Abstract
We isolated and sequenced two complete endogenous feline leukemia viruses (enFeLVs), designated enFeLV-AGTT and enFeLV-GGAG. In enFeLV-AGTT, the open reading frames are reminiscent of a functioning FeLV genome, and the 5′ and 3′ long terminal repeat sequences are identical. Neither endogenous provirus is genetically fixed in cats but polymorphic, with 8.9 and 15.2% prevalence for enFeLV-AGTT and enFeLV-GGAG, respectively, among a survey of domestic cats. Neither provirus was found in the genomes of related species of the Felis genus, previously shown to harbor enFeLVs. The absence of mutational divergence, polymorphic incidence in cats, and absence in related species suggest that these enFeLVs may have entered the germ line more recently than previously believed, perhaps coincident with domestication, and reopens the question of whether some enFeLVs might be replication competent.
Endogenous feline leukemia virus (enFeLV) sequences are present in the genome of the domestic cat, Felis catus, with an estimated 6 to 12 copies per haploid genome (4, 17, 28, 30, 31). These sequences are homologous to exogenous FeLVs (exFeLVs), which are horizontally transmitted oncogenic retroviruses capable of inducing both proliferative and degenerative diseases (12, 27). Endogenous feline leukemia proviruses are part of the germ line and are transmitted from parent to offspring as integral components of chromosomes (4, 17). enFeLV sequences do not produce infectious virus, and attempts to rescue or induce endogenous virus by cocultivation in cell lines have failed (3, 19). Restriction enzyme mapping has revealed the presence of large deletions in some enFeLVs, which would render them incapable of producing an exogenous virus (41, 42). Molecular copies of enFeLVs without major deletions were found in DNA transfection studies also to be noninfectious, presumably owing either to alterations in sequences regulating gene expression or to coding sequence mutation (42). Both frameshift and nonsense mutations have been identified in the gag and env regions of full-length enFeLVs (5, 27).
Although they do not produce infections on their own, enFeLV sequences readily recombine with exFeLVs (32, 37, 43). Transmissible exFeLVs lack recombinant enFeLV segments and are classified as subgroup A (12, 15). The two other exFeLV subgroups, B and C, result from recombination between enFeLV segments and exFeLVs (27, 32, 35, 43). Recombinant viruses may exhibit altered biological activity and pathogenicity (13, 33, 37, 39, 47); for example, the recombinant subgroup C viruses have been found to induce aplastic anemia (14). Additionally, segments of enFeLVs are transcribed and translated in lymphoma and other cell lines: a truncated enFeLV envelope protein has been detected that inhibits infection by subgroup B exFeLVs (24). Transcription and translation of enFeLV genes have also been demonstrated in tissues from healthy cats, including lymphoid tissue, raising the prospect of a protective role for enFeLVs in vivo (6, 24). In contrast, a protein derived from an enFeLV env region was found to facilitate infection by a T-cell-tropic exFeLV (1).
Despite their possible role in protecting against infection by exFeLVs, and their established capacity to recombine with exFeLVs to produce new strains, the genomic structure and variation of enFeLVs have not been well characterized. Although sequences of endogenous long terminal repeats (LTRs) (5, 18), env (18), pol (33), and part of gag (5) have been determined, the full sequence of a complete enFeLV has not been reported. We therefore generated a probe from a 7-kb gag-pol-env segment (pKHR2-gpe; see Appendix) of a recombinant subgroup B exFeLV (pKHR-2/λHF60) (11, 26) and screened a domestic cat lambda FIX II genomic library (9- to 23-kb insert size; Stratagene) (36). Two previously undescribed full-length enFeLVs, designated enFeLV-AGTT and enFeLV-GGAG (the distinguishing label is the unique 4-bp segment of host DNA duplicated during viral integration), were isolated and sequenced. The proviral genome was 8,695 bp long for enFeLV-AGTT and 8667 bp long for enFeLV-GGAG. These are longer than the 8,440- to 8,448-bp genomes of the two nonrecombinant exFeLVs whose complete sequences are available (GenBank accession numbers M18247 [10] and AF052723 [8]). They are also longer than the 8.2 kb previously estimated by restriction fragment analysis for a full-length enFeLV (42). The gag, pol, and env regions of the two novel proviruses were closer in sequence to enFeLVs than to exFeLVs. For example, enFeLV-AGTT pol had 98.3% nucleotide sequence identity to endogenous L06140 pol but only 95.4% identity to exFeLV M18247 pol. The sequences of enFeLV-AGTT and enFeLV-GGAG were remarkably similar, differing by only a single substitution in the 1,512-bp gag region and by eight substitutions in the 3,630-bp pol region. The length of the env region was 2,009 bp in enFeLV-AGTT and 2,010 bp in enFeLV-GGAG, with two nucleotide substitutions (including one in the region of overlap between pol and env) and one indel distinguishing them. The cat genomic DNA flanking the enFeLV proviral genomes was also sequenced (600 bp each for the 5′ and 3′ flanks), and physical mapping (with a radiation hybrid cell panel) of the unique cellular flanks adjacent to each proviral integration site has determined that the two proviral integrations are on different domestic cat chromosomes (A. L. Roca, W. G. Nash, J. C. Menninger, W. J. Murphy, and S. J. O'Brien, unpublished data).
