DR (Fig. 7A, lane 7). This interaction was greatly reduced upon TGF-β treatment in HKc/HPV16 (Fig. 7A, lane 6) and was also reduced, but to a lesser extent, in HKc/DR (Fig. 7A, lane 8). Our data demonstrate that Ski interacts with NFI in HPV16-immortalized HKc, and this interaction decreases upon TGF-β treatment. Taken together, these results suggest that NFI transcriptional activation of the HPV16 URR is likely mediated by Ski, and thus, a decrease in Ski levels reduces NFI activity upon TGF-β treatment.
Overexpression of both NFI and Ski eliminates TGF-β inhibition of the HPV16 URR.
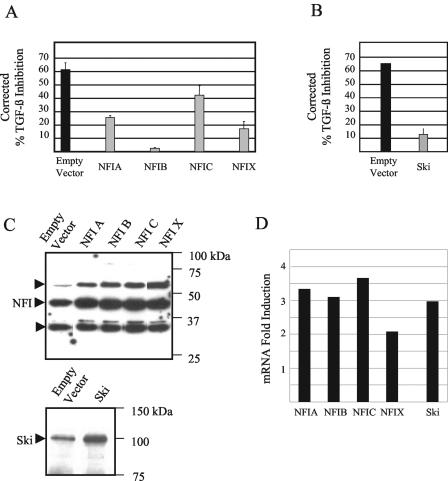
To further confirm that NFI and Ski contribute to TGF-β modulation of HPV16 early gene expression, we cotransfected each of the NFI family members (NFIA, NFIB, NFIC, and NFIX) or Ski with pGL3/URR in HKc/HPV16 and treated the cells with or without TGF-β. Overexpression of each NFI family member reduced TGF-β inhibition, although to various degrees (Fig. 8A). Ski overexpression also resulted in the elimination of TGF-β inhibition of the HPV16 URR reporter construct (Fig. 8B). The overexpression of Ski and each NFI family member was confirmed both at the protein level, by immunoblot analysis (Fig. 8C), and at the RNA level, by real-time PCR analysis (Fig. 8D). These data demonstrate that overexpression of either NFI or Ski can interfere with TGF-β inhibition of the HPV16 URR, further supporting the conclusion that each plays an important role in TGF-β modulation of HPV16 early gene expression.
FIG. 8.
TGF-β inhibition of the HPV16 URR is overcome by overexpression of either NFI or Ski. (A) Effects of NFI family member overexpression on TGF-β modulation of the HPV16 URR. pGL3/URR and pCMV vectors expressing each of the NFI family members were cotransfected into TGF-β-sensitive HKc/HPV16 using Transfast (Promega). Luciferase activity was determined after treatment with and without 40 pM TGF-β for 42 h, 68 h posttransfection. TGF-β inhibition resulting from transfection of an empty vector (black bar) is compared with the percent TGF-β inhibition upon overexpression of each NFI family member (gray bars). (B) Effects of Ski overexpression on TGF-β modulation of the HPV16 URR. pGL3/URR and a pcDNA3.1 vector expressing Ski were cotransfected and analyzed as described above (A). (C) NFI and Ski protein levels were determined by Western analysis. Lysates from TGF-β-sensitive HKc/HPV16 cotransfected with pGL3/URR and pCMV vectors expressing each NFI family member, empty pCMV vector, Ski, or an empty pcDNA3.1 vector were separated on an SDS-12% polyacrylamide gel, transferred to nitrocellulose, and probed with an anti-NFI or an anti-Ski antibody. Molecular mass markers are shown on the right with arrows pointing to three NFI bands (top panel) or Ski (bottom panel). (D) mRNA expression of each NFI family member and Ski was determined by real-time PCR analysis. RNA was collected using RNeasy columns (Qiagen) from TGF-β-sensitive HKc/HPV16 cotransfected with pGL3/URR and pCMV vectors expressing each NFI family member or a pcDNA vector expressing Ski. Expression was determined using primers specific for each NFI family member or Ski and is given as fold induction over empty vector.
