Abstract
Bacteriophage Mu replicates as a transposable element, exploiting host enzymes to promote initiation of DNA synthesis. The phage-encoded transposase MuA, assembled into an oligomeric transpososome, promotes transfer of Mu ends to target DNA, creating a fork at each end, and then remains tightly bound to both forks. In the transition to DNA synthesis, the molecular chaperone ClpX acts first to weaken the transpososome's interaction with DNA, apparently activating its function as a molecular matchmaker. This activated transpososome promotes formation of a new nucleoprotein complex (prereplisome) by yet unidentified host factors [Mu replication factors (MRFα2)], which displace the transpososome in an ATP-dependent reaction. Primosome assembly proteins PriA, PriB, DnaT, and the DnaB–DnaC complex then promote the binding of the replicative helicase DnaB on the lagging strand template of the Mu fork. PriA helicase plays an important role in opening the DNA duplex for DnaB binding, which leads to assembly of DNA polymerase III holoenzyme to form the replisome. The MRFα2 transition factors, assembled into a prereplisome, not only protect the fork from action by nonspecific host enzymes but also appear to aid in replisome assembly by helping to activate PriA's helicase activity. They consist of at least two separable components, one heat stable and the other heat labile. Although the MRFα2 components are apparently not encoded by currently known homologous recombination genes such as recA, recF, recO, and recR, they may fulfill an important function in assembling replisomes on arrested replication forks and products of homologous strand exchange.
Bacteriophage Mu's characteristics as a transposable element play a critical part in the establishment of lysogeny as well as in lytic development. On injection of phage DNA into the host cell, it is integrated at a random site of the host chromosome (1) by the phage-encoded transposition apparatus (2–4), a process that can lead to the establishment of lysogeny. Although the initial integration event is conservative (5) (i.e., not involving replication of Mu), lytic development involves many replicative transposition events (6–8) that exploit the host replication apparatus to form multiple integrated copies of Mu (8–12). Long considered a mechanism distinct from homologous recombination, Mu transposition may nevertheless have much in common with this process during the transition from strand exchange to DNA replication.
The Mu Transposition Apparatus.
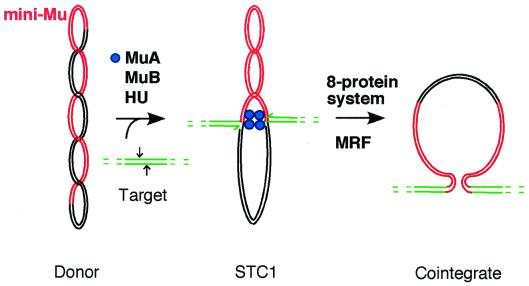
The establishment by Mizuuchi (8) of a crude extract system that catalyzes replicative Mu transposition has led to a detailed understanding of both the strand exchange reaction and the key steps involved in the initiation of DNA synthesis (for reviews, see refs. 13–16). This system uses a supercoiled donor substrate that bears a miniature version of the Mu genome (mini-Mu) and a target plasmid that contains no Mu DNA sequence. The strand exchange step that forms the template for Mu DNA synthesis can be catalyzed with three proteins (17): the phage-encoded transposase MuA, a second transposition protein MuB, and the host-encoded protein HU (see Fig. 1). HU aids in the assembly of MuA into an oligomeric transpososome tightly bound to both Mu ends (18–22), and the transpososome promotes integration of Mu ends into target DNA that is bound by MuB (23).
Figure 1.
Replication of Mu by transposition. In the first stage, the phage-encoded transposition proteins aided by the histone-like protein HU promote transfer of 3′-OH ends of miniMu (red) to each strand of target DNA (green). Two sites, 5 bp apart on target DNA, that will be subjected to a nucleophilic attack by each Mu end are indicated by arrows. Strand exchange produces a fork at each Mu end, the target providing 3′-OH ends (indicated by half arrows) that can potentially serve a primers for leading strand synthesis. MuA transposase, which has been assembled into an oligomeric transpososome, remains tightly bound to both Mu ends in the strand exchange product (strand transfer complex, STC1). Host factors then initiate Mu DNA synthesis from one end to duplicate Mu and form the final cointegrate product. The DNA synthesis phase was initially reconstituted in an eight-protein system supplemented with partially purified host factors (MRF), as described in the text.
In this process, the tetrameric core of the transpososome (24–26) produces a nick at each Mu end (Fig. 1) and promotes the transfer of the resulting 3′-OH ends to target DNA (19, 21, 27). The resulting strand exchange product (28) has at each Mu end a forked structure that can become the initiation site for Mu DNA synthesis. Host replication proteins will initiate semidiscontinuous DNA synthesis at one of these forks to duplicate Mu DNA (Fig. 1) and form the final cointegrate product (12, 29). However, the transpososome remains very tightly bound to both ends after strand exchange has been completed (21). Although this transpososome appears to pose an impediment to DNA replication, it plays an important role in promoting transition to DNA synthesis (11, 30).
Host Factors Involved in Mu DNA Replication.
Before the development of an in vitro Mu transposition system, Escherichia coli functions found to be required for bacteriophage Mu DNA replication included dnaE, dnaX, dnaB, dnaC, dnaG, gyrA, and gyrB (9, 10, 31, 32). The dnaE and dnaX genes encode subunits of the DNA polymerase (pol) III holoenzyme (33–36), the replicase at the replication fork. The DnaB protein is the major helicase at the fork, translocating 5′ to 3′ along the lagging strand template to unwind the helix for the propagating fork (37), and it can attract primase (38), encoded by dnaG (39), for initiation of lagging strand synthesis. DnaC protein forms a 1:1 complex with DnaB (40–42), acting as a molecular matchmaker (43) to promote loading of DnaB onto the replication fork. The gyrA and gyrB genes encode the two subunits of DNA gyrase (44–46), and this requirement may in part reflect the need for a supercoiled donor substrate for the strand exchange reaction (47, 48). As suggested by in vivo requirements for Mu DNA replication, we have found that cointegrate formation in the in vitro system requires DnaB–DnaC complex and the DNA polymerase III holoenzyme (11), confirming that the replisome involved in replicating the bacterial chromosome replicates Mu DNA during transposition.
