Abstract
We report the use of a cDNA microarray to monitor global transcriptional responses of the chestnut blight fungus, Cryphonectria parasitica, to infection by mild and severe isolates of virulence-attenuating hypoviruses that share 87 to 93% and 90 to 98% identity at the nucleotide and amino acid levels, respectively. Infection by the mild hypovirus isolate CHV1-Euro7 resulted in differential expression of 166 of the ca. 2,200 genes represented on the microarray (90 upregulated and 76 downregulated). This is roughly half the number of genes scored as differentially expressed after infection by the severe isolate, CHV1-EP713 (295 genes; 132 upregulated and 163 downregulated). Comparison of the lists of genes responsive to infection by the two hypovirus isolates revealed 80 virus-common responsive genes. Infection by CHV1-EP713 also caused changes in gene transcript accumulation that were, in general, of greater magnitude than those observed with CHV1-Euro7 infections. Thus, the host transcriptional response to infection by severe hypovirus CHV1-EP713 appears to be considerably more dynamic than the response to infection by the mild isolate CHV1-Euro7. Real-time reverse transcription-PCR was performed on 39 different clones, with false-positive rates of 3 and 8% observed for the microarray-predicted list of genes responsive to CHV1-EP713 and CHV1-Euro7 infections, respectively. This analysis has allowed an initial assignment for ca. 2,200 unique C. parasitica-expressed genes as being unresponsive to hypovirus infection, selectively responsive to a specific hypovirus, or generally responsive to hypovirus infection.
Members of the RNA virus family Hypoviridae persistently alter phenotypic traits, modulate gene expression, and attenuate virulence of their fungal host, the chestnut blight fungus Cryphonectria parasitica (2, 9). Infectious cDNA clones have been constructed for two hypovirus isolates, CHV1-EP713 (5) and CHV1-Euro7 (4), that cause quite different changes in host phenotypic traits while sharing a high level of sequence identity at both the nucleotide (87 to 93%) and amino acid (90 to 98%) levels. Hypovirus CHV1-EP713, classified as a severe isolate, significantly reduces the ability of C. parasitica to invade and produce asexual spore-forming bodies on chestnut tissue. In contrast, C. parastica strains infected with CHV1-Euro7, classified as a mild hypovirus isolate, aggressively invade chestnut tissue early after colonization and then abruptly cease expansion when the canker reaches a size three- to fourfold larger than that attained by CHV1-EP713-infected strains. Moreover, the surface of these cankers is covered with a significant level of spore-forming bodies (4). Virus-free C. parasitica strains continue to expand until the tree is girdled and produce copious amounts of asexual spores on the surface of colonized bark tissue (2, 9).
Fungal strains infected with the two hypoviruses also exhibit a number of distinguishing phenotypic traits when cultured under defined laboratory conditions. CHV1-EP713-infected C. parasitica strains exhibit reduced growth rates on solid media and produce colonies characterized by reduced production of aerial hyphae, irregular margins, and the general absence of asexual spores (4). C. parasitica strains infected with the mild hypovirus CHV1-Euro7 actually grow faster than the corresponding virus-free strain, and produce colonies with abundant aerial hyphae and regular margins and that produce conidia at levels intermediate between those of virus-free and CHV1-EP713-infected strains (4). Both hypoviruses suppress production of orange pigment, resulting in white colonies, and cause loss of female fertility.
Efforts to understand the mechanisms underlying the differences in symptom expression by the mild and severe hypovirus isolates revealed differential modulation of cyclic AMP-regulated signaling and laccase production by the mild and severe isolates (25). These results suggested the possibility that the two hypoviruses, even though closely related at the nucleotide and amino acid sequence levels, may have quite different effects on host gene expression. However, caution must be exercised in making such predictions since these studies relied on single gene readouts or differences in the activity of a specific family of enzymes.
Allen et al. (1) recently used a spotted cDNA microarray containing ca. 2,200 C. parastica genes to monitor the change in host transcriptional profile after infection with severe hypovirus isolate CHV1-EP713. That analysis confirmed that CHV1-EP713 infection results in a persistent reprogramming of a significant portion (estimated at 13.4%) of the C. parasitica transcriptome and expanded the number of identified C. parasitica genes that respond to hypovirus infection from less than 20 (9, 17) to nearly 300. We have taken advantage of the C. parasitica cDNA microarray platform to determine here whether the mild and severe hypovirus isolates cause similar or dissimilar changes in the host transcriptional profile and to identify virus-common and virus-specific host-responsive genes.
MATERIALS AND METHODS
Strains and media.
Hypovirus-free C. parasitica strain EP155 (ATCC 38755), isogenic strain EP155/CHV1-EP713 (ATCC 52571) infected with the prototypic hypovirus CHV1-EP713 (28), and EP155/CHV1-Euro7 generated through transfection of virus-free EP155 with infectious CHV1-Euro7 RNA (4) were maintained on potato dextrose agar (PDA; Difco, Detroit, Mich.) at a temperature between 22 and 24°C with a 12-h light-dark cycle at 1,300 to 1,600 lx. Cultures used for RNA preparations were grown under similar conditions on cellophane membranes overlaying PDA (cellophane-PDA).
Total RNA isolation.
Cultures used for RNA isolation were grown on PDA-cellophane for 6 days and harvested by freezing the mycelia in liquid nitrogen with immediate grinding of the mycelia into a fine powder by using a mortar and pestle. RNA isolation was performed as described by Allen et al. (1).
Microarray analysis.