DISCUSSION
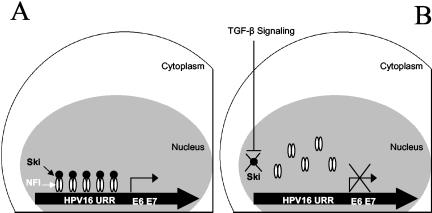
These studies, initiated to explore the molecular details of TGF-β modulation of HPV16 URR activity, yielded the following important observations: (i) NFI binding sites within the HPV16 URR are critical for TGF-β modulation of URR activity; (ii) TGF-β treatment of HKc/HPV16 reduces NFI binding to selected NFI sites within the HPV16 URR, without changing overall NFI protein or mRNA levels; (iii) Ski interacts with NFI, and Ski protein levels decrease dramatically following TGF-β treatment of HKc/HPV16 and HKc/DR; and (iv) overexpression of either NFI or Ski in HKc/HPV16 interferes with TGF-β inhibition of HPV16 URR activity. Overall, our data support a model whereby TGF-β inhibition of HPV16 early gene expression, including E6 and E7 expression, results from a decrease in Ski levels, which in turn reduces NFI transcriptional activity (Fig. 9).
FIG. 9.
Proposed model of TGF-β inhibition of HPV16 URR activity through NFI-Ski interactions in TGF-β-sensitive HKc/HPV16. (A) In the absence of TGF-β NFI-Ski complexes bind to and activate the HPV16 URR promoter. (B) In the presence of TGF-β signaling, Ski is degraded and no longer available to complex with NFI to induce promoter activity of the HPV16 URR.
The KDE region of the URR contains the majority of the transcription factor binding sites, which contribute either positively or negatively to overall URR activity in keratinocytes. The KDE includes seven NFI binding half-sites of the sequence 5′-TTGGC-3′, making it one of the most abundant transcription factor binding sites within the URR. Previous DNase I footprinting experiments support NFI binding to these seven putative NFI motifs (17). Furthermore, deletions and mutations of the URR, as well as EMSAs, have demonstrated an essential role for NFI in transcriptional activation of the HPV16 enhancer and suggest an important role for NFI in the determination of the viral enhancer's epithelial cell specificity (1, 8). Our DNase I footprinting studies (Fig. 2) found that, upon TGF-β treatment of HKc/HPV16, protection by nuclear protein(s) was decreased around several NFI sites within the HPV16 URR. However, EMSAs with oligonucleotides spanning all seven NFI half-sites found that binding to all NFI sites was not equal, suggesting that nucleotide sequences outside the NFI binding motif must also contribute to NFI binding. NFI sites 2 and 3 showed the most dramatic loss of NFI binding following TGF-β treatment (Fig. 3).
Our finding that TGF-β had no effect on the levels of NFI protein or NFI mRNA (Fig. 6) suggested to us that TGF-β modulation of NFI binding to the URR might occur through proteins that interact with NFI. The possibility that NFI requires a coactivator(s) for maximal URR activity has been previously proposed (1), and a study by Terapore et al. (54) demonstrates that the oncoprotein Ski interacts with NFI and results in enhanced NFI transcriptional activity. These observations prompted us to explore a potential role of Ski in TGF-β modulation of the URR. Our data suggest that Ski is necessary for efficient URR activity through interaction with NFI. This is demonstrated by significant inhibition of URR activity upon TGF-β treatment, which results in Ski degradation and a loss of binding of NFI to the URR. Our conclusion that Ski acts as a coactivator of NFI in positive transcriptional regulation of the HPV16 URR is also supported by results showing that ectopic Ski overexpression eliminated TGF-β inhibition of the URR (Fig. 8B).
Endogenous Ski is expressed at low levels in TGF-β-sensitive HKc/HPV16 (Fig. 7A, lane 1) but shows a dramatic increase in HKc/DR (Fig. 7A, lane 3). This is consistent with observations that endogenous Ski is expressed at relatively low levels in normal tissues but is abundant in transformed cells (15, 30, 45). Our data demonstrating a decrease in Ski levels upon TGF-β treatment in HKc/DR were initially puzzling, since HKc/DR have reduced expression of the TGF-β receptor type I and are refractory to growth inhibition by TGF-β (4). However, using a Smad reporter construct, our laboratory has shown that TGF-β signaling is reduced but not absent in HKc/DR (unpublished data). Therefore, it appears that sufficient TGF-β signaling through the Smads is retained, resulting in Ski degradation in HKc/DR in response to TGF-β treatment. This observation points to a potentially important concept: growth control by TGF-β appears to require intact or almost intact TGF-β signaling, while other TGF-β-mediated responses can be elicited even when TGF-β signaling pathways are partially disrupted (3). Our results here indicate that enough TGF-β signaling is retained in HKc/DR to still modulate the levels of Ski. However, given the considerable increase in total Ski levels in HKc/DR, even when reduced by TGF-β treatment, enough Ski is present to still mediate NFI activation of the URR, and thus TGF-β inhibition of URR activity is lost. In other words, we propose that, following TGF-β treatment, Ski is limiting in TGF-β-sensitive, low-passage-number cells but not any more in the TGF-β-resistant HKc/DR that have such high levels of Ski.
