Abstract
Wild-type human respiratory syncytial virus (HRSV) is a poor inducer of alpha/beta interferons (IFN-α/β). However, recombinant HRSV lacking the NS1 and NS2 genes (ΔNS1/2) induced high levels of IFN-α and -β in human pulmonary epithelial cells (A549) as well as in macrophages derived from primary human peripheral blood monocytes. Results with NS1 and NS2 single- and double-gene-deletion viruses indicated that the two proteins function independently as well as coordinately to achieve the full inhibitory effect, with NS1 having a greater independent role. The relative contributions of the individual NS proteins were the converse of that recently described for bovine RSV (J. F. Valarcher, J. Furze, S. Wyld, R. Cook, K. K. Conzelmann, and G. Taylor, J. Virol. 77:8426-8439, 2003). This pattern of inhibition by HRSV NS1 and NS2 also extended to the newly described antiviral cytokines IFN-λ1, -2 and -3.
Human respiratory syncytial virus (HRSV) is the most common cause of viral bronchiolitis and pneumonia in infants and children worldwide, and a vaccine is needed (9, 10). HRSV belongs to the genus Pneumovirus of the family Paramyxoviridae and has single-stranded, negative-sense RNA as its genome (10). One of the differences between the members of the genus Pneumovirus and other members of Paramyxoviridae is that Pneumovirus species express two putative nonstructural proteins, NS1 and NS2, from separate mRNAs encoded by the first two genes in the viral gene order. Recombinant HRSVs in which the NS1 and/or NS2 genes have been deleted singly or in combination (ΔNS1, ΔNS2, and ΔNS1/2 viruses) exhibit reduced replication in cultured cells that are competent to produce alpha interferon (IFN-α) and IFN-β, as well as in mice, monkeys, and chimpanzees, but replicate more like wild-type (wt) HRSV in Vero cells that lack the IFN-α/β genes (15, 16, 21-23, 25, 28). Clinical trials of recombinant HRSV (rHRSV) vaccine candidates lacking NS1 or NS2 are under way or in preparation. Bovine RSV (BRSV) is an animal counterpart of HRSV that exhibits a strong host range specificity in vivo and differs from HRSV by up to 71% with regard to the amino acid sequences of the various individual proteins (5). Studies with BRSV provided evidence that its NS1 and NS2 proteins can function independently as well as cooperatively to reduce the effectiveness of the IFN α/β-mediated antiviral state (3, 19). How this occurs is unclear, although it does not appear to involve an inhibition of either intracellular signaling or the expression of IFN-stimulated genes (4, 27).
In the present study, we describe inhibition of the induction of IFN-α and -β by the NS1 and NS2 proteins of HRSV. In addition, we investigated the effect of HRSV infection and of deleting NS1 and/or NS2 on the expression of the newly described antiviral cytokines IFN-λ1, -2 and -3 (alternatively designated interleukin 29 (IL-29), -28A, and -28B, respectively) (17, 20). These cytokines differ genetically and structurally from INF-α/β but share the following characteristics. They appear to be broadly expressed, are inducible by double-stranded RNA (dsRNA) or virus infection, bind to a heterodimeric receptor (albeit one distinct from that of IFN-α/β), activate the same JAK/STAT signal transduction pathway, up-regulate genes that render cells resistant to infection, and potentially augment the subsequent adaptive immune response. Thus, IFN-λ appears to be an IFN-α/β-like cytokine with pleiotropic effects that include potent antiviral activities.
The rHRSV ΔNS1, ΔNS2, and ΔNS1/2 viruses were constructed in previous work and are derivatives of the recombinant version of the wt A2 strain (rA2) (21-23, 25). All of the viruses used in this study were grown in Vero cells and purified by centrifugation through a sucrose gradient to ensure the absence of IFN in the viral inocula. The identities and purity of the specific virus stocks used in these experiments were confirmed by reverse transcription (RT)-PCR. The expression levels of IFN-α/βs were evaluated in A549 cells, an established line of human type II alveolar epithelial cells, and in human macrophages. The macrophages were prepared by incubating human peripheral blood monocytes in Iscove's medium containing 20 ng of recombinant human granulocyte-monocyte colony-stimulating factor/ml, 10% human serum, and 1% sodium pyruvate for 1 week, after which residual nonadherent cells were removed by washing.