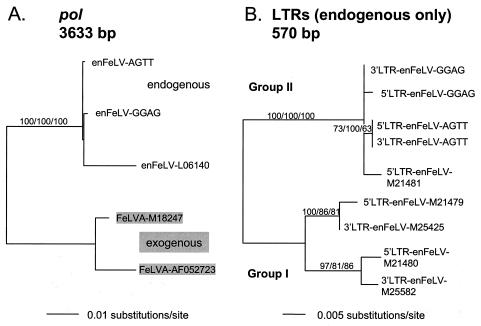
The novel endogenous proviral sequences were aligned versus previously characterized enFeLV and exFeLV sequences with CLUSTALX (45), and phylogenetic analyses were implemented in PAUP*4.0b4 (44) with three different methods (neighbor joining [NJ], maximum parsimony [MP], and maximum likelihood [ML]), each of which yielded similar tree topologies. The ML tree for pol is shown in Fig. 1A, and it reflects the closer relationship of the novel proviral segments to endogenous rather than exogenous sequences (also true for gag and env; not shown, see Appendix).
The gag, pol, and env viral coding regions are flanked by noncoding LTRs. Unlike the rest of the proviral enFeLV genome, the U3 region of the LTR is not homologous between enFeLVs and exFeLVs (5). The U3 region forms the 5′ end of each proviral LTR (in enFeLV-AGTT, the 568-bp LTR includes a 423-bp U3 region). The U3 region has been used as a straightforward means of distinguishing between exFeLV and enFeLV sequences in hybridization studies (7, 29), and the U3 sequences of enFeLV-AGTT and enFeLV-GGAG readily identified them as enFeLVs and not exFeLVs. Because of the absence of homology, exFeLV LTRs were not included in the LTR phylogenetic analyses, which revealed that enFeLV LTRs cluster into two groups (Fig. 1B; Appendix). The enFeLV-AGTT and enFeLV-GGAG LTRs group with enFeLV-M21481 (5), while the other four previously described endogenous LTRs (Fig. 1B) (5, 18) form a separate clade (Fig. 1B). It is uncertain whether these two groups of endogenous LTRs represent two separate integrations of FeLVs into the feline genome.
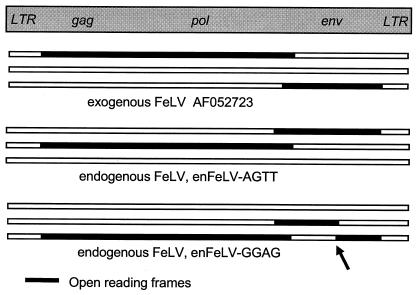
An analysis of reading frame structure revealed large open reading frames (ORFs) in enFeLV-AGTT similar to the ORFs observed in pathogenic exFeLVs (Fig. 2). Unlike other enFeLVs (41, 42), enFeLV-AGTT includes no major deletions or frameshift mutations. Although recombination with exogenous viruses could restore the ORFs of an ancient endogenous provirus, the dissimilarity in sequence between enFeLV-AGTT and exFeLVs rules this out as an explanation for the intact ORF in enFeLV-AGTT. Selective pressure could maintain the ORF in env, since intact endogenous env protects lymphocytes from infection by exFeLVs (24). However, there is no obvious selective advantage in maintaining the integrity of enFeLV sequences in toto. Thus, it seems likely that enFeLV-AGTT represents integration following an evolutionarily recent infection. In enFeLV-GGAG, the ORF of env is disrupted by a frameshift mutation in the coding region for the gp70 protein (arrow in Fig. 2) (10). This site (residue 200) contains a succession of nine cytosines in the undisrupted enFeLV-AGTT coding sequence. In enFeLV-GGAG, a 10th cytosine is present in the poly(C) region, presumably resulting after strand slippage during DNA replication. Another mutation disrupts the putative start codon for env in enFeLV-GGAG.
