Abstract
Influenza A virus mutants expressing C-terminally deleted forms of the NS1 protein (NS1-81 and NS1-110) were generated by plasmid rescue. These viruses were temperature sensitive and showed a small plaque size at the permissive temperature. The accumulation of virion RNA in mutant virus-infected cells was reduced at the restrictive temperature, while the accumulation of cRNA or mRNA was not affected, indicating that the NS1 protein is involved in the control of transcription versus replication processes in the infection. The synthesis and accumulation of late virus proteins were reduced in NS1-81 mutant-infected cells at the permissive temperature and were essentially abolished for both viruses at the restrictive temperature, while synthesis and accumulation of nucleoprotein (NP) were unaffected. Probably as a consequence, the nucleocytoplasmic export of virus NP was strongly inhibited at the restrictive temperature. These results indicate that the NS1 protein is essential for nuclear and cytoplasmic steps during the virus cycle.
The genome of influenza A virus consists of eight single-stranded RNA molecules of negative polarity associated with nucleoprotein (NP) molecules and the polymerase in the form of ribonucleoprotein (RNP) complexes (for reviews, see references 40, 43, and 66). The first step in viral gene expression is primary transcription from the incoming viral RNPs (28). The expression of virus proteins, at least NP, leads to the shift from transcription to the synthesis of complete positive-polarity RNAs (cRNAs) (29, 73), which serve as templates for the synthesis of virion RNAs (vRNAs). Transcription and replication of vRNA take place in the nucleus of the infected cell (30, 34) and require at least the activity of the three subunits of the polymerase (PB1, PB2, and PA) and the NP (9, 31, 38, 55, 64). The syntheses of the various vRNAs are not simultaneous during the infection cycle. Thus, NS1 or NP vRNAs are replicated earlier than M1 or hemagglutinin (HA) vRNAs (72). Since transcription is coupled to replication at the beginning of vRNA synthesis, the NS1 protein and NP are expressed earlier than the M1 protein and HA (72). However, later in the process of vRNA synthesis, transcription is discontinued and viral protein synthesis rests on previously synthesized mRNAs (72). In the course of the infection, viral gene expression takes over the cell machinery, leading to the shutoff phenomenon. Several alterations induced by the virus in the infected cell may be connected to shutoff: nuclear retention and degradation of polymerase II transcripts in the nucleus (35), inhibition of cellular pre-mRNA cleavage and polyadenylation (56, 74), cytoplasmic degradation of preexisting cellular mRNAs (3, 32), and preferential utilization of the translation machinery by the virus-specific mRNAs (36).
Influenza A virus encodes a nonstructural protein (NS1) that is translated from the unspliced transcript of segment 8 (33, 44). NS1 is a nuclear protein, both in the infected cell (5, 41) and when expressed from cDNA (23, 45, 65). It accumulates in the nucleus early in the infection, but later in the infectious cycle it is found in the nucleus and the cytoplasm (58), where it is associated with polysomes (8, 16, 41). The NS1 protein can bind various types of RNA: vRNA, poly(A)-containing RNAs, U6 snRNA, and viral mRNA (26, 27, 51, 63, 68, 69). Its expression from cDNA in cultured cells leads to a variety of alterations in cellular processes that involve RNA. These include nuclear retention of poly(A)-containing mRNAs (17, 68), inhibition of cellular pre-mRNA cleavage and polyadenylation (56), alterations in the splicing of pre-mRNAs (17, 18, 47, 68), and enhancement of the translation of viral, but not cellular, mRNAs (8, 14).
Several studies have addressed the phenotypes of mutant NS1 cDNAs. Site-directed mutagenesis at various positions along the NS1 gene led to the proposal that the NS1 protein contains two separate functional domains: an N-proximal domain (amino acids 19 to 38) involved in RNA binding and a C-proximal domain (amino acids 134 to 161) presumably acting as effector for the functions of nuclear retention of poly(A)-containing RNA and inhibition of splicing (47, 67). On the other hand, deletion from the C terminus indicated that the N-terminal half of the NS1 protein is sufficient for nuclear retention of poly(A)-containing RNA and enhancement of viral mRNA translation (51). The phenotypes of temperature-sensitive virus mutants with alterations in the NS1 gene are diverse. Some mutants show a transcriptional block at the nonpermissive temperature (ts412 [mutation R25K]) (39, 49), while others are affected in viral gene expression at a posttranscriptional level (SPC45 [mutation K62N] or ICR1629 [mutation A132T]) (24). For the latter type, a correlation has been established between their ability to bind RNA and their capacity to activate PKR (25). In addition, reverse genetics has been used to construct mutant viruses containing C-terminal deletions of the NS1 protein (12, 15, 71). Their efficiency of replication in Vero cells was similar to that of wild-type (wt) virus, suggesting that the NS1 protein is not essential for virus replication in these cultures (12). In other cell types, in which the replication of these NS1 mutant viruses was severely compromised, the NS1 protein could act as a virulence factor responsible for counteracting the interferon (IFN) response, since Vero cells are deficient in IFN genes (10). In line with this interpretation, a PR8-based mutant virus lacking the NS1 open reading frame (delNS1 virus) was able to replicate in cultured cells devoid of the IFN response but not in normal cells (20). Accordingly, delNS1 virus was attenuated in normal mice but pathogenic in STAT−/− mice (20) or PKR−/− knockout mice (4). It was proposed that the NS1 protein would mediate this function by sequestering double-stranded RNA (dsRNA) during viral infection (48). In agreement with this proposal, it has been shown that the NS1 protein could counteract in trans the delNS1-induced activation of IRF-3 and that the dsRNA-binding domain is essential for such blocking activity (75). In fact, the NS1 protein can block the signaling pathway mediated by JNK (50) and down-regulates apoptosis (81). However, the phenotype of NS1 virus mutants affected in the effector domain (37, 59) has led to an alternative hypothesis about the mode of action of the NS1 protein as a virulence factor (42). The fact that delNS1 mutant virus replicates almost normally in Vero cells (20, 71) does not preclude the possibility that the NS1 protein may play other roles in the virus infection cycle, in addition to the above-mentioned function as a virulence factor. In fact, delNS1 virus was generated by using a Vero cell-adapted PR8 strain and may carry genetic alterations that could make it not representative of a true field strain.
In this study we have generated mutant influenza viruses encoding C-terminally deleted NS1 proteins in the genetic background of a field strain. These viruses are thermosensitive and are defective in virus RNA replication, translation of late virus mRNAs, and nucleocytoplasmic export of NP, indicating that the NS1 protein plays essential roles in both nuclear and cytoplasmic steps in the virus replication cycle.
MATERIALS AND METHODS
Biological materials.
The COS-1 cell line (22) was provided by Y. Gluzman, and 293T HEK cells (11) were obtained from A. Portela. The MDCK cell line was purchased from the American Type Culture Collection. The cells were cultivated as described previously (62). Plasmids containing the cDNAs of WSN virus RNAs under control of the polI promoter, as well as expression plasmids encoding the WSN virus polymerase and NP genes (57), were gifts of Y. Kawaoka. Plasmid vector pHH21 (57) was kindly provided by G. Hobom. Antisera specific for NP and the NS1 protein were generated by hyperimmunization of rabbits or rats with purified MBP-NP or HisNS1 protein, respectively. Monoclonal antibody E10, recognizing both the M1 and M2 proteins was a gift of A. García-Sastre.
Plasmid constructions.
To generate plasmids pHHVicPB1, pHHVicPB2, pHHVicPA, pHHVicNP, and pHHVicNS, their viral cDNAs were amplified from the corresponding pGEM plasmids (17, 55) by using the Expand PCR system from Roche and cloned into plasmid pHH21 as described previously (57). Plasmids expressing the A/Victoria/3/75 polymerase and NP genes under control of the cytomegalovirus promoter (pCMVPB1, pCMVPB2, pCMVPA, and pCMVNP) were constructed by subcloning the cDNAs derived from the corresponding pGEM plasmids (17, 55) into the pCMV vector. For the construction of mutant plasmids pHHVicNS1-81 and pHHVicNS1-110, the mutant cDNAs were amplified by using the corresponding pGEM constructs as templates (51) and cloned into the pHH21 vector as indicated above.