Microarray slide printing, fluorescent probe generation, microarray hybridization, scanning, and data analyses were performed as described by Allen et al. (1). Integrated pixel intensity values for each spot were calculated by using TIGR Spotfinder software and saved in tab-delimited format for use by MIDAS software (http://www.tigr.org/software) from The Institute for Genomic Research (Rockville, Md.). All hybridization data among three sets of dye-swap experiments were normalized simultaneously in MIDAS to correct for experimental error within a specific hybridization and between repeated hybridizations.
Selection of differentially expressed clones in each hybridization was performed in the Functional Genomics module of Spotfire DecisionSite 7.0 (Spotfire, Inc., Somerville, Mass.) by calculating the log2 (cy3/cy5) ratio for each clone. The clones were divided into “groups” based on the number of standard deviations that the corresponding log2 ratio varied from the data sets' average log2 ratio. Clones with log2 ratios of ≥2 standard deviations in a minimum of four of six hybridizations were considered differentially expressed. For experiments comparing RNA isolated from EP155/CHV1-EP713 to that from EP155/CHV1-Euro7, a clone was considered differentially expressed if its log2 ratio was ≥2 standard deviations in a minimum of three of five hybridizations. Genes identified as differentially expressed were subjected to hierarchical cluster analysis in Spotfire DecisionSite 7.0.
Microarray data management.
The arrayed EST (AEST) library used to construct the cDNA microarray chip consisted of 3,864 EST clones representing ca. 2,200 unique C. parasitica genes, of which roughly 1,600 were estimated to be present as a single clone (1, 8). Thus, ca. 27% of the genes present on the microarray are represented by multiple independent EST clones. Consequently, a subset of the genes on the primary list of differentially expressed genes generated in a microarray experiment will also be represented by more than one related AEST clone (8). To reduce the redundancy of the primary lists for publication purposes (1), the scored AEST clones are sorted according to NCBI gene index (gi) numbers assigned by basic local alignment search tool (BLAST) analysis. The AEST clone with the strongest BLAST hit is subsequently chosen to represent all of the scored AEST clones with the same gi number. This level of redundancy can present some minor cross-referencing problems when two published lists are compared. That is, if the representative AEST of a gi family for some technical reason fails to be scored, it will be replaced by the related EST with the next strongest BLAST hit. This can result in the same gene being represented by different related AEST clones on two different published, nonredundant lists of differentially expressed genes. A list of related AEST clones sorted according to gi numbers is available to facilitate cross-referencing (http://www.umbi.umd.edu/∼cbr/AESTRedundant.pdf). Nonredundant lists for hybridizations involving EP155/CHV1-EP713 versus EP155, EP155/CHV1-Euro7 versus EP155, and EP155/CHV1-EP713 versus EP155/CHV1-Euro7 are available at http://www.umbi.umd.edu/∼cbr/155-713NonRedundant.pdf, http://www.umbi.umd.edu /∼cbr/155-Euro7NonRedundant.pdf, and http://www.umbi.umd.edu/∼cbr/713-Euro7NonRedundant.pdf, respectively.
It is important that primary lists of responsive genes and not the published, nonredundant lists are used in all list comparison analyses. For example, host genes responsive to both mild and severe hypoviruses were identified by comparing the primary CHV1-EP713- and CHV1-Euro7-responsive lists in Microsoft Excel by using the MATCH command. The resulting common list was reduced in redundancy as described above for presentation in Table 1. Occasionally, members of a gi family are found to be present on two primary lists, but the representative of the family with the strongest BLAST hit is absent on one or both of the lists. In such cases, the family members common to both lists are sorted according to BLAST scores, and the AEST clone with the strongest BLAST hit is included in the published, nonredundant common list to represent the gene corresponding to that gi family.
TABLE 1.
C. parasitica genes responsive to infection by both CHV1-EP713 and CHV1-Euro7a
| Category | 713vs155
|
AEST ID | Euro7vs155
|
E-value | Description (organism) | ||
|---|---|---|---|---|---|---|---|
| Direction | Fold change | Direction | Fold change | ||||
| Amino acid metabolism | UP | 23.69 | [13] AEST-08-F-10 | UP | 5.80 | 5.00E-95 | SAMS (Neurospora crassa) |
| DN | 4.95 | AEST-22-H-07 | DN | 2.24 | 1.00E-20 | Putative tyrosinase (Gibberella zeae) | |
| Carbohydrate metabolism | DN | 3.43 | AEST-19-G-03 | DN | 2.22 | 1.00E-74 | Hypothetical protein (Neurospora crassa) |