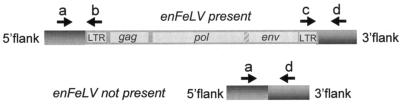
A polymorphic distribution (i.e., integrated virus versus empty chromosomal sites) of enFeLVs has been suggested by Southern blotting (17). We determined the distribution among cats of enFeLV-AGTT and enFeLV-GGAG by using one PCR primer based on the genomic DNA flanks unique to each individual enFeLV, with a second primer based on the proviral sequence (Fig. 3 and Appendix). Cats from different genetic backgrounds and geographic origins, including individuals from nine recognized breeds, were screened for the presence of enFeLV-AGTT and enFeLV-GGAG (Table 1 and Appendix). Among the 79 domestic cats screened, enFeLV-AGTT was present in 7 individuals (8.9% of the cats, 4.4% of the chromosomes) (Table 1). Two Egyptian Mau cats and two of three Turkish Van cats had enFeLV-AGTT. Three nonbreed cats, including a feral cat from Australia, were also found to have enFeLV-AGTT. All seven of the cats were heterozygous for the presence of enFeLV-AGTT, since a PCR designed to span the proviral integration site was also successful, indicating that enFeLV-AGTT was present in only one of the two sister chromosomes. Because enFeLV-AGTT is found in a small minority of domestic cats, it may have been absent from the cell lines used in induction studies, in which FeLVs could not be induced from enFeLVs (3, 19, 42). While this result was attributed to deletions or mutations in enFeLVs, induction studies have not been attempted with cell lines screened for the presence of the undisrupted enFeLV-AGTT provirus, which reopens the question of whether some enFeLVs might be replication competent.
Among the 79 domestic cats (Table 1 and Appendix), enFeLV-GGAG was present in 12 (15.2% of the cats, 8.2% of the chromosomes), including 4 of the 6 Persian cats screened and 1 of 2 Siamese cats (Table 1). The other seven enFeLV-GGAG-positive cats were nonbreed cats or of unknown lineage. One cat, Fca 215, was homozygous for the presence of the provirus, as repeated attempts to amplify an unintegrated site, with different combinations of primers corresponding to the 5′ and 3′ sequences flanking enFeLV-GGAG, were unsuccessful. No cats carried both enFeLV-AGTT and enFeLV-GGAG, although both were discovered in the original cat used to construct the genomic library.
We also screened for the presence of enFeLV-AGTT and enFeLV-GGAG in individuals from wild Felis species of the domestic cat lineage, which are known to carry enFeLVs (Appendix) (3, 21, 23). The presence of enFeLVs in only these species of felids had suggested that enFeLVs entered the germ line of a common ancestor of the domestic cat lineage before the lineage radiated (2, 3, 17, 27), i.e., millions of years ago (23). Neither enFeLV-AGTT nor enFeLV-GGAG was found to be present in any of the wild cats tested with multiple primer pairs, although primers spanning the proviral integration site readily amplified both in Felis species and in more distantly related Felidae species (Appendix). The absence of enFeLV-AGTT among wild cats and lack of fixation among domestic cats raise the possibility that integration of enFeLV-AGTT occurred subsequent to the domestication of cats. The viruses that produced enFeLVs were not thought to have persisted except as molecular “fossils” in the genome of the ancestor of the domestic cat lineage (27). The enFeLV-AGTT provirus suggests more recent persistence of these FeLVs among domestic cats or related Felis species. Since enFeLVs may derive from rodent viruses (2, 3), the possibility that viruses emerged from rodents on multiple occasions also cannot be excluded.
Retroviral 5′ and 3′ LTRs are identical in sequence at the time of integration, although random mutation would causes proviral 5′ and 3′ LTR sequences to gradually drift apart after incorporation into the host germ line (16). The 5′ and 3′ LTRs of enFeLV-GGAG were different from each other at two nucleotide sites, while the 5′ and 3′ LTR sequences of enFeLV-AGTT are identical, which suggests that integration of the latter provirus occurred relatively recently in the evolutionary history of cats. The substitution rate for endogenous LTRs in humans, apes, and Old World monkeys has been estimated to be 2.28 to 5.00 substitutions per site per 109 years (16). This rate was used to estimate the length of time after integration in which an initial difference would be expected to appear between 5′ and 3′ LTRs (9, 16, 22, 38, 40, 46). Adjusting for the longer LTRs in feline (1,136-bp combined LTRs in enFeLV-AGTT) versus human (HERV-K113; 970-bp combined LTRs) endogenous retroviruses, the first difference between enFeLV LTRs would be expected within 170,000 to 385,000 years after proviral integration (16, 46), although this estimate does not adjust for shorter generation times in cats versus primates. If we consider a divergence rate estimate for noncoding regions of the domestic cat genome of 1.2% per 106 years (20), then the LTRs would diverge, on average, by 74,000 years. Both estimates suggest that enFeLV-AGTT integration occurred after the radiation of species within the domestic cat lineage began more than two million years ago (21, 23).