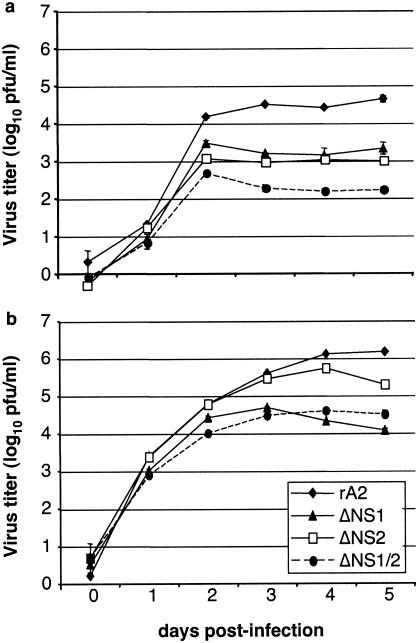
We compared the growth of the mutant viruses to that of wt rA2 in A549 cells (Fig. 1a), which are competent for the expression of IFN-α/β, unlike Vero cells (Fig. 1b), which lack the structural genes for these cytokines. Cells were infected at an input multiplicity of infection (MOI) of 0.01 PFU per cell for 2 h at 37°C, and aliquots of the medium were harvested daily. Virus titers were determined by plaque assay. The ΔNS1/2 double mutant was severely restricted (>100-fold reduced) in growth in A549 cells compared to wt rA2, whereas the growth of ΔNS1 and ΔNS2 was intermediate (20- to 45-fold reduced) between that of the double-deletion mutant and wt rA2. The NS deletion viruses also replicated less efficiently than wt rA2 in Vero cells, although the difference was less dramatic; for example, the ΔNS1/2 double deletion virus replicated 20-fold less efficiently than wt rA2. Thus, the attenuation phenotypes of the NS deletion viruses can be only partly accounted for by IFN-α/β effects, particularly for viruses in which the NS1 gene was deleted. Whether the residual restriction is due to IFN-λ or to some other aspect of NS1/2 function is not yet known. We next examined the expression of IFN-α, -β, and -λ mRNA in A549 epithelial cells by quantitative RT-PCR (qRT-PCR). Cells were infected with the ΔNS1, ΔNS2, or ΔNS1/2 mutant viruses in parallel with wt rA2, UV-inactivated wt rA2, and human parainfluenza virus type 3 (HPIV3) at an input MOI of 3 PFU or its equivalent per cell, adsorbed for 2 h at 37°C, washed, and harvested at 2-, 6-, 10-, 14-, 18-, 26-, 38-, and 50-h postinfection (p.i.) in independent experiments. Total RNA was extracted from cell pellets by using TRIzol reagent (Invitrogen) and the RNeasy total RNA isolation kit (QIAGEN). Prior to qRT-PCR, RNA was treated with DNase 1 (QIAGEN) to remove any contaminating DNA, the absence of which was confirmed in control experiments in which the reverse transcriptase enzyme was omitted (data not shown). qRT-PCR was performed by using reagents from the Brilliant two-step qRT-PCR kit (Stratagene). RT was performed with random primers and total intracellular RNA. PCR primers and TaqMan probes for the single species of IFN-β, the multiple species of IFN-α, the several species of IFN-λ, and the housekeeping mRNA β-actin were designed by using Primer 3 (www-genome.wi.mit.edu/cgi-bin/primer/primer3_www.cgi) software (Table 1). Human IFN-α exists as at least 13 species; based on their high degree of sequence relatedness, we designed two sets of primers and probes representing 10 species. IFN-α primer set 1 was specific to IFN-α1, -6, and -13, and IFN-α primer set 2 was specific to IFN-α4, -5, -8, -10, -14, -17, and -21 (Table 1). Three species of IFN-λ have been described, and the primers and probes used in this study differentiated between those for IFN-λ1 (IL-29) and those for IFN-λ2 and -3 combined (IL-28A and B, which have 96% amino acid sequence identity). The probe for β-actin was labeled with the reporter dye 5-carboxyfluorescein (FAM) at the 5′ end and black-hole quencher (BHQ1) at the 3′ end. All IFN probes were labeled with the reporter dye 5′HEX at the 5′ end and BHQ1 at the 3′ end. Reactions contained a passive reference dye, which is not involved in amplification and which served to normalize the probe reporter dye signals. The use of differential reporter dyes (FAM and HEX) allowed qRT-PCRs to be done for each IFN mRNA of interest and the β-actin housekeeping mRNA to be duplexed (performed simultaneously in the same tube), and thus each result was normalized to the internal housekeeping-mRNA control. In addition, each set of PCRs was run in parallel with a single-standard cDNA preparation made from RNA from A549 cells infected with ΔNS1/2 and known to contain significant levels of IFN mRNA; this cDNA was run as a 10-fold dilution series that was used to make a curve for the purposes of quantitation and comparison between runs and for confirming the linearity of each PCR.
FIG. 1.
Growth of NS gene deletion viruses in A549 and Vero cells. Triplicate cultures of A549 (a) or Vero (b) cells were infected at an MOI of 0.01 PFU/cell with wt recombinant HRSV (rA2), ΔNS1, ΔNS2, or ΔNS1/2. The supernatants were harvested daily, and viral titers were determined by plaque assay using the cell line from which the supernatants were derived. Plaques were visualized by immunostaining with anti-F monoclonal antibodies (21), and the mean titers (log10 PFU/ml) are shown.
TABLE 1.
β-Actin and IFN-α/β primers and probes used for quantitative RT-PCR
| Target gene | Primera | Nucleotide sequence | GenBank accession no. |
|---|---|---|---|
| β-actin | F | 5′-GGCATCCACGAAACTACCTT-3′ | NM_001101 |
| R | 5′-AGCACTGTGTTGGCGTACAG-3′ | ||
| P | 5′-ATCATGAAGTGTGACGTGGACATCCG-3′ | ||
| IFN-β | F | 5′-CGCCGCATTGACCATCTA-3′ | M28622 |
| R | 5′-GACATTAGCCAGGAGGTTCTCA-3′ | ||
| P | 5′-TCAGACAAGATTCATCTAGCACTGGCTGGA-3′ | ||
| IFN-α set 1b | F | 5′-CAGAGTCACCCATCTCAGCA-3′ | NM_024013 |
| R | 5′-CACCACCAGGACCATCAGTA-3′ | ||
| P | 5′-ATCTGCAATATCTACGATGGCCTCGCC-3′ | ||
| IFN-α set 2c | F | 5′-CTGGCACAAATGGGAAGAAT-3′ | NM_021068 |
| R | 5′-CTTGAGCCTTCTGGAACTGG-3′ | ||
| P | 5′-TTTCTCCTGCCTGAAGGACAGACATGA-3′ | ||
| IFN-λ1 | F | 5′-CTGGAGGCATCTGTCACCTT-3′ | AY184372 |
| R | 5′-TGGGTTGACGTTCTCAGACA-3′ | ||
| P | 5′-CCTCCTCACGCGAGACCTCAAATATGT-3′ | ||
| IFN-λ2/3 | F | 5′-GCCTCTGTCACCTTCAACCTC-3′ | AY184373/4 |
| R | 5′-GGAGGGTCAGACACACAGGT-3′ | ||
| P | 5′-CTCCTCACGCGAGACCTGAATTGTGTT-3′ |
The primers are identified as forward or positive-sense (F), reverse (R), or probe (P).