In our URR reporter construct experiments, we found that a complete loss of TGF-β inhibition of the HPV16 URR was not observed until all seven NFI sites present in the URR were mutated. This might be explained by the fact that, even though certain NFI sites demonstrate more NFI binding than others (Fig. 3B) and are probably more important to basal HPV16 URR activity (Fig. 5), some compensation could occur by binding to the remaining NFI sites when others have been mutated. The various degrees of NFI binding imply that negative regulatory elements may exist near NFI binding sites. This possibility is supported by our NFI binding data demonstrating that binding to NFI sites 1 and 5 is minimal (Fig. 3B, lanes 1 and 9) and that mutation of those two sites produces only a small decrease in the basal activity of the HPV16 URR (Fig. 5). One explanation for variation in NFI binding may lie in the fact that YY1 binding sites are present adjacent to five of the NFI binding sites in the URR (Fig. 1). The two NFI sites not adjacent to YY1 sites are located next to Tef-1 sites (NFI sites 4 and 6, Fig. 1), and YY1 has been shown to compete with Tef-1 for binding to the Tef-1 sites. Consequently, YY1 could affect binding to all seven NFI binding sites in the HPV16 URR. While our mutational analysis of YY1 did not demonstrate an effect on TGF-β modulation of the URR, we cannot completely rule out the possibility that YY1 may play some role in TGF-β modulation of the URR.
Although it has been reported that NFIX is not expressed in epithelial cells (1), we have found that all NFI family members are expressed in HKc/HPV16 at the RNA level (Fig. 5B). This discrepancy could be explained by differences in the epithelial cell lines used, as our studies were performed in HPV16-immortalized human foreskin keratinocytes, whereas the former study was performed in HeLa cells (1). We also demonstrate that ectopic expression of each family member can reduce TGF-β inhibition of URR activity (Fig. 7), demonstrating that each NFI family member is active.
The Smads, which have been shown to interact with numerous transcription factors to transduce TGF-β signaling (55), have not been shown to directly bind NFI. TGF-β upregulation of gene expression through NFI, however, has been documented for several genes including those for cyclooxygenase 2 (60), rat bone sialoprotein (42), and glial fibrillary acidic protein (26). This is the first report of negative transcriptional regulation by TGF-β through NFI, and we describe for the first time a mechanism whereby TGF-β signaling is transduced through NFI-Ski interactions. Our results demonstrate that NFI-Ski interactions modulate TGF-β inhibition of HPV16 early gene expression.
Acknowledgments
Antiserum to NFI was kindly provided by Naoko Tanese. We also thank Edward Stavnezer for kindly proving the Ski expression vectors and Richard M. Gronostajski for the gift of the NFI expression vectors. We thank Lubomir Turek for many helpful discussions on this topic over the years and Karl Munger for critically reading the manuscript.
This work was supported by grant R01-CA89502 from the National Institutes of Health to K.E.C., by a Research Development Award from the USC School of Medicine, and by the South Carolina Endowment for Children's Cancer Research.