Because Mu replication depends on host factors, we have been using this system to better understand the host apparatus needed to make the transition from recombination to replication. The studies were undertaken on the basis of the rationale that the Mu transposition system may be invaluable not only for dissecting the transition process in transposition but also for identifying cellular factors that play crucial roles in linking homologous recombination and replication. We initially set up a system of eight purified host proteins to identify host factors needed to initiate DNA synthesis on the product of Mu strand exchange. This system includes the DNA pol III holoenzyme, DnaB helicase, DnaC protein, primase, and DNA gyrase, the factors originally implicated in Mu replication in vivo. It also includes the single strand-binding protein, which would be needed at a propagating fork, as well as DNA pol I and DNA ligase, which would be needed for removing RNA primers from lagging strand synthesis and for forming covalently closed circular cointegrates, respectively. The template used for this reaction is the Mu strand exchange product formed by MuA, MuB, and HU with supercoiled donor and target substrates, the transpososome remaining very tightly bound to the two forks of this template [Fig. 1; strand transfer complex 1 (STC1)]. MuB and HU have also been found to be loosely bound to this template (49); however, we have been able to strip these proteins off STC1 without producing any apparent changes in the way it is replicated (11). Not surprisingly, the eight-protein system is not sufficient to catalyze any amount of Mu DNA synthesis on STC1. However, if the transpososome is removed from the STC1 template by phenol extraction, some of the replication proteins could gain access to the fork. On the deproteinized template, DNA pol I catalyzes limited strand displacement synthesis (11, 50). The deproteinized template can also be converted to a cointegrate in the reaction system even when DnaB is absent, provided that the DNA pol III holoenzyme preparation also contains helicase II (J.M.J. and H.N., unpublished results).
STC1 is converted to a cointegrate if the eight-protein system is supplemented with a partially purified host enzyme fraction [Mu replication factors (MRF)] (11). No cointegrates are formed when DNA pol III holoenzyme, DnaB helicase, or DnaC protein is omitted from the reaction system. If any one of these replication proteins or MRF is missing from the reaction system, we are not able to detect DnaB-independent cointegrate formation or the low levels of strand displacement DNA synthesis that can be catalyzed by DNA pol I on the deproteinized template. This indicates that the transpososome bound to the template imposes a strict requirement for both MRF and the specific replication proteins for initiation of Mu DNA synthesis. MRF was originally separated further into two fractions, MRFα and MRFβ (see Fig. 2), which can be functionally distinguished (30). MRFα removes the transpososome in an ATP-dependent reaction, and when the resulting template is isolated free of unbound proteins by gel filtration, it is converted to a cointegrate in the presence of MRFβ and the eight-protein system. MRFβ and the specific replication proteins are essential for converting this isolated template to cointegrate; however, if this template is stripped of bound proteins by phenol extraction, these factors are no longer essential for Mu DNA synthesis (30). This result implies that a new nucleoprotein complex (a prereplisome) takes the place of the transpososome and imposes specific requirements for MRFβ, DnaB, DnaC, and DNA pol III holoenzyme.
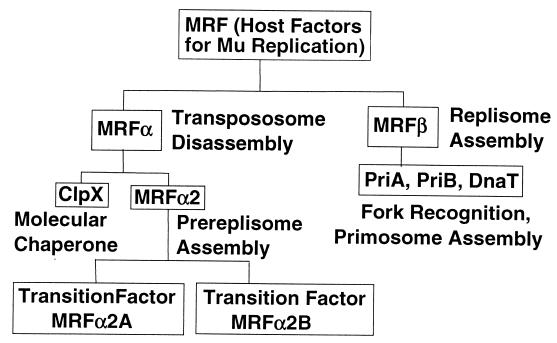
Figure 2.
Components of the MRF. MRF was originally identified as host factors needed in addition to the eight-protein system to convert STC1 to cointegrates. Resolution of MRF into enzyme fractions distinguishable by function and into pure components (ClpX, PriA, PriB, and DnaT) is indicated.
MRFα and MRFβ have each been found to consist of multiple components (see Fig. 2). The MRFα group is made up of the molecular chaperone ClpX and yet unidentified factors (MRFα2), and MRFβ is composed of primosomal constituents PriA, PriB, and DnaT (12). ClpX can play a distinct role at two different stages of the Mu life cycle (51). Together with the ClpP protein, it constitutes a chaperone-linked protease (52, 53) that can degrade the Mu immunity repressor (51, 54, 55). This process leads to derepression of Mu transposition, promoting exit out of lysogeny and induction of lytic development. ClpX, but not the protease component ClpP, is also required for Mu DNA replication in vivo (51), and as discussed below, it is one of the factors needed to promote transition from transpososome to replisome. The critical function of MRFβ factors in vivo was confirmed by the demonstration that Mu cannot undergo lytic development and cannot be replicated by transposition in a priA knockout mutant (12). In addition, a dnaT knockout mutant has recently been constructed, and it does not support Mu development (S. J. Sandler, personal communication). Although priB knockout mutants do support Mu development, this is consistent with the finding that priB has an essential cellular function that is redundant with priC (56). PriA, PriB, PriC, and DnaT proteins were first characterized as primosomal components needed to prime complementary DNA synthesis on single-stranded phage φX174 DNA (57, 58). The role of these proteins in initiating Mu DNA synthesis was consistent with their function in homologous recombination envisioned by Kogoma (59, 60). He hypothesized that these primosomal components promote assembly of a replisome at the site of homologous strand exchange. Biochemical evidence that these proteins can direct replisome assembly at D-loops (61) as well as other branched structures such as the Mu fork has supported their role in initiation of DNA synthesis on recombination intermediates and in the restart of arrested replication forks (62).
Role of the Transpososome in Mu DNA Replication.
A major strength of the in vitro Mu transposition system is that its essential components include the host factors required for Mu replication in vivo and that alternate pathways that initiate Mu DNA synthesis without one of these factors are prevented. As discussed so far, the transpososome plays a critical role in maintaining specificity for host factors involved in Mu DNA replication. When the transpososome is removed by phenol extraction, initiation of Mu DNA synthesis without MRF, DnaB, DnaC, or even DNA pol III holoenzyme can be detected (11, 30, 50, 63). The critical step promoted by the transpososome is the assembly of the prereplisome with MRFα2 at the Mu forks.
In the transition to DNA synthesis (Fig. 3), the molecular chaperone catalyzes the first step by acting on the transpososome in an ATP-dependent process (63, 64). Levchenko et al. (64) demonstrated that ClpX can cause the transpososome to disassemble, MuA dissociating from DNA in monomeric form. The released MuA could catalyze strand exchange, indicating that the cycle of transpososome assembly and ClpX-promoted disassembly produced no apparent alteration of MuA. These results indicate that ClpX can promote changes in transpososome conformation, altering MuA quaternary interactions and its interaction with DNA.