The date estimates refer to the appearance of an initial difference between LTRs; the proviral integration could be considerably more recent. A lower limit for the integration date of enFeLV-AGTT may be suggested by its presence in the genomes of seven domestic cats, including Turkish Van cats derived from the Near East (25), nonbreed cats from the United States, and a feral cat from Australia. Similarly, the enFeLV-GGAG provirus was found among the Persian breed derived from Middle Eastern cats, Siamese cats derived from Southeast Asia (25), and nonbreed cats from the United States. If the proviruses entered the genome once in a common ancestor of the cats in which they are now found, sufficient time must have elapsed for gene flow or population expansion across the broad geographic areas represented. The alternative, that the seven cats represent more recent multiple independent viral integrations into the same site on the genome, is unlikely given nonspecific integrations of retroviruses into the genome (34) and the presence of only 6 to 12 enFeLV copies per haploid genome (4, 17, 28, 30, 31).
The presence of enFeLV-AGTT and -GGAG in only 8.9 and 15.2%, respectively, of domestic cats also suggests that the genomic location and distribution of enFeLVs in general may be quite diverse. The total number of enFeLV integration sites may be larger than previously reported, with only a small proportion present in any individual cat. Identification of additional enFeLV integration sites will determine whether other enFeLVs are found in a greater proportion of cats, whether some are restricted to a subset of the domestic cat lineage species, and whether any enFeLVs are present universally within the domestic cat lineage. Since the presence of enFeLVs at the same genomic location in two individuals is an indication of common ancestry, enFeLVs may prove useful as genetic markers for establishing relationships among individuals, lineages, and species within the genus Felis.
Nucleotide sequence accession numbers.
The sequences of the novel enFeLVs described here have been deposited in the GenBank database (accession numbers AY364318 and AY364319).
Acknowledgments
We thank E. R. Wilson for outstanding assistance. We also thank W. J. Murphy, E. Eizirik, M. Menotti-Raymond, R. Benveniste, D. Blair, A. Brandt, S. Cevario, V. David, B. Gough, M. Gough, J. Martenson, G. Pei, A. Robert, and A. Snyder.
This publication was funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract N01-CO-12400.
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Appendix
For each of the previously published sequences used in phylogenetic analyses, the accession number is included as part of the name.
Felids with neither enFeLV-AGTT nor enFeLV-GGAG present were as follows: domestic cat, Felis catus, by breed or locale, Abyssinian breed, Fca 567, 618, and 641; American Shorthair, Fca 326, 327, 329, and 391; Birman, Fca 620 and 626; Burmese breed, Fca 9, 364, 368, 376, 379, 381, 382, 382, 384, 385, 386, 387, 388, 389, and 390; Havana Brown, Fca 792; Japanese Bobtail, Fca 599, 600, and 603; Persian breed, Fca 1061 and 1067; Russian Blue, Fca 1094 and 1095; Siamese breed, Fca 559; Turkish Van, Fca 583; Argentina, Fca 157; Australia (feral), Fca 168 and 169; Britain, Fca GWK, GW1, and TB4; Costa Rica, Fca 150; Russia, Fca 140; United States, Fca 12, 17, 18, 21, 23, 24, 39, 52, 122, 123, 132, 133, 136, 186, 223, 264, 265, and FAS13. Wild species of the domestic cat lineage: Felis bieti, Chinese mountain cat, Fbi 2; Felis chaus, jungle cat, Fch 1, 2, 4, and 5; Felis lybica, African wild cat, Fli 3; Felis margarita, sand cat, Fma 5, 8, 10, 11, and 13; Felis nigripes, black-footed cat, Fni 3, 4, 5, 6, and 14; Felis silvestris, European wild cat, Fsi 1, 6, 7, 9, 13, 18, 21, and 25. Other wild felid species: Herpailurus yagouaroundi, jaguarundi, Hya 12; Leopardus wiedii, margay, Lwi 19 and 70; Lynx pardinus, Iberian lynx, Lpa 11; Lynx rufus, bobcat, Lru 38 and 43; Otocolobus manul, Pallas cat, Oma 3, 4, 5, 10, 14, and 15; Panthera leo, lion, Ple 7; Panthera uncia, snow leopard, Pun 13; Puma concolor, puma, Pco 333.
