Abstract
Vascular endothelial cells in vivo are exposed to multiple biophysical cues provided by the basement membrane, a specialized extracellular matrix through which vascular endothelial cells are attached to the underlying stroma. The importance of biophysical cues has been widely reported, but the signaling pathways that mediate cellular recognition and response to these cues remain poorly understood. Anisotropic topographically patterned substrates with nano- through microscale feature dimensions were fabricated to investigate cellular responses to topographic cues. The present study focuses on early events following exposure of human umbilical vein endothelial cells (HUVECs) to these patterned substrates. In serum-free medium and on substrates without protein coating, HUVECs oriented parallel to the long axis of underlying ridges in as little as 30 min. Immunocytochemistry showed clear differences in the localization of the focal adhesion proteins Src, p130Cas, and focal adhesion kinase (FAK) in HUVECs cultured on topographically patterned surfaces and on planar surfaces, suggesting involvement of these proteins in mediating the response to topographic features. Knockdown experiments demonstrated that FAK was not necessary for HUVEC alignment in response to topographic cues, although FAK knockdown did modulate HUVEC migration. These data identify key events early in the cellular response to biophysical stimuli.
Keywords: surface topography, focal adhesion kinase, vascular endothelial cells, cell signaling
cardiovascular disease (CVD) remains a leading cause of death within the United States, as well as globally, causing over one-third of all deaths in 2007 alone (35). Despite extensive research efforts, there remain knowledge gaps regarding CVD onset and progression. In the past, most studies focused on the influence of soluble signaling factors on vascular endothelial cell behavior; more recently, the impact of biophysical cues on cell biology has become apparent. The microenvironment of cells in vivo is characterized by a rich, “felt-like” topography that induces a cascade of signaling events that ultimately participate in determining cell phenotype. The importance of biophysical cues in fundamental cell behaviors, including shape (22), adhesion (20), proliferation (22, 25, 33), migration (9, 27, 34), cell alignment (1, 43–45), and survival (33), has been widely reported. These studies demonstrate that biophysical cues from the tissue microenvironment modulate cellular functions that are distinct from biochemical stimuli. However, the signaling processes that enable cells to identify and respond to topographic cues remain largely unknown. Topographically patterned substrates with anisotropically ordered features of parallel ridges and grooves in the biomimetic nano- through microscale range have been used to study the impact of topographic cues on a variety of cell types (20).
Vascular endothelial cells in vivo are intimately associated with a basement membrane (BM). The BM is a specialized extracellular matrix (ECM) possessing a complex three-dimensional meshwork consisting of pores and fibers in the submicroscale (100–1,000 nm) and nanoscale (1–100 nm) range, as shown for the BM from a variety of species and anatomic sites (1–3, 24, 26). The ECM is linked to the cytoskeleton by focal adhesions, multiprotein complexes that are formed upon binding of integrin family transmembrane receptors to extracellular ligands. Aside from their structural roles, focal adhesions are suggested to be involved in the transmission of extracellular cues to intracellular signals. Binding of integrin family transmembrane receptors to extracellular ligands upon contact with the cell surface initiates integrin clustering, which leads to the recruitment of focal adhesion kinase (FAK) to these focal adhesions. FAK then binds indirectly to integrins through interactions with other integrin-associated proteins (16, 39).
FAK, encoded by the protein tyrosine kinase (PTK) 2 (PTK2) gene, is an intracellular nonreceptor PTK that is activated through autophosphorylation at Tyr397 (38). This phosphorylation induces several signaling cascades (41) by initiating the recruitment of multiple structural and signaling proteins, including vinculin (4), paxillin (1, 17, 40, 46), talin (7), p130Cas (6), and Src family kinases (8). The known cellular functions of FAK include control of the cell cycle (52, 53), survival (28), and proliferation (33). Fibroblasts isolated from FAK-null mice displayed decreased motility, indicating involvement of FAK in cell migration (19, 41). Additionally, FAK has been proposed to act as a mechanosensor on the basis of reports documenting reduced responses of FAK-null fibroblasts to both traction forces applied to flexible substrates (49) and differences in response to substrate stiffness compared with wild-type fibroblasts (33). The goal of this study was to describe early responses of human umbilical vein endothelial cells (HUVECs) to topographically patterned surfaces and to investigate initial cell-signaling processes that enable HUVECs to recognize and react to biologically relevant topographic cues.
MATERIALS AND METHODS
Fabrication of Topographically Patterned Substrates With Anisotropic Features
Patterned silicon chips containing ridge and groove features were fabricated using X-ray lithography, as previously described (20, 27). The silicon masters were fabricated containing either an array of six (2 × 2 mm) ridge and groove areas with pitches of 400, 800, 1,200, 1,600, 2,000, and 4,000 nm (pitch = groove + ridge width) separated by planar control areas (i.e., “6-packs”) or a single 6.5-cm2 area with a single pitch (planar 400-, 1,400-, and 4,000-nm pitch, i.e., “monotypic”). The dimensions of the various topographies were of equal ridge and groove width, with a groove depth of 300 nm. These silicon masters were used as templates for replication of the patterns through soft lithography using poly(dimethylsiloxane) stamps (20). The pattern was stamped into a spun-coat layer of NOA81 optical adhesive (Norland Products, Cranbury, NJ) (32) onto 35-mm (for 6-packs) or 60-mm (for monotypics) tissue culture plates and cured in a UV cross-linker (model XL-1500, Dymax, Torrington, CT) under 365-nm light for 100 min. NOA81, a proprietary mercaptoester compound that cures to a rigid polymer upon exposure to UV light, has previously been demonstrated by our research group to be a suitable material for cell culture (25, 27). In preparation for tissue culture, all polyurethane surfaces were sterilized by 20 min of exposure to UV (280-nm) light.
Cell Culture
Primary HUVECs (Lonza, Walkersville, MD) were cultured in endothelial basal medium-2 supplemented with the EGM-2 SingleQuot Kit containing GA-1000, human EGF, 2% fetal bovine serum, heparin, ascorbic acid, R3-IGF-1, VEGF, human FGF-B, and hydrocortisone (Lonza). Cell cultures were incubated at 37°C and 5% CO2 and used between passages 4 and 7 for all experiments.
Gene Downregulation by Small Interfering RNA Transfection
At 60–80% confluence of HUVECs, small interfering RNA (siRNA) transfections were performed using the DharmaFect 4 transfection reagent (Dharmacon, Lafayette, CO) following the manufacturer's instructions, with final concentrations of 28.5 nM FAK siRNA (catalog no. Hs_PTK2_11, Qiagen, Valencia, CA) or control siRNA (ON-TARGETplus Nontargeting siRNA #3, Dharmacon). At 48–120 h posttransfection, the cells were harvested for RNA isolation. Knockdown to expression levels <20% was achieved as determined by real-time quantitative PCR (qPCR) analyses.
RNA Isolation and Real-Time qPCR Analysis
HUVECs were seeded at 2.5 × 105 cells/6.5 cm2 on topographically patterned substrates (400-, 1,400-, and 4,000-nm pitch) and planar control surfaces. After 12 h of incubation, the cells were harvested for RNA isolation using the RNeasy kit (Qiagen) following the manufacturer's instructions. The expression levels of FAK in cells cultured on topographically patterned surfaces were determined by real-time qPCR analyses using a StepOne qPCR machine (Applied Biosystems, Carlsbad, CA). Sixty nanograms of RNA were used to determine expression levels of FAK and 18S rRNA using the TaqMan One-Step PCR kit and aptamers specific to FAK and 18S rRNA (catalog nos. Hs00178587_m1 and Hs99999901_m1, respectively, Applied Biosystems). The reverse transcription reaction was performed for 30 min at 50°C followed by PCR enzyme activation for 10 min at 95°C. Forty cycles of 60°C for 1 min followed by 95°C for 15 s were performed. Relative expression levels of the genes of interest were normalized to the expression of 18S rRNA. The experiment was performed in triplicate and repeated three times.