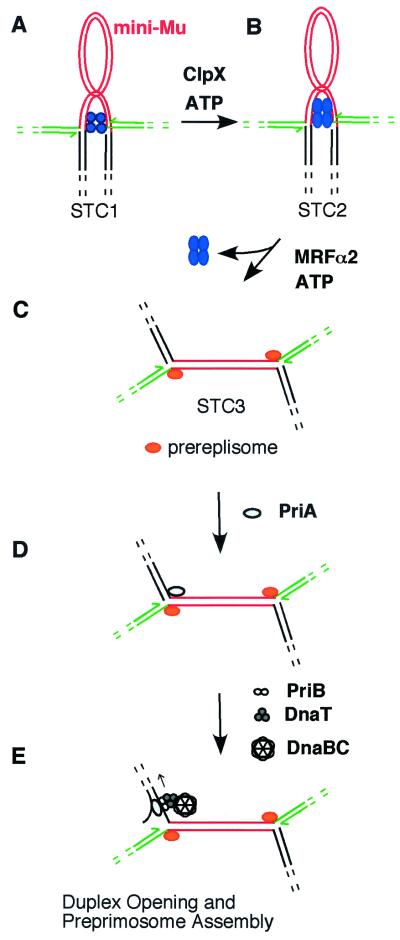
Figure 3.
Transition from transpososome to replisome. The molecular chaperone ClpX converts STC1 (A) to STC2 (B), altering the conformation of the transpososome. MRFα2 then displaces the transpososome to assemble the prereplisome at the Mu forks, forming STC3 (C). PriA binds to the forked DNA structure created by strand exchange (D) and begins the process of assembling a replisome at one Mu end. The mechanism that determines which Mu end is used to initiate DNA synthesis is not yet clear. PriA assembles a preprimosome complex by recruiting PriB, DnaT, and the DnaB-DnaC complex (E). In this process, DnaB must be bound to single-stranded lagging strand template. To create this binding site, PriA unwinds duplex DNA by translocating 3′ to 5′ along this template. Once bound to DNA, DnaB attracts primase to form a primosome, which catalyzes primer synthesis for lagging strand synthesis, and DnaB promotes binding of the DNA pol III holoenzyme to complete replisome assembly.
Although we also found that ClpX acts on the transpososome for the transition to DNA synthesis, our studies indicated that the transpososome must remain bound to DNA to promote the transition to DNA synthesis (63). When STC1 (the template with bound transpososome) was treated with ClpX under reaction conditions used for the in vitro Mu replication system, the resulting nucleoprotein complex (STC2) could be isolated free of unbound proteins (including ClpX) and then converted to a cointegrate in the eight-protein system supplemented with MRFα2 (no ClpX) and MRFβ. In isolated STC2, oligomeric MuA still holds the two Mu ends together in a synaptic complex (Fig. 3B). Initiation of Mu DNA synthesis strictly required MRFα2, MRFβ (or PriA, PriB, and DnaT), DnaB, DnaC, and the DNA pol III holoenzyme. If any one of these components was missing, not even partial replication of Mu DNA could be detected. However, if the transpososome was removed from DNA by phenol extraction or by high ionic strength as described below, initiation of Mu DNA synthesis did not absolutely depend on each of these components.
Even though the transpososome in STC2 remains bound to the two Mu ends, this complex is not as stable as the transpososome in STC1 (63). Intact STC2 could be isolated free of unbound proteins by gel filtration in the presence of 60 mM KCl or 200 mM potassium glutamate, ionic conditions used in the in vitro Mu replication system. Under conditions of higher ionic strength (300 mM NaCl), MuA dissociated from DNA. Levchenko et al. (64) had used such conditions to separate DNA from released MuA. This suggested to us that an intact STC2 transpososome can be isolated after ClpX treatment, so long as it is kept under conditions of lower ionic strength, which is required for the Mu replication system, and that this transpososome dissociates under conditions of higher ionic strength.
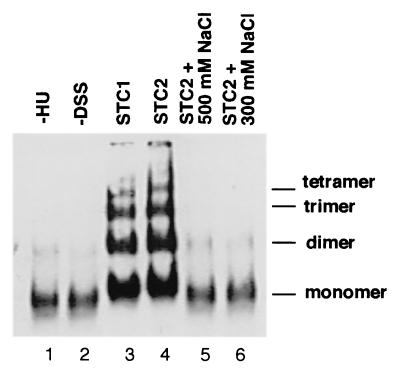
Crosslinking analysis confirmed that the oligomeric structure of the STC2 transpososome can be disrupted at 300 mM NaCl (Fig. 4). The oligomeric nature of the transpososome was originally established by chemical crosslinking (25, 27). MuA in solution is a monomer (65), and intermolecular crosslinking between MuA monomers does not readily occur. But once assembled into a transpososome, MuA protomers are crosslinked to form tetramers and even higher-order oligomers. Because the STC1 transpososome is extremely stable and remains intact even at 2 M NaCl (21), MuA protomers in the transpososome have readily been crosslinked at 0.5 M NaCl (25, 27). We performed crosslinking analysis at conditions of lower ionic strength (60 mM KCl) that allow isolation of intact STC2. Under our reaction conditions, there was very little crosslinking of MuA monomers by disuccinimidyl suberate (DSS), even when MuA was added to donor DNA without HU (conditions that do not allow the transpososome to be assembled; Fig. 4, lane 1). When STC1 isolated free of unbound proteins was treated with DSS, MuA was crosslinked to a ladder of dimers, trimers, tetramers, and higher order oligomers (lane 3). In previous studies (25, 27), the transpososome was crosslinked predominantly to tetramers by using dithio-bis(succinimidyl propionate), which can be cleaved by using a reducing agent. Because we used reaction conditions that largely reflect optimal conditions for Mu transposition and replication in vitro, we used the crosslinking agent DSS, which cannot be cleaved, and these conditions yielded somewhat less efficient crosslinking than in previous analysis. The pattern of MuA crosslinking was not apparently changed when the isolated STC1 was converted to STC2 with ClpX (lane 4). Although no major differences in MuA crosslinking between STC1 and STC2 can be discerned, subtle changes in crosslinking between MuA protomers cannot be ruled out. Treatment of STC2 at 0.3–0.5 M NaCl resulted in the loss of MuA crosslinking, indicating that the transpososome was disassembled back to monomers (lanes 5 and 6). The resulting template isolated free of unbound proteins has all of the characteristics of the deproteinized strand exchange product: the requirement for the specific host factors in initiating Mu DNA synthesis is lost (63).