IFN-α set 1 includes IFN-α1, -6, and -13.
IFN-α set 2 includes IFN-α4, -5, -8, -10, -14, -17, and -21.
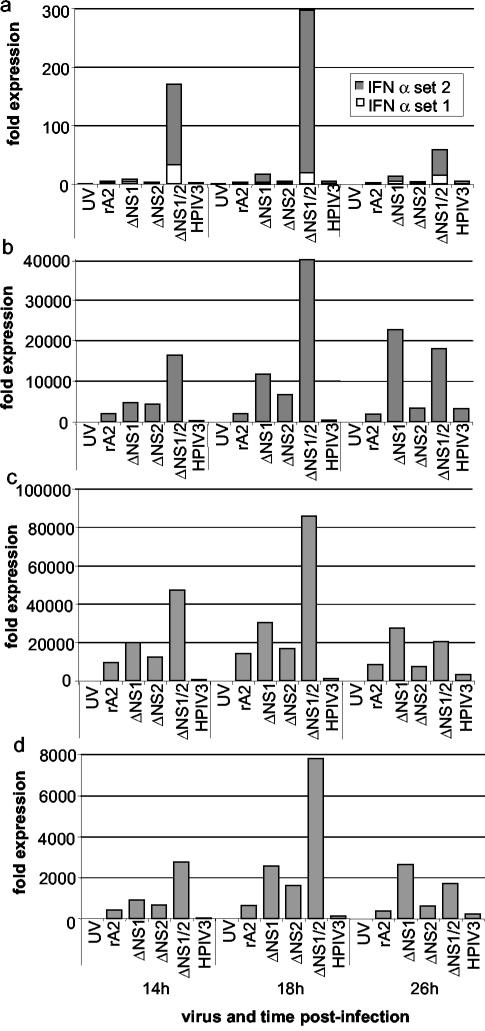
The response of IFN-α to infection with wt rA2 and the NS deletion mutants and controls is shown in Fig. 2a for time points 14-, 18-, and 26-h p.i.; at the other time points there was only minimal expression, and there is no data shown. Infection with the ΔNS1/2 virus resulted in a 300-fold increase in the expression of the combined species at 18-h p.i. compared to that for mock-treated cells, which diminished thereafter. Infection with ΔNS1 resulted in an 18-fold increase that peaked at 26 h, whereas the remaining viruses were associated with levels that were threefold or less that of the mock-infected control. In particular, infection with UV-inactivated wt rA2 as a control did not induce a detectable IFN-α response, implying a need for viral RNA synthesis. Infection with wt HPIV3 as another control was also inefficient in inducing IFN-α, implying that HPIV3 also strongly inhibits the induction of IFN-α. Similar results with UV-HRSV and HPIV3 were observed with IFN-β (Fig. 2b) and -λ (Fig. 2c and d).
FIG. 2.
Expression levels of mRNA encoding human IFN-α (a), -β (b), -λ1 (c), or -λ2/3 (d) in A549 cells infected with wt rA2, ΔNS1, ΔNS2, ΔNS1/2, or HPIV3 or in mock-infected A549 cells. mRNA was measured by qRT-PCR using specific primers and Taqman probes (Table 1). The IFN-α primers were designed as two sets, each of which detected multiple species due to the high degree of sequence relatedness. Set 1 detected IFN-α1, -6, and -13, and set 2 detected IFN-α4, -5, -8, -10, -14, -17 and -21 (Table 1). For each sample, the expression level of each IFN mRNA was calculated in relation to the expression level of β-actin and expressed as a fold increase compared to that for the mock-infected sample. Representative data from one of two independent experiments that gave similar results are shown. There was minimal expression at the 2-, 6-, 10-, 38-, and 50-h time points, and no datum is shown for these time points.
In the case of IFN-β (Fig. 2b), infection of A549 cells with the ΔNS1/2 virus resulted in a 40,000-fold increase in the expression level of IFN-β mRNA compared to that for mock-infected cells. The accumulation of IFN-β mRNA peaked at 18-h p.i. and diminished thereafter. This pattern of a sharp peak followed by rapid decay is consistent with the induction of IFN-β mRNA in response to dsRNA, where the half-life of the mRNA was calculated to be 18 min (6, 12, 19). The peak induction of IFN-β in response to dsRNA occurs within 1.5 h (6), and the considerably longer time frame in response to HRSV infection presumably reflects a requirement for extensive HRSV RNA replication and transcription. In comparison, the peak values of wt rA2, HPIV3, ΔNS1, and ΔNS2 were 2010, 3220, 22,600, and 6,635 times those of the mock-infected control, and thus each of these viruses induced a substantial response, even if it was much less than that of ΔNS1/2. This finding is consistent with the reported induction of IFN-β in epithelial cells by wt HRSV (14). The kinetics of accumulation of intracellular IFN-β mRNA was generally the same for each of the HRSVs, although the peak for the ΔNS1 virus was at 26 h rather than 18 h in this particular experiment (Fig. 2b), reflecting experimental variability. Thus, the highest induction of IFN-β mRNA was associated with deletion of the NS1 and NS2 genes together, followed, respectively, by deletion of NS1 and NS2.