Virus rescue.
To rescue infectious virus from cDNAs, 105 293T HEK cells were cotransfected with a mixture of 12 plasmid DNAs (100 ng each) including (i) 8 genomic plasmids each carrying a viral segment cDNA under the control of the polI promoter and (ii) 4 expression plasmids encoding the three polymerase subunits and the NP. Transfection was carried out at 32.5°C with Lipofectamine Plus (Gibco) under the conditions recommended by the manufacturer. At 16 h posttransfection, the cells were washed with Dulbecco's modified Eagle's medium (DMEM) and further incubated with DMEM containing 2% fetal bovine serum. Alternatively, transfected cells were plated onto an excess of MDCK cells and cultivated as described previously (21). When a cytopathic effect was apparent, the supernatant medium was collected and used for plaque assay on MDCK cells at 32.5°C. A well-isolated plaque was picked and used to produce a virus stock at a low multiplicity of infection. For rescue of viruses with the A/Victoria/3/75 genetic background, the corresponding genomic plasmids for PB1, PB2, PA, NP, and NS were mixed with those for the HA, NA, and M segments from the WSN virus and complemented with the A/Victoria/3/75 expression plasmids. The identity of rescued mutant viruses was ascertained by sequencing of DNAs derived from the NS RNA segment by reverse transcription-PCR (RT-PCR) amplification.
Protein analyses.
Protein labeling in vivo was carried out as described before (82). At various times postinfection, cultures were washed, incubated for 1 h in DMEM lacking Met-Cys, and labeled with [35S]Met-Cys to a final concentration of 200 μCi/ml. After incubation for 1 h, total extracts were prepared in Laemmli sample buffer and processed by polyacrylamide gel electrophoresis and autoradiography. Immunofluorescence analysis was performed as follows. Infected or mock-infected MDCK cells were washed with phosphate-buffered saline (PBS), fixed for 20 min in methanol at −20°C, and washed with PBS. The cells were pretreated with PBS-3% bovine serum albumin (BSA) and incubated with anti-NP rabbit serum and/or with rat anti-NS1 serum (each at a 1:200 dilution in PBS-0.5% BSA) for 1 h at room temperature. After washing with PBS-0.5% BSA, the bound antibodies were revealed with fluorescein isothiocyanate-conjugated goat anti-rabbit antibody and/or Texas Red-conjugated goat anti-rat antibody (each at a 1:200 dilution) by incubation for 1 h at room temperature. Finally, the cells were washed with PBS and the preparations were mounted with Mowiol. Optical sections were obtained with a Zeiss Axiophot fluorescence microscope equipped with a Bio-Rad confocal unit. Western blotting were carried out as follows. Total cell extracts were separated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis and transferred to Immobilon filters that were saturated with 3% BSA in PBS for 1 h at room temperature. The filters were incubated with the primary antibodies diluted in 1% BSA in PBS for 1 h at room temperature. After being washed two times for 30 min each with PBS containing 0.25% Tween 20, the filters were incubated with a 1:10,000 dilution of goat anti-rabbit or anti-mouse immunoglobulin G conjugated to horseradish peroxidase. Finally, the filters were washed two times for 30 min each as described above and developed by enhanced chemiluminescence.
RNA analyses.
The accumulation of vRNAs and cRNAs was determined by real-time RT-PCR with oligonucleotide primers specific for the NP and M RNA segments, whose sequences are available upon request. The isolation of total RNA from infected or mock-infected cells was carried out as described previously (64). Appropriate dilutions of the samples were boiled for 5 min and rapidly cooled to 0°C. The RT step was carried out for 10 min at 50°C in the presence of the reverse transcriptase primer. After inactivation of the reverse transcriptase for 10 min at 95°C and treatment with 100 ng of RNase A for 15 min at 37°C, the PCR step was performed in the presence of both primers. Control experiments in which the reverse transcriptase primer was omitted in the first step verified that no fluorescence could be obtained. Quantitation was carried out with the Sybr Green RT-PCR kit from Applied Biosystems as recommended by the manufacturer. A standard curve was obtained with RNA isolated at late times after high-multiplicity infection with influenza virus. Only determinations that led to a standard curve with a fit of better than 0.99 were considered. Denaturation curves and agarose gel electrophoresis of each reaction product verified that it contained a single amplification fragment of the expected melting temperature and length. The accumulation of the various mRNAs was determined by dot blot hybridization. Infected or mock-infected cells were fractionated into nuclear and cytoplasmic fractions, and total RNA from each one was isolated as described previously (64). For detection of NP mRNA, a full-gene riboprobe of negative polarity was used. Detection of M1 mRNA was carried out with a 200-nucleotide-long riboprobe located within the intron of segment 7. For M2 mRNA and mRNA3 quantitation, the cDNA probes were used complementary to the exon-exon junctions (76). These probes contained an additional T12-13 tail and were labeled with [32P]dTTP by asymmetric PCR. For control of RNA loading, each filter was further hybridized with a 200-nucleotide-long 28S-specific rRNA probe.
RESULTS
Mutant influenza viruses with C-terminal deletions in the NS1 protein are thermosensitive.
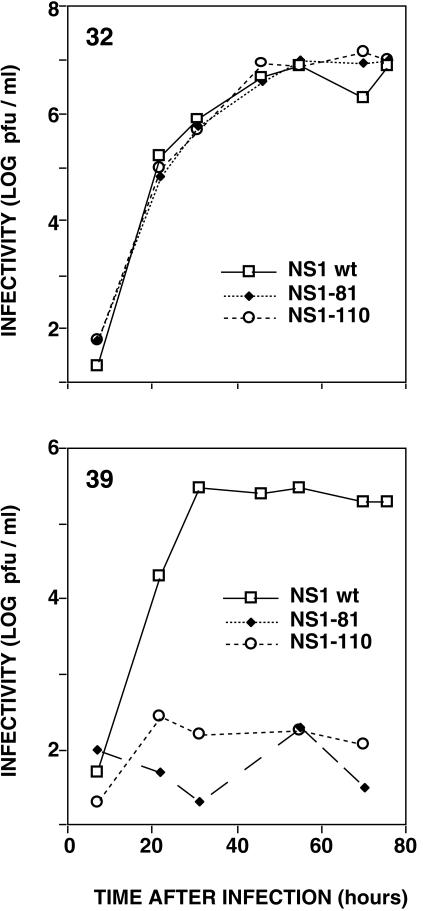
In order to test for possible roles of the NS1 protein other than as a virulence factor, we have constructed mutant viruses containing C-terminal deletions of the NS1 protein in the genetic background of the A/Victoria/3/75 strain (Fig. 1). For easier handling, the mutants were generated by using the membrane protein genes of WSN strain, but the polymerase, NP, and NS genes were derived from low-passage strain A/Victoria/3/75. These mutant viruses should behave like the wt in regard to the inhibition of the IFN response, since their NS1 gene-encoded proteins contain an intact dsRNA-binding domain and indeed bind RNA as wt NS1 does (51). Mutant and wt recombinant viruses were rescued from cDNA at 32.5°C (permissive temperature), plaque purified, and tested for growth at both permissive and restrictive (39.5°C) temperatures. The mutant NS1-81 and NS1-110 viruses showed small-plaque phenotype at the permissive temperature and very marked titer reductions when plated at the restrictive temperature (data not shown), even compared to the wt recombinant virus, which shows reduced plating efficiency at the restrictive temperature (probably due to its particular gene constellation). Multiple-cycle growth curves indicated that NS1-81 and NS1-110 mutant viruses are temperature sensitive (Fig. 2). In contrast, neither of these mutations led to a temperature-sensitive phenotype when assayed in the genetic background of WSN virus (reference 15 and data not shown).