Figure 4.
Fragile property of the STC2 transpososome. Formation of STC1, its conversion to STC2, crosslinking of the transpososome with DSS, and detection of crosslinked MuA by Western blot analysis was conducted as previously described (63). Lane 1: As control, the strand exchange reaction mixture was incubated without HU protein, conditions which do not permit transpososome assembly, and then MuA was subjected to crosslinking with DSS. Lanes 2–6: STC1 was isolated free of unbound proteins by filtration through a Bio-Gel (Bio-Rad) A-15 m column equilibrated with 25 mM Hepes-KOH (pH 7.5), 12 mM magnesium acetate, and 60 mM KCl. Isolated complexes were incubated at 37°C for 30 min in the presence of ATP, ClpX being included for conversion to STC2. The reaction mixture was adjusted to 300 or 500 mM NaCl, as indicated, and allowed to stand at room temperature for 15 min before addition of DSS. For lane 2, STC1 was not subjected to DSS treatment as control.
The MRFα2 Transition Factors.
Although the dissociation of the STC2 transpososome at 300 mM NaCl results in loss of the requirement for the specific host factors, disassembly of this transpososome by MRFα2 forms a template that maintains this specificity (63). MRFα2 is able to disassemble the transpososome from isolated STC2 but not from STC1. This is an ATP-dependent process that results in the release of the transpososome in oligomeric form, indicated by our ability to crosslink released MuA after it has been separated from DNA by gel filtration. Whereas the STC2 transpososome can be disassembled from DNA and dissociated to monomers at 300 mM NaCl or higher, it is not yet clear how the oligomeric transpososome is dissociated from the Mu ends by MRFα2. The transpososome may simply be displaced by assembly of MRFα2 components at the Mu ends and may be fully capable of rebinding to Mu ends. Alternatively, the dissociated transpososome may be bound to an MRFα2 component that does not remain bound to the template, or it may assume an inactive conformation such that it may no longer bind to Mu ends. Once MuA is displaced from the template, a new nucleoprotein complex (STC3) is apparently formed. STC3 isolated free of MuA retains the strict requirement for MRFβ, DnaB, DnaC, and DNA pol III holoenzyme for its conversion to a cointegrate (30, 63). The importance of proteins bound to STC3 is indicated by the loss of host factor specificity when they are removed from the template by phenol extraction. Because components of the replisome will assemble on STC3, we have referred to this nucleoprotein complex as a prereplisome that prepares the template for replisome assembly. And because we have not been able to form STC3 by incubating MRFα with the deproteinized template, we conclude that the STC2 transpososome plays a crucial role in promoting assembly of the prereplisome.
The transpososome's role in maintaining specificity for host factors is therefore associated with promoting prereplisome assembly. ClpX apparently activates a molecular matchmaker function of the transpososome (63). The STC2 transpososome allows assembly of a prereplisome in an ATP-dependent process at the Mu forks, the transpososome being displaced from DNA in the process. The interaction between prereplisome components and the Mu forks is apparently not established without the help of the transpososome. Although the transpososome remains stably bound to the Mu ends after ClpX treatment under reaction conditions used to catalyze Mu DNA replication, the presence of 300 mM NaCl or higher causes the dissociation of MuA as monomers and thus prevents prereplisome assembly. Formation of the prereplisome may commit the template to be replicated by allowing only the specific host proteins PriA, PriB, DnaT, DnaB–DnaC complex, and DNA pol III holoenzyme to gain access to the fork. If the transpososome disassembles without promoting prereplisome assembly, other cellular enzymes could compete for access to the Mu fork. The deproteinized strand transfer product can be converted to a cointegrate when introduced into a crude extract (28), but as we have discussed, this template can be converted to a cointegrate by a pathway not dependent on the specific replication factors. Thus, significant levels of alternate and aberrant products such as partially replicated and degraded templates can also accumulate.
The protein content and organization of the prereplisome complex are not yet clear. Although two identical prereplisomes are shown to be assembled at the two Mu forks in Fig. 3C, it is also possible that distinct complexes are assembled at these forks. In induced lysogens, replication of full-length (37-kb) Mu DNA proceeds semidiscontinuously from one end to the other, and Mu DNA synthesis initiates 80–90% of the time from the Mu left end (29, 66–68). DNA synthesis in the reconstituted system also initiates at one end and proceeds semidiscontinuously to the other end, and preference for the Mu left end can be detected (12). These results indicate that PriA promotes assembly of a replisome predominantly at one Mu end, preferentially the left end, and not both (Fig. 3D). One possible mechanism for the choice of Mu ends used for replisome assembly would be the asymmetric assembly of nucleoprotein complexes at the two Mu ends during transition to STC3. The two ends are composed of different sequences, and the transpososome is bound differently to each end (25, 27). It is therefore possible that the transpososome promotes asymmetric prereplisome assembly such that replisome assembly at the left end is favored.
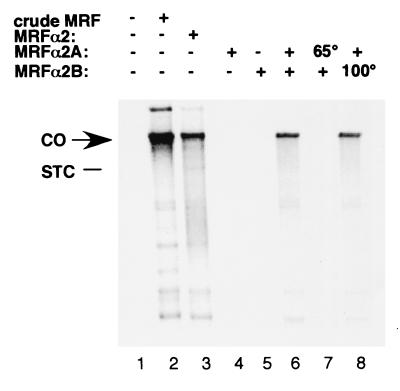
We are currently purifying MRFα2 to identify the remaining host factors needed to replicate Mu DNA during transposition. We have so far determined that MRFα2 can be resolved into two components (MRFα2A and MRFα2B). MRFα2 activity is assayed by using a reaction system that includes STC1 as template, molecular chaperone ClpX, and purified replication proteins (the original eight-protein system plus PriA, PriB, PriC, and DnaT). This assay system cannot promote cointegrate formation unless supplemented with both MRFα2 components (Fig. 5, compare lane 6 with lanes 4 and 5). The conclusion that these are two distinct factors is confirmed by the finding that one is heat labile, whereas the other is heat stable. MRFα2A is readily inactivated by heating at 65°C for 10 min (lane 7), whereas MRFα2B remains active even after heating at 100°C for 10 min (lane 8).
Figure 5.