We also examined the expression of IFN-λ by using primers that differentiated between IFN-λ1 (Fig. 2c) and -λ2/3 (Fig. 2d). In each case, ΔNS1/2 induced an increase in the accumulation of IFN-λ1 and -2/3 that peaked at 18-h p.i. and declined thereafter. Cells infected with ΔNS1 also displayed modestly elevated levels of IFN-λ, peaking at 26 h. Thus, the time frame of the increase and decrease of mRNA was similar for all of the IFN-α/βs.
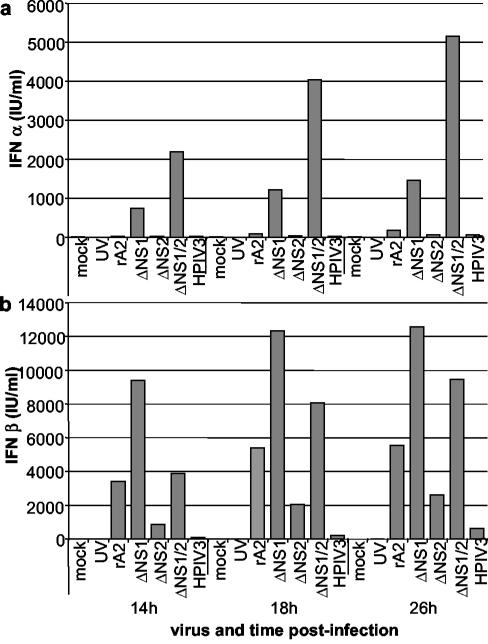
The secretion of IFN-α and -β from infected A549 cells was quantified by antigen-capture enzyme-linked immunosorbent assay (ELISA) of the medium supernatants (Fig. 3). The secretion of IFN-α faithfully reflected the results of the qRT-PCR and was the greatest for ΔNS1/2, reaching a level of nearly 5,000 IU of IFN-α/ml, followed by that for ΔNS1 (Fig. 3a). In the case of IFN-β (Fig. 3b), the pattern of secretion did not completely match the results of qRT-PCR. In particular, the peak level of secretion in response to ΔNS1 was somewhat higher than that of ΔNS1/2, and the amount of secretion in response to wt rA2 was unexpectedly high. Similar results were obtained in independent experiments. The reason for the inconsistent correspondence between the IFN-β qRT-PCR and the ELISA is not known. One possible factor is that the level of secretion of IFN-β was very high, up to 6,000 to 12,000 IU/ml, and it might be that its production or secretion by A549 cells was not completely responsive to intracellular mRNA levels. Another possibility is that some of the mRNA measured by qRT-PCR was not translatable, reflecting the rapid posttranscriptional repression of IFN-β synthesis in which translatability of the mRNA is lost before the body of the mRNA is degraded (11, 18). A previous study with Sendai virus indicated that viral factors can influence IFN-β mRNA translatability (11), which might cause a further difference between the various viruses studied here. Nonetheless, the IFN-β ELISA did confirm the most important point, that the ΔNS1 and ΔNS1/2 mutants were associated with the highest levels of IFN-β secretion. The secretion of IFN-λ1, -2 and -3 was not monitored due to the lack of an established, specific, functional or antibody-based assay.
FIG. 3.
Expression levels of IFN-α (a) and -β (b) protein in A549 cells infected with wt rA2, ΔNS1, ΔNS2, ΔNS1/2, or HPIV3. Expression levels of IFN-α and -β were measured by using, respectively, the Human Interferon-α Multi-Species ELISA kit (PBL Biomedical Laboratories) or the Human Interferon-β ELISA kit (Biosource International) and were calibrated by using standard curves generated from international standards for IFN-α or -β, as designated by the National Institutes of Health (PBL Biomedical Laboratories).
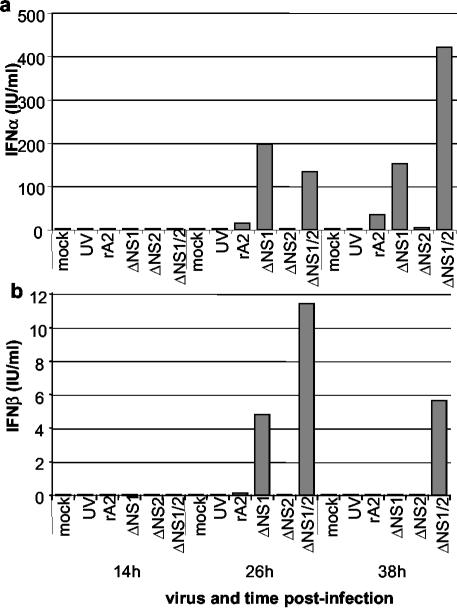
We also investigated the expression of IFN-α/βs in response to wt and mutant HRSVs in macrophages, immune cells that are exposed to HRSV during pulmonary infection. Monocyte-derived macrophages were infected with the NS mutants or UV-rA2 in the same manner as the A549 cells, and total cellular RNA was extracted 2-, 14-, 26-, and 38-h p.i. in three independent experiments by using cells from two donors. The expression of IFN-α was strongly induced in cells infected with the ΔNS1/2 virus and, to a lesser extent, with ΔNS1 (Fig. 4a). A similar pattern was observed for IFN-β; in the experiment shown, the expression level in response to the ΔNS1 virus was nearly equal to that of ΔNS1/2 (Fig. 4b), whereas in other experiments the response to ΔNS1 was intermediate between that of ΔNS1/2 and the control viruses. A similar pattern was observed in the case of IFN-λ1 (Fig. 4c) and -λ2/3 (Fig. 4d), with the response being the greatest to the ΔNS1/2 virus followed by that for the ΔNS1 virus. For each of the IFNs, the peak of mRNA accumulation in the macrophages was 26-h p.i., with some experiment-to-experiment variability. Specifically, in the particular experiment shown in Fig. 4c, the peak level of IFN-λ1 was at 38-h rather than 26-h p.i., but in other experiments it was at 26-h p.i.(data not shown). There was also experiment-to-experiment variability in the background levels of mRNA, which were high in this experiment (Fig. 4c and d) but not in other experiments. However, the general pattern was consistent. At the protein level (Fig. 5), the pattern of secretion of IFN-α and -β was consistent with the qRT-PCR data, although the maximum levels, 410 IU of IFN-α/ml and 11.5 IU of IFN-β/ml, were much lower than that observed with A549 cells. Also, the balance between IFN-α set 1 and set 2 in the qRT-PCR was the converse of that observed for A549 cells. In each case, expression levels were the greatest for ΔNS1/2, followed by those for ΔNS1, with the other viruses inducing little IFN.