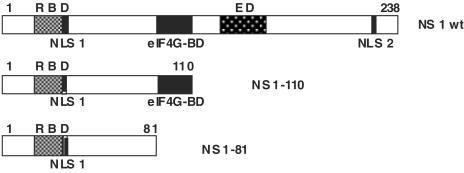
FIG. 1.
Structures of the NS1 protein and its deletion mutants. The diagram shows the structure of the NS1 protein, including the locations of nuclear localization signals (NLSs) (23), the RNA-binding domain (RBD), the effector domain (ED) (67), and the region that recognizes the eIF4G translation initiation factor (2), as well as the structures of the NS1-81 and NS1-110 deletion mutants.
FIG. 2.
Kinetics of replication of wt or deletion mutant NS1 viruses. Cultures of MDCK cells were infected with either wt, NS1-81, or NS1-110 virus at 10−2 PFU/cell. Replicate infections were incubated at the permissive (32°C) or restrictive (39°C) temperature. At the indicated times after infection, aliquots of the culture supernatants were withdrawn and analyzed by plaque assay at the permissive temperature on MDCK cells.
Replication, but not transcription, of vRNA is thermosensitive in mutant viruses with C-terminal deletions in the NS1 protein.
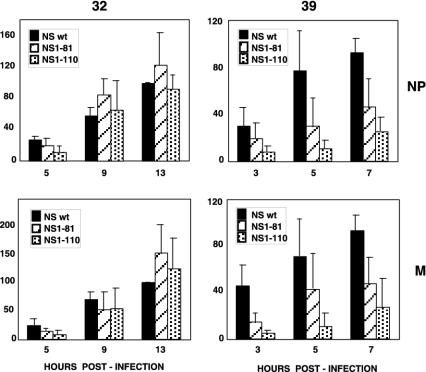
To determine the molecular basis of the temperature-sensitive phenotype of these NS1 mutant viruses, we first analyzed the accumulation of vRNA at the permissive and restrictive temperatures. Total cell RNA was isolated at various times after high-multiplicity infection, and the concentrations of NP and M vRNAs were determined by real-time RT-PCR. The results are shown in Fig. 3. Compared to that in wt recombinant virus-infected cells, the accumulations of vRNA obtained for the NS1-81 and NS1-110 mutant viruses were normal at the permissive temperature but decreased at the restrictive temperature, especially for NS1-110, which showed around a four- to fivefold vRNA reduction for both NP and M vRNAs. To analyze whether this reduction was a consequence of a previous defect in cRNA production, the accumulations of NP and M cRNAs were determined at both temperatures in wt and mutant virus-infected cells. The results are presented in Fig. 4 and do not show a temperature-dependent defect in cRNA accumulation. These results indicate that the NS1 protein plays a role in the process of virus RNA replication, probably in the generation of vRNA from cRNA, and that a defect in this function may contribute to the temperature-sensitive phenotype observed.
FIG. 3.
Accumulation of vRNAs in cells infected with wt or deletion mutant NS1 viruses. Cultures of MDCK cells were infected with either wt, NS1-81, or NS1-110 virus at 5 to 10 PFU/cell or mock infected. Replicate infections were incubated at the permissive (32°C) or restrictive (39°C) temperature. At the times indicated, total cell RNA was isolated and aliquots of each sample were used to determine the concentration of NP or M vRNA by real-time quantitative RT-PCR as described in Materials and Methods. Averages and standard deviations from at least four determinations of the kinetics of infection are shown. The results are presented as percentages of the maximal accumulation for wt virus-infected cells in each kinetic experiment.
FIG. 4.
Accumulation of cRNAs in cells infected with wt or deletion mutant NS1 viruses. Cultures of MDCK cells were infected with either wt, NS1-81, or NS1-110 virus at 5 to 10 PFU/cell or mock infected. Replicate infections were incubated at the permissive (32°C) or restrictive (39°C) temperature. At the times indicated, total cell RNA was isolated and aliquots of each sample were used to determine the concentration of NP or M cRNA by real-time quantitative RT-PCR as described in Materials and Methods. Averages and standard deviations from three determinations of the kinetics of infection are shown. The results are presented as percentages of the maximal accumulation for wt virus-infected cells in each kinetic experiment.
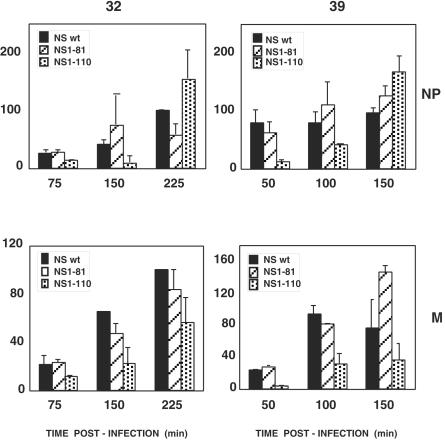
To check whether the reduction in vRNA accumulation at the restrictive temperature would have a reflection in viral transcription, the accumulations of various mRNAs were determined. As the real-time RT-PCR technique was not reliable for this purpose, probably due to the excess accumulation of vRNAs at late times after infection, we used dot blot hybridization as an alternative. The results obtained for NP mRNA are presented in Fig. 5. In contrast to the inhibition of vRNA accumulation (Fig. 3), NP mRNA accumulation was not diminished at the restrictive temperature in mutant virus-infected cells. Similar results were obtained for total accumulation of mRNAs derived from segment 7 (data not shown). Taken together, the accumulations of vRNAs and mRNAs in mutant virus-infected cells at the restrictive temperature indicate that the NS1 protein plays a role as regulator of the transcription versus replication activities of the viral RNPs.
FIG. 5.
Accumulation of mRNAs in cells infected with wt or deletion mutant NS1 viruses. Cultures of MDCK cells were infected with either wt, NS1-81, or NS1-110 virus at 5 to 10 PFU/cell or mock infected. Replicate infections were incubated at the permissive (32°C) or restrictive (39°C) temperature. At the times indicated, total cell RNA was isolated and aliquots of each sample were used to determine the concentration of NP mRNA by dot blot hybridization as described in Materials and Methods. Averages and standard deviations from at least three determinations of kinetics of infection are shown. The results are presented as percentages of the maximal accumulation for wt virus-infected cells in each kinetic experiment.
The accumulations of virus mRNAs in both nuclear and cytoplasmic fractions were assayed to ascertain possible alterations in nucleocytoplasmic transport. No significant change was observed in the distributions of viral mRNAs in the nuclei and cytoplasm of wt- or mutant-infected cells at either temperature (data not shown). As previous experiments using transfection of NS1 cDNAs showed nuclear retention of mRNAs upon NS1 expression (17, 67, 68), these results might be interpreted to indicate that the NS1 protein does not play a role in the transport of viral mRNAs during infection. However, it is possible that the deletions introduced did not affect such a potential function. In fact, we have previously shown that the first 113 amino acids of the NS1 protein are sufficient for nuclear mRNA retention (51).
Synthesis of M1 protein is reduced at the permissive temperature in mutant virus NS1-81.