MRFα2 consists of at least two distinct components. The reconstituted Mu DNA replication reaction (50 μl) with [α-32P]dNTPs was assembled with ClpX, the 12-protein system, and the indicated MRFα2 components, and products were resolved by alkaline agarose electrophoresis as previously described (75). Crude MRF (fraction II) (30) and MRFα2 (fraction III) (63) were prepared as described. Resolution of MRFα2 into two components, MRFα2A and MRFα2B, will be described in a future publication (V.D. and H.N., unpublished work). Approximately 10 units (63) of the indicated MRFα2 components were added. Where indicated, MRFα2A and MRFα2B were heated at 65 and 100°C, respectively, for 10 min. In the reaction catalyzed with crude MRF, greater than 95% of STC1 was converted to cointegrate. When MRFα2 was supplied as two components, typically 50–95% of STC1 was converted to a cointegrate. CO, position of the cointegrate; STC, position of the strand exchange product (not radiolabeled and therefore not visible).
Genetic analysis has not yet suggested the possible identity of MRFα2 components. The critical role PriA plays in both homologous recombination and Mu replication has suggested that Mu may exploit host homologous recombination functions for the transition from recombination to replication. We have examined a number of homologous recombination functions, including those that play a critical role in restart of chromosomal replication (for reviews, see refs. 62 and 69–72), for a possible role in Mu replication. So far, we have not found mutants that are as defective in Mu development as the priA and dnaT knockout mutants. We have examined recA, recF, recO, recR, recJ, recG, and ruvA mutants, most of which have knockout mutations (exceptions are recF143 and recF4101 strains provided to us by S. Sandler, University of Massachusetts, Amherst, MA), and all were able to support Mu lytic development. S. Lamrani and G. Maenhaut-Michel (personal communication) have also found that knockout mutations in the following genes still allow Mu lytic development to proceed: recA, recB, recD, recG, ruvA, ruvC, ruvABC, and rusA. We cannot rule out the unlikely possibility that more than one host factor can provide the same MRFα2 function and that one of these recombination genes provides such a redundant function. On the other hand, MRFα2 components may turn out to be factors not yet implicated in any step in recombination or replication. Just as MRFα2 components may act at a stage when the transpososome function is being completed and the replisome function is about to begin, they may act at a corresponding stage in cellular recombination-dependent replication, having a function distinct from that of the currently known homologous recombination proteins.
Role of PriA's Helicase Activity.
The involvement of host factors such as PriA in Mu DNA replication has helped us better understand their role in cellular chromosome replication and recombination. For example, the essential role of PriA in Mu DNA replication in vitro and in vivo has demonstrated the critical function it can play in assembling replisomes on recombination intermediates (12). In addition, PriA's 3′ to 5′ helicase activity (73, 74) has been found to play an important role in initiating Mu DNA replication (75). PriA mutants defective in helicase activity such as PriA K230R are proficient in assembling a primosome on the single-stranded template of phage φX174, and they are able to reverse characteristics of slow growth, low viability, and filamentous morphology (76) characteristic of priA knockout strains (77, 78). Although the cellular phenotype of priA helicase mutants suggests no apparent function for the helicase activity, studies of the role of PriA helicase in Mu DNA replication indicate a general function in duplex opening for replisome assembly (13, 75).
Mutants expressing PriA K230R support Mu DNA replication at greatly reduced rates (less than 20% the rate of wild-type cells). Infected cells exhibit delayed lysis and a reduced burst, and when Mu is plated with these mutants, minute plaques are formed. In the reconstituted system, which consists of the purified protein system supplemented with partially purified MRFα2, little to no cointegrates can be formed if PriA K230R is used instead of wild-type PriA. Unlike the potential replication fork at a D-loop created by homologous strand exchange, the Mu fork created by the transposase has no single-stranded DNA on the lagging strand arm of the fork to load the DnaB helicase: there is a gap of only five bases on the leading strand side of the fork. To assemble a replisome at the fork, DnaB must be bound to the lagging strand template of the fork, and it occupies 20 nucleotides of single-stranded DNA (79). PriA binds to the forked DNA structures found at D-loops, arrested replication forks, and the Mu fork (75, 80, 81), and then PriA can translocate 3′ to 5′ along the lagging strand template as it promotes preprimosome assembly with PriB, DnaT, and the DnaB-DnaC complex (75) (Fig. 3E). Translocation of PriA along that template is tightly coupled to the binding of DnaB to the same DNA strand. Thus, PriA helicase can function to create the single-stranded template needed for primosome assembly when there is an insufficient amount needed to load DnaB.
Although inactivation of PriA helicase greatly reduces the rate of Mu DNA replication in vivo, it does not eliminate Mu DNA replication entirely. This is most likely because of the action of other host factors such as other helicases and exonucleases that may also serve to create a duplex opening on the lagging strand side of the fork. Even though Mu DNA replication is catalyzed poorly in the reconstituted system by using PriA K230R, addition of a crude host enzyme fraction can complement this deficiency to promote higher levels of cointegrate formation (75). For DNA synthesis at a D-loop, PriA helicase activity may not be required because the template already has a duplex opening for loading DnaB. But other pathways for restarting DNA replication, such as the regression of the replication fork, could very well require duplex opening (13). Even if such pathways requiring duplex opening are critical for cell viability, mutations that inactivate PriA helicase may exhibit no severe phenotype because other host enzymes may also carry out this function. At least in the case of Mu, however, duplex opening by other factors is inefficient because helicase deficiency impairs Mu replication in vivo. For its role in cellular DNA recombination and replication, PriA may require its helicase activity to function optimally, and this may be reflected by the observation that the helicase motif of priA genes identified in various species is highly conserved (13).
In the initiation of chromosomal replication at the bacterial origin oriC, a critical point of regulation is duplex opening catalyzed by the initiator protein DnaA (82). At the Mu fork, the prereplisome may influence whether PriA helicase can open the DNA duplex. The deproteinized strand exchange product is not so readily converted to a cointegrate in the reconstituted system (no helicase II present), especially when DNA pol I is omitted so that limited strand displacement synthesis, which can create a duplex opening, is not catalyzed. At optimal levels, only about 30–40% of the deproteinized strand exchange products are converted to cointegrates in the absence of DNA pol I (12); greater than 95% of STC1 can be converted to cointegrate in the reconstituted system. Examining PriA helicase activity by using oligonucleotide substrates, we have found that oligonucleotides with the structure of the Mu fork are not good substrates for PriA helicase even though they are bound with high affinity by PriA (ref. 75; J. M. Jones and H.N., unpublished work). Two alterations of the Mu fork substrate increase unwinding of the duplex lagging strand arm. Significant amounts of unwinding can be detected if the leading strand arm of the fork is rendered single stranded. A similar amount of unwinding can be detected if a five-base gap is introduced at the fork on the lagging strand arm. The two modifications together increase unwinding ≈10-fold over the DNA substrate that has only one of the modifications.