REFERENCES
- 1.Apt, D., T. Chong, Y. Liu, and H. U. Bernard. 1993. Nuclear factor I and epithelial cell-specific transcription of human papillomavirus type 16. J. Virol. 67:4455-4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Armendariz-Borunda, J., C. P. Simkevich, N. Roy, R. Raghow, A. H. Kang, and J. M. Seyer. 1994. Activation of Ito cells involves regulation of AP-1 binding proteins and induction of type I collagen gene expression. Biochem. J. 304:817-824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berger, D. H., X. H. Feng, J. Yao, D. Saha, R. D. Beauchamp, and X. Lin. 2002. Resistance to transforming growth factor-beta occurs in the presence of normal Smad activation. Surgery 132:310-316. [DOI] [PubMed] [Google Scholar]
- 4.Borger, D. R., Y. Mi, G. Geslani, L. L. Zyzak, A. Batova, T. S. Engin, L. Pirisi, and K. E. Creek. 2000. Retinoic acid resistance at late stages of human papillomavirus type 16-mediated transformation of human keratinocytes arises despite intact retinoid signaling and is due to a loss of sensitivity to transforming growth factor-beta. Virology 270:397-407. [DOI] [PubMed] [Google Scholar]
- 5.Butz, K., and F. Hoppe-Seyler. 1993. Transcriptional control of human papillomavirus (HPV) oncogene expression: composition of the HPV type 18 upstream regulatory region. J. Virol. 67:6476-6486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan, W. K., T. Chong, H. U. Bernard, and G. Klock. 1990. Transcription of the transforming genes of the oncogenic human papillomavirus-16 is stimulated by tumor promotors through AP1 binding sites. Nucleic Acids Res. 18:763-769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chaudhry, A. Z., G. E. Lyons, and R. M. Gronostajski. 1997. Expression patterns of the four nuclear factor I genes during mouse embryogenesis indicate a potential role in development. Dev. Dyn. 208:313-325. [DOI] [PubMed] [Google Scholar]
- 8.Chong, T., D. Apt, B. Gloss, M. Isa, and H. U. Bernard. 1991. The enhancer of human papillomavirus type 16: binding sites for the ubiquitous transcription factors oct-1, NFA, TEF-2, NF1, and AP-1 participate in epithelial cell-specific transcription. J. Virol. 65:5933-5943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chong, T., W. K. Chan, and H. U. Bernard. 1990. Transcriptional activation of human papillomavirus 16 by nuclear factor I, AP1, steroid receptors and a possibly novel transcription factor, PVF: a model for the composition of genital papillomavirus enhancers. Nucleic Acids Res. 18:465-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chung, K. Y., A. Agarwal, J. Uitto, and A. Mauviel. 1996. An AP-1 binding sequence is essential for regulation of the human α2(I) collagen (COL1A2) promoter activity by transforming growth factor-beta. J. Biol. Chem. 271:3272-3278. [DOI] [PubMed] [Google Scholar]
- 11.Cripe, T. P., A. Alderborn, R. D. Anderson, S. Parkkinen, P. Bergman, T. H. Haugen, U. Pettersson, and L. P. Turek. 1990. Transcriptional activation of the human papillomavirus-16 P97 promoter by an 88-nucleotide enhancer containing distinct cell-dependent and AP-1-responsive modules. New Biol. 2:450-463. [PubMed] [Google Scholar]
- 12.Dollard, S. C., T. R. Broker, and L. T. Chow. 1993. Regulation of the human papillomavirus type 11 E6 promoter by viral and host transcription factors in primary human keratinocytes. J. Virol. 67:1721-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dong, X. P., F. Stubenrauch, E. Beyer-Finkler, and H. Pfister. 1994. Prevalence of deletions of YY1-binding sites in episomal HPV 16 DNA from cervical cancers. Int. J. Cancer 58:803-808. [DOI] [PubMed] [Google Scholar]
- 14.Frederick, J. P., and X. F. Wang. 2002. Smads “Freeze” when they ski. Structure (Cambridge) 10:1607-1611. [DOI] [PubMed] [Google Scholar]
- 15.Fumagalli, S., L. Doneda, N. Nomura, and L. Larizza. 1993. Expression of the c-ski proto-oncogene in human melanoma cell lines. Melanoma Res. 3:23-27. [DOI] [PubMed] [Google Scholar]