The PCR primers used to generate the pKHR2-gpe DNA for library screening were GA-GAG-F1 (ATGGGCCAAACTATAACTACCC) and GA-ENV-R1 (TGGTCGGTCCGGATCGTATTGC). The long PCR used to isolate each LTR on a separate DNA fragment used one primer based on the left phage arm (FIXII-LA; GCGGCCGCGAGCTCTAATACGA) or the right phage arm (FIXII-RA; GCGGCCGCGAGCTCAATTAACC) and a second primer based on the enFeLV pol sequence, in either the forward (POL-F8XL; ACCRAGGRAAAACTATAATGCCTGA) or the reverse (POL-R8XL; GCCCAGCCAGAGAAGGTGTCTAT) direction. PCR screening for the presence of enFeLV-AGTT was done with primers 6FL5-F1 (CCTTGATTAGAAGGTAAGGT) and LTR-R4 (CTCAGCAAAGACTTGCGC), primers LTR-F8 (AAACAGGATATCTGTGGTCA) and 6FL3-R4 (ATTCCTTACTAACACTGGAT), primers 6-5F11 (CCCCRGGTTGTGAGGAAAT) and LTR-R21 (CRGGTGGCTGACCACAGATA), or primers LTR-F22 (GCGCAAGTCTTTGCTGAG) and 6-3R11 (TGAAACTCAGAAAGAAGCAGAGG). For absence of enFeLV-AGTT, the primer combinations used were 6FL5-F3 (CTTCAGTGCATACAACAGG) and 6FL3-R3 (TTCAGATTTGAAAGATTAGTCA), 6FL5-F3 and 6FL3-R4 (ATTCCTTACTAACACTGGAT), 6FL5-F4 (TTCTCAGTGGGGCAGTGT) and 6FL3-R3, and 6-5F11 and 6-3R11. For the presence of enFeLV-GGAG, the primers used were LTR-F8 and 16FL3-R4 (CAACTCCTTTGTACGTCG), 16FL5-F3 (TGGCAGAACAGTGATTGAA) and LTR-R4, 16-5F11 (TTTCCAGAGACAGACYGTGA) and LTR-R21, and LTR-F22 and 16-3R11 (AAGGAGACCTCTAAGGTGAAGC). The primers used to test for the absence of enFeLV-GGAG were 16FL5-F1 (AACACAAACCACAGTACAA) and 16FL3-R4, 16FL5-F3 and 16FL3-R4, 16FL5-F4 (CTCTCCCACTTGTGCTCT) and 16FL3-R3 (CCTCAGCTTTGTTCTACG), and 16-5F11 and 16-3R11. All results were verified by sequencing and by repetition of screens with a second pair of primers.
For phylogenetic analyses, exhaustive searches were performed in all cases except ML analysis for env and LTRs, which used 50 replicates of heuristic searches with random taxon addition and tree bisection-reconnection (TBR) branch swapping. NJ analyses were performed with Kimura-2 parameter distances. All analyses of gag used a partial sequence consisting of an alignment of 322 characters at the 5′ end of the gag region (of which 46 were parsimony informative in the MP analysis). Full-length sequences were used for pol (3,633 characters, 153 parsimony informative), for env (2,059 characters, 538 parsimony informative), and for LTRs (570 characters, 82 parsimony informative). MP analyses treated gaps as a fifth state. ML analyses used empirical base frequencies, with program estimation of transition/transversion ratios (estimates: gag = 5.740359, pol = 8.161742, env = 3.265367, LTRs = 1.720779), of proportion invariable sites (estimates: gag = 0.0801995, pol 0.587492, env = 0.272717, LTRs = 0.74054), and of the α parameter for the γ distribution of the rate variation among sites (estimates: gag = infinity, pol = 0.885470, env = 1.288299, LTRs = 0.750109). Bootstrap resampling support was based on 100 (ML) or 1,000 (NJ and MP) iterations, with heuristic searches and TBR branch swapping (ML and MP), with starting trees generated by NJ (for ML bootstrap) or by simple stepwise addition (MP). Sequence alignments and further details of the methods used are available at http://home.ncifcrf.gov/ccr/lgd.
FIG. 1.
Phylogenetic analyses of proviral regions from enFeLV-AGTT and enFeLV-GGAG and sequences in the GenBank database. ML trees are depicted, drawn by midpoint rooting, with bootstrap support (100 iterations) listed above branches for nodes supported by all three methods: NJ (left), MP (middle), and ML (right). The novel sequences enFeLV-AGTT and -GGAG are compared to previously published sequences labeled with their GenBank accession numbers. FeLVA, exFeLV subgroup A. (A) Analyses of sequences from the pol viral region demonstrate that enFeLV-AGTT and enFeLV-GGAG are more closely related to enFeLV than to exFeLV sequences. The full-length sequence was used to generate the tree for pol (3,633 bp). The score (−ln likelihood) of the best ML tree was 6,421.51427; the same tree topology was produced by NJ and MP (best tree found by MP: length = 277, consistency index [CI] = 0.968, retention index [RI] = 0.941). (B) ML tree for the full-length (570-bp) proviral LTRs of enFeLVs reveals their subdivision into two sets of sequences, designated groups I and II. The U3 regions of the LTRs of exFeLVs are too dissimilar for alignment with those of endogenous LTRs; thus, these were excluded from this analysis. ML tree −ln likelihood = 1,004.08144. Subdivision of endogenous LTRs into two groups was also supported by NJ and MP analyses, which generated the same tree topology (best tree found by MP: length = 102, CI = 0.941, RI = 0.969).