Protein Isolation and Western Blotting
Protein from HUVECs was isolated by addition of M-PER buffer (Thermo Scientific, Rockford, IL) with Halt Protease Inhibitor Cocktail (Thermo Scientific) for 10 min on ice. Cell debris was removed by centrifugation at 14,000 rpm for 5 min at 4°C. The protein concentration was determined using the DC Protein Assay (Bio-Rad, Hercules, CA). The samples were prepared for electrophoresis by incubation at 95°C for 5 min after addition of 5× Laemmli buffer. Equal amounts of protein (10 μg/lane) were loaded on a 4–12% NuPAGE Bis-Tris gel (Life Technologies, Carlsbad, CA), subjected to electrophoresis, and blotted onto a nitrocellulose membrane (Life Technologies). The blot was blocked in milk diluent (KPL, Gaithersburg, MD) and then incubated overnight with primary antibodies specific for total FAK (mouse anti-FAK, 1:50 dilution; BD Biosciences, Franklin Lakes, NJ) and β-actin (chicken anti-β-actin, 1:1,000 dilution; Abcam, Cambridge, MA) diluted in 10% milk diluent (KPL). Secondary antibodies (peroxidase-labeled goat anti-mouse or goat anti-chicken, KPL) were used at a 1:20,000 dilution. Protein bands of interest were detected using the chemiluminescent ECL Plus Western blotting reagent (GE Healthcare, Pittsburgh, PA) and a charge-coupled device camera (ImageQuant 350, GE Healthcare).
Time-Lapse Microscopy and Analysis of Migration
For the migration assay, 35-mm dishes with two separate chambers, each containing one 6-pack, were fabricated to allow simultaneous imaging of control and FAK siRNA-transfected cells. At 48 h after transfection, 3 × 104 cells were seeded into each chamber and allowed to adhere in an incubator for 3–4 h at 37°C and 5% CO2. Plates were transferred to the incubated stage of a Zeiss microscope to maintain cell culture conditions.
Areas containing each topographically patterned region (400- to 4,000-nm pitch) and planar control were identified and marked using AxioVision software (version 4.6, Carl Zeiss) and an automated stage. Sequential phase-contrast images of each marked area were taken every 10 min over a 10-h period at ×10 magnification (0.4 numerical aperture objective) using a Zeiss Axiovert 200 M microscope. Only cells that remained within the field of view and did not come into contact with another cell or undergo cell division during the course of the experiment were analyzed. The center of mass of each cell was identified, and the x and y coordinates were recorded using a cell tracker module in AxioVision 4.6. From the resulting data, cell trajectories, total distance, and the rate of migration were calculated for a total of ≥40 FAK or control siRNA-transfected cells per topographic feature or planar control from ≥3 separate experiments. As a control, the analyses were checked in two experiments with untreated cells on topographic surfaces or planar control areas (≥32 cells per topography or planar control). Results of these experiments showed that the control siRNA-treated cells had migration characteristics similar to those of untreated cells (data not shown).
Analysis of Cell Orientation to Topographic Features
HUVECs were plated at a density of 1.5 × 104 cells/cm2 in 35-mm plates containing 6-pack polyurethane substrates with topographic features ranging from 400- to 4,000-nm pitch, as well as intervening planar regions. After 4 h, cells were fixed for 20 min with 2% paraformaldehyde in PBS, permeabilized with 0.1% Triton X-100 (Fisher Scientific, Waltham, MA) in PBS, and stained with Alexa Fluor 568-phalloidin (Invitrogen, Carlsbad, CA) and 4′,6-diamidino-2-phenylindole (BioGenex, San Ramon, CA). Images were taken at ×10 magnification with an Axiovert 200 M inverted microscope.
Orientation of individual cells was analyzed from fluorescent images using AxioVision 4.6 software, which allowed us to define the boundary of the cell and the orientation of the cell in relation to the underlying pitch. Only cells that were fully contained within the border of the image, not in physical contact with other cells, and not undergoing mitosis were included for analysis. The orientation of the cells was based on the angle between the major axis of the object and the underlying feature. Cells were considered aligned parallel with the ridges and grooves when this angle was between 0° and 10° and perpendicular when the angle was between 80° and 90°. Cell elongation is defined as the ratio of the length to the width of each cell. Cells were considered elongated if this factor was >1.3. A minimum of 50 cells for a given pitch size were analyzed in four replicates.
To ensure that the transfection method had no impact on the ability of the cell to orient to topographic cues, we additionally included untreated cells and mock-transfected cells, i.e., cells that were transfected with water instead of siRNA. No significant differences were found in these groups in their response compared with control siRNA-transfected cells (data not shown).
Immunocytochemistry and Fluorescence Microscopy
HUVECs were fixed with 3% paraformaldehyde and permeabilized in 0.05% Triton X-100 (Fisher Scientific) for 3.5 min. Nonspecific binding sites were blocked with 1 mg/ml bovine serum albumin overnight at 4°C. The cells were incubated with primary antibody for 2 h at room temperature and then with secondary fluorophore-conjugated antibodies (Alexa 488- and/or Alexa 555-conjugated goat anti-mouse or goat anti-rabbit, Invitrogen). Primary antibodies were anti-phosphotyrosine (Millipore, Billerica, MA), monoclonal rabbit anti-phosphorylated (Tyr397) FAK (Invitrogen), anti-phosphorylated (Tyr416) Src (Millipore), and anti-phosphorylated (Tyr165) p130Cas (Cell Signaling, Danvers, MA). A ×63 objective (1.4 numerical aperture) was used to acquire immunofluorescent z stacks of HUVECs on topographically patterned and planar surfaces. The z stacks were iteratively deconvolved with theoretical point-spread functions using the AxioVision Deconvolution Module. Other images were acquired as standard epifluorescent images with use of a ×40 objective as indicated.
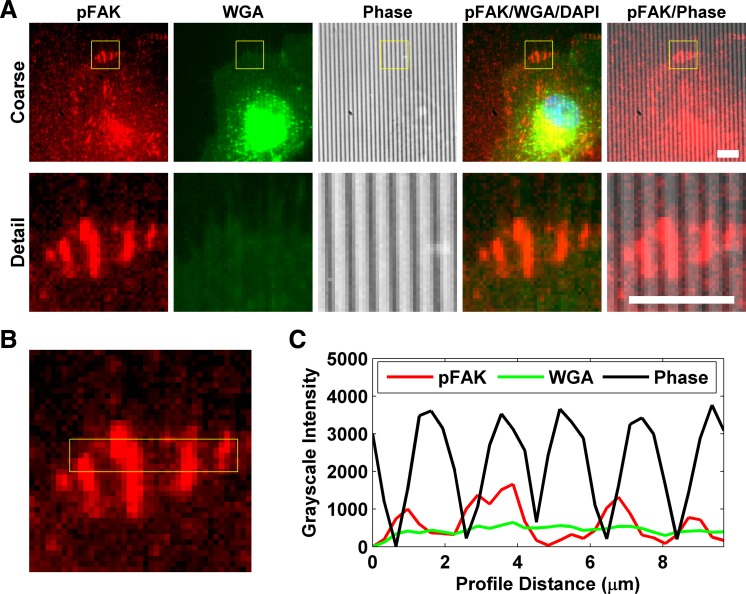
To ensure that the topographic surfaces did not induce an imaging artifact, cells grown on patterned and planar substrates were exposed to a general cell stain; specifically, they were costained with 4′,6-diamidino-2-phenylindole, phosphorylated FAK, and wheat germ agglutinin (WGA; Fig. 1A). Cells were fixed in 4% paraformaldehyde for 20 min and washed three times in PBS. WGA conjugated to Alexa Fluor 488 (Life Technologies) was added to cells at 5 μg/ml in PBS for 30 min. The cells were washed three times in PBS, permeabilized with 0.2% Triton X-100 in PBS for 5 min, washed three times, and stained as described above.