- 16.Garcia-Carranca, A., F. Thierry, and M. Yaniv. 1988. Interplay of viral and cellular proteins along the long control region of human papillomavirus type 18. J. Virol. 62:4321-4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gloss, B., T. Chong, and H. U. Bernard. 1989. Numerous nuclear proteins bind the long control region of human papillomavirus type 16: a subset of 6 of 23 DNase I-protected segments coincides with the location of the cell-type-specific enhancer. J. Virol. 63:1142-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gronostajski, R. M. 2000. Roles of the NFI/CTF gene family in transcription and development. Gene 249:31-45. [DOI] [PubMed] [Google Scholar]
- 19.Inoue, N., R. C. Venema, H. S. Sayegh, Y. Ohara, T. J. Murphy, and D. G. Harrison. 1995. Molecular regulation of the bovine endothelial cell nitric oxide synthase by transforming growth factor-beta 1. Arterioscler. Thromb. Vasc. Biol. 15:1255-1261. [DOI] [PubMed] [Google Scholar]
- 20.Iozzo, R. V., J. Pillarisetti, B. Sharma, A. D. Murdoch, K. G. Danielson, J. Uitto, and A. Mauviel. 1997. Structural and functional characterization of the human perlecan gene promoter. Transcriptional activation by transforming growth factor-beta via a nuclear factor 1-binding element. J. Biol. Chem. 272:5219-5228. [DOI] [PubMed] [Google Scholar]
- 21.Ishiji, T., M. J. Lace, S. Parkkinen, R. D. Anderson, T. H. Haugen, T. P. Cripe, J. H. Xiao, I. Davidson, P. Chambon, and L. P. Turek. 1992. Transcriptional enhancer factor (TEF)-1 and its cell-specific co-activator activate human papillomavirus-16 E6 and E7 oncogene transcription in keratinocytes and cervical carcinoma cells. EMBO J. 11:2271-2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kannius-Janson, M., E. M. Johansson, G. Bjursell, and J. Nilsson. 2002. Nuclear factor 1-C2 contributes to the tissue-specific activation of a milk protein gene in the differentiating mammary gland. J. Biol. Chem. 277:17589-17596. [DOI] [PubMed] [Google Scholar]
- 23.Keeton, M. R., S. A. Curriden, A. J. van Zonneveld, and D. J. Loskutoff. 1991. Identification of regulatory sequences in the type 1 plasminogen activator inhibitor gene responsive to transforming growth factor beta. J. Biol. Chem. 266:23048-23052. [PubMed] [Google Scholar]
- 24.Khan, M. A., G. R. Jenkins, W. H. Tolleson, K. E. Creek, and L. Pirisi. 1993. Retinoic acid inhibition of human papillomavirus type 16-mediated transformation of human keratinocytes. Cancer Res. 53:905-909. [PubMed] [Google Scholar]
- 25.Khan, M. A., W. H. Tolleson, J. D. Gangemi, and L. Pirisi. 1993. Inhibition of growth, transformation, and expression of human papillomavirus type 16 E7 in human keratinocytes by alpha interferons. J. Virol. 67:3396-3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krohn, K., I. Rozovsky, P. Wals, B. Teter, C. P. Anderson, and C. E. Finch. 1999. Glial fibrillary acidic protein transcription responses to transforming growth factor-β1 and interleukin-1β are mediated by a nuclear factor-1-like site in the near-upstream promoter. J. Neurochem. 72:1353-1361. [DOI] [PubMed] [Google Scholar]
- 27.Kulkarni, S., and R. M. Gronostajski. 1996. Altered expression of the developmentally regulated NFI gene family during phorbol ester-induced differentiation of human leukemic cells. Cell Growth Differ. 7:501-510. [PubMed] [Google Scholar]
- 28.Lafont, J., M. Laurent, H. Thibout, F. Lallemand, Y. Le Bouc, A. Atfi, and C. Martinerie. 2002. The expression of novH in adrenocortical cells is down-regulated by TGFβ 1 through c-Jun in a Smad-independent manner. J. Biol. Chem. 277:41220-41229. [DOI] [PubMed] [Google Scholar]
- 29.Lin, C. J., J. W. Martens, and W. L. Miller. 2001. NF-1C, Sp1, and Sp3 are essential for transcription of the human gene for P450c17 (steroid 17alpha-hydroxylase/17,20 lyase) in human adrenal NCI-H295A cells. Mol. Endocrinol. 15:1277-1293. [DOI] [PubMed] [Google Scholar]