FIG. 2.
Structure of enFeLV proviruses enFeLV-AGTT and enFeLV-GGAG compared to that of exFeLV (GenBank accession no. AF052723). The three horizontal segments represent the three reading frames for each FeLV sequence; long ORFs are highlighted in black, with corresponding gag, pol, and env regions displayed above. While enFeLV-AGTT (middle) has intact ORFs reminiscent of exFeLV (top), a frameshift mutation (arrow) disrupts the ORF of env in enFeLV-GGAG (bottom).
FIG. 3.
PCR screening strategy for detecting enFeLV proviruses enFeLV-AGTT and enFeLV-GGAG in cats. Primers were designed on the basis of sequences within the enFeLV (primers b and c) or in cat genomic sequence flanking the proviral integration site (primers a and d). If the enFeLV was present (top), primers a and b or primers c and d would amplify short DNA segments but primers a and d would not. If the enFeLV was not present (bottom), then primers a and d would amplify but the other combinations would not. If only one of the two chromosome homologues contained the enFeLV, then all of the primers would amplify a PCR product.
TABLE 1.
Domestic cats with enFeLV-AGTT or -GGAGa
| Reference no. | enFeLV-AGTT | enFeLV-GGAG | Breed or locale |
|---|---|---|---|
| Fca 138 | +/− | −/− | Egyptian Mau |
| Fca 146 | +/− | −/− | Egyptian Mau |
| Fca 569 | +/− | −/− | Turkish Van |
| Fca 828 | +/− | −/− | Turkish Van |
| Fca 171 | +/− | −/− | Australia, feral |
| Fca 84 | +/− | −/− | United States |
| Fca 165 | +/− | −/− | United States |
| Fca 215 | −/− | +/+ | Persian |
| Fca 997 | −/− | +/− | Persian |
| Fca 1082 | −/− | +/− | Persian |
| Fca 1093 | −/− | +/− | Persian |
| Fca 761 | −/− | +/− | Siamese |
| Fca BW7 | −/− | +/− | Britain |
| Fca 38 | −/− | +/− | United States |
| Fca 42 | −/− | +/− | United States |
| Fca 127 | −/− | +/− | United States |
| Fca 129 | −/− | +/− | United States |
| Fca FAS11 | −/− | +/− | United States |
| Fca FAS5 | −/− | +/− | United States |
+/+, copy present on both chromosome homologues; +/−, copy present on one of two homologues; −/−, no copies present.
REFERENCES
- 1.Anderson, M. M., A. S. Lauring, C. C. Burns, and J. Overbaugh. 2000. Identification of a cellular cofactor required for infection by feline leukemia virus. Science 287:1828-1830. [DOI] [PubMed] [Google Scholar]
- 2.Benveniste, R. E. 1985. The contributions of retroviruses to the study of mammalian evolution, p. 359-417. In R. J. MacIntyre (ed.), Molecular evolutionary genetics. Plenum Press, New York, N.Y.
- 3.Benveniste, R. E., C. J. Sherr, and G. J. Todaro. 1975. Evolution of type C viral genes: origin of feline leukemia virus. Science 190:886-888. [DOI] [PubMed] [Google Scholar]
- 4.Benveniste, R. E., and G. J. Todaro. 1975. Segregation of RD-114 and FeLV-related sequences in crosses between domestic cat and leopard cat. Nature 257:506-508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berry, B. T., A. K. Ghosh, D. V. Kumar, D. A. Spodick, and P. Roy-Burman. 1988. Structure and function of endogenous feline leukemia virus long terminal repeats and adjoining regions. J. Virol. 62:3631-3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Busch, M. P., B. G. Devi, L. H. Soe, B. Perbal, M. A. Baluda, and P. Roy-Burman. 1983. Characterization of the expression of cellular retrovirus genes and oncogenes in feline cells. Hematol. Oncol. 1:61-75. [DOI] [PubMed] [Google Scholar]
- 7.Casey, J. W., A. Roach, J. I. Mullins, K. B. Burck, M. O. Nicolson, M. B. Gardner, and N. Davidson. 1981. The U3 portion of feline leukemia virus DNA identifies horizontally acquired proviruses in leukemic cats. Proc. Natl. Acad. Sci. USA 78:7778-7782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, H., M. K. Bechtel, Y. Shi, A. Phipps, L. E. Mathes, K. A. Hayes, and P. Roy-Burman. 1998. Pathogenicity induced by feline leukemia virus, Rickard strain, subgroup A plasmid DNA (pFRA). J. Virol. 72:7048-7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dangel, A. W., B. J. Baker, A. R. Mendoza, and C. Y. Yu. 1995. Complement component C4 gene intron 9 as a phylogenetic marker for primates: long terminal repeats of the endogenous retrovirus ERV-K(C4) are a molecular clock of evolution. Immunogenetics 42:41-52. [DOI] [PubMed] [Google Scholar]
- 10.Donahue, P. R., E. A. Hoover, G. A. Beltz, N. Riedel, V. M. Hirsch, J. Overbaugh, and J. I. Mullins. 1988. Strong sequence conservation among horizontally transmissible, minimally pathogenic feline leukemia viruses. J. Virol. 62:722-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elder, J. H., and J. I. Mullins. 1983. Nucleotide sequence of the envelope gene of Gardner-Arnstein feline leukemia virus B reveals unique sequence homologies with a murine mink cell focus-forming virus. J. Virol. 46:871-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hardy, W. D., Jr. 1992. Feline oncoretroviruses, p. 109-180. In J. A. Levy (ed.), The Retroviridae, vol. 2. Plenum Press, New York, N.Y.