Fig. 1.
Analysis of staining specificity. A: representative human umbilical vein endothelial cells (HUVECs) on the edge of a topographically patterned region labeled with phosphorylated FAK (pFAK; red) and wheat germ agglutinin (WGA; green). 4′,6-Diaminido-2-phenylindole (DAPI) was used as nuclear counterstain. The 2,000-nm pitch ridges are clearly visible on phase and can be seen as a faint nonspecific signal on the fluorescent channels. Yellow box on the coarse view corresponds to the region in the detail view. Scale bars, 10 μm. B: enlarged “detail” view of the pFAK stain. Yellow rectangle denotes a 3-pixel-wide strip that was used to generate a profile of grayscale intensities of the pFAK signal (red line in C). The 3 focal adhesions captured by the strip are clearly visible as peaks in the profile. C: similar profiles of the same strip were generated for the WGA and phase signals. The pFAK signal intensifies at the rising of the bright phase peaks, which indicates ridges; in contrast, the WGA signal shows only faint variation across the ridges. All grayscale values were zeroed to the minimum value of the profile.
The WGA label is clearly visible in areas of ridges and grooves, consistent with previous reports showing that the membrane spans the grooves and that the primary points of cell attachment are on the ridges (21, 29, 30, 36, 45). This further demonstrates that there is minimal fluorescent artifact from the underlying topography. We used ImageJ software (42) to measure the grayscale intensity of WGA, phosphorylated FAK, and the phase image along the ridges and grooves of a representative area of the image (yellow box in Fig. 1B), and a graphical representation of this measurement was prepared (Fig. 1C). The resulting profile shows bright peaks of the phase grayscale intensities corresponding to the ridges of the underlying patterned substrate. The three focal adhesions captured in this image section appear as peaks in the profile that coincide with the phase peaks at the ridges. In contrast, the membrane staining intensity remains relatively stable along the ridges and grooves, reflecting the consistent presence of the membrane along the pattern.
Statistical Analysis
All statistics were performed using SigmaPlot 11 (Systat Software, San Jose, CA). Two-way ANOVAs were used to determine the differences of two varying factors (topography and time or silencing), unless stated otherwise. The Holm-Sidak post hoc test was used to determine significant difference within a factor from control (e.g., planar surfaces and transfections with control siRNA).
RESULTS AND DISCUSSION
Cellular Responses to Topographic Cues Do Not Depend on Adsorbed Proteins
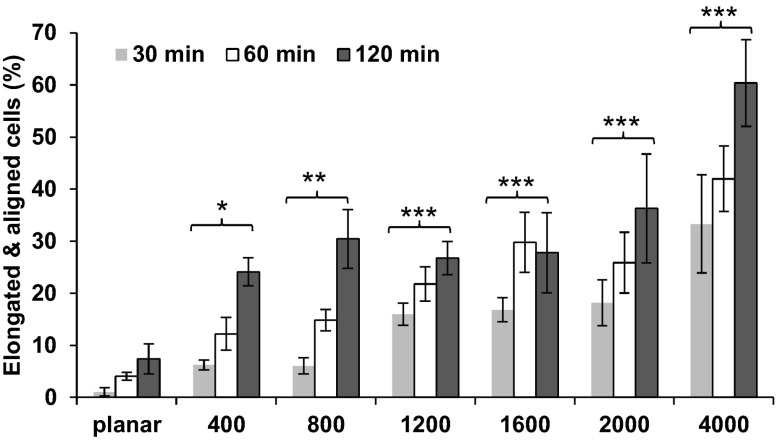
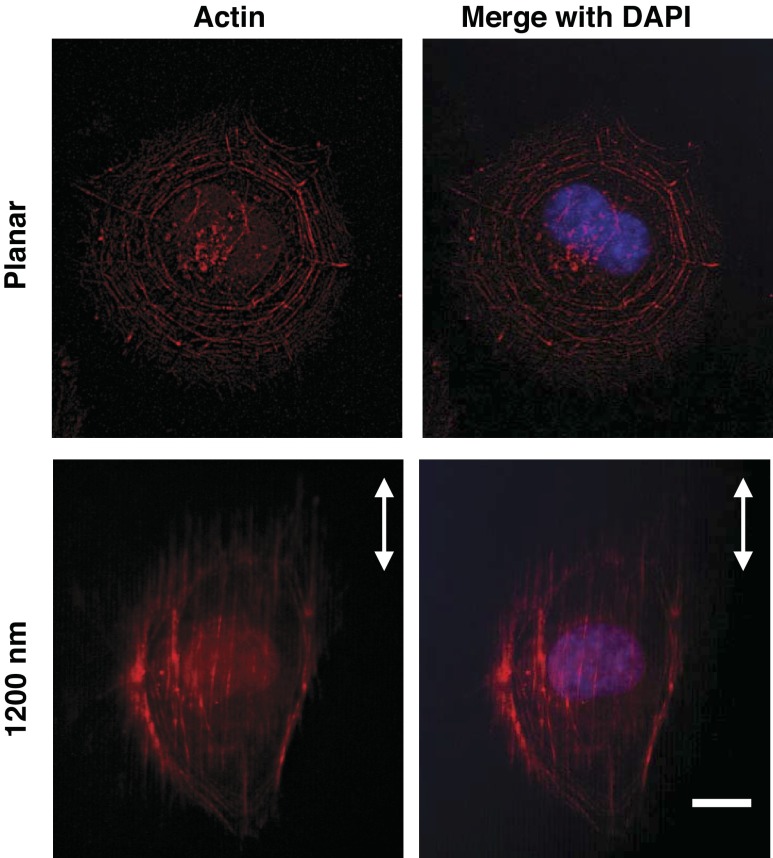
Vascular endothelial cells have long been known to respond to various biophysical cues (11a, 14, 27, 29, 50), including topographic features. It remains unknown, however, if the cells were responding specifically to the underlying topographic features or to protein patterns created through deposition on the features. We therefore tested the response of HUVECs to topographic surfaces in the absence of adsorbed protein. After trypsinization, HUVECs were washed thoroughly with PBS and then plated in protein-free basal medium without additives on 6-pack topographically patterned surfaces, as well as on planar control areas. Morphology was imaged in cells fixed at 30, 60, 90, and 120 min postplating and stained with fluorescent phalloidin. Even within 30 min of exposure to the patterned surface, HUVECs responded to the topographic cues by preferentially elongating and aligning along the ridges and grooves, in the absence of added protein (Fig. 2). As time progressed, an increasing number of cells elongated and aligned. We observed a significant difference between 30 and 120 min, but not between 30 and 60 min, postplating (Fig. 2), demonstrating the temporal component of the cell alignment. At <30 min postplating, we observed no substantial cell alignment (data not shown). All pitches significantly increased the percentage of elongated and aligned cells. Within 90 min postplating, the phalloidin-stained actin stress fibers (red) aligned parallel to the ridges and grooves on patterned substrates while being oriented in random directions on the planar substrate (Fig. 3). These data demonstrate that HUVECs do not require prior protein deposition to respond to topographic cues. This result was consistent with previous reports that serum proteins were not a requirement for the alignment of corneal epithelial cells along topographic surface features (12, 43). The data suggest that cells can respond directly to the physical shape of the surface, instead of indirectly to the pattern of protein that had previously been deposited on the topographic ridges.
Fig. 2.