FIG. 4.
Expression levels of mRNA encoding IFN-α (a), -β (b), -λ1 (c), or -λ2/3 (d) in monocyte-derived macrophages infected with wt rA2, ΔNS1, ΔNS2, ΔNS1/2, UV-inactivated rA2 (UV), or mock infected. Expression levels were measured by qRT-PCR as described in the legend to Fig. 2. Representative data from one of three independent experiments involving two different donors, which yielded comparable results, are shown. Minimal expression occurred at the 2-h time point and is not shown.
FIG. 5.
Expression levels of IFN-α (a) and IFN-β (b) protein in peripheral blood-derived human macrophages infected with wt rA2, ΔNS1, ΔNS2, ΔNS1/2, or UV-inactivated rA2 (UV). IFN-α and IFN-β levels were measured as described in the legend to Fig. 3.
Since it is likely that viral dsRNA is important in inducing the IFN responses observed here, we compared the magnitude of intracellular viral RNA synthesis by wt rA2 and the NS gene deletion viruses. Analysis of the intracellular accumulation of viral genomic RNA and the viral mRNAs and proteins during single-cycle growth in both IFN-competent HEp-2 and IFN-lacking Vero cells revealed at most only modest reductions for the NS deletion viruses compared to those for wt rA2, differences that did not appear to be cell type-specific (15; M. N. Teng, K. M. Spann, and P. L. Collins, unpublished data). In addition, we measured the intracellular accumulation of viral genomic RNA in A549 cells and macrophages 14-h p.i. by using TaqMan probes and primers specific to the N and F regions of the RSV genome (data not shown). The reaction was made specific to genomic RNA by using a positive-sense primer for RT. Genomic RNA was chosen for measurement, since it presumably would be the duplex partner that would be limiting in the formation of intracellular viral dsRNA. For both cell types, the accumulations of genomic RNA by wt rA2, ΔNS1, ΔNS2, and ΔNS1/2 were within a twofold range and thus were very similar and should not be a factor in these virus-to-virus comparisons.
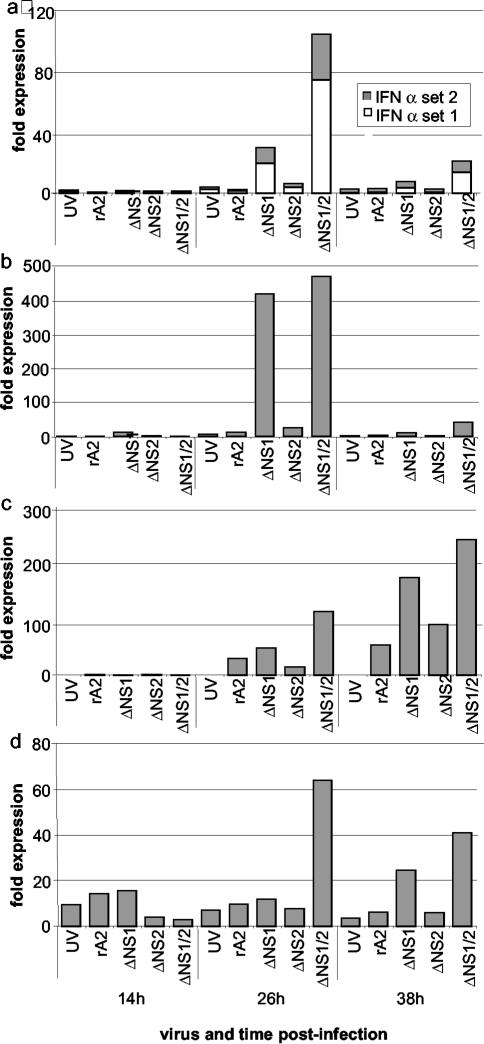
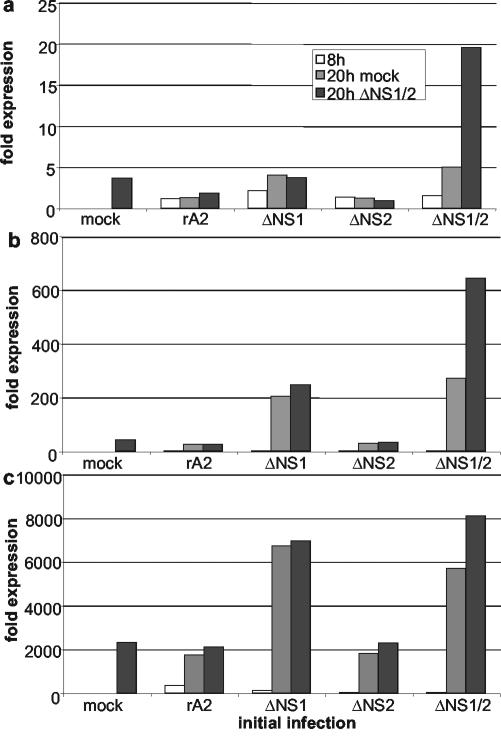
We then tested the ability of wt rA2 and the ΔNS1 and ΔNS2 single-gene-deletion mutants to inhibit the induction of IFN by the ΔNS1/2 double-gene-deletion mutant. A549 cells were infected with wt rA2, ΔNS1, ΔNS2, or ΔNS1/2 or were mock infected. The cells were incubated for 8 h in order to allow the NS1 and NS2 proteins to begin to accumulate at significant levels, which were confirmed by Western blot analysis (data not shown). The cells were then superinfected with ΔNS1/2 or mock superinfected and incubated for 12 h more to allow the possible induction or superinduction of IFN by ΔNS1/2. While a longer chase would have provided for a greater possible response to the superinfection, based on the kinetics shown in Fig. 2, we chose this shorter period in order to avoid the cytopathic effects of a long exposure to virus. The cells were then harvested and processed for qRT-PCR to measure intracellular levels of IFN-α set 1 (Fig. 6a), IFN-α set 2 (Fig. 6b), or IFN-β (Fig. 6c). Consistent with the results shown in Fig. 2, the initial infection with ΔNS1 or ΔNS1/2 followed by the mock superinfection induced substantially elevated levels of mRNA for IFN-α set 2 and IFN-β, while the induction of IFN-α set 1 was less substantial. Cells infected with the other viruses followed by mock superinfection had much lower levels of each IFN mRNA. Superinfection with ΔNS1/2 induced a further increase in levels of IFN-α set 1 (Fig. 6a), IFN-α set 2 (Fig. 6b), or IFN-β (Fig. 6c) in cells that had originally been mock infected or infected with ΔNS1/2 and not in cells that had originally been infected with wt rA2, ΔNS1, or ΔNS2. Thus, superinfection by ΔNS1/2 failed to induce a further induction of IFN in cells expressing NS1 and/or NS2, supporting the idea that each of these proteins, together or alone, inhibits the induction of IFN.