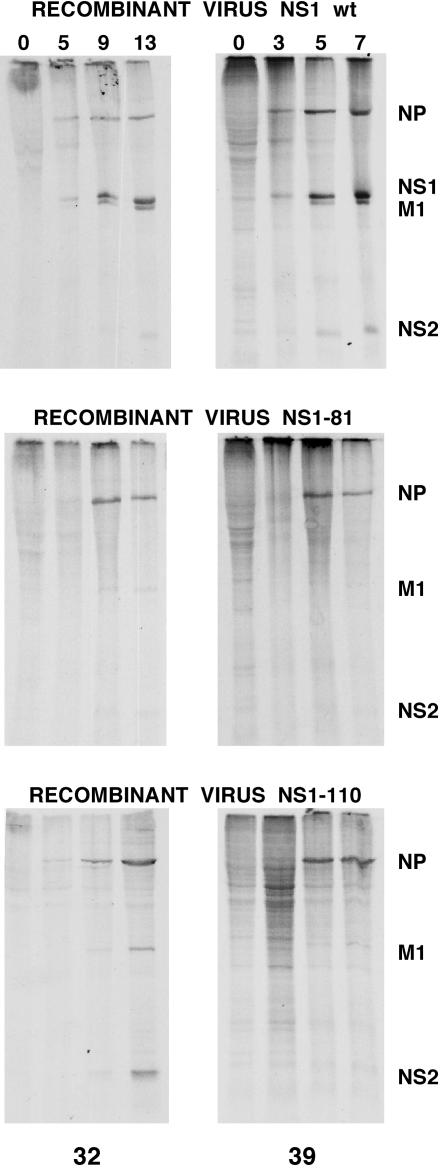
To determine the consequences of NS1 mutations at the protein level, cells infected with wt or mutant recombinant viruses were pulse-labeled at various times after infection at either the permissive or restrictive temperature and the labeled proteins were separated by polyacrylamide gel electrophoresis. The results are presented in Fig. 6. The synthesis of the main viral proteins (NP, M1, NS1, and NS2) was clearly apparent in wt recombinant-infected cells at either temperature (Fig. 6, top panels). In contrast, in cells infected with the NS1-81 mutant virus, the synthesis of late proteins (M1 and NS2) was reduced at the permissive temperature and was not detectable at the restrictive temperature, while the synthesis of NP was essentially normal (Fig. 6, middle panels). The situation in cells infected with the NS1-110 mutant virus was different. While synthesis of late proteins at the permissive temperature was only slightly affected, M1 and NS2 were essentially not detectable at the restrictive temperature. Since viral mRNA accumulation is not affected at the permissive temperature in mutant virus-infected cells (Fig. 5 and data not shown), a possible explanation for the absence of a correct M1 protein synthesis in NS1-81 mutant-infected cells at the permissive temperature is that this mutant NS1 protein is unable to stimulate late viral protein synthesis, in contrast to the NS1-110 mutant. To test an alternative possibility, namely, that splicing of the M segment is very efficient in NS1-81 mutant-infected cells at the restrictive temperature, the accumulations of M1 and M2 proteins were determined at various times after infection at either temperature. The results are presented in Fig. 7. The M1/M2 ratio, which should be changed if the splicing of the segment 7 transcript were altered by the NS1 mutations, remained essentially constant for wt or mutant viruses at either temperature. Furthermore, the accumulations of the three mRNAs derived from viral segment 7 (M1 and M2 mRNAs and mRNA3) were determined by dot blot hybridization with intronic and splice junction probes, and their ratios were not altered in mutant virus-infected cells compared to wt-infected cells at either temperature (data not shown).
FIG. 6.
Kinetics of viral protein synthesis in wt and deletion mutant NS1 viruses. Cultures of MDCK cells were infected with either wt, NS1-81, or NS1-110 virus at 5 to 10 PFU/cell. Replicate infections were incubated at the permissive (32°C) or restrictive (39°C) temperature. At the times (in hours) indicated at the top of each lane, the cultures were labeled with [35S]Met-Cys for 1 h in a Met-Cys-free medium. Total cell extracts were prepared in Laemmli sample buffer and analyzed by electrophoresis on an SDS-15% polyacrylamide gel and autoradiography. The positions of relevant virus proteins are indicated on the right.
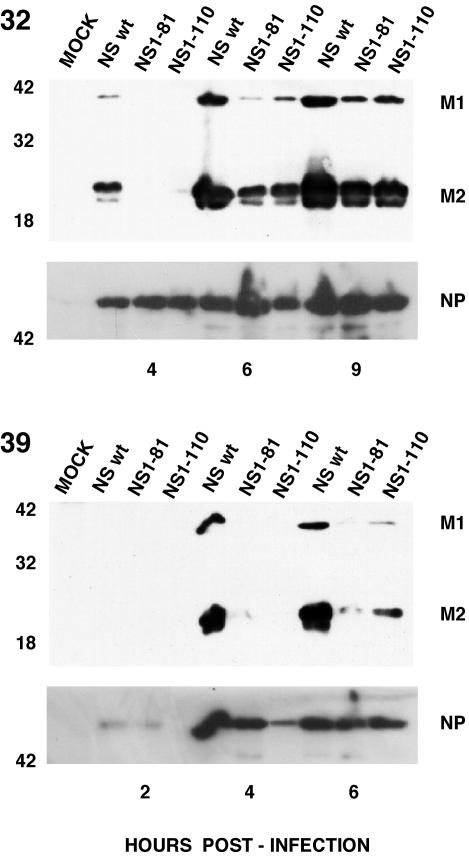
FIG. 7.
Accumulation of M1 and M2 proteins in wt and deletion mutant NS1 viruses. Cultures of MDCK cells were infected with either wt, NS1-81, or NS1-110 virus at 5 to 10 PFU/cell. Replicate infections were incubated at the permissive (32°C) or restrictive (39°C) temperature. At the times (in hours) indicated at the bottom of each panel, total cell extracts were prepared in Laemmli sample buffer and analyzed by electrophoresis on SDS-polyacrylamide gels and Western blotting with the E10 monoclonal antibody, which recognizes both the M1 and M2 proteins.
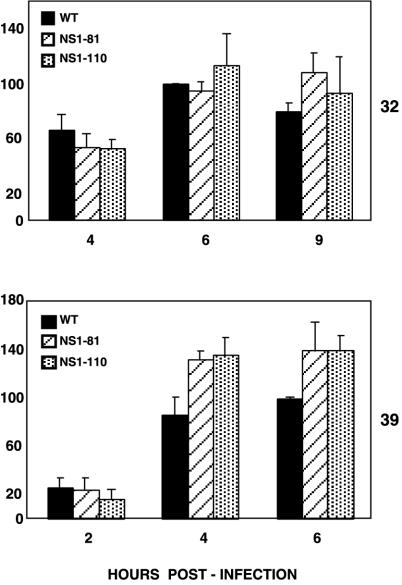
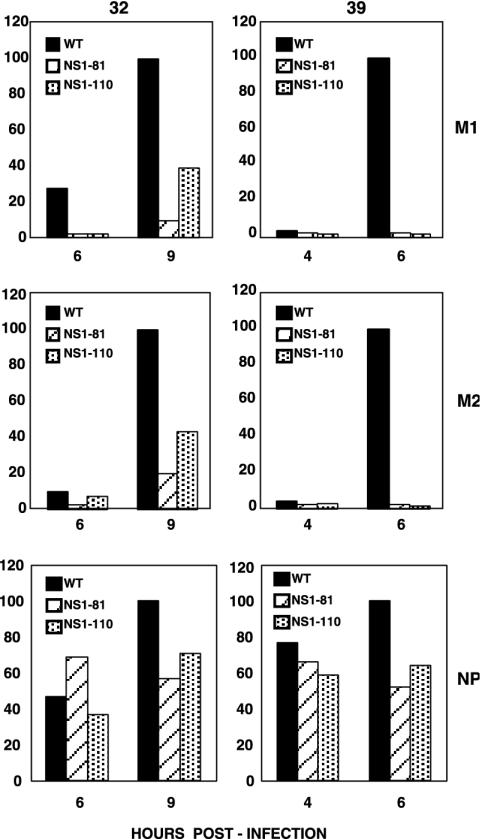
On the other hand, the absolute accumulations of the M1 and M2 proteins were affected by the NS1-81 and NS1-110 mutations, particularly at the restrictive temperature, while the accumulation of NP was not affected (Fig. 7), in agreement with the results for protein synthesis presented in Fig. 6. These mutant phenotypes are best represented by the ratio of protein to cytoplasmic mRNA accumulations, as a measure of the efficiency of translation of the corresponding messengers (Fig. 8). The translation of the M1 and M2 proteins was partly affected in mutant-infected cells at the permissive temperature and became almost undetectable at the restrictive temperature, but translation of NP was essentially unaltered.