| 5.00E-71 | 1,2-α-Mannosidase (Aspergillus phoenicis) | ||||||
| DN | 4.28 | AEST-30-G-01 | DN | 1.76 | 3.00E-56 | Hypothetical protein (Neurospora crassa) | |
| 9.00E-44 | Acid-stable alpha-amylase (Aspergillus kawachii) | ||||||
| DN | 4.92 | AEST-37-F-03 | DN | 2.13 | 1.00E-52 | β-Glucosidase homolog (Cochliobolus heterostrophus) | |
| Cell wall or growth | DN | 5.53 | AEST-20-G-02 | DN | 1.94 | 2.00E-27 | Predicted protein (Neurospora crassa) |
| 4.00E-10 | Hard surface-induced protein 3 (Glomerella cingulata) | ||||||
| Development | DN | 3.94 | AEST-30-F-01 | DN | 1.95 | 6.00E-23 | Clock-controlled protein 6 (Neurospora crassa) |
| Electron transport | UP | 2.11 | AEST-33-E-05 | DN | 2.46 | 2.00E-32 | Hypothetical protein (Neurospora crassa) |
| 1.00E-17 | Oxidoreductase, MmyG [Streptomyces coelicolor A3(2)] | ||||||
| UP | 8.14 | AEST-33-G-12 | UP | 3.67 | 2.00E-17 | Hypothetical protein (Burkholderia fungorum) | |
| 1.00E-15 | Oxidoreductase (Pseudomonas syringae) | ||||||
| UP | 3.20 | AEST-35-E-02 | DN | 2.86 | 9.00E-23 | Hypothetical protein (Microbulbifer degradans 2-40) | |
| 4.00E-19 | Oxidoreductase (Clostridium acetobutylicum) | ||||||
| Lipid metabolism | UP | 3.69 | AEST-11-C-02 | UP | 3.81 | 7.00E-84 | Hypothetical protein (Neurospora crassa) |
| 1.00E-61 | Similar to LTA4 hydrolase (Homo sapiens) | ||||||
| Metabolism related | DN | 6.19 | [11] AEST-06-B-03 | DN | 3.16 | 2.00E-32 | Unnamed protein product (Podospora anserina) |
| 3.00E-30 | Possible CGI-83 protein (Leishmania major) | ||||||
| UP | 5.36 | [26] AEST-17-H-10 | DN | 2.02 | 3.00E-32 | Isoamyl alcohol oxidase (Neurospora crassa) | |
| UP | 18.83 | AEST-20-A-03 | UP | 5.23 | 3.00E-57 | FK506-binding protein (Neurospora crassa) | |
| UP | 3.99 | AEST-28-C-01 | UP | 2.03 | 3.00E-20 | Hypothetical protein (Neurospora crassa) | |
| 4.00E-12 | Putative acetyltransferase (Clostridium tetani E88) | ||||||
| UP | 3.11 | AEST-28-D-08 | UP | 2.02 | 4.00E-22 | Short-chain dehydrogenase (Schizosaccharomyces pombe) | |
| DN | 1.79 | AEST-36-F-03 | DN | 1.88 | 2.00E-22 | Hypothetical protein (Neurospora crassa) | |
| 3.00E-13 | Aflatoxin B1 aldehyde reductase 1 (Mus musculus) | ||||||
| Nucleic acid metabolism | DN | 6.20 | [9] AEST-05-C-02 | DN | 2.31 | 3.00E-27 | Hydrogenase regulation HoxX (Aquifex aeolicus) |
| UP | 11.45 | AEST-08-E-11 | UP | 5.13 | 5.00E-47 | Hypothetical protein (Neurospora crassa) | |
| 8.00E-19 | ATP synthase/rho (Brucella melitensis) | ||||||
| UP | 3.11 | [17] AEST-11-E-06 | UP | 2.91 | 4.00E-29 | Hypothetical protein (Neurospora crassa) | |
| 7.00E-22 | Transcriptional regulator (Clostridium tetani E88) | ||||||
| UP | 2.02 | [31] AEST-24-D-01 | UP | 2.92 | 2.00E-32 | SnodProt1 precursor (Neurospora crassa) | |
| UP | 11.04 | AEST-28-F-06 | UP | 2.00 | 7.00E-48 | Hypothetical protein (Neurospora crassa) | |
| 2.00E-14 | Probable AAA family ATPase (Campylobacter jejuni) | ||||||
| Oxygen/radical metabolism | UP | 8.90 | [38] AEST-31-F-09 | UP | 4.67 | 3.00E-39 | Hypothetical protein (Neurospora crassa) |
| 5.00E-35 | Fum12p (Gibberella moniliformis) | ||||||
| Protein metabolism | DN | 4.73 | [8] AEST-05-A-09 | DN | 2.08 | 2.00E-53 | Aspergillopepsin II precursor (Aspergillus niger) |
| UP | 9.86 | AEST-05-E-01 | UP | 5.44 | 2.00E-66 | Probable zinc metallo-protease (Neurospora crassa) | |
| DN | 3.19 | AEST-08-C-09 | DN | 2.19 | 5.00E-92 | Acid proteinase (Cryphonectria parasitica) | |
| DN | 3.91 | AEST-11-D-08 | DN | 2.63 | 1.00E-109 | Endothiapepsin precursor (Cryphonectria parasitica) | |
| DN | 2.99 | [24] AEST-17-C-03 | UP | 2.15 | 1.00E-33 | Aspartic protease precursor (Botryotinia fuckeliana) | |
| DN | 3.09 | AEST-18-C-11 | DN | 2.01 | 4.00E-72 | Carboxypeptidase S1 (Penicillium janthinellum) | |
| UP | 5.51 | AEST-38-C-10 | UP | 2.48 | 5.00E-54 | Ubiquitin-conjugating enzyme E2 (Magnaporthe grisea) | |
| Stress response | UP | 5.61 | AEST-10-H-10 | UP | 2.73 | 2.00E-77 | Heat shock 70-kDa protein (Ajellomyces capsulatus) |
| UP | 5.39 | [21] AEST-12-G-04 | DN | 4.04 | 3.00E-67 | Hypothetical protein (Neurospora crassa) | |
| 2.00E-38 | GST (Schizosaccharomyces pombe) | ||||||
| DN | 4.00 | AEST-38-C-04 | DN | 3.91 | 1.00E-108 | HSP70 (Neurospora crassa) | |
| Transport | UP | 9.31 | AEST-01-G-12 | UP | 3.24 | 4.00E-61 | Hypothetical protein (Neurospora crassa) |
| 1.00E-40 | Nontransporter ABC protein (Dictyostelium discoideum) | ||||||
| UP | 3.76 | [27] AEST-22-A-11 | UP | 1.90 | 2.00E-68 | Stomatin-like protein (Gibberella fujikuroi) | |
| DN | 3.44 | AEST-27-E-05 | DN | 3.03 | 3.00E-25 | Acetylcholine transporter (Schizosaccharomyces pombe) | |
| Unknown | UP | 5.05 | AEST-01-B-11 | UP | 2.51 | ||
| UP | 8.78 | AEST-01-C-10 | UP | 4.20 | 4.00E-05 | Predicted protein (Neurospora crassa) | |