FIG. 6.
Ability of the NS1 and NS2 proteins to independently block the induction of expression of mRNA encoding IFN-α set 1 (a), IFN-α set 2 (b), or IFN-β (c) in A549 cells in response to superinfection by the ΔNS1/2 virus. Triplicate sets of cells were infected with wt rA2, ΔNS1, ΔNS2, or ΔNS1/2 or were mock infected and were incubated for 8 h. One culture from each set was processed to purify intracellular RNA, and the remaining cultures were mock superinfected or superinfected with ΔNS1/2. The cultures were incubated for an additional 12 h and processed to purify intracellular RNA. The expression levels of IFN mRNA were measured by qRT-PCR.
The production of IFN-α/βs is an early response to virus infection. IFN-α/βs mediate the autocrine and paracrine induction of a wide array of genes. Some of these gene products, including 2-5A synthetase, RNA-dependent protein kinase PKR, adenosine deaminase, and Mx, are potent antiviral effectors that establish an intracellular antiviral state (14). Other IFN-stimulated gene products, such as major histocompatibility complex proteins and antigen-processing proteins, have the potential to enhance the adaptive immune response (13, 14). IFN-α/βs are also potent cytokines that enhance cell-mediated responses of both innate and adaptive immunity, including Th1 helper cells, dendritic cells, NK cells, and cytotoxic T cells (1, 2, 12). The BRSV NS1 and NS2 proteins had previously been shown to counteract the IFN-induced intracellular antiviral state (3, 19). Also, while this manuscript was in preparation, Valarcher et al. and Bossart et al. (4, 24) showed that the BRSV NS1 and NS2 proteins together inhibit the induction of IFN-α/β and that BRSV NS2 had the greater independent inhibitory activity. In the case of HRSV, the two NS proteins also cooperate to inhibit the induction of IFN-α/βs. Each also appeared to have a significant independent role, but NS1 rather than NS2 had the greater independent role, reflecting an apparent species-specific difference. Previous studies of the rHRSV mutants in chimpanzees showed that the ΔNS1 virus was significantly more attenuated than the ΔNS2 virus (23, 25), and similar results were observed with BALB/c mice (M. N. Teng and P. L. Collins, unpublished data). This finding is consistent with the interpretation that IFN is responsible for the greater attenuation of ΔNS1 in vivo.
In the present paper, deletion of the HRSV NS proteins also resulted in the induction of the newly described cytokines IFN-λ1 and -λ2/3. While the full range of their biological properties remains to be described, IFN-λ has been shown to have an antiviral potency comparable to that of IFN-α (17, 20). IFN-λ had been shown to be inducible by dsRNA or by infection with several different viruses, and the present paper shows that IFN-λ was comparable to IFN-α/β with regard to the kinetics of induction, the transient nature of induction, the apparent short half-life of the mRNA, induction in both epithelial cells and macrophages, and inhibition by the HRSV NS proteins.