HUVECs elongate and align parallel to topographic features in the absence of serum. HUVECs were plated in serum-free medium on topographically patterned or planar surfaces and fixed 30, 60, or 120 min later. Values (means ± SE of 3 independent experiments) represent percentage of cells that elongated and aligned parallel to the features of the underlying topography. A 2-way ANOVA was performed against topography and time. *P < 0.05, **P < 0.01, ***P < 0.001 vs. planar (by Holm-Sidak post hoc test). There was also a significant difference between data obtained at 30 and 120 min (P < 0.001), but not between data obtained at 30 and 60 min (P < 0.056).
Fig. 3.
Actin fibers of the cytoskeleton align along the topographic features of the underlying substrate within 60–90 min postplating in serum-free medium. Representative images show HUVECs on a planar or a topographically patterned surface (1,200-nm pitch). The actin cytoskeleton, stained with phalloidin (red), has an isotropic orientation on the planar surface; on the patterned substrate, the cell, as well as the actin fibers, orient along the ridges and grooves. Nuclei are stained with DAPI (blue). Arrows indicate orientation of the ridges and grooves of the patterned surface. Scale bar, 10 μm; ×63 objective.
Early Cell-Signaling Events Triggered by Topographic Cues
Activation of the focal adhesion proteins p130Cas and Src in cells on patterned surfaces.
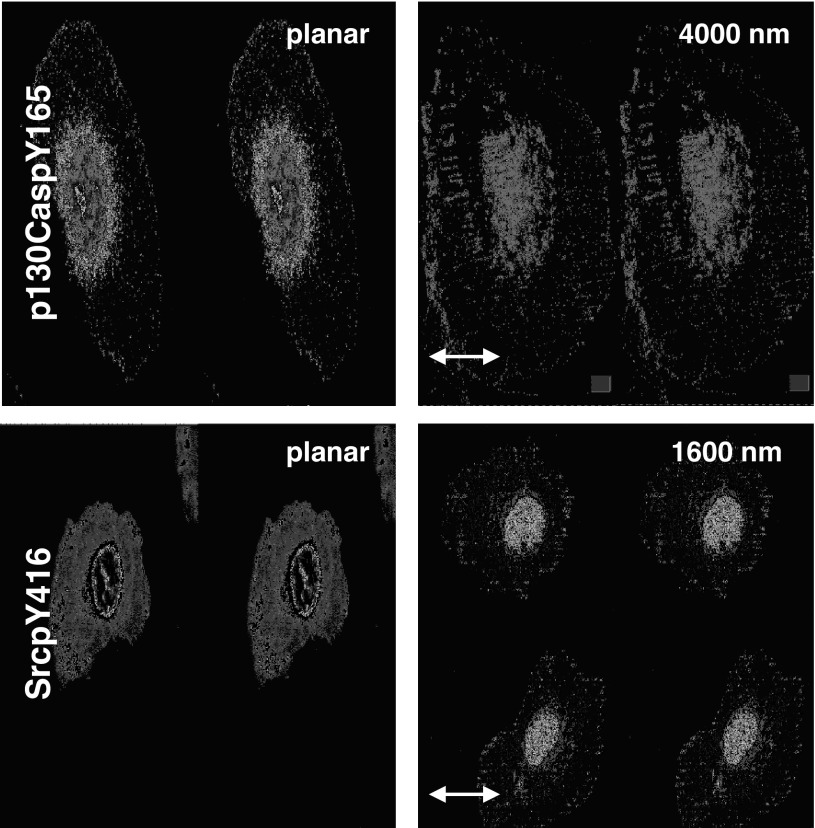
Focal adhesions have been documented to play an essential role in the transduction of extracellular cues into intracellular signals. To study signaling pathways that may be involved in the cellular responses to topography, we focused on phosphorylation changes in focal adhesion proteins at the sites of surface contact. Upon activation, focal adhesion proteins localize to focal adhesions, which can be visualized by immunofluorescent staining as characteristic freckle-like structures. The focal adhesion proteins p130Cas and Src have been reported to be involved in sensing of mechanical forces (23, 37). Src becomes activated when phosphorylated at Tyr416 and Tyr418, while p130Cas is activated by phosphorylation at multiple sites, including Tyr165. Using immunocytochemistry, we stained for the activated focal adhesion proteins phosphorylated (Tyr165) p130Cas and phosphorylated (Tyr416) Src in HUVECs fixed 30 min postplating on 6-packs in protein-free media and found differential distributions of both proteins on patterned surfaces compared with planar controls. Within 30 min of plating, phosphorylated (Tyr165) p130Cas localized to the edges of the topographic ridges in linear structures parallel to the surface patterns (Fig. 4, top). Similarly, Src was activated within long linear focal adhesion structures on topographically patterned surfaces (Fig. 4, bottom). In contrast, immunofluorescent staining of cells on the planar substrates revealed a random or radial orientation of the focal adhesions, which were mainly confined to the periphery of the cell. The distinctive localization of these activated proteins on the patterned surfaces was observed on all pitches (data not shown) within 30 min, indicating an involvement of these proteins in the early cellular response to topographic cues.
Fig. 4.
Focal adhesion proteins [phosphorylated (Tyr165) p130Cas (p130CaspY165) and phosphorylated (Tyr416) Src (SrcpY416)] respond to topographic cues. HUVECs were fixed 30 min after plating on topographically patterned or planar surfaces. Representative immunofluorescent images show that p130Cas (top) and Src (bottom), downstream targets of focal adhesion kinase (FAK) signaling cascades, are phosphorylated and orient in linear structures that align with the ridges and grooves of the underlying patterned surface. Arrows indicate orientation of the ridges and grooves. Scale bar, 10 μm; ×63 objective.
The specificity of this staining pattern on the patterned substrates was previously reported (21, 29, 30, 36, 45) and confirmed in methods (Fig. 1).
FAK activation in response to topography.
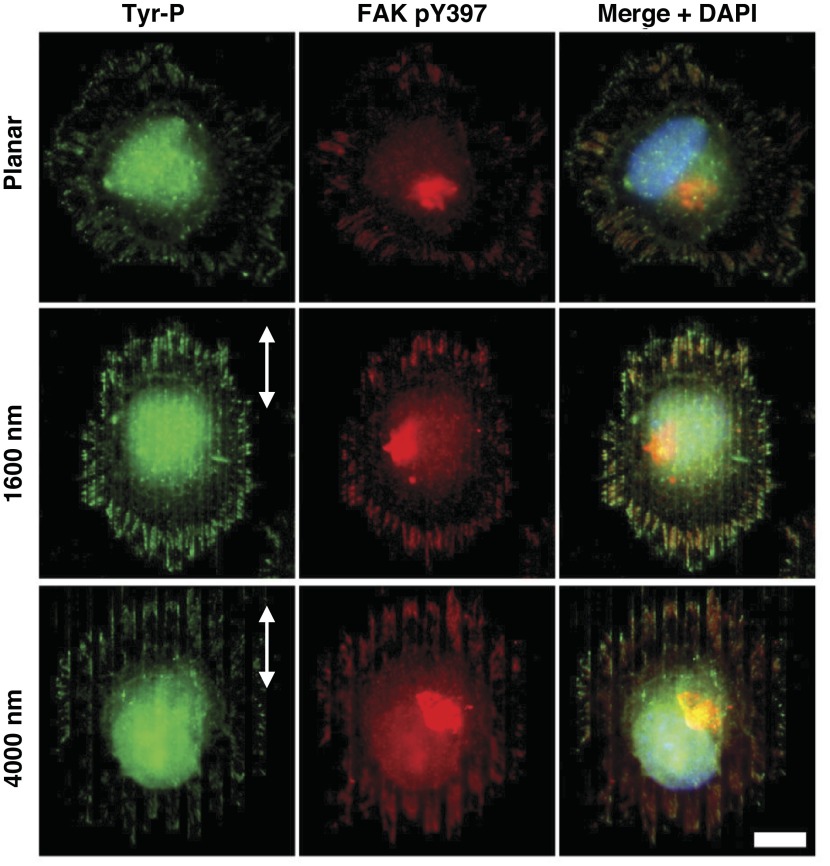
FAK, a signaling molecule upstream of Src and p130Cas, might be responsible for the localization and activation of those proteins upon culture on topographic surfaces. Phosphorylation of FAK at Tyr397 initiates signaling cascades that can affect cell motility and morphology, among other behaviors, and has also been reported to play a potential role in mechanotransduction (33, 49). We therefore chose to observe FAK activation and localization in HUVECs interacting with topography. Immunocytochemistry using a monoclonal antibody recognizing general tyrosine phosphorylation events (green) and an antibody against phosphorylated (Tyr397) FAK (red) was performed to specifically detect activated FAK in HUVECs fixed 30 min after plating on 6-packs. Deconvolved immunofluorescent images showed that FAK is activated within 30 min of exposure to the topography and forms strong linear structures that localize to the edges of the ridges. In contrast, on the planar control substrate, phosphorylated (Tyr397) FAK is localized in random speckles at the cell periphery (Fig. 5). These results were consistent with the results of p130Cas and Src localization. We analyzed FAK activation at the earliest time point of cell elongation and observed that the activation and localization of FAK within 30 min of exposure to the topographically patterned surfaces coincide with the time required for cell alignment to the ridges and grooves. This intriguing observation suggests that FAK activation might play a role during the cell's early response to topography and led us to further investigate if FAK might be a trigger for cellular responses to biophysical cues.