| UP | 3.96 | AEST-01-E-12 | UP | 2.45 | 1.00E-36 | Unnamed protein product (Podospora anserina) | |
| UP | 4.55 | AEST-02-F-06 | DN | 1.91 | 3.00E-12 | Hypothetical protein (Rhodopseudomonas palustris)/PICK> | |
| DN | 8.98 | [7] AEST-04-D-04 | DN | 5.12 | |||
| DN | 6.78 | [10] AEST-05-D-11 | DN | 5.60 | |||
| DN | 3.90 | AEST-07-A-11 | DN | 2.01 | |||
| UP | 7.09 | AEST-09-B-02 | UP | 2.04 | 1.00E-04 | Hypothetical protein (Neurospora crassa) | |
| UP | 6.19 | AEST-09-G-04 | UP | 3.62 | 2.00E-04 | Glu-Asp-rich protein (Dictyostelium discoideum) | |
| DN | 5.73 | AEST-09-G-11 | DN | 2.74 | |||
| UP | 7.89 | AEST-09-H-05 | UP | 3.94 | |||
| UP | 6.22 | AEST-09-H-07 | UP | 3.66 | 1.00E-05 | Hypothetical protein (Neurospora crassa) | |
| DN | 5.39 | AEST-11-A-09 | DN | 2.30 | 2.00E-05 | Probable aldolase (Nostoc sp. strain PCC 7120) | |
| DN | 9.68 | AEST-12-C-05 | DN | 2.04 | 1.00E-09 | Membrane glycoprotein (Equine herpesvirus 1) | |
| UP | 7.88 | AEST-13-E-03 | UP | 3.55 | 9.00E-30 | Predicted protein (Neurospora crassa) | |
| DN | 3.11 | AEST-15-C-06 | DN | 2.41 | 3.00E-36 | Predicted protein (Neurospora crassa) | |
| UP | 2.38 | AEST-15-F-06 | UP | 2.08 | 3.00E-27 | Predicted protein (Neurospora crassa) | |
| DN | 3.30 | AEST-16-A-02 | DN | 1.94 | 8.00E-05 | Hypothetical protein (Neurospora crassa) | |
| DN | 5.44 | AEST-16-A-07 | DN | 2.32 | 8.00E-03 | Cholesterol oxidase precursor (Neurospora crassa) | |
| DN | 4.58 | AEST-16-A-09 | DN | 1.77 | 3.00E-17 | Predicted protein (Neurospora crassa) | |
| UP | 3.79 | AEST-16-C-12 | UP | 2.34 | 4.00E-03 | Hypothetical protein L1177.03 (Leishmania major) | |
| UP | 5.81 | AEST-17-G-12 | UP | 3.21 | 1.00E-06 | Predicted protein (Neurospora crassa) | |
| UP | 15.68 | AEST-18-C-02 | UP | 4.66 | |||
| UP | 6.56 | AEST-18-C-10 | UP | 2.01 | |||
| UP | 5.51 | AEST-19-A-08 | UP | 3.59 | 2.00E-27 | Predicted protein (Neurospora crassa) | |
| DN | 5.76 | AEST-19-A-11 | DN | 2.18 | |||
| DN | 3.05 | AEST-19-C-02 | DN | 1.77 | 1.00E-07 | Polyketide synthase (Cochliobolus heterostrophus) | |
| UP | 5.54 | AEST-19-G-01 | UP | 3.29 | 2.00E-04 | Class A calcium channel variant (Rattus norvegicus) | |
| DN | 3.77 | AEST-19-H-08 | DN | 2.19 | |||
| DN | 3.22 | AEST-20-C-06 | DN | 1.79 | 2.00E-10 | Acid phosphatase precursor (Yarrowia lipolytica) | |
| DN | 5.87 | AEST-20-D-11 | DN | 3.26 | 5.00E-03 | Predicted protein (Neurospora crassa) | |
| DN | 3.29 | AEST-20-E-08 | DN | 2.21 | |||
| DN | 11.26 | AEST-21-A-03 | DN | 2.08 | 2.00E-08 | Hypothetical protein XP_211670 (Homo sapiens) | |
| DN | 3.88 | AEST-21-D-04 | DN | 2.26 | 4.00E-09 | Hypothetical protein (Neurospora crassa) | |
| DN | 10.79 | AEST-21-E-08 | DN | 2.39 | 8.00E-07 | YFW1 gene (Saccharomyces cerevisiae) | |
| UP | 4.92 | AEST-21-E-10 | UP | 2.14 | 2.00E-24 | Predicted protein (Neurospora crassa) | |
| UP | 4.61 | AEST-21-H-05 | UP | 2.50 | 9.00E-05 | Proteophosphoglycan (Leishmania major) | |
| UP | 3.08 | AEST-22-G-02 | UP | 1.81 | 1.00E-07 | Hypothetical protein XP_209582 (Homo sapiens) | |
| DN | 4.75 | AEST-23-B-10 | DN | 2.67 | 2.00E-05 | Keratin-associated protein 4.12 (Mus musculus) | |
| UP | 3.94 | [33] AEST-26-B-03 | DN | 1.81 | 1.00E-36 | Predicted protein (Neurospora crassa) | |
| UP | 6.06 | AEST-27-E-11 | UP | 4.29 | |||
| UP | 10.48 | AEST-30-B-02 | UP | 3.72 | |||
| DN | 4.38 | AEST-36-C-10 | DN | 3.70 | |||
| DN | 3.06 | AEST-39-H-07 | DN | 2.04 | 2.00E-06 | Glycoprotein gp2 precursor (Caenorhabditis elegans) | |
The AEST library identification (AEST ID [1]) for each gene is indicated in column 4. Clones for which real-time RT-PCR confirmations were performed are preceded by a number in brackets that refers to the corresponding clone in Table 3. Two clone IDs, [17] AEST-11-E-06 and [33] AEST-26-B-03, indicate a discrepancy between the microarray and real-time RT-PCR results. The direction and magnitude (average fold change for six hybridizations) of change in transcript accumulation for each gene after CHV1-EP713 infection are indicated in columns 2 and 3, respectively. Similarly, the direction and magnitude of change in transcript accumulation for each gene after CHV1-Euro7 infection are indicated in columns 5 and 6, respectively. As for column 3, the values in column 6 represent the average fold change for six hybridizations. Column 7 indicates the strength of the BLAST hit corresponding to the biological process description and source organism of the matched sequence in column 8. In cases in which a clone was grouped by using a secondary BLAST hit, the strongest biological process description is listed first, followed by the biological process description used to group the clone. In addition, a few clone IDs listed under the “unknown” ontological category returned no BLAST information and are left blank. Direction: UP, up; DN, down.