A vaccine for HRSV is presently not available, but live attenuated viruses, including some involving deletion of the NS1 or NS2 gene (16, 23, 25), appear to be promising candidates and are under development and in clinical trials (26). Ideally, an HRSV vaccine should be given to infants of 1 month of age or less, given the very young age at which infants become susceptible to HRSV disease (9, 10). Immune responses in this age group are reduced due to immunologic immaturity and the immunosuppressive effects of maternally derived serum antibodies, posing a major obstacle to HRSV immunoprophylaxis. The immunologic immaturity of the young infant extends to IFN-α/β responses (7), and a pediatric HRSV vaccine that does not further inhibit these responses might be particularly advantageous. It has also been hypothesized that wt HRSV infection suppresses the adaptive immune response, as suggested by the recent observation that virus-specific CD8+ T cells induced during HRSV infection of BALB/c mice exhibited reduced cytolytic activity, cytokine secretion, and development of memory cells (8). It would be interesting to determine whether these defects are mitigated by an increased IFN-α/β response, such as during infection by ΔNS1/2 or ΔNS1. In addition, although wt HRSV is a poor inducer of IFN-α/β, the IFN that is produced appears to play a regulatory role that minimizes pathogenic, Th2-associated components of the immune response in the BALB/c mouse model (12). Thus, an HRSV vaccine virus that has a reduced ability to block the synthesis and signaling of IFN-α/βs has the potential for both increased immunogenicity and decreased reactogenicity. Indeed, the BRSV ΔNS2 virus appeared to be more immunogenic than the BRSV ΔNS1 virus when administered to the respiratory tracts of calves, suggesting that increased synthesis of IFN-α/β indeed correlated with improved immunogenicity (24). Comparison of the HRSV ΔNS1 and ΔNS2 viruses in studies with chimpanzees did not reveal a difference in immunogenicity (23, 25), but this comparison was complicated by the difference in replication efficiency between the two viruses and by the small number and outbred nature of the animals. The double-deletion ΔNS1/2 virus would be the best choice for a robust IFN-α/β response, but a BRSV ΔNS1/2 virus was overattenuated in calves (24), and an HRSV ΔNS1/2 virus was overattenuated in African Green monkeys (16). However, the poor replication observed in these studies might have been exaggerated due to the use of a laboratory strain in the case of BRSV and a semipermissive animal model in the case of HRSV. Thus, these studies might not accurately predict the level of attenuation of the HRSV ΔNS1/2 virus in infants, particularly since the attenuation might be reduced due to the reduced IFN-α/β response in that age group. Thus, the HRSV ΔNS1 and ΔNS1/2 viruses are attractive vaccine candidates.
Acknowledgments
The views in this article are those of the authors and do not reflect the official policy or position of the Food and Drug Administration or the United States Government.
While this work was in progress, the NIAID laboratory received support from Wyeth for the development of live-attenuated vaccines against HRSV.
REFERENCES
- 1.Biron, C. A. 1998. Role of early cytokines, including alpha and beta interferons (IFN-α/β), in innate and adaptive immune responses to viral infections. Semin. Immunol. 10:383-390. [DOI] [PubMed] [Google Scholar]
- 2.Bogdan, C. 2000. The function of type I interferons in antimicrobial immunity. Curr. Opin. Immunol. 12:419-424. [DOI] [PubMed] [Google Scholar]
- 3.Bossert, B., and K.-K. Conzelmann. 2002. Respiratory syncytial virus (RSV) nonstructural (NS) proteins as host range determinants: a chimeric bovine RSV with NS genes from human RSV is attenuated in interferon-competent bovine cells. J. Virol. 76:4187-4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bossert, B., S. Marozin, and K. K. Conzelmann. 2003. Nonstructural proteins NS1 and NS2 of bovine respiratory syncytial virus block activation of interferon regulatory factor 3. J. Virol. 77:8661-8668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buchholz, U. J., H. Granzow, K. Schuldt, S. S. Whitehead, B. R. Murphy, and P. L. Collins. 2000. Chimeric bovine respiratory syncytial virus with glycoprotein gene substitutions from human respiratory syncytial virus (HRSV): effects on host range and evaluation as a live-attenuated HRSV vaccine. J. Virol. 74:1187-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cavalieri, R. L., E. A. Havell, J. Vilcek, and S. Pestka. 1977. Induction and decay of human fibroblast interferon. Proc. Natl. Acad. Sci. USA 74:4415-4419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cederblad, B., T. Riesenfeld, and G. V. Alm. 1990. Deficient herpes simplex virus-induced interferon-alpha production by blood leukocytes of preterm and term newborn infants. Pediatr. Res. 27:7-10. [DOI] [PubMed] [Google Scholar]
- 8.Chang, J., and T. J. Braciale. 2002. Respiratory syncytial virus infection suppresses CD8+ T-cell effector activity and peripheral CD8+ T-cell memory in the respiratory tract. Nat. Med. 8:54-60. [DOI] [PubMed] [Google Scholar]
- 9.Collins, P. L., and B. R. Murphy. 2002. Respiratory syncytial virus: reverse genetics and vaccine strategies. Virology 296:204-211. [DOI] [PubMed] [Google Scholar]