Fig. 5.
Activation of focal adhesion kinase (FAK) in HUVECs on topographically patterned surfaces. HUVECs were plated on topographically patterned and planar surfaces and fixed 30 min later. Immunocytochemistry was used to detect all tyrosine-phosphorylated proteins (Tyr-P, green) or, specifically, FAK phosphorylated at Tyr397 (FAK pY397, red). Nuclei were stained with DAPI (blue). On all patterned surfaces, phosphorylated and, thus, activated FAK forms large linear patterns that align with the ridges and grooves. Arrows indicate direction of the ridges and grooves. Scale bar, 10 μm; ×63 objective.
Topography Does Not Modulate Inherent FAK Expression
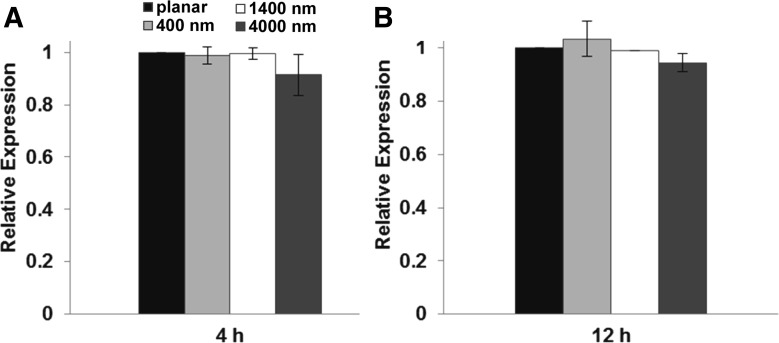
We were interested in whether biophysical cues alter the gene expression levels of FAK. A previously performed gene chip analysis showed no differences in FAK expression between HUVECs cultured on the 400-nm pitch and those cultured on planar surfaces (14). To analyze additional pitch sizes, we performed a qPCR using RNA isolated from HUVECs cultured on topographically patterned surfaces and planar control substrates. At 4 and 12 h postplating, FAK expression was not altered on any of our topographically patterned surfaces (Fig. 6).
Fig. 6.
Substratum topography does not alter the gene expression levels of protein tyrosine kinase 2 (PTK2), coding for FAK, in HUVECs. Expression levels of PTK2 in HUVECs at 4 h (A) and 12 h (B) postplating on topographically patterned and planar surfaces were detected by quantitative PCR. Values (means ± SD of technical triplicates from 3 experiments) were normalized to expression levels on planar surfaces, which were arbitrarily set as “1.”. No statistically significant differences were detected between expression levels on planar and patterned surfaces.
Effect of FAK Knockdown
Successful siRNA-mediated knockdown of FAK in HUVECs.
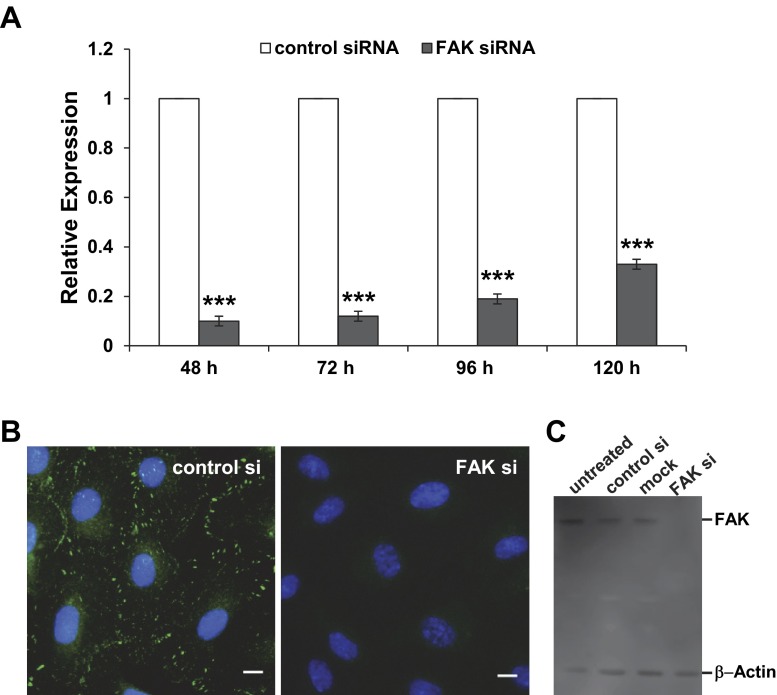
To investigate the importance of FAK as a mechanosensor in HUVECs, FAK mRNA was downregulated by siRNA transfection. Time points from 48 to 120 h posttransfection were analyzed. As determined by qPCR analysis, knockdown to expression levels <20% compared with control siRNA-transfected cells was achieved at 48–96 h posttransfection. At 120 h posttransfection, the mRNA levels started to increase and exceeded 30% of gene expression (Fig. 7A). We therefore chose to perform our experiments during the first 72 h posttransfection to ensure consistency throughout our experiments. In contrast to the knockdown experiment, similar FAK gene expression levels were detected in untreated or mock-transfected cells and in cells transfected with control siRNA (data not shown).
Fig. 7.
Successful small interfering RNA (siRNA)-mediated FAK knockdown in HUVECs. HUVECs were transfected with control or FAK siRNA and 48–120 h later either harvested for RNA or protein isolation or fixed for detection of phosphorylated (Tyr397) FAK by immunocytochemistry. A: transfection with FAK siRNA led to downregulation of FAK mRNA expression. At 48–96 h posttransfection, residual FAK expression levels were <20% of FAK expression in control siRNA-transfected cells as detected by quantitative PCR. At 120 h posttransfection, expression of FAK increased to ∼30% compared with expression of FAK in control cells, which was arbitrarily set as 1. Values are means ± SD of technical triplicates from 3 experiments. ***P < 0.001 vs. control siRNA (by 1-way ANOVA). B: in control cells, phosphorylated (Tyr397) FAK (green) is detectable by immunocytochemistry as punctate structures predominantly at the periphery of the cells. These structures could not be detected in cells fixed 96 h posttransfection with FAK siRNA. Nuclei are stained with DAPI (blue). Scale bar, 10 μm; ×40 objective. C: Western blot confirming no detectable FAK at the protein level in HUVECs 48 h posttransfection with FAK siRNA (FAK si). In untreated or mock-transfected HUVECs, as well as in HUVECs transfected with control siRNA (control si), FAK was detected as a 125-kDa band. As a loading control, β-actin was detected as a ∼46-kDa protein in all lanes.