Validation of differentially expressed clones through real-time RT-PCR.
A total of 39 clones (32 predicted by microarray to be differentially expressed and 7 predicted not to be differentially expressed) were tested by quantitative reverse transcription-PCR (RT-PCR) by using an Applied Biosystems (Foster City, Calif.) GeneAMP 5700 sequence detection system and an Applied Biosystems TaqMan RT kit. Reaction conditions were as described by Allen et al. (1). Transcript abundance was calculated by using the comparative ΔCt method (16) relative to the amount of 18S rRNA in the sample with primers and conditions as described by Parsley et al. (25). Differential expression based on real-time RT-PCR measurements was defined as a change in transcript accumulation of ≥2-fold.
RESULTS
Transcriptional profiling and data analysis.
A custom microarray chip (1) containing EST clones representing ca. 2,200 unique C. parasitica genes (8), which was successfully used to monitor host transcriptional responses to persistent infection by severe hypovirus isolate CHV1-EP713 (1), was used here to determine whether the mild hypovirus isolate CHV1-Euro7 causes a similar change in the host transcriptional profile. Reciprocal (dye-swap) hybridizations were performed for each of three RNA preparations (a total of six hybridizations) obtained from parallel cultures of virus-free C. parasitica strain EP155 and CHV1-Euro7-infected strain EP155 (designated EP155/CHV1-Euro7). An EST clone (gene) was scored as being differentially expressed if the log ratio of the relative probe signal intensities was at least two standard deviations from the experimental average log ratio in at least four of six hybridizations.
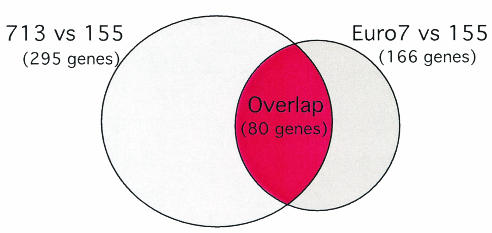
Of the 2,200 C. parasitica genes represented on the microarray, only 166 were scored as being differentially expressed in the CHV1-Euro7-infected strain (90 upregulated and 76 downregulated [available at http://www.umbi.umd.edu/∼cbr/155-Euro7NonRedundant.pdf]). This is roughly one-half the number of genes found to be altered in transcript accumulation after infection by the severe isolate CHV1-EP713 (295 genes, with 132 upregulated and 163 downregulated [1]). That is, CHV1-Euro7 caused only half as many changes in the transcriptional profile as CHV1-EP713. Comparison of the 295 genes differentially expressed after CHV1-EP713 infection (EP155/CHV1-EP713 versus EP155) with the 166 genes differentially expressed after CHV1-Euro7 infection (EP155/CHV1-Euro7 versus EP155) indicated an overlap of 80 genes that were altered in transcript accumulation after infection of EP155 independently by CHV1-EP713 and CHV1-Euro7 (Fig. 1). The list of virus-common genes is presented in Table 1 under headings of putative biological processes as assigned by Dawe et al. (8), according to the classification guidelines outlined by the Gene Ontology Consortium (http://www.genontology.org) and as previously reported for CHV1-EP713-responsive genes by Allen et al. (1).
FIG. 1.
Venn diagram illustrating the total number of differentially expressed genes identified in hybridizations between EP155/CHV1-EP713 versus EP155 (1) and EP155/CHV1-Euro7 versus EP155 (the present study). A total of 80 genes were found on both lists of differentially expressed clones, and these genes are described in Table 1.
Comparison of the expression change profiles for the EP155/CHV1-EP713 versus EP155 and EP155/CHV1-Euro7 versus EP155 hybridizations also suggested that, in addition to causing altered transcript accumulation for a greater number of genes, infection by CHV1-EP713 caused changes that were, in general, of greater magnitude than found for the CHV1-Euro7-infected strain. This was further tested experimentally by performing hybridizations with probes specific for RNA isolated from strain EP155 infected with CHV1-EP713 and CHV1-Euro7, respectively. The results of these hybridizations reflected a number of differences predicted from the lists of differentially expressed genes observed for the EP155/CHV1-EP713 versus EP155 and EP155/CHV1-Euro7 versus EP155 hybridizations. These included the disappearance of genes that were regulated in the same direction by both viruses but that differed in magnitude by <2-fold and in the appearance of new genes that were regulated in different directions by the two viruses, but at levels below the cutoff value in the hybridizations comparing virus-infected strains with uninfected strains (Fig. 2). A total of 411 differentially expressed clones were identified for the EP155/CHV1-EP713 versus EP155/CHV1-Euro7 hybridizations (available at http://www.umbi.umd.edu/∼cbr/713-Euro7NonRedundant.pdf). Of these, 195 were increased in transcript abundance in EP155/CHV1-EP713 relative to EP155/CHV1-Euro7; 216 were decreased.