- 30.Liu, X., Y. Sun, R. A. Weinberg, and H. F. Lodish. 2001. Ski/Sno and TGF-beta signaling. Cytokine Growth Factor Rev. 12:1-8. [DOI] [PubMed] [Google Scholar]
- 31.Massague, J. 2000. How cells read TGF-beta signals. Nat. Rev. Mol. Cell Biol. 1:169-178. [DOI] [PubMed] [Google Scholar]
- 32.Massague, J., and D. Wotton. 2000. Transcriptional control by the TGF-beta/Smad signaling system. EMBO J. 19:1745-1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matrisian, L. M., G. L. Ganser, L. D. Kerr, R. W. Pelton, and L. D. Wood. 1992. Negative regulation of gene expression by TGF-beta. Mol. Reprod. Dev. 32:111-120. [DOI] [PubMed] [Google Scholar]
- 34.May, M., X. P. Dong, E. Beyer-Finkler, F. Stubenrauch, P. G. Fuchs, and H. Pfister. 1994. The E6/E7 promoter of extrachromosomal HPV16 DNA in cervical cancers escapes from cellular repression by mutation of target sequences for YY1. EMBO J. 13:1460-1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Misawa, H., and M. Yamaguchi. 2002. Identification of transcription factor in the promoter region of rat regucalcin gene: binding of nuclear factor I-A1 to TTGGC motif. J. Cell. Biochem. 84:795-802. [DOI] [PubMed] [Google Scholar]
- 36.Moustakas, A., K. Pardali, A. Gaal, and C. H. Heldin. 2002. Mechanisms of TGF-beta signaling in regulation of cell growth and differentiation. Immunol. Lett. 82:85-91. [DOI] [PubMed] [Google Scholar]
- 37.Moustakas, A., S. Souchelnytskyi, and C. H. Heldin. 2001. Smad regulation in TGF-beta signal transduction. J. Cell Sci. 114:4359-4369. [DOI] [PubMed] [Google Scholar]
- 38.Nakshatri, H., M. M. Pater, and A. Pater. 1990. Ubiquitous and cell-type-specific protein interactions with human papillomavirus type 16 and type 18 enhancers. Virology 178:92-103. [DOI] [PubMed] [Google Scholar]
- 39.Nicol, R., and E. Stavnezer. 1998. Transcriptional repression by v-Ski and c-Ski mediated by a specific DNA binding site. J. Biol. Chem. 273:3588-3597. [DOI] [PubMed] [Google Scholar]
- 40.Nurnberg, W., M. Artuc, G. Vorbrueggen, F. Kalkbrenner, K. Moelling, B. M. Czarnetzki, and D. Schadendorf. 1995. Nuclear proto-oncogene products transactivate the human papillomavirus type 16 promoter. Br. J. Cancer 71:1018-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.O'Connor, M. J., S. H. Tan, C. H. Tan, and H. U. Bernard. 1996. YY1 represses human papillomavirus type 16 transcription by quenching AP-1 activity. J. Virol. 70:6529-6539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ogata, Y., N. Niisato, S. Furuyama, S. Cheifetz, R. H. Kim, H. Sugiya, and J. Sodek. 1997. Transforming growth factor-beta 1 regulation of bone sialoprotein gene transcription: identification of a TGF-beta activation element in the rat BSP gene promoter. J. Cell. Biochem. 65:501-512. [DOI] [PubMed] [Google Scholar]
- 43.Pirisi, L., S. Yasumoto, M. Feller, J. Doniger, and J. A. DiPaolo. 1987. Transformation of human fibroblasts and keratinocytes with human papillomavirus type 16 DNA. J. Virol. 61:1061-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rafty, L. A., F. S. Santiago, and L. M. Khachigian. 2002. NF1/X represses PDGF A-chain transcription by interacting with Sp1 and antagonizing Sp1 occupancy of the promoter. EMBO J. 21:334-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reed, J. A., E. Bales, W. Xu, N. A. Okan, D. Bandyopadhyay, and E. E. Medrano. 2001. Cytoplasmic localization of the oncogenic protein Ski in human cutaneous melanomas in vivo: functional implications for transforming growth factor beta signaling. Cancer Res. 61:8074-8078. [PubMed] [Google Scholar]