The successful silencing of FAK was confirmed on a protein level by immunocytochemistry on HUVECs 48 h posttransfection, which showed a signal in control siRNA-transfected cells, but not in FAK siRNA-transfected cells (Fig. 7B). Similarly, the amount of FAK protein was below the level of detection by Western blotting in cell lysate obtained from FAK siRNA-transfected cells (Fig. 7C) compared with untreated cells and HUVECs transfected with control siRNA or mock-transfected HUVECs. β-Actin was also measured and detected in all samples, confirming that equal amounts of protein were loaded.
FAK-depleted cells remain sensitive to topographic cues.
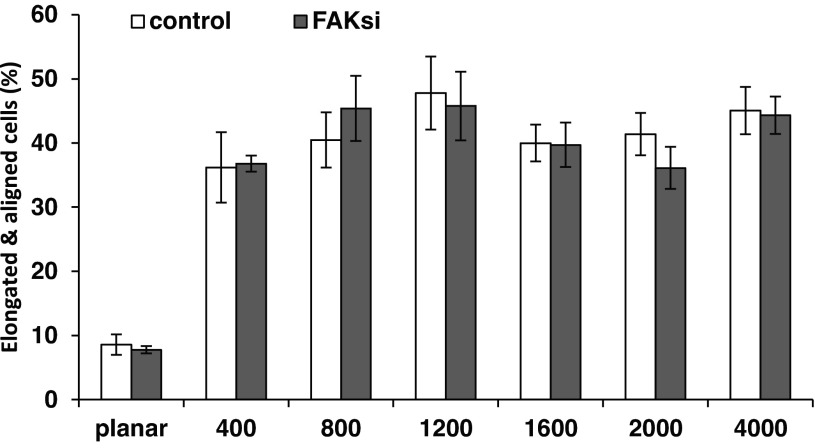
At ∼48 h posttransfection with control and FAK siRNA, HUVECs were plated on topographically patterned surfaces of different pitch sizes, as well as planar control areas, and left to adhere for 4 h. After fixation and staining of nuclei and the actin cytoskeleton, the percentage of cells aligned with the topographic features was determined. On all pitch sizes, HUVECs preferentially aligned parallel to the ridges and grooves of the underlying surface (Fig. 8). No statistically significant differences could be detected between cells transfected with control and cells transfected with FAK siRNA, indicating that, in HUVECs, FAK is not required for the cell orientation along the ridges and grooves. Similar values were obtained with untreated or mock-transfected cells (data not shown).
Fig. 8.
HUVECs remain responsive to topographic cues after FAK knockdown. HUVECs were plated on topographically patterned surfaces 48 h posttransfection with control or FAK siRNA. Cells were fixed 4 h postplating, and the percentage of cells that were elongated and aligned along the ridges and grooves was determined. Values are means ± SE of 4 independent experiments. On all pitches, HUVECs aligned preferentially along the ridges and grooves of the underlying surface. A 2-way ANOVA was performed against topography and transfection conditions. Elongation and alignment of cells on all pitches were significantly different from cells on planar surfaces (P < 0.001, by Holm-Sidak post hoc test). There was no significant difference with FAK knockdown (P < 0.803).
Previous work in FAK-null fibroblasts showed that these cells were unable to respond to mechanical forces, durotaxis (49), or the topographic cues of micrometer-sized pillars (13). Our observation that FAK knockdown had little or no impact on the ability of HUVECs to align along the ridges and grooves counters the hypothesis that FAK is a fundamental mechanosensor in topographic cueing. In agreement with our data, we recently observed that, in corneal epithelial cells, FAK knockdown led to an increased cellular response to topographic cues (10). The differences in the impact of FAK knockdown on different cell types suggest altered sensing modalities for the specific tissue microenvironments to which each cell type is exposed in vivo. Like most cell types, vascular endothelial cells are exposed to topographic cues provided by the ECM on their basal surface. In addition, they are exposed to several hemodynamic cues, such as pressure, stretch, and shear (29, 47). The dramatic difference in the biophysical microenvironment could influence the importance of specific mechanotransduction pathways. Interestingly, reports that the cellular responses to substrate stretch can be independent of FAK (18, 31) provide further evidence that FAK is not always essential for mechanotransduction. Therefore, FAK being dispensable for vascular endothelial cell mechanotransduction is not without precedent. Thus, FAK may be involved in the reaction to biophysical cues in some cell types and conditions but is not necessary in HUVECs to induce cell alignment and elongation in response to topographic cues.
Migration of HUVECs on topographically patterned surfaces after knockdown of FAK.
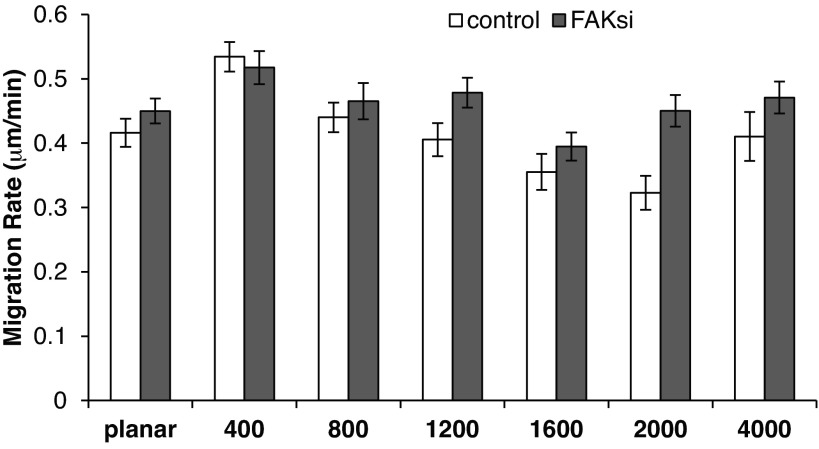
At ∼48 h posttransfection with control and FAK siRNA, HUVECs were plated on 6-pack substrates and left to adhere for ∼4 h. During the following 10 h, cell migration was tracked by time-lapse microscopy to analyze the impact of FAK knockdown on the motility of HUVECs. The migration rate of control and FAK siRNA-transfected cells was significantly increased on the 400-nm pitch (Fig. 9). Additionally, there was a statistically significant difference between the migration rate of control and FAK siRNA-transfected cells with FAK knockdown, resulting in an increase in cell migration (Fig. 9). This result was contrary to our expectations, since FAK-null fibroblasts, as well as inactive FAK-mutant fibroblasts, have been reported to show a reduced migration rate (19, 49). Also, overexpression of FAK resulted in an increased migration rate of Chinese hamster ovary cells (5). In addition, a recent study from our laboratory documented a decreased migration rate of human corneal epithelial cells after downregulation of FAK by siRNA transfection (10). While it is possible that the migration was influenced by the residual FAK mRNA expression, we consider this unlikely, as FAK was below detection limits at the protein level. Together with previously published literature, these data point to variability in the mechanotransduction pathways active within a specific cell type. In HUVECs, it is likely that other signaling molecules may compensate for the loss of FAK upon FAK downregulation. Recent publications report an important role of Yes-associated protein in mechanotransduction (11, 15, 48) in a FAK-independent pathway (51). Congruent with this hypothesis, Yes-associated protein mRNA expression in HUVECs is not affected in our FAK-knockdown experiments (data not shown). Therefore, this pathway should remain fully functional and may compensate for the loss of FAK signaling in HUVECs.
Fig. 9.
Topography and FAK knockdown modulate the migration rate of HUVECs. HUVECs transfected with control or FAK siRNA were plated on topographically patterned and planar surfaces 48 h posttransfection. Values are means ± SE from ≥40 individual cells in ≥4 experiments. A 2-way ANOVA was performed against topography and transfection conditions. Migration on the 400-nm pitch was significantly elevated from planar (P < 0.001, by Holm-Sidak post hoc test), and there is a statistically significant difference between the migration rate of control and FAK siRNA-transfected cells, except on the 400-nm pitch substrates (P < 0.001, by Holm-Sidak post hoc test).