- 13.Jarrett, O. 1992. Pathogenicity of feline leukemia virus is commonly associated with variant viruses. Leukemia 6:153S-154S. [PubMed] [Google Scholar]
- 14.Jarrett, O., M. C. Golder, S. Toth, D. E. Onions, and M. F. Stewart. 1984. Interaction between feline leukaemia virus subgroups in the pathogenesis of erythroid hypoplasia. Int. J. Cancer. 34:283-288. [DOI] [PubMed] [Google Scholar]
- 15.Jarrett, O., and P. H. Russell. 1978. Differential growth and transmission in cats of feline leukaemia viruses of subgroups A and B. Int. J. Cancer 21:466-472. [DOI] [PubMed] [Google Scholar]
- 16.Johnson, W. E., and J. M. Coffin. 1999. Constructing primate phylogenies from ancient retrovirus sequences. Proc. Natl. Acad. Sci. USA 96:10254-10260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koshy, R., R. C. Gallo, and F. Wong-Staal. 1980. Characterization of the endogenous feline leukemia virus-related DNA sequences in cats and attempts to identify exogenous viral sequences in tissues of virus-negative leukemic animals. Virology 103:434-445. [DOI] [PubMed] [Google Scholar]
- 18.Kumar, D. V., B. T. Berry, and P. Roy-Burman. 1989. Nucleotide sequence and distinctive characteristics of the env gene of endogenous feline leukemia provirus. J. Virol. 63:2379-2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Livingston, D. M., and G. J. Todaro. 1973. Endogenous type C virus from a cat cell clone with properties distinct from previously described feline type C virus. Virology 53:142-151. [DOI] [PubMed] [Google Scholar]
- 20.Lopez, J. V., M. Culver, J. C. Stephens, W. E. Johnson, and S. J. O'Brien. 1997. Rates of nuclear and cytoplasmic mitochondrial DNA sequence divergence in mammals. Mol. Biol. Evol. 14:277-286. [DOI] [PubMed] [Google Scholar]
- 21.Lopez, J. V., N. Yuhki, R. Masuda, W. Modi, and S. J. O'Brien. 1994. Numt, a recent transfer and tandem amplification of mitochondrial DNA to the nuclear genome of the domestic cat. J. Mol. Evol. 39:174-190. [DOI] [PubMed] [Google Scholar]
- 22.Mager, D. L., and J. D. Freeman. 1995. HERV-H endogenous retroviruses: presence in the New World branch but amplification in the Old World primate lineage. Virology 213:395-404. [DOI] [PubMed] [Google Scholar]
- 23.Masuda, R., J. V. Lopez, J. P. Slattery, N. Yuhki, and S. J. O'Brien. 1996. Molecular phylogeny of mitochondrial cytochrome b and 12S rRNA sequences in the Felidae: ocelot and domestic cat lineages. Mol. Phylogenet. Evol. 6:351-365. [DOI] [PubMed] [Google Scholar]
- 24.McDougall, A. S., A. Terry, T. Tzavaras, C. Cheney, J. Rojko, and J. C. Neil. 1994. Defective endogenous proviruses are expressed in feline lymphoid cells: evidence for a role in natural resistance to subgroup B feline leukemia viruses. J. Virol. 68:2151-2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morris, D. 1999. Cat breeds of the world. Viking, New York, N.Y.
- 26.Mullins, J. I., J. W. Casey, M. O. Nicolson, K. B. Burck, and N. Davidson. 1981. Sequence arrangement and biological activity of cloned feline leukemia virus proviruses from a virus-productive human cell line. J. Virol. 38:688-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mullins, J. I., and E. A. Hoover. 1990. Molecular aspects of feline leukemia virus pathogenesis, p. 87-116. In R. C. Gallo and F. Wong-Staal (ed.), Retrovirus biology and human disease. Dekker, New York, N.Y.