- 46.Riccio, A., P. V. Pedone, L. R. Lund, T. Olesen, H. S. Olsen, and P. A. Andreasen. 1992. Transforming growth factor beta 1-responsive element: closely associated binding sites for USF and CCAAT-binding transcription factor-nuclear factor I in the type 1 plasminogen activator inhibitor gene. Mol. Cell. Biol. 12:1846-1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ritzenthaler, J. D., R. H. Goldstein, A. Fine, A. Lichtler, D. W. Rowe, and B. D. Smith. 1991. Transforming-growth-factor-beta activation elements in the distal promoter regions of the rat alpha 1 type I collagen gene. Biochem. J. 280:157-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ritzenthaler, J. D., R. H. Goldstein, A. Fine, and B. D. Smith. 1993. Regulation of the alpha 1(I) collagen promoter via a transforming growth factor-beta activation element. J. Biol. Chem. 268:13625-13631. [PubMed] [Google Scholar]
- 49.Rousse, S., F. Lallemand, D. Montarras, C. Pinset, A. Mazars, C. Prunier, A. Atfi, and C. Dubois. 2001. Transforming growth factor-beta inhibition of insulin-like growth factor-binding protein-5 synthesis in skeletal muscle cells involves a c-Jun N-terminal kinase-dependent pathway. J. Biol. Chem. 276:46961-46967. [DOI] [PubMed] [Google Scholar]
- 50.Sen, E., J. L. Bromberg-White, and C. Meyers. 2002. Genetic analysis of cis regulatory elements within the 5′ region of the human papillomavirus type 31 upstream regulatory region during different stages of the viral life cycle. J. Virol. 76:4798-4809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shih, S. C., and K. P. Claffey. 2001. Role of AP-1 and HIF-1 transcription factors in TGF-beta activation of VEGF expression. Growth Factors 19:19-34. [DOI] [PubMed] [Google Scholar]
- 52.Sibbet, G. J., and M. S. Campo. 1990. Multiple interactions between cellular factors and the non-coding region of human papillomavirus type 16. J. Gen. Virol. 71:2699-2707. [DOI] [PubMed] [Google Scholar]
- 53.Tahara, T., J. P. Kraus, and L. E. Rosenberg. 1990. Direct DNA sequencing of PCR amplified genomic DNA by the Maxam-Gilbert method. BioTechniques 8:366-368. [PubMed] [Google Scholar]
- 54.Tarapore, P., C. Richmond, G. Zheng, S. B. Cohen, B. Kelder, J. Kopchick, U. Kruse, A. E. Sippel, C. Colmenares, and E. Stavnezer. 1997. DNA binding and transcriptional activation by the Ski oncoprotein mediated by interaction with NFI. Nucleic Acids Res. 25:3895-3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ten Dijke, P., M. J. Goumans, F. Itoh, and S. Itoh. 2002. Regulation of cell proliferation by Smad proteins. J. Cell. Physiol. 191:1-16. [DOI] [PubMed] [Google Scholar]
- 56.Thierry, F., G. Spyrou, M. Yaniv, and P. Howley. 1992. Two AP1 sites binding JunB are essential for human papillomavirus type 18 transcription in keratinocytes. J. Virol. 66:3740-3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wakefield, L. M., and A. B. Roberts. 2002. TGF-beta signaling: positive and negative effects on tumorigenesis. Curr. Opin. Genet. Dev. 12:22-29. [DOI] [PubMed] [Google Scholar]
- 58.Woodworth, C. D., V. Notario, and J. A. DiPaolo. 1990. Transforming growth factors beta 1 and 2 transcriptionally regulate human papillomavirus (HPV) type 16 early gene expression in HPV-immortalized human genital epithelial cells. J. Virol. 64:4767-4775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xie, Y., V. Madelian, J. Zhang, G. Ling, and X. Ding. 2001. Activation of the NPTA element of the CYP2A3 gene by NFI-A2, a nasal mucosa-selective nuclear factor 1 isoform. Biochem. Biophys. Res. Commun. 289:1225-1228. [DOI] [PubMed] [Google Scholar]
- 60.Yang, X., F. Hou, L. Taylor, and P. Polgar. 1997. Characterization of human cyclooxygenase 2 gene promoter localization of a TGF-beta response element. Biochim. Biophys. Acta 1350:287-292. [DOI] [PubMed] [Google Scholar]
- 61.Zhang, J., and X. Ding. 1998. Identification and characterization of a novel tissue-specific transcriptional activating element in the 5′-flanking region of the CYP2A3 gene predominantly expressed in rat olfactory mucosa. J. Biol. Chem. 273:23454-23462. [DOI] [PubMed] [Google Scholar]
- 62.zur Hausen, H. 2002. Papillomaviruses and cancer: from basic studies to clinical application. Nat. Rev. Cancer 2:342-350. [DOI] [PubMed] [Google Scholar]