Conclusions
The present data document that prior coating of substrate surfaces is not a requirement for the ability of HUVECs to align along the ridges and grooves of the underlying topographically patterned substrates. Even without the addition of protein in the culture medium, HUVECs responded to topographic cues within 30 min of plating by orienting along the various pitches. At this early time point, we found activated FAK, p130Cas, and Src localized to long linear structures along the edges of the topography, in sharp contrast to the random peripheral organization on planar substrates. Nevertheless, depletion of FAK did not inhibit cell orientation to topography, suggesting the existence of parallel signaling pathways. We theorize that FAK activation is a prime event in the cellular response to topographic cues but that parallel pathways exist in HUVECs, emphasizing that cellular mechanotransduction systems are not generalizable between cell types. The cell type-specific differences may reflect the particular roles and microenvironments of each cell type. On the basis of these data and current literature, we hypothesize that several signaling pathways are involved in the cellular responses to biophysical cues that allow the cells to respond to a variety of biophysical stimuli in their specific microenvironment.
GRANTS
This project was supported by National Heart, Lung, and Blood Institute Grant 1R01 HL-079012-01A and National Eye Institute Grants 1R01 EY-017367-01A, 1R01 EY-019475, and P30 EY-12576.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
B.D., J.Z.G., P.F.N., P.R., and C.J.M. conceived and designed the research; B.D., J.Z.G., and J.T.M. performed the experiments; B.D., J.Z.G., and J.T.M. analyzed the data; B.D., J.Z.G., J.T.M., P.R., and C.J.M. interpreted the results of the experiments; B.D., J.Z.G., and J.T.M. prepared the figures; B.D. and J.Z.G. drafted the manuscript; B.D., J.Z.G., J.T.M., P.R., and C.J.M. edited and revised the manuscript; B.D., J.Z.G., J.T.M., P.F.N., P.R., and C.J.M. approved the final version of the manuscript.
ACKNOWLEDGMENTS
The authors thank Kunal Patel for help with the image analysis and Sara Liliensiek for helpful discussions.
REFERENCES
- 1. Abrams GA, Bentley E, Nealey PF, Murphy CJ. Electron microscopy of the canine corneal basement membranes. Cells Tissues Organs 170: 251–257, 2002 [DOI] [PubMed] [Google Scholar]
- 2. Abrams GA, Goodman SL, Nealey PF, Franco M, Murphy CJ. Nanoscale topography of the basement membrane underlying the corneal epithelium of the rhesus macaque. Cell Tissue Res 299: 39–46, 2000 [DOI] [PubMed] [Google Scholar]
- 3. Abrams GA, Murphy CJ, Wang ZY, Nealey PF, Bjorling DE. Ultrastructural basement membrane topography of the bladder epithelium. Urol Res 31: 341–346, 2003 [DOI] [PubMed] [Google Scholar]
- 4. Brown MC, Perrotta JA, Turner CE. Identification of LIM3 as the principal determinant of paxillin focal adhesion localization and characterization of a novel motif on paxillin directing vinculin and focal adhesion kinase binding. J Cell Biol 135: 1109–1123, 1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cary LA, Chang JF, Guan JL. Stimulation of cell migration by overexpression of focal adhesion kinase and its association with Src and Fyn. J Cell Sci 109: 1787–1794, 1996 [DOI] [PubMed] [Google Scholar]
- 6. Cary LA, Han DC, Polte TR, Hanks SK, Guan JL. Identification of p130Cas as a mediator of focal adhesion kinase-promoted cell migration. J Cell Biol 140: 211–221, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen HC, Appeddu PA, Parsons JT, Hildebrand JD, Schaller MD, Guan JL. Interaction of focal adhesion kinase with cytoskeletal protein talin. J Biol Chem 270: 16995–16999, 1995 [DOI] [PubMed] [Google Scholar]
- 8. Cobb BS, Schaller MD, Leu TH, Parsons JT. Stable association of pp60src and pp59fyn with the focal adhesion-associated protein tyrosine kinase, pp125FAK. Mol Cell Biol 14: 147–155, 1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Diehl KA, Foley JD, Nealey PF, Murphy CJ. Nanoscale topography modulates corneal epithelial cell migration. J Biomed Mater Res A 75: 603–611, 2005 [DOI] [PubMed] [Google Scholar]
- 10. Dreier B, Raghunathan VK, Russell P, Murphy CJ. Focal adhesion kinase knockdown modulates the response of human corneal epithelial cells to topographic cues. Acta Biomater 8: 4285–4294, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dupont S, Morsut L, Aragona M, Enzo E, Giulitti S, Cordenonsi M, Zanconato F, Le Digabel J, Forcato M, Bicciato S, Elvassore N, Piccolo S. Role of YAP/TAZ in mechanotransduction. Nature 474: 179–183, 2011 [DOI] [PubMed] [Google Scholar]
- 11a. Franco D, Klingauf M, Bednarzik M, Cecchini M, Kurtcuoglu V, Gobrecht J, Poulikakos D, Ferrari A. Control of initial endothelial spreading by topographic activation of focal adhesion kinase. Soft Matter 7: 7313–7324, 2011 [Google Scholar]
- 12. Fraser SA, Ting YH, Mallon KS, Wendt AE, Murphy CJ, Nealey PF. Sub-micron and nanoscale feature depth modulates alignment of stromal fibroblasts and corneal epithelial cells in serum-rich and serum-free media. J Biomed Mater Res A 86: 725–735, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Frey MT, Tsai IY, Russell TP, Hanks SK, Wang YL. Cellular responses to substrate topography: role of myosin II and focal adhesion kinase. Biophys J 90: 3774–3782, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gasiorowski JZ, Liliensiek SJ, Russell P, Stephan DA, Nealey PF, Murphy CJ. Alterations in gene expression of human vascular endothelial cells associated with nanotopographic cues. Biomaterials 31: 8882–8888, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Halder G, Dupont S, Piccolo S. Transduction of mechanical and cytoskeletal cues by YAP and TAZ. Nat Rev Mol Cell Biol 13: 591–600, 2012 [DOI] [PubMed] [Google Scholar]
- 16. Hanks SK, Calalb MB, Harper MC, Patel SK. Focal adhesion protein-tyrosine kinase phosphorylated in response to cell attachment to fibronectin. Proc Natl Acad Sci USA 89: 8487–8491, 1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hildebrand JD, Schaller MD, Parsons JT. Paxillin, a tyrosine phosphorylated focal adhesion-associated protein binds to the carboxyl terminal domain of focal adhesion kinase. Mol Biol Cell 6: 637–647, 1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hsu HJ, Lee CF, Locke A, Vanderzyl SQ, Kaunas R. Stretch-induced stress fiber remodeling and the activations of JNK and ERK depend on mechanical strain rate, but not FAK. PLos One 5: e12470, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ilic D, Furuta Y, Kanazawa S, Takeda N, Sobue K, Nakatsuji N, Nomura S, Fujimoto J, Okada M, Yamamoto T. Reduced cell motility and enhanced focal adhesion contact formation in cells from FAK-deficient mice. Nature 377: 539–544, 1995 [DOI] [PubMed] [Google Scholar]
- 20. Karuri NW, Liliensiek S, Teixeira AI, Abrams G, Campbell S, Nealey PF, Murphy CJ. Biological length scale topography enhances cell-substratum adhesion of human corneal epithelial cells. J Cell Sci 117: 3153–3164, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Karuri NW, Nealey PF, Murphy CJ, Albrecht RM. Structural organization of the cytoskeleton in SV40 human corneal epithelial cells cultured on nano- and microscale grooves. Scanning 30: 405–413, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Karuri NW, Porri TJ, Albrecht RM, Murphy CJ, Nealey PF. Nano- and microscale holes modulate cell-substrate adhesion, cytoskeletal organization, and β1-integrin localization in SV40 human corneal epithelial cells. IEEE Trans Nanobiosci 5: 273–280, 2006 [DOI] [PubMed] [Google Scholar]