FIG. 8.
Translation efficiencies of late virus mRNAs in wt and deletion mutant NS1 viruses. The relative translation efficiencies of M1 and M2 mRNAs were estimated as the ratios of protein to cytoplasmic mRNAs present in the infected cells at various times after infection at either the permissive (32°C) or restrictive (39°C) temperature. The accumulations of protein were determined by Western blotting and those of mRNAs were determined by blot hybridization, as described in Materials and Methods. The results are presented as percentages of the maximal ratios obtained for wt infections.
Nuclear export of virus NP is defective at the restrictive temperature in mutant viruses with C-terminal deletions in the NS1 protein.
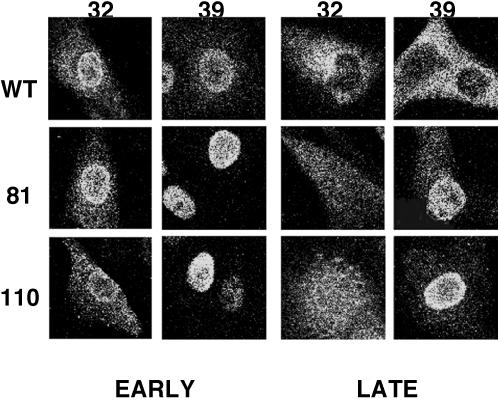
Mutant NS1 proteins might have altered the normal viral gene expression processes by improper intracellular localization. In fact, the existence of a nuclear export signal sequence at positions 138 to 147 in NS1 has been reported (46), and such a structure would be absent in the NS1-81 and NS1-110 mutants. It is known that wt NS1 accumulates within the nucleus early in the infection and is present also in the cytoplasm at later times. To determine whether mutant NS1-81 and NS1-110 proteins behave like wt NS1, we carried out immunofluorescence experiments at various times after infection at both the permissive and restrictive temperatures, using anti-NP antibodies as a control for the infection kinetics. The intracellular localization of mutant NS1 proteins was not substantially different from that of wt protein. Thus, both the NS1-81 and NS1-110 proteins accumulated in the nucleus early in the infection and later became cytoplasmic proteins, although these intracellular translocations were delayed compared to that of wt protein at the restrictive temperature (data not shown). However, the most striking results came from the localization of NP in mutant virus-infected cells (Fig. 9). At the permissive temperature the kinetics of NP nucleocytoplasmic export during the infection, a measure of the transport to the cytoplasm of progeny RNPs, was essentially normal, but a clear block in NP export was observed at the restrictive temperature. This was most apparent late in the infection (5 h postinfection), at a time when NP is completely localized in the cytoplasm of wt virus-infected cells but still essentially nuclear in mutant virus-infected cells. We conclude that deletion of the C-terminal half of the NS1 protein results in thermosensitive export of viral NP, at least in part representing virus RNPs, from the nucleus. Since the M1 and NS2 proteins are responsible for viral RNP transport to the cytoplasm (13, 54, 60, 79, 80), the simplest explanation of this result is that the expression of the M1 and NS2 proteins is so severely diminished at the restrictive temperature that viral RNP export is basically abolished.
FIG. 9.
Intracellular localization of NP in wt and deletion mutant NS1 viruses. Cultures of MDCK cells on coverslips were infected with either wt, NS1-81, or NS1-110 virus at 5 to 10 PFU/cell. Replicate infections were incubated at the permissive (32°C) or restrictive (39°C) temperature and fixed at various times after infection. The cultures were stained for immunofluorescence with anti-NP rabbit serum and fluorescein isothiocyanate-labeled goat anti-rabbit immunoglobulin G, and optical sections were obtained by confocal microscopy. Representative patterns are shown for each mutant and infection time.
DISCUSSION
In the last few years, a number of transfection and in vitro experiments by various groups have contributed to the idea that the influenza virus NS1 protein is a viral multifunctional regulator of gene expression (for reviews, see references 7 and 61). Recently, the application of reverse genetics to the RNA segment encoding the NS1 protein has put in perspective the implication of this small protein in virus multiplication. Thus, virus mutants lacking large C-terminal portions of the NS1 protein, or lacking the NS1 protein altogether, are viable in cell systems devoid of the IFN response but do not replicate in its presence (4, 12, 20). These results and other evidence (75, 78; reviewed in reference 19) indicate that NS1 plays a role as a virulence factor in the influenza virus infection. In this study we have tested the relevance of the NS1 protein for the replication of influenza virus. The phenotype caused by long C-terminal deletions of the NS1 protein in the genetic background of a nonadapted virus strain indicates that these mutants are thermosensitive (Fig. 2) and show altered virus RNA replication (Fig. 3) and late gene expression (Fig. 6 to 8). As a result of these changes, the nucleocytoplasmic export of viral NP is severely retarded (Fig. 9). There are partial disagreements between the phenotypes reported here and those previously reported elsewhere for similar mutant viruses (15, 71). These differences may be the result of using high-yield laboratory strains that had been adapted to grow in mice (15) or chicken embryo and Vero cells (71) rather than a recombinant virus containing the RNP genes from a field strain, which was used in the present study. It could be considered that the phenotypes we observed for the NS1-81 and NS1-110 mutant viruses are indirect consequences of an incomplete block of the IFN response, but several points argue against such possibility: (i) the deleted NS1-81 and NS1-110 proteins bind dsRNA as the wt NS1 protein does (51); (ii) the IFN response is effective primarily in the cells surrounding an infected one, and it is improbable that it alters the result of a high-multiplicity infection; and (iii) there is not a general translation block, since translation of NP mRNA occurs normally at the restrictive temperature in mutant virus-infected cells, even at late times in the infection (Fig. 6).
At the permissive temperature, the main phenotype observed was a reduced capacity for late protein synthesis in mutant NS1-81-infected cells (Fig. 6 to 8). Earlier experiments using cDNA transfection have indicated that the NS1 protein is involved in the specific stimulation of viral mRNA translation (8, 14). The phenotype of the mutant NS1-81 virus conforms to these conclusion. Furthermore, the different behaviors of the NS1-81 and NS1-110 mutants at the permissive temperature are in full agreement with earlier experiments carried out by transfection. Indeed, they established that the NS1-110 protein was able to enhance the translation of a co expressed virus gene while the NS1-81 protein was not (51). The activity of the NS1 protein to stimulate virus translation has been correlated to its capacity to interact with elements of the translation preinitiation complex (eIF4GI and PABPI) and to associate specifically with viral mRNAs during the infection (2, 6). Thus, the NS1-110 protein was shown to interact with eIF4GI, but the NS1-81 protein was unable to do so (2). Similarly, NS1-81 cannot interact with human Staufen protein, a dsRNA-binding protein involved in the localization and efficient translation of mRNAs, but longer NS1 proteins do (16, 52). Therefore, we conclude that a protein determinant located between positions 81 and 110 in the NS1 protein is important to allow an efficient translation of viral late mRNAs during the infection.