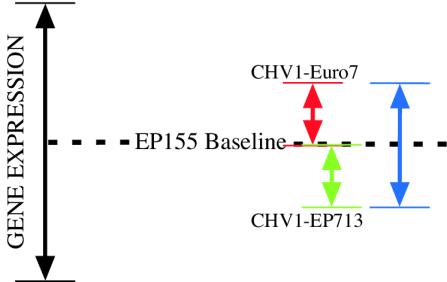
FIG. 2.
Illustration of the differences predicted in the list of differentially regulated genes generated from EP155/CHV1-EP713 versus EP155/CHV1-Euro7 hybridizations from those lists generated from EP155/CHV1-EP713 versus EP155 and EP155/CHV1-Euro7 versus EP155 hybridizations. The length of each colored arrow represents transcript abundance measurements of various magnitude relative to virus-free EP155. The red and green arrows represent transcript changes too small to be reliably detected by microarray profiling for EP155/CHV1-Euro7 versus EP155 and EP155/CHV1-EP713 versus EP155 hybridizations, respectively. The blue arrow represents the same transcript that can be reliably detected by microarray profiling for EP155/CHV1-EP713 versus EP155/CHV1-Euro7 hybridizations. Clones identified in this manner include clones [35], [23], and [12] highlighted in Fig. 3 and Table 3.
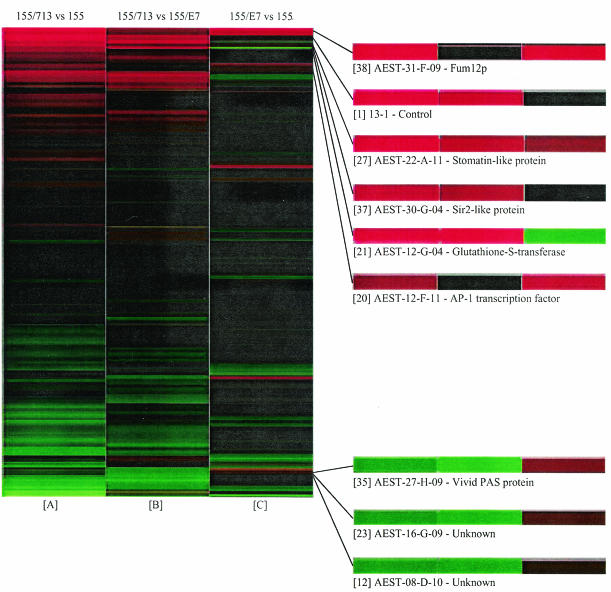
It was reasoned that if the changes in transcript accumulation were of a significantly higher magnitude for CHV1-EP713 infection than for CHV1-Euro7 infection, then the shape of the expression change profile for the EP155/CHV1-EP713 versus EP155/CHV1-Euro7 hybridizations would show a high similarity to that generated for the EP155/CHV1-EP713 versus EP155 hybridizations. This prediction was confirmed by cluster analysis of the average log2 (cy3/cy5) ratios for each cDNA clone on the spotted microarray chip across the three hybridization data sets as visualized in Fig. 3. In columns A and C, red lines indicate an increase in transcript abundance in EP155 after infection with either CHV1-EP713 or CHV1-Euro7, respectively. Green lines indicate a decrease in transcript abundance. In column B, red lines indicate an increase in transcript abundance for EP155/CHV1-EP713 relative to EP155/CHV1-Euro7, whereas green lines indicate a decrease. In all cases, black lines indicate no significant change in transcript abundance was noted between biological samples. It is apparent that the shapes of the expression change profiles for the EP155/CHV1-EP713 versus EP155 and the EP155/CHV1-EP713 versus EP155/CHV1-Euro7 hybridizations are much more similar to each other than either is to the shape of the expression change profile obtained for the EP155/CHV1-Euro7 versus EP155 hybridizations.
FIG. 3.
Visual representation of hierarchically clustered hybridization data sorted according to similarities in gene expression patterns. Column A represents the average log2 (cy3/cy5) ratio for each cDNA clone measured in six hybridizations of EP155/CHV1-EP713 versus EP155. Column B represents the average log2 (cy3/cy5) ratio for each cDNA clone measured in five hybridizations of EP155/CHV1-EP713 versus EP155/CHV1-Euro7. Column C represents the average log2 (cy3/cy5) ratio for each cDNA clone measured in six hybridizations of EP155/CHV1-Euro7 versus EP155. In columns A and C, red lines indicate an increase in transcript abundance in hypovirus-infected strains relative to virus-free EP155. Green lines indicate a decrease in transcript accumulation. In column B, red lines indicate an increase in transcript abundance in EP155/CHV1-EP713 relative to EP155/CHV1-Euro7. Green lines indicate a decrease. In all columns, black lines indicate no significant change in transcript accumulation between biological samples. Clones of interest are highlighted to the right of the cluster diagram. Each clone is preceded by a number in brackets, which refers the reader to the real-time RT-PCR data in Table 3.