Nurse et al. (80) have characterized two distinct modes of PriA binding to DNA. One mode is reflected by binding of PriA to duplexes with 3′ single strand extensions. This is thought to reflect recognition of DNA by the helicase domain. In the second mode, PriA binds to forked substrates, recognizing bent DNA in three-arm junctions. Our recent results (J. M. Jones and H.N., unpublished work) indicate that the fork-specific binding mode leads to translocation of PriA 3′ to 5′ along the lagging strand template, suggesting that this mode of binding orients the helicase domain to bind to this strand. Alterations of the Mu fork that promote PriA helicase action facilitate access of the single strand to which the helicase domain binds. The assembly of the prereplisome at the Mu fork may hold the fork in a conformation that allows activation of the PriA helicase without these alterations.
Potential Role of Transition Mechanisms in Cellular DNA Replication.
The transition mechanisms involved in Mu DNA replication promote the assembly of a replisome at the Mu fork with specific host factors, apparently excluding access of the fork to nonspecific host enzymes that can lead to inefficient Mu replication or that may damage the template. Such mechanisms may also play a very important role in cellular DNA replication linked to homologous recombination. MRFα potentially consists of two types of components. One type comprises host factors that normally do not function in cellular recombination-dependent replication. That is, the transposase has evolved to exploit these factors to promote transition to DNA replication. For example, the molecular chaperone ClpX may not play any function in linking cellular recombination and replication; its role in linking recombination and replication may be limited to activating the transposase's molecular matchmaker function. A second type of MRFα component may be factors that have evolved to function in the transition between recombination and replication, preparing DNA recombination intermediates for replisome assembly. Components of the prereplisome in STC3 most likely belong to this class. Presumably, they did not evolve only to promote the transition process in transposition.
Assembly of a prereplisome on a template created by homologous recombination proteins may commit the pathway to the assembly of a replisome and the establishment of a replication fork. The prereplisome assembly step may distinguish this pathway from other processes associated with homologous strand exchange such as double-strand break repair or replication-dependent recombination (69), which may require only a limited amount of DNA synthesis linked to recombination and not a highly processive replisome needed for chromosomal replication. In pathways that repair DNA lesions at an arrested replication fork, the prereplisome may play a critical function to assure re-establishment of the replication fork by requiring that DnaB and DNA pol III holoenzyme be engaged on that template.
If the prereplisome components evolved to promote transition from homologous recombination to replication, what processes might be involved in assembling the prereplisome at the site of homologous strand exchange? In transposition, the transpososome may aid in prereplisome assembly in one of two ways. First, the transpososome may interact specifically with prereplisome components to recruit them to the Mu fork. Such a function may be assumed by one of the homologous recombination proteins such as RecA at a D-loop. Alternatively, the transpososome may hold the DNA together in a conformation that mimics a structure created by homologous recombination proteins as they promote restart of chromosomal replication. This type of strategy is used in recruiting PriA, which binds to the branched DNA structure created by Mu strand exchange (75), a structure resembling D-loops and arrested forks. The STC2 transpososome still holds the two Mu forks together in a synaptic complex (63), but unlike the STC1 transpososome, this complex is fragile. ClpX's role in weakening the transpososome's interaction with DNA (63, 64) may be the key feature of activating the apparent molecular matchmaker function of the transpososome. As the prereplisome assembles on the DNA structure maintained on STC2, the transpososome disassembles, completing the handoff of the Mu forks from phage-encoded transposase to host factors. How cellular recombination functions trigger prereplisome assembly may be a key to understanding what regulates replisome assembly during homologous recombination. The identification and characterization of MRFα2 transition factors promise to provide better understanding of the transition between recombination and replication for the transposable element as well as its host.
Acknowledgments
We thank Robert Kruklitis, who performed the initial work to fractionate host factors for Mu DNA replication, including early experiments to characterize the role of ClpX and MuA in initiation. Supplies of E. coli replication proteins for this study have been maintained as a collaboration with Kirsten Skarstad, Elliott Crooke, and Nick Dixon. J.M.J. was a recipient of a predoctoral training grant from the Department of Defense Breast Cancer Research Program (DAMD 17–98-1–8090). This work was supported by grants from the National Institutes of Health (GM49649 and GM58265).
Abbreviations
- MRF
Mu replication factors
- pol
polymerase
- STC
strand transfer complex
- DSS
disuccinimidyl suberate
Footnotes
This paper results from the National Academy of Sciences colloquium, “Links Between Recombination and Replication: Vital Roles of Recombination,” held November 10–12, 2000, in Irvine, CA.