At the restrictive temperature, the ratio of replication to transcription activities of viral RNPs was considerably altered (three- to eightfold; compare Fig. 3 and 5). These results, together with the phenotypes of other mutant viruses previously characterized, support the notion of the NS1 protein being a multifunctional factor involved in virus multiplication at various levels. First, NS1 would play a role in the regulation of the transcription and replication processes of virus RNPs in the cell nucleus. As NS1 mutants show a diminished vRNA, but not cRNA, accumulation under nonpermissive conditions (Fig. 3 and 4), it can be suggested that wt NS1 would stimulate virus RNA replication at the cRNA-to-vRNA step. This is in agreement with the phenotype of mutant tsmN3 (1, 70), but another temperature-sensitive mutant virus (ts412) showed reduced mRNA synthesis (39, 49). These phenotypes are in line with the reported association of the NS1 protein with viral RNPs during virus infection (53). The expression of the NS1 protein has also been implicated in the inhibition of cellular pre-mRNA cleavage and polyadenylation, by its interaction with cleavage and polyadenylation specificity factor (CPSF) (56), and a mutant virus affected in the NS1 gene shows a phenotype compatible with such a role in infection (74). However, the deletion of the domain responsible for the interaction of the NS1 protein with the 30-kDa subunit of CPSF, which is absent in both the NS1-81 and NS1-110 mutants described here, did not lead to a significant reduction in virus multiplication at the permissive temperature, although it could be at the basis of the reduction in plaque size in the mutants. A potential role for the NS1 protein in other nuclear events, such as alterations in the nucleocytoplasmic transport or splicing of mRNAs, could not be verified with the mutations analyzed here. Thus, no temperature-sensitive changes in the nucleocytoplasmic distribution of viral mRNAs or the proportions of the various mRNAs derived from segment 7 could be detected (Fig. 7 and data not shown).
The translation phenotype of the NS1-81 mutant was augmented at the restrictive temperature and became clearly apparent also for mutant NS1-110 (Fig. 6 to 8). This could be the consequence of conformational changes in the NS1-110 protein or deficiencies in its dimerization capacity (77) at the restrictive temperature, but it is not due to thermal instability (data not shown). Decreases in the synthesis of the M1 protein have been also observed in other virus mutants containing deleted NS1 proteins (12, 15, 71), but these viruses did not show a temperature-sensitive phenotype. Moreover, some temperature-sensitive virus mutants, such as viruses SPC45 or ICR1629 (24), also showed a posttranscriptional defect in late gene expression.
In summary, NS1 is a key regulatory factor for virus multiplication that affects replication and transcription in the nucleus as well as translation steps in the cytoplasm. Its mutation leads to less-fit viruses affected in one or more gene expression steps, depending on the nature of the mutation. Its ability to alter cellular gene expression and/or its capacity to bind dsRNA enables the NS1 protein to act also as an essential virulence factor to counteract the IFN response in vivo.
Acknowledgments
We thank A. García-Sastre, G. Hobom, and Y. Kawaoka for providing biological materials. The technical assistance of Y. Fernández and J. Fernández is gratefully acknowledged.
A. M. Falcón was a fellow of the Programa de Formación de Personal Investigador, Ministerio de Ciencia y Tecnología. T. Zürcher was a fellow of the Swiss National Science Foundation. R. M. Marión was a fellow of the Comunidad de Madrid. This work was supported by the Programa Sectorial de Promoción General del Conocimiento (grants PB97-1160 and BMC01-1223) and the Comunidad de Madrid (grants 08.2/0025/98 and 08.2/0012.1/2001).
REFERENCES
- 1.Almond, J. W., D. McGeoch, and R. D. Barry. 1979. Temperature-sensitive mutants of fowl plague virus: isolation and genetic characterization. Virology 92:416-427. [DOI] [PubMed] [Google Scholar]
- 2.Aragón, T., S. de la Luna, I. Novoa, L. Carrasco, J. Ortín, and A. Nieto. 2000. Translation factor eIF4GI is a cellular target for NS1 protein, a translational activator of influenza virus. Mol. Cell. Biol. 20:6259-6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beloso, A., C. Martínez, J. Valcárcel, J. Fernández-Santarén, and J. Ortín. 1992. Degradation of cellular mRNA during influenza virus infection: its possible role in protein synthesis shutoff. J. Gen. Virol. 73:575-581. [DOI] [PubMed] [Google Scholar]
- 4.Bergmann, M., A. García-Sastre, E. Carnero, H. Pehamberger, K. Wolff, P. Palese, and T. Muster. 2000. Influenza virus NS1 protein counteracts PKR-mediated inhibition of replication. J. Virol. 74:6203-6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Briedis, D. J., G. Conti, E. A. Munn, and B. W. J. Mahy. 1981. Migration of influenza virus-specific polypeptides from cytoplasm to nucleus of infected cells. Virology 111:154-164. [DOI] [PubMed] [Google Scholar]
- 6.Burgui, I., T. Aragón, J. Ortín, and A. Nieto. 2003. PABP1 and eIF4GI associate with influenza virus NS1 protein in viral mRNA translation initiation complexes. J. Gen. Virol. 84:3263-3274. [DOI] [PubMed] [Google Scholar]
- 7.Chen, Z., and R. M. Krug. 2000. Selective nuclear export of viral mRNAs in influenza-virus-infected cells. Trends Microbiol. 8:376-383. [DOI] [PubMed] [Google Scholar]
- 8.de la Luna, S., P. Fortes, A. Beloso, and J. Ortín. 1995. Influenza virus NS1 protein enhances the rate of translation initiation of viral mRNAs. J. Virol. 69:2427-2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de la Luna, S., J. Martín, A. Portela, and J. Ortín. 1993. Influenza virus naked RNA can be expressed upon transfection into cells co-expressing the three subunits of the polymerase and the nucleoprotein from SV40 recombinant viruses. J. Gen. Virol. 74:535-539. [DOI] [PubMed] [Google Scholar]
- 10.Diaz, M. O., S. Ziemin, M. M. Le Beau, P. Pitha, S. D. Smith, R. R. Chilcote, and J. D. Rowley. 1988. Homozygous deletion of the alpha- and beta 1-interferon genes in human leukemia and derived cell lines. Proc. Natl. Acad. Sci. USA 85:5259-5263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DuBridge, R. B., P. Tang, H. C. Hsia, P. M. Leong, J. H. Miller, and M. P. Calos. 1987. Analysis of mutation in human cells by using an Epstein-Barr virus shuttle system. Mol. Cell. Biol. 7:379-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Egorov, A., S. Brandt, S. Sereinig, J. Romanova, B. Ferko, D. Katinger, A. Grassauer, G. Alexandrova, H. Katinger, and T. Muster. 1998. Transfectant influenza A viruses with long deletions in the NS1 protein grow efficiently in Vero cells. J. Virol. 72:6437-6441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Enami, K., Y. Qiao, R. Fukuda, and M. Enami. 1993. An influenza virus temperature-sensitive mutant defective in the nuclear-cytoplasmic transport of the negative-sense viral RNAs. Virology 194:822-827. [DOI] [PubMed] [Google Scholar]
- 14.Enami, K., T. A. Sato, S. Nakada, and M. Enami. 1994. Influenza virus NS1 protein stimulates translation of the M1 protein. J. Virol. 68:1432-1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Enami, M., and K. Enami. 2000. Characterization of influenza virus NS1 protein by using a novel helper-virus-free reverse genetic system. J. Virol. 74:5556-5561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Falcón, A. M., P. Fortes, R. M. Marión, A. Beloso, and J. Ortín. 1999. Interaction of influenza virus NS1 protein and the human homologue of Staufen in vivo and in vitro. Nucleic Acids Res. 27:2241-2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fortes, P., A. Beloso, and J. Ortín. 1994. Influenza virus NS1 protein inhibits pre-mRNA splicing and blocks RNA nucleocytoplasmic transport. EMBO J. 13:704-712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fortes, P., A. I. Lamond, and J. Ortín. 1995. Influenza virus NS1 protein alters the subnuclear localization of cellular splicing components. J. Gen. Virol. 76:1001-1007. [DOI] [PubMed] [Google Scholar]