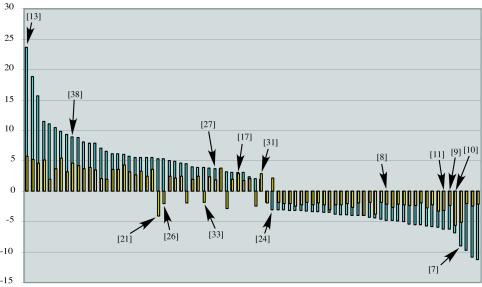
Inspection of the magnitude changes for the 80 virus-common responsive genes (Table 1) further supports the conclusion that host transcriptional response to infection by severe isolate CHV1-EP713 is considerably more dynamic than the response to infection by the mild isolate CHV1-Euro7. This is represented graphically in Fig. 4. It is striking that 73 of the 80 virus-common genes are changed in the same direction by both viruses but almost invariably to a greater magnitude by the severe virus. Moreover, the upregulated and downregulated genes are nearly evenly divided, with 36 upregulated and 37 downregulated. Only seven of the virus-common genes are regulated in opposite directions by the two viruses.
FIG. 4.
Graphical representation of the relative magnitudes of microarray-predicted changes in transcript accumulation for the 80 identified virus-common C. parasitica-responsive genes (from Table 1). Blue-shaded bars indicate the magnitude of transcript accumulation change following CHV1-EP713 infection (fold change [y axis]), whereas the magnitude of change for the same genes after CHV1-Euro7 infection is indicated by the yellow-shaded bars. Specific genes tested by real-time RT-PCR are indicated by bracketed numbers that refer the reader to data listed in Table 3.
Validation of microarray results.
Several control cDNAs were included on the microarray chip to help monitor the consistency between hybridizations (1). These included cDNAs corresponding to CHV1-EP713-encoded proteins p29, p48, and several other portions of open reading frame B (28), the CHV1-EP713-inducible C. parasitica gene, 13-1 (25), and all four exons of the C. parasitica gene epn-1 that encode the aspartyl protease endothiapepsin (1). As indicated in Table 2, a significant increase in transcript abundance was observed for the CHV1-EP713 coding domains p29, p48, and open reading frame B in the CHV1-EP713-infected strain, whereas smaller, but still substantial, increases were observed for the CHV1-Euro7-infected strain due to weaker hybridization by the heterologous CHV1-Euro7 probes to the CHV1-EP713 target sequences (ca. 90% identity between the two viral nucleotide sequences) (4). Consistency was also observed across the different hybridization experiments for changes in transcript abundance for the control C. parasitica cDNAs. Gene 13-1 displayed strong upregulation in the presence of CHV1-EP713 (5-fold [Table 2]), whereas it displayed little or no change in the presence of CHV1-Euro7 (1.5-fold [Table 2]), a finding consistent with previous reports based on real-time RT-PCR measurements (25). It is noteworthy that the 1.5-fold increase in 13-1 transcript accumulation in the CHV1-Euro7-infected strain would not be scored as differentially expressed because it is below the two-standard-deviation cutoff. All four exons of epn-1 (endothiapepsin) exhibited a reduction in transcript accumulation in all three lists.
TABLE 2.
Performance of control spots under three different hybridization conditionsa
| AEST ID | Avg fold change (direction)
|
||
|---|---|---|---|
| 713/155 | Euro7/155 | 713/Euro7 | |
| p48 | 64.08 (UP) | 28.84 (UP) | 3.34 (UP) |
| p29 | 62.29 (UP) | 35.51 (UP) | 2.99 (UP) |
| ORFB | 14.47 (UP) | 4.47 (UP) | 3.92 (UP) |
| 13-1 | 5.04 (UP) | 1.47 (UP) | 2.84 (UP) |
| epn1 (exon 1) | 3.40 (DN) | 2.07 (DN) | 2.22 (DN) |
| epn1 (exon 2) | 4.18 (DN) | 5.13 (DN) | 1.46 (DN) |
| epn1 (exon 3) | 3.69 (DN) | 2.16 (DN) | 2.17 (DN) |
| epn1 (exon 4) | 3.02 (DN) | 2.22 (DN) | 1.92 (DN) |
A list of control spots on the C. parasitica cDNA chip and their relative average abundance measurements across three hybridization conditions is given. Column 1 indicates the clone identification (ID). Column 2 indicates the average fold change up (UP) or down (DN) in transcript abundance measured for EP155/CHV1-EP713 relative to EP155. Column 3 indicates the average fold change up or down in transcript abundance measured for EP155/CHV1-Euro7 relative to EP155. Column 4 indicates the average fold change up or down in transcript abundance measured for EP155/CHV1-EP713 relative to EP155/CHV1-Euro7. ORFB, open reading frame B.
An external quality-control check on the lists of differentially expressed clones generated through microarray profiling was provided by analyzing a subset of 39 clones by quantitative RT-PCR (Table 3). Thirty-two clones that appeared on one or more of the differentially expressed clone lists and seven clones that were absent on all three lists were selected based on putative functions or magnitude of the predicted change in transcript accumulation. Fourteen of the clones were also on the virus-common list presented in Table 1. Transcript accumulation was measured for each of the 39 clones in triplicate for each of two independent RNA isolations in virus-free EP155, EP155/CHV1-EP713, and EP155/CHV1-Euro7. As indicated in Table 3, there were very few cases in which real-time RT-PCR analysis failed to support a microarray-predicted increase (false positive [boldface]) or decrease (false negative [underscored]) in transcript accumulation. These included one false-positive (3%) and two false-negative (5%) results for the EP155/CHV1-EP713 versus EP155 list, three false-positive (8%) and two false-negative (5%) results for the EP155/CHV1-Euro7 versus EP155 list, and two false-positive (3%) and six false-negative (15%) results for the EP155/CHV1-EP713 versus EP155/CHV1-Euro7 list. Microarray-predicted changes in transcript accumulation were confirmed for 12 of the 14 clones found on the virus-common list. These included four that were predicted to be upregulated by both viruses, five that were predicted to be downregulated by both viruses, and three that were predicted to be regulated in opposite directions by the two viruses. False positives were recorded for each of the two virus-common clones that were not supported by real-time RT-PCR: AEST-11-E-06 and AEST-26-B-03. The former was predicted to be upregulated by 3.1-fold in EP155/CHV1-EP713 and was found to be slightly downregulated, whereas the latter was predicted to be upregulated in EP155/CHV1-Euro7 by 1.8-fold and was found to be downregulated by 1.2-fold. Significantly, all seven clones that were selected because of their absence on all three differentially expressed clone lists were also found not to be differentially expressed by real-time RT-PCR. Taken together, the low percentage of false positives and false negatives revealed by real-time RT-PCR (Table 3) provides a high level of confidence in the full lists of differentially expressed clones presented in the present study.