- 23. Kostic A, Sheetz MP. Fibronectin rigidity response through Fyn and p130Cas recruitment to the leading edge. Mol Biol Cell 17: 2684–2695, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Last JA, Liliensiek SJ, Nealey PF, Murphy CJ. Determining the mechanical properties of human corneal basement membranes with atomic force microscopy. J Struct Biol 167: 19–24, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liliensiek SJ, Campbell S, Nealey PF, Murphy CJ. The scale of substratum topographic features modulates proliferation of corneal epithelial cells and corneal fibroblasts. J Biomed Mater Res A 79: 185–192, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liliensiek SJ, Nealey P, Murphy CJ. Characterization of endothelial basement membrane nanotopography in rhesus macaque as a guide for vessel tissue engineering. Tissue Eng A 15: 2643–2651, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Liliensiek SJ, Wood JA, Yong J, Auerbach R, Nealey PF, Murphy CJ. Modulation of human vascular endothelial cell behaviors by nanotopographic cues. Biomaterials 31: 5418–5426, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lu Q, Rounds S. Focal adhesion kinase and endothelial cell apoptosis. Microvasc Res 83: 56–63, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Morgan JT, Wood JA, Shah NM, Hughbanks ML, Russell P, Barakat AI, Murphy CJ. Integration of basal topographic cues and apical shear stress in vascular endothelial cells. Biomaterials 33: 4126–4135, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Myrna KE, Mendonsa R, Russell P, Pot SA, Liliensiek SJ, Jester JV, Nealey PF, Brown D, Murphy CJ. Substratum topography modulates corneal fibroblast to myofibroblast transformation. Invest Ophthalmol Vis Sci 53: 811–816, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ngu H, Feng Y, Lu L, Oswald SJ, Longmore GD, Yin FC. Effect of focal adhesion proteins on endothelial cell adhesion, motility and orientation response to cyclic strain. Ann Biomed Eng 38: 208–222, 2010 [DOI] [PubMed] [Google Scholar]
- 32. Odom TW, Love JC, Wolfe DB, Paul KE, Whitesides GM. Improved pattern transfer in soft lithography using composite stamps. Langmuir 18: 5314–5320, 2002 [Google Scholar]
- 33. Owen KA, Abshire MY, Tilghman RW, Casanova JE, Bouton AH. FAK regulates intestinal epithelial cell survival and proliferation during mucosal wound healing. PLos One 6: e23123, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pot SA, Liliensiek SJ, Myrna KE, Bentley E, Jester JV, Nealey PF, Murphy CJ. Nanoscale topography-induced modulation of fundamental cell behaviors of rabbit corneal keratocytes, fibroblasts, and myofibroblasts. Invest Ophthalmol Vis Sci 51: 1373–1381, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Roger VL, Go AS, Lloyd-Jones DM, Adams RJ, Berry JD, Brown TM, Carnethon MR, Dai S, de Simone G, Ford ES, Fox CS, Fullerton HJ, Gillespie C, Greenlund KJ, Hailpern SM, Heit JA, Ho PM, Howard VJ, Kissela BM, Kittner SJ, Lackland DT, Lichtman JH, Lisabeth LD, Makuc DM, Marcus GM, Marelli A, Matchar DB, McDermott MM, Meigs JB, Moy CS, Mozaffarian D, Mussolino ME, Nichol G, Paynter NP, Rosamond WD, Sorlie PD, Stafford RS, Turan TN, Turner MB, Wong ND, Wylie-Rosett J. Heart disease and stroke statistics—2011 update: a report from the American Heart Association. Circulation 123: e18–e209, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Russell P, Gasiorowski JZ, Nealy PF, Murphy CJ. Response of human trabecular meshwork cells to topographic cues on the nanoscale level. Invest Ophthalmol Vis Sci 49: 629–635, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sawada Y, Tamada M, Dubin-Thaler BJ, Cherniavskaya O, Sakai R, Tanaka S, Sheetz MP. Force sensing by mechanical extension of the Src family kinase substrate p130Cas. Cell 127: 1015–1026, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schaller MD, Hildebrand JD, Shannon JD, Fox JW, Vines RR, Parsons JT. Autophosphorylation of the focal adhesion kinase, pp125FAK, directs SH2-dependent binding of pp60src. Mol Cell Biol 14: 1680–1688, 1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Schaller MD, Otey CA, Hildebrand JD, Parsons JT. Focal adhesion kinase and paxillin bind to peptides mimicking β-integrin cytoplasmic domains. J Cell Biol 130: 1181–1187, 1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schaller MD, Parsons JT. pp125FAK-dependent tyrosine phosphorylation of paxillin creates a high-affinity binding site for Crk. Mol Cell Biol 15: 2635–2645, 1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Schlaepfer DD, Mitra SK, Ilic D. Control of motile and invasive cell phenotypes by focal adhesion kinase. Biochim Biophys Acta 1692: 77–102, 2004 [DOI] [PubMed] [Google Scholar]
- 42. Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9: 671–675, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Teixeira AI, Abrams GA, Bertics PJ, Murphy CJ, Nealey PF. Epithelial contact guidance on well-defined micro- and nanostructured substrates. J Cell Sci 116: 1881–1892, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Teixeira AI, McKie GA, Foley JD, Bertics PJ, Nealey PF, Murphy CJ. The effect of environmental factors on the response of human corneal epithelial cells to nanoscale substrate topography. Biomaterials 27: 3945–3954, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Teixeira AI, Nealey PF, Murphy CJ. Responses of human keratocytes to micro- and nanostructured substrates. J Biomed Mater Res A 71: 369–376, 2004 [DOI] [PubMed] [Google Scholar]
- 46. Turner CE, Miller JT. Primary sequence of paxillin contains putative SH2 and SH3 domain binding motifs and multiple LIM domains: identification of a vinculin and pp125Fak-binding region. J Cell Sci 107: 1583–1591, 1994 [DOI] [PubMed] [Google Scholar]
- 47. Uttayarat P, Chen M, Li M, Allen FD, Composto RJ, Lelkes PI. Microtopography and flow modulate the direction of endothelial cell migration. Am J Physiol Heart Circ Physiol 294: H1027–H1035, 2008 [DOI] [PubMed] [Google Scholar]
- 48. Wada K, Itoga K, Okano T, Yonemura S, Sasaki H. Hippo pathway regulation by cell morphology and stress fibers. Development 138: 3907–3914, 2011 [DOI] [PubMed] [Google Scholar]
- 49. Wang HB, Dembo M, Hanks SK, Wang Y. Focal adhesion kinase is involved in mechanosensing during fibroblast migration. Proc Natl Acad Sci USA 98: 11295–11300, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wood JA, Shah NM, McKee CT, Hughbanks ML, Liliensiek SJ, Russell P, Murphy CJ. The role of substratum compliance of hydrogels on vascular endothelial cell behavior. Biomaterials 32: 5056–5064, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhao B, Li L, Wang L, Wang CY, Yu J, Guan KL. Cell detachment activates the Hippo pathway via cytoskeleton reorganization to induce anoikis. Genes Dev 26: 54–68, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhao J, Zheng C, Guan J. Pyk2 and FAK differentially regulate progression of the cell cycle. J Cell Sci 113: 3063–3072, 2000 [DOI] [PubMed] [Google Scholar]
- 53. Zhao JH, Reiske H, Guan JL. Regulation of the cell cycle by focal adhesion kinase. J Cell Biol 143: 1997–2008, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]