- 19.Garcia-Sastre, A. 2001. Inhibition of interferon-mediated antiviral responses by influenza A viruses and other negative-strand RNA viruses. Virology 279:375-384. [DOI] [PubMed] [Google Scholar]
- 20.García-Sastre, A., A. Egorov, D. Matassov, S. Brandt, D. E. Levy, J. E. Durbin, P. Palese, and T. Muster. 1998. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology 252:324-330. [DOI] [PubMed] [Google Scholar]
- 21.Gastaminza, P., B. Perales, A. M. Falcón, and J. Ortín. 2003. Influenza virus mutants in the N-terminal region of PB2 protein are affected in virus RNA replication but not transcription. J. Virol. 76:5098-5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gluzman, Y. 1981. SV40 transformed simian cells support the replication or early SV40 mutants. Cell 23:175-182. [DOI] [PubMed] [Google Scholar]
- 23.Greenspan, D., P. Palese, and M. Krystal. 1988. Two nuclear location signals in the influenza virus NS1 nonstructural protein. J. Virol. 62:3020-3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hatada, E., M. Hasegawa, K. Shimizu, M. Hatanaka, and R. Fukuda. 1990. Analysis of influenza A virus temperature-sensitive mutants with mutations in RNA segment 8. J. Gen. Virol. 71:1283-1292. [DOI] [PubMed] [Google Scholar]
- 25.Hatada, E., S. Saito, and R. Fukuda. 1999. Mutant influenza viruses with a defective NS1 protein cannot block the activation of PKR in infected cells. J. Virol. 73:2425-2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hatada, E., S. Saito, N. Okishio, and R. Fukuda. 1997. Binding of the influenza virus NS1 protein to model genome RNAs. J. Gen. Virol. 78:1059-1063. [DOI] [PubMed] [Google Scholar]
- 27.Hatada, E., T. Takizawa, and R. Fukuda. 1992. Specific binding of influenza virus NS1 protein to the virus minus-sense RNA in vitro. J. Gen. Virol. 73:17-25. [DOI] [PubMed] [Google Scholar]
- 28.Hay, A. J., B. Lomniczi, A. R. Bellamy, and J. J. Skehel. 1977. Transcription of the influenza virus genome. Virology 83:337-355. [DOI] [PubMed] [Google Scholar]
- 29.Hay, A. J., J. J. Skehel, and J. McCauley. 1982. Characterization of influenza virus RNA complete transcripts. Virology 116:517-522. [DOI] [PubMed] [Google Scholar]
- 30.Herz, C., E. Stavnezer, R. M. Krug, and T. Gurney. 1981. Influenza virus, an RNA virus, synthesizes its messenger RNA in the nucleus of infected cells. Cell 26:391-400. [DOI] [PubMed] [Google Scholar]
- 31.Huang, T. S., P. Palese, and M. Krystal. 1990. Determination of influenza virus proteins required for genome replication. J. Virol. 64:5669-5673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Inglis, S. C. 1982. Inhibition of host protein synthesis and degradation of cellular mRNAs during infection by influenza and herpes simplex virus. Mol. Cell. Biol. 2:1644-1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Inglis, S. C., T. Barret, C. M. Brown, and J. W. Almond. 1979. The smallest genome RNA segment of influenza virus contains two genes that may overlap. Proc. Natl. Acad. Sci. USA 76:3790-3794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jackson, D. A., A. J. Caton, S. J. McCready, and P. R. Cook. 1982. Influenza virus RNA is synthesized at fixed sites in the nucleus. Nature 296:366-368. [DOI] [PubMed] [Google Scholar]
- 35.Katze, M. G., Y. T. Chen, and R. M. Krug. 1984. Nuclear-cytoplasmic transport and VAI RNA-independent translation of influenza viral messenger RNAs in late adenovirus-infected cells. Cell 37:483-490. [DOI] [PubMed] [Google Scholar]
- 36.Katze, M. G., D. DeCorato, and R. M. Krug. 1986. Cellular mRNA translation is blocked at both initiation and elongation after infection by influenza virus or adenovirus. J. Virol. 60:1027-1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim, M. J., A. G. Latham, and R. M. Krug. 2002. Human influenza viruses activate an interferon-independent transcription of cellular antiviral genes: outcome with influenza A virus is unique. Proc. Natl. Acad. Sci. USA 99:10096-100101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kimura, N., M. Mishida, K. Nagata, A. Ishihama, K. Oda, and S. Nakada. 1992. Transcription of a recombinant influenza virus RNA in cells that can express the influenza virus RNA polymerase and nucleoprotein genes. J. Gen. Virol. 73:1321-1328. [DOI] [PubMed] [Google Scholar]
- 39.Koennecke, I., C. B. Boschek, and C. Scholtissek. 1981. Isolation and properties of a temperature-sensitive mutant (ts412) of the influenza virus recombinant with a lesion in the gene coding for the nonstructural protein. Virology 110:16-25. [DOI] [PubMed] [Google Scholar]
- 40.Krug, R. M., F. V. Alonso-Kaplen, I. Julkunen, and M. G. Katze. 1989. Expression and replication of the influenza virus genome, p. 89-152. In R. M. Krug (ed.), The influenza viruses. Plenum Press, New York, N.Y.
- 41.Krug, R. M., and P. R. Etkind. 1973. Cytoplasmic and nuclear specific proteins in influenza virus-infected MDCK cells. Virology 56:334-348. [DOI] [PubMed] [Google Scholar]
- 42.Krug, R. M., W. Yuan, D. L. Noah, and A. G. Latham. 2003. Intracellular warfare between human influenza viruses and human cells: the roles of the viral NS1 protein. Virology 309:181-189. [DOI] [PubMed] [Google Scholar]
- 43.Lamb, R. A. 1989. The genes and proteins of influenza viruses, p. 1-87. In R. M. Krug (ed.), The influenza viruses. Plenum Press, New York, N.Y.
- 44.Lamb, R. A., and P. W. Choppin. 1979. Segment 8 of the influenza virus genome is unique in coding for two polypeptides. Proc. Natl. Acad. Sci. USA 76:4908-4912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lamb, R. A., and C. J. Lai. 1984. Expression of unspliced NS1 mRNA, spliced NS2 mRNA, and a spliced chimera mRNA from cloned influenza virus NS DNA in an SV40 vector. Virology 135:139-147. [DOI] [PubMed] [Google Scholar]
- 46.Li, Y., Y. Yamakita, and R. M. Krug. 1998. Regulation of a nuclear export signal by an adjacent inhibitory sequence: the effector domain of the influenza virus NS1 protein. Proc. Natl. Acad. Sci. USA 95:4864-4869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lu, Y., X. Y. Qian, and R. M. Krug. 1994. The influenza virus NS1 protein: a novel inhibitor of pre-mRNA splicing. Genes Dev. 8:1817-1828. [DOI] [PubMed] [Google Scholar]
- 48.Lu, Y., M. Wambach, M. G. Katze, and R. M. Krug. 1995. Binding of the influenza virus NS1 protein to double-stranded RNA inhibits the activation of the protein kinase that phosphorylates the elF-2 translation initiation factor. Virology 214:222-228. [DOI] [PubMed] [Google Scholar]
- 49.Ludwig, S., U. Vogel, and C. Scholtissek. 1995. Amino acid replacements leading to temperature-sensitive defects of the NS1 protein of influenza A virus. Arch Virol. 140:945-950. [DOI] [PubMed] [Google Scholar]