- 28.Niman, H. L., M. Akhavi, M. B. Gardner, J. R. Stephenson, and P. Roy-Burman. 1980. Differential expression of two distinct endogenous retrovirus genomes in developing tissues of the domestic cat. J. Natl. Cancer Inst. 64:587-594. [PubMed] [Google Scholar]
- 29.Okabe, H., J. DuBuy, R. V. Gilden, and M. B. Gardner. 1978. A portion of the feline leukaemia virus genome is not endogenous in cat cells. Int. J. Cancer 22:70-78. [DOI] [PubMed] [Google Scholar]
- 30.Okabe, H., J. DuBuy, M. Hatanaka, and R. V. Gilden. 1978. Reiteration frequency of feline type C viral genomes in homologous and heterologous host cell DNA. Intervirology 9:253-260. [DOI] [PubMed] [Google Scholar]
- 31.Okabe, H., E. Twiddy, R. V. Gilden, M. Hatanaka, E. A. Hoover, and R. G. Olsen. 1976. FeLV-related sequences in DNA from a FeLV-free cat colony. Virology 69:798-801. [DOI] [PubMed] [Google Scholar]
- 32.Overbaugh, J., N. Riedel, E. A. Hoover, and J. I. Mullins. 1988. Transduction of endogenous envelope genes by feline leukaemia virus in vitro. Nature 332:731-734. [DOI] [PubMed] [Google Scholar]
- 33.Pandey, R., A. K. Ghosh, D. V. Kumar, B. A. Bachman, D. Shibata, and P. Roy-Burman. 1991. Recombination between feline leukemia virus subgroup B or C and endogenous env elements alters the in vitro biological activities of the viruses. J. Virol. 65:6495-6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pryciak, P. M., A. Sil, and H. E. Varmus. 1992. Retroviral integration into minichromosomes in vitro. EMBO J. 11:291-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Riedel, N., E. A. Hoover, R. E. Dornsife, and J. I. Mullins. 1988. Pathogenic and host range determinants of the feline aplastic anemia retrovirus. Proc. Natl. Acad. Sci. USA 85:2758-2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roca, A. L., C. Godson, D. R. Weaver, and S. M. Reppert. 1996. Structure, characterization, and expression of the gene encoding the mouse Mel1a melatonin receptor. Endocrinology 137:3469-3477. [DOI] [PubMed] [Google Scholar]
- 37.Roy-Burman, P. 1995. Endogenous env elements: partners in generation of pathogenic feline leukemia viruses. Virus Genes 11:147-161. [DOI] [PubMed] [Google Scholar]
- 38.SanMiguel, P., B. S. Gaut, A. Tikhonov, Y. Nakajima, and J. L. Bennetzen. 1998. The paleontology of intergene retrotransposons of maize. Nat. Genet. 20:43-45. [DOI] [PubMed] [Google Scholar]
- 39.Sheets, R. L., R. Pandey, W. C. Jen, and P. Roy-Burman. 1993. Recombinant feline leukemia virus genes detected in naturally occurring feline lymphosarcomas. J. Virol. 67:3118-3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shih, A., E. E. Coutavas, and M. G. Rush. 1991. Evolutionary implications of primate endogenous retroviruses. Virology 182:495-502. [DOI] [PubMed] [Google Scholar]
- 41.Soe, L. H., B. G. Devi, J. I. Mullins, and P. Roy-Burman. 1983. Molecular cloning and characterization of endogenous feline leukemia virus sequences from a cat genomic library. J. Virol. 46:829-840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Soe, L. H., R. W. Shimizu, J. R. Landolph, and P. Roy-Burman. 1985. Molecular analysis of several classes of endogenous feline leukemia virus elements. J. Virol. 56:701-710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stewart, M. A., M. Warnock, A. Wheeler, N. Wilkie, J. I. Mullins, D. E. Onions, and J. C. Neil. 1986. Nucleotide sequences of a feline leukemia virus subgroup A envelope gene and long terminal repeat and evidence for the recombinational origin of subgroup B viruses. J. Virol. 58:825-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Swofford, D. L. 1998. PAUP*4.0b2: phylogenetic analysis using parsimony. Sinauer, Sunderland, Mass.
- 45.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Turner, G., M. Barbulescu, M. Su, M. I. Jensen-Seaman, K. K. Kidd, and J. Lenz. 2001. Insertional polymorphisms of full-length endogenous retroviruses in humans. Curr. Biol. 11:1531-1535. [DOI] [PubMed] [Google Scholar]
- 47.Tzavaras, T., M. Stewart, A. McDougall, R. Fulton, N. Testa, D. E. Onions, and J. C. Neil. 1990. Molecular cloning and characterization of a defective recombinant feline leukaemia virus associated with myeloid leukaemia. J. Gen. Virol. 71:343-354. [DOI] [PubMed] [Google Scholar]