- 10.Collins, P. L., R. M. Chanock, and B. R. Murphy. 2001. Respiratory syncytial virus, p. 1443-1485. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 11.Dehlin, E., A. von Gabain, G. Alm, R. Dingelmaier, and O. Resnekov. 1996. Repression of beta interferon gene expression in virus-infected cells is correlated with a poly(A) tail elongation. Mol. Cell. Biol. 16:468-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Durbin, J. E., T. R. Johnson, R. K. Durbin, S. E. Mertz, R. A. Morotti, R. S. Peebles, and B. S. Graham. 2002. The role of IFN in respiratory syncytial virus pathogenesis. J. Immunol. 168:2944-2952. [DOI] [PubMed] [Google Scholar]
- 13.Goodbourn, S., L. Pidcock, and R. E. Randall. 2000. Interferons: cell signalling, immune modulation, antiviral responses and virus countermeasures. J. Gen. Virol. 81:2341-2364. [DOI] [PubMed] [Google Scholar]
- 14.Jamaluddin, M., S. Wang, R. P. Garofalo, T. Elliott, A. Casola, S. Baron, and A. R. Brasier. 2001. IFN-β mediates coordinate expression of antigen-processing genes in RSV-infected pulmonary epithelial cells. Am. J. Physiol. Lung Cell. Mol. Physiol. 280:L248-L257. [DOI] [PubMed] [Google Scholar]
- 15.Jin, H., H. Zhou, X. Cheng, R. Tang, M. Munoz, and N. Nguyen. 2000. Recombinant respiratory syncytial viruses with deletions in the NS1, NS2, SH, and M2-2 genes are attenuated in vitro and in vivo. Virology 273:210-218. [DOI] [PubMed] [Google Scholar]
- 16.Jin, H., H. Zhou, X. Cheng, R. Tang, M. Munoz, and N. Nguyen. 2003. Evaluation of recombinant respiratory syncytial virus gene deletion mutants in African green monkeys for their potential as live attenuated vaccine candidates. Vaccine 21:3647-3652. [DOI] [PubMed] [Google Scholar]
- 17.Kotenko, S. V., G. Gallagher, V. V. Baurin, A. Lewis-Antes, M. Shen, N. K. Shah, J. A. Langer, F. Sheikh, H. Dickensheets, and R. P. Donnelly. 2003. IFN-λs mediate antiviral protection through a distinct class II cytokine receptor complex. Nat. Immunol. 4:69-77. [DOI] [PubMed] [Google Scholar]
- 18.Paste, M., G. Huez, and V. Kruys. 2003. Deadenylation of interferon-β mRNA is mediated by both the AU-rich element in the 3′-untranslated region and an instability sequence in the coding region. Eur. J. Biochem. 270:1590-1597. [DOI] [PubMed] [Google Scholar]
- 19.Schlender, J., B. Bossert, U. Buchholz, and K-K. Conzelmann. 2000. Bovine respiratory syncytial virus nonstructural proteins NS1 and NS2 cooperatively antagonize alpha/beta interferon-induced antiviral response. J. Virol. 74: 8234-8242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sheppard, P., W. Kindsvogel, W. Xu, K. Henderson, S. Schlutsmeyer, T. E. Whitmore, R. Kuestner, U. Garrigues, C. Birks, J. Roraback, C. Ostrander, D. Dong, J. Shin, S. Presnell, B. Fox, B. Haldeman, E. Cooper, D. Taft, T. Gilbert, F. J. Grant, M. Tackett, W. Krivan, G. McKnight, C. Clegg, D. Foster, and K. M. Klucher. 2003. IL-28, IL-29 and their class II cytokine receptor IL-28R. Nat. Immunol. 4:63-68. [DOI] [PubMed] [Google Scholar]
- 21.Spann, K. M., P. L. Collins, and M. N. Teng. 2003. Genetic recombination during coinfection of two mutants of human respiratory syncytial virus. J. Virol. 77:11201-11211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Teng, M. N., and P. L. Collins. 1999. Altered growth characteristics of recombinant respiratory syncytial viruses which do not produce NS2 protein. J. Virol. 73:466-473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Teng, M. N., S. S. Whitehead, A. Bermingham, M. St. Claire, W. R. Elkins, B. R. Murphy, and P. L. Collins. 2000. Recombinant respiratory syncytial virus that does not express the NS1 or M2-2 protein is highly attenuated and immunogenic in chimpanzees. J. Virol. 74:9317-9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Valarcher, J. F., J. Furze, S. Wyld, R. Cook, K. K. Conzelmann, and G. Taylor. 2003. Role of alpha/beta interferons in the attenuation and immunogenicity of recombinant bovine respiratory syncytial viruses lacking NS proteins. J. Virol. 77:8426-8439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Whitehead, S. S., A. Bukreyev, M. N. Teng, C. Y. Firestone, M. St. Claire, W. R. Elkins, P. L. Collins, and B. R. Murphy. 1999. Recombinant respiratory syncytial virus bearing a deletion of either the NS2 or SH gene is attenuated in chimpanzees. J. Virol. 72:4467-4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wright, P. F., R. A. Karron, R. B. Belshe, J. Thompson, J. E. Crowe, Jr., T. G. Boyce, L. L. Halburnt, G. W. Reed, S. S. Whitehead, E. L. Anderson, A. E. Wittek, R. Casey, M. Eichelberger, B. Thumar, V. B. Randolph, S. A. Udem, R. M. Chanock, and B. R. Murphy. 2000. Evaluation of a live, cold-passaged, temperature-sensitive, respiratory syncytial virus vaccine candidate in infancy. J. Infect. Dis. 182:1331-1342. [DOI] [PubMed] [Google Scholar]
- 27.Young, D. F., L. Pidcock, S. Goodbourne, and R. E. Randall. 2000. Paramyxoviridae use distinct virus-specific mechanisms to circumvent the interferon response. Virology 269:383-390. [DOI] [PubMed] [Google Scholar]
- 28.Young, D. F., L Andrejeve, A. Livingstone, S. Goodbourn, R. A. Lamb, P. L. Collins, R. M. Elliott, and R. E. Randall. 2002. Virus replication in engineered human cells that do not respond to interferons. J. Virol. 77:2174-2181. [DOI] [PMC free article] [PubMed] [Google Scholar]