- 50.Ludwig, S., X. Wang, C. Ehrhardt, H. Zheng, N. Donelan, O. Planz, S. Pleschka, A. Garcia-Sastre, G. Heins, and T. Wolff. 2002. The influenza A virus NS1 protein inhibits activation of Jun N-terminal kinase and AP-1 transcription factors. J. Virol. 76:11166-11171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Marión, R. M., T. Aragón, A. Beloso, A. Nieto, and J. Ortín. 1997. The N-terminal half of the influenza virus NS1 protein is sufficient for nuclear retention of mRNA and enhancement of viral mRNA translation. Nucleic Acids Res. 25:4271-4277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marión, R. M., P. Fortes, A. Beloso, C. Dotti, and J. Ortín. 1999. A human sequence homologue of Staufen is an RNA-binding protein that localizes to the polysomes of the rough endoplasmic reticulum. Mol. Cell. Biol. 19:2212-2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Marión, R. M., T. Zürcher, S. de la Luna, and J. Ortín. 1997. Influenza virus NS1 protein interacts with viral transcription-replication complexes in vivo. J. Gen. Virol. 78:2447-2451. [DOI] [PubMed] [Google Scholar]
- 54.Martin, K., and A. Helenius. 1991. Nuclear transport of influenza virus ribonucleoproteins: the viral matrix protein (M1) promotes export and inhibits import. Cell 67:117-130. [DOI] [PubMed] [Google Scholar]
- 55.Mena, I., S. de la Luna, C. Albo, J. Martín, A. Nieto, J. Ortín, and A. Portela. 1994. Synthesis of biologically active influenza virus core proteins using a vaccinia-T7 RNA polymerase expression system. J. Gen. Virol. 75:2109-2114. [DOI] [PubMed] [Google Scholar]
- 56.Nemeroff, M. E., S. M. Barabino, Y. Li, W. Keller, and R. M. Krug. 1998. Influenza virus NS1 protein interacts with the cellular 30 kDa subunit of CPSF and inhibits 3′ end formation of cellular pre-mRNAs. Mol. Cell 1:991-1000. [DOI] [PubMed] [Google Scholar]
- 57.Neumann, G., T. Watanabe, H. Ito, S. Watanabe, H. Goto, P. Gao, M. Hughes, D. R. Perez, R. Donis, E. Hoffmann, G. Hobom, and Y. Kawaoka. 1999. Generation of influenza A viruses entirely from cloned cDNAs. Proc. Natl. Acad. Sci. USA 96:9345-9350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nieto, A., S. de la Luna, J. Bárcena, A. Portela, J. Valcárcel, J. A. Melero, and J. Ortín. 1992. Nuclear transport of influenza virus polymerase PA protein. Virus Res. 24:65-75. [DOI] [PubMed] [Google Scholar]
- 59.Noah, D. L., K. Y. Twu, and R. M. Krug. 2003. Cellular antiviral responses against influenza A virus are countered at the posttranscriptional level by the viral NS1A protein via its binding to a cellular protein required for the 3′ end processing of cellular pre-mRNAS. Virology 307:386-395. [DOI] [PubMed] [Google Scholar]
- 60.O'Neill, R. E., J. Talon, and P. Palese. 1998. The influenza virus NEP (NS2 protein) mediates the nuclear export of viral ribonucleoproteins. EMBO J. 17:288-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ortín, J. 1998. Multiple levels of post-transcriptional regulation of Influenza virus gene expression. Semin. Virol. 3:335-342. [Google Scholar]
- 62.Ortín, J., R. Nájera, C. López, M. Dávila, and E. Domingo. 1980. Genetic variability of Hong Kong (H3N2) influenza viruses: spontaneous mutations and their location in the viral genome. Gene 11:319-331. [DOI] [PubMed] [Google Scholar]
- 63.Park, Y. W., and M. G. Katze. 1995. Translational control by influenza virus. Identification of cis-acting sequences and trans-acting factors which may regulate selective viral mRNA translation. J. Biol. Chem. 270:28433-28439. [DOI] [PubMed] [Google Scholar]
- 64.Perales, B., and J. Ortín. 1997. The influenza A virus PB2 polymerase subunit is required for the replication of viral RNA. J. Virol. 71:1381-1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Portela, A., J. A. Melero, C. Martinez, E. Domingo, and J. Ortin. 1985. A primer vector system that allows temperature dependent gene amplification and expression in mammalian cells: regulation of the influenza virus NS1 gene expression. Nucleic Acids Res. 13:7959-7977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Portela, A., T. Zürcher, A. Nieto, and J. Ortín. 1999. Replication of Orthomyxoviruses. Adv. Virus Res. 54:319-348. [DOI] [PubMed] [Google Scholar]
- 67.Qian, X.-Y., F. Alonso-Caplen, and R. M. Krug. 1994. Two functional domains of the influenza virus NS1 protein are required for regulation of nuclear export of mRNA. J. Virol. 68:2433-2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Qiu, Y., and R. M. Krug. 1994. The influenza virus NS1 protein is a poly(A)-binding protein that inhibits nuclear export of mRNAs containing poly(A). J. Virol. 68:2425-2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Qiu, Y., M. Nemeroff, and R. M. Krug. 1995. The influenza virus NS1 protein binds to a specific region in human U6 snRNA and inhibits U6-U2 and U6-U4 snRNA interactions during splicing. RNA 1:304-316. [PMC free article] [PubMed] [Google Scholar]
- 70.Robertson, J. S., E. Robertson, I. Roditi, J. W. Almond, and S. C. Inglis. 1983. Sequence analysis of fowl plague virus mutant ts47 reveals a nonsense mutation in the NS1 gene. Virology 126:391-394. [DOI] [PubMed] [Google Scholar]
- 71.Salvatore, M., C. F. Basler, J. P. Parisien, C. M. Horvarth, S. Bourmakina, H. Zheng, T. Muster, P. Palese, and A. García-Sastre. 2002. Effects of influenza A virus NS1 protein on protein expression: the NS1 protein enhances translation and is not required for shutoff of host protein synthesis. J. Virol. 76:1206-1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shapiro, G. I., T. J. Gurney, and R. M. Krug. 1987. Influenza virus gene expression: control mechanisms at early and late times of infection and nuclear-cytoplasmic transport of virus-specific RNAs. J. Virol. 61:764-773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shapiro, G. I., and R. M. Krug. 1988. Influenza virus RNA replication in vitro: synthesis of viral template RNAs and virion RNAs in the absence of an added primer. J. Virol. 62:2285-2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shimizu, K., A. Iguchi, R. Gomyou, and Y. Ono. 1999. Influenza virus inhibits cleavage of the HSP70 pre-mRNAs at the polyadenylation site. Virology 254:213-219. [DOI] [PubMed] [Google Scholar]
- 75.Talon, J., C. M. Horvath, R. Polley, C. F. Basler, T. Muster, P. Palese, and S. A. Garcia. 2000. Activation of interferon regulatory factor 3 is inhibited by the influenza A virus NS1 protein. J. Virol. 74:7989-7996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Valcárcel, J., P. Fortes, and J. Ortin. 1993. Splicing of influenza virus matrix protein mRNA expressed from a simian virus 40 recombinant. J. Gen. Virol. 74:1317-1326. [DOI] [PubMed] [Google Scholar]
- 77.Wang, X., C. F. Basler, B. R. Williams, R. H. Silverman, P. Palese, and A. Garcia-Sastre. 2002. Functional replacement of the carboxy-terminal two-thirds of the influenza A virus NS1 protein with short heterologous dimerization domains. J. Virol. 76:12951-12962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang, X., M. Li, H. Zheng, T. Muster, P. Palese, A. A. Beg, and A. Garcia Sastre. 2000. Influenza A virus NS1 protein prevents activation of NF-κB and induction of alpha/beta interferon. J. Virol. 74:11566-11573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Whittaker, G., M. Bui, and A. Helenius. 1996. Nuclear trafficking of influenza virus ribonuleoproteins in heterokaryons. J. Virol. 70:2743-2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yasuda, J., S. Nakada, A. Kato, T. Toyoda, and A. Ishihama. 1993. Molecular assembly of influenza virus: association of the NS2 protein with virion matrix. Virology 196:249-255. [DOI] [PubMed] [Google Scholar]
- 81.Zhirnov, O. P., T. E. Konakova, T. Wolff, and H. D. Klenk. 2002. NS1 protein of influenza A virus down-regulates apoptosis. J. Virol. 76:1617-1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zürcher, T., R. M. Marión, and J. Ortín. 2000. The protein synthesis shutoff induced by influenza virus infection is independent of PKR activity. J. Virol. 74:8781-8784. [DOI] [PMC free article] [PubMed] [Google Scholar]