References
- 1.Taylor A L. Proc Natl Acad Sci USA. 1963;50:1043–1051. doi: 10.1073/pnas.50.6.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ljungquist E, Khatoon H, DuBow M, Ambrosio L, de Bruijn F, Bukhari A I. Cold Spring Harbor Symp Quant Biol. 1979;43:1151–1158. doi: 10.1101/sqb.1979.043.01.130. [DOI] [PubMed] [Google Scholar]
- 3.Chaconas G, Giddens E B, Miller J L, Gloor G. Cell. 1985;41:857–865. doi: 10.1016/s0092-8674(85)80066-0. [DOI] [PubMed] [Google Scholar]
- 4.O'Day K J, Schultz D W, Howe M M. In: Microbiology 1978. Schlessinger D, editor. Washington, DC: Am. Soc. Microbiol.; 1978. pp. 48–51. [Google Scholar]
- 5.Harshey R M. Nature (London) 1984;311:580–581. doi: 10.1038/311580a0. [DOI] [PubMed] [Google Scholar]
- 6.Wijffelman C, Lotterman B. Mol Gen Genet. 1977;151:169–174. doi: 10.1007/BF00338691. [DOI] [PubMed] [Google Scholar]
- 7.Faelen M, Huisman O, Toussaint A. Nature (London) 1978;271:580–582. doi: 10.1038/271580a0. [DOI] [PubMed] [Google Scholar]
- 8.Mizuuchi K. Cell. 1983;35:785–794. doi: 10.1016/0092-8674(83)90111-3. [DOI] [PubMed] [Google Scholar]
- 9.Toussaint A, Faelen M. Mol Gen Genet. 1974;131:209–214. doi: 10.1007/BF00267960. [DOI] [PubMed] [Google Scholar]
- 10.Résibois A, Pato M, Higgins P, Toussaint A. In: Proteins Involved in DNA Replication. Hubscher U, Spadari S, editors. New York: Plenum; 1984. pp. 69–76. [Google Scholar]
- 11.Kruklitis R, Nakai H. J Biol Chem. 1994;269:16469–16477. [PubMed] [Google Scholar]
- 12.Jones J M, Nakai H. EMBO J. 1997;16:6886–6895. doi: 10.1093/emboj/16.22.6886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jones J M, Nakai H. Mol Microbiol. 2000;36:519–527. doi: 10.1046/j.1365-2958.2000.01888.x. [DOI] [PubMed] [Google Scholar]
- 14.Mizuuchi K. Annu Rev Biochem. 1992;61:1011–1051. doi: 10.1146/annurev.bi.61.070192.005051. [DOI] [PubMed] [Google Scholar]
- 15.Chaconas G, Lavoie B D, Watson M A. Curr Biol. 1996;6:817–820. doi: 10.1016/s0960-9822(02)00603-6. [DOI] [PubMed] [Google Scholar]
- 16.Lavoie B D, Chaconas G. Curr Top Microbiol Immunol. 1996;204:83–102. doi: 10.1007/978-3-642-79795-8_4. [DOI] [PubMed] [Google Scholar]
- 17.Craigie R, Arndt-Jovin D J, Mizuuchi K. Proc Natl Acad Sci USA. 1985;82:7570–7574. doi: 10.1073/pnas.82.22.7570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lavoie B D, Chaconas G. Genes Dev. 1993;7:2510–2519. doi: 10.1101/gad.7.12b.2510. [DOI] [PubMed] [Google Scholar]
- 19.Craigie R, Mizuuchi K. Cell. 1987;51:493–501. doi: 10.1016/0092-8674(87)90645-3. [DOI] [PubMed] [Google Scholar]
- 20.Lavoie B D, Chaconas G. J Biol Chem. 1994;269:15571–15576. [PubMed] [Google Scholar]
- 21.Surette M G, Buch S J, Chaconas G. Cell. 1987;49:253–262. doi: 10.1016/0092-8674(87)90566-6. [DOI] [PubMed] [Google Scholar]
- 22.Surette M G, Chaconas G. Cell. 1992;68:1101–1108. doi: 10.1016/0092-8674(92)90081-m. [DOI] [PubMed] [Google Scholar]
- 23.Adzuma K, Mizuuchi K. Cell. 1988;53:257–266. doi: 10.1016/0092-8674(88)90387-x. [DOI] [PubMed] [Google Scholar]
- 24.Baker T A, Mizuuchi M, Savilahti H, Mizuuchi K. Cell. 1993;74:723–733. doi: 10.1016/0092-8674(93)90519-v. [DOI] [PubMed] [Google Scholar]
- 25.Lavoie B D, Chan B S, Allison R G, Chaconas G. EMBO J. 1991;10:3051–3059. doi: 10.1002/j.1460-2075.1991.tb07856.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu Z G, Chaconas G. J Mol Biol. 1997;267:132–141. doi: 10.1006/jmbi.1996.0854. [DOI] [PubMed] [Google Scholar]
- 27.Mizuuchi M, Baker T A, Mizuuchi K. Cell. 1992;70:303–311. doi: 10.1016/0092-8674(92)90104-k. [DOI] [PubMed] [Google Scholar]
- 28.Craigie R, Mizuuchi K. Cell. 1985;41:867–876. doi: 10.1016/s0092-8674(85)80067-2. [DOI] [PubMed] [Google Scholar]
- 29.Higgins N P, Moncecchi D, Manlapaz-Ramos P, Olivera B M. J Biol Chem. 1983;258:4293–4297. [PubMed] [Google Scholar]
- 30.Nakai H, Kruklitis R. J Biol Chem. 1995;270:19591–19598. doi: 10.1074/jbc.270.33.19591. [DOI] [PubMed] [Google Scholar]
- 31.Ross W, Shore S H, Howe M M. J Bacteriol. 1986;167:905–919. doi: 10.1128/jb.167.3.905-919.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Toussaint A, Résibois A. In: Mobile Genetic Elements. Shapiro J A, editor. New York: Academic; 1983. pp. 105–158. [Google Scholar]
- 33.Maki S, Kornberg A. J Biol Chem. 1988;263:6547–6554. [PubMed] [Google Scholar]
- 34.Gefter M L, Hirota Y, Kornberg T, Wechsler J A, Barnoux C. Proc Natl Acad Sci USA. 1971;68:3150–3153. doi: 10.1073/pnas.68.12.3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hübscher U, Kornberg A. J Biol Chem. 1980;255:11698–11703. [PubMed] [Google Scholar]
- 36.Wickner W, Schekman R, Geider K, Kornberg A. Proc Natl Acad Sci USA. 1973;70:1764–1767. doi: 10.1073/pnas.70.6.1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.LeBowitz J H, McMacken R. J Biol Chem. 1986;261:4738–4748. [PubMed] [Google Scholar]
- 38.Tougo K, Peng H, Marians K J. J Biol Chem. 1994;269:4675–4682. [PubMed] [Google Scholar]
- 39.Rowen L, Kornberg A. J Biol Chem. 1978;253:758–764. [PubMed] [Google Scholar]
- 40.Kobori J A, Kornberg A. J Biol Chem. 1982;257:13770–13775. [PubMed] [Google Scholar]
- 41.Lanka E, Schuster H. Nucleic Acids Res. 1983;11:987–997. doi: 10.1093/nar/11.4.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wickner S, Hurwitz J. Proc Natl Acad Sci USA. 1975;72:921–925. doi: 10.1073/pnas.72.3.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sancar A, Hearst J E. Science. 1993;259:1415–1420. doi: 10.1126/science.8451638. [DOI] [PubMed] [Google Scholar]
- 44.Gellert M, Mizuuchi K, O'Dea M H, Itoh T, Tomizawa J. Proc Natl Acad Sci USA. 1977;74:4772–4776. doi: 10.1073/pnas.74.11.4772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gellert M, O'Dea M H, Itoh T, Tomizawa J. Proc Natl Acad Sci USA. 1976;73:4474–4478. doi: 10.1073/pnas.73.12.4474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sugino A, Peebles C L, Kreuzer K N, Cozarelli N R. Proc Natl Acad Sci USA. 1977;74:4767–4771. doi: 10.1073/pnas.74.11.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pato M L, Karlok M, Wall C, Higgins N P. J Bacteriol. 1995;177:5937–5942. doi: 10.1128/jb.177.20.5937-5942.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Craigie R, Mizuuchi K. Cell. 1986;45:793–800. doi: 10.1016/0092-8674(86)90554-4. [DOI] [PubMed] [Google Scholar]
- 49.Lavoie B D, Chaconas G. J Biol Chem. 1990;265:1623–1627. [PubMed] [Google Scholar]
- 50.Jones J M, Welty D J, Nakai H. J Biol Chem. 1998;273:459–465. doi: 10.1074/jbc.273.1.459. [DOI] [PubMed] [Google Scholar]
- 51.Mhammedi-Alaoui A, Pato M, Gama M-J, Toussaint A. Mol Microbiol. 1994;11:1109–1116. doi: 10.1111/j.1365-2958.1994.tb00387.x. [DOI] [PubMed] [Google Scholar]
- 52.Gottesman S, Clark W P, de Crécy-Lagard V, Maurizi M R. J Biol Chem. 1993;268:22618–22626. [PubMed] [Google Scholar]
- 53.Woo K M, Chung W J, Ha D B, Goldberg A L, Chung C H. J Biol Chem. 1989;264:2088–2091. [PubMed] [Google Scholar]
- 54.Geuskens V, Mhammedi-Alaoui A, Desmet L, Toussaint A. EMBO J. 1992;11:5121–5127. doi: 10.1002/j.1460-2075.1992.tb05619.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Welty D J, Jones J M, Nakai H. J Mol Biol. 1997;272:31–41. doi: 10.1006/jmbi.1997.1193. [DOI] [PubMed] [Google Scholar]
- 56.Sandler S J, Marians K J, Zavitz K H, Coutu J, Prent M A, Clark A J. Mol Microbiol. 1999;34:91–101. doi: 10.1046/j.1365-2958.1999.01576.x. [DOI] [PubMed] [Google Scholar]
- 57.Schekman R, Weiner J H, Weiner A, Kornberg A. J Biol Chem. 1975;250:5859–5865. [PubMed] [Google Scholar]
- 58.Wickner S, Hurwitz J. Proc Natl Acad Sci USA. 1974;71:4120–4124. doi: 10.1073/pnas.71.10.4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Asai T, Kogoma T. J Bacteriol. 1994;176:1807–1812. doi: 10.1128/jb.176.7.1807-1812.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kogoma T. Cell. 1996;85:625–627. doi: 10.1016/s0092-8674(00)81229-5. [DOI] [PubMed] [Google Scholar]
- 61.Liu J, Marians K J. J Biol Chem. 1999;274:25033–25041. doi: 10.1074/jbc.274.35.25033. [DOI] [PubMed] [Google Scholar]
- 62.Marians K J. Trends Biochem Sci. 2000;25:185–189. doi: 10.1016/s0968-0004(00)01565-6. [DOI] [PubMed] [Google Scholar]
- 63.Kruklitis R, Welty D J, Nakai H. EMBO J. 1996;15:935–944. [PMC free article] [PubMed] [Google Scholar]
- 64.Levchenko I, Luo L, Baker T A. Genes Dev. 1995;9:2399–2408. doi: 10.1101/gad.9.19.2399. [DOI] [PubMed] [Google Scholar]
- 65.Kuo C-F, Zou A, Jayaram M, Getzoff E, Harshey R. EMBO J. 1991;10:1585–1591. doi: 10.1002/j.1460-2075.1991.tb07679.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wijffelman C, van de Putte P. In: DNA Insertion Elements, Plasmids, and Episomes. Bukhari A I, Shapiro J A, Adhya S L, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1977. pp. 329–333. [Google Scholar]
- 67.Goosen T. In: DNA Synthesis: Present and Future. Molineux I, Kohiyama M, editors. New York: Plenum; 1978. pp. 121–126. [Google Scholar]
- 68.Pato M L, Waggoner B T. In: Phage Mu. Symonds N, Toussaint A, van de Putte P, Howe M M, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1987. pp. 177–189. [Google Scholar]
- 69.Kowalczykowski S C. Trends Biochem Sci. 2000;25:156–165. doi: 10.1016/s0968-0004(00)01569-3. [DOI] [PubMed] [Google Scholar]
- 70.Michel B. Trends Biochem Sci. 2000;25:173–178. doi: 10.1016/s0968-0004(00)01560-7. [DOI] [PubMed] [Google Scholar]
- 71.Kuzminov A. Microbiol Mol Biol Rev. 1999;63:751–813. doi: 10.1128/mmbr.63.4.751-813.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cox M M. Genes Cells. 1998;3:65–78. doi: 10.1046/j.1365-2443.1998.00175.x. [DOI] [PubMed] [Google Scholar]
- 73.Lasken R S, Kornberg A. J Biol Chem. 1988;263:5512–5518. [PubMed] [Google Scholar]
- 74.Lee M S, Marians K J. Proc Natl Acad Sci USA. 1987;84:8345–8349. doi: 10.1073/pnas.84.23.8345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jones J M, Nakai H. J Mol Biol. 1999;289:503–515. doi: 10.1006/jmbi.1999.2783. [DOI] [PubMed] [Google Scholar]
- 76.Zavitz K H, Marians K J. J Biol Chem. 1992;267:6933–6940. [PubMed] [Google Scholar]
- 77.Lee E H, Kornberg A. Proc Natl Acad Sci USA. 1991;88:3029–3032. doi: 10.1073/pnas.88.8.3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nurse P, Zavitz K H, Marians K J. J Bacteriol. 1991;173:6686–6693. doi: 10.1128/jb.173.21.6686-6693.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bujalowski W, Jezewska M J. Biochemistry. 1995;34:8513–8519. doi: 10.1021/bi00027a001. [DOI] [PubMed] [Google Scholar]
- 80.Nurse P, Liu J, Marians K J. J Biol Chem. 1999;274:25026–25032. doi: 10.1074/jbc.274.35.25026. [DOI] [PubMed] [Google Scholar]
- 81.McGlynn P, Al-Deib A A, Liu J, Marians K J, Lloyd R G. J Mol Biol. 1997;270:212–221. doi: 10.1006/jmbi.1997.1120. [DOI] [PubMed] [Google Scholar]
- 82.Bramhill D, Kornberg A. Cell. 1988;52:743–755. doi: 10.1016/0092-8674(88)90412-6. [DOI] [PubMed] [Google Scholar]