TABLE 3.
Validation of microarray measurements using real-time RT-PCRa
| Clone ID | Hyb condition | Microarray (fold change) | RT-PCR (fold change) | Putative ID (organism) | E-value |
|---|---|---|---|---|---|
| [1] 13-1 | 713/155 | 5.04 | 53.8 (6.6) | Positive control; Cryphonectria parasitica gene | |
| Euro7/155 | ND | 1.5 (0.1) | |||
| 713/Euro7 | 2.84 | 35.87 | |||
| [2] AEST-02-A-07 (AEST-22-B-11)b | 713/155 | ND | 4.3 (1.1) | SAHH (Saccharomyces cerevisiae) | 7.00E-50 |
| Euro7/155 | 2.35 | 2.6 (0.17) | |||
| 713/Euro7 | ND | 1.65 | |||
| [3] AEST-02-B-01 | 713/155 | ND | 1.4 (0.2) | Heterokaryon incompatibility protein (Saccharomyces cerevisiae) | 1.00E-11 |
| Euro7/155 | ND | −1.0 (0.2) | |||
| 713/Euro7 | ND | 1.4 | |||
| [4] AEST-02-B-12 | 713/155 | ND | 1.3 (0.2) | ABC transporter-like protein (Botryotinia fuckeliana) | 1.00E-36 |
| Euro7/155 | ND | −1.0 (0.1) | |||
| 713/Euro7 | ND | 1.3 | |||
| [5] AEST-02-D-01 | 713/155 | ND | 1.4 (0.3) | DNA replication licensing factor (Aspergillus nidulans) | 3.00E-08 |
| Euro7/155 | ND | −1.1 (0.2) | |||
| 713/Euro7 | ND | 1.54 | |||
| [6] AEST-04-C-01 | 713/155 | ND | 2.0 (0.3) | NADH-cytochrome b5 reductase (Saccharomyces cerevisiae) | 4.00E-34 |
| Euro7/155 | ND | −1.6 (0.2) | |||
| 713/Euro7 | ND | 3.2 | |||
| [7] AEST-04-D-04 | 713/155 | −8.98 | −39.9 (9.2) | Unknown | >E-02 |
| Euro7/155 | −5.12 | −8.0 (2.2) | |||
| 713/Euro7 | −2.52 | −4.9 | |||
| [8] AEST-05-A-09 | 713/155 | −4.73 | −5.5 (1.2) | Aspergillopepsin II precursor (Aspergillus niger) | 2.00E-39 |
| Euro7/155 | −2.08 | −3.1 (0.1) | |||
| 713/Euro7 | ND | −1.77 | |||
| [9] AEST-05-C-02 | 713/155 | −6.20 | −16.9 (5.9) | Hydrogenase regulation HoxX (Aquifex aeolicus) | 3.00E-27 |
| Euro7/155 | −2.30 | −7.0 (0.7) | |||
| 713/Euro7 | ND | −2.4 | |||
| [10] AEST-05-D-11 | 713/155 | −6.78 | −891 (297) | Unknown | >E-02 |
| Euro7/155 | −5.62 | −566 (170) | |||
| 713/Euro7 | −9.02 | −1.57 | |||
| [11] AEST-06-B-03 | 713/155 | −6.19 | −62.5 (20.9) | Possible CGI-83 protein (Leishmania major) | 3.00E-30 |
| Euro7/155 | −3.16 | −17.5 (0.9) | |||
| 713/Euro7 | ND | −3.57 | |||
| [12] AEST-08-D-10 | 713/155 | ND | −2.2 (0.4) | Sperm chromatin HMrBNP/H1 (Pseudopleuronectes americanus) | 1.00E-03 |
| Euro7/155 | ND | 3.6 (1.3) | |||
| 713/Euro7 | −2.18 | −7.92 | |||
| [13] AEST-08-F-10 | 713/155 | 23.68 | 4.2 (0.6) | SAMS (Neurospora crassa) | 3.00E-95 |
| Euro7/155 | 5.8 | 2.1 (0.1) | |||
| 713/Euro7 | 2.37 | 2.0 | |||
| [14] AEST-09-C-08 | 713/155 | ND | 1.2 (0.2) | Proteasome regulatory subunit (Schizosaccharomyces pombe) | 1.00E-55 |
| Euro7/155 | −2.72 | −1.3 (0.2) | |||
| 713/Euro7 | ND | 1.56 | |||
| [15] AEST-09-G-11 | 713/155 | −5.73 | −61.7 (21.7) | Unknown | >E-02 |
| Euro7/155 | −2.74 | −17.5 (2.8) | |||
| 713/Euro7 | ND | −3.53 | |||
| [16] AEST-11-B-12 | 713/155 | −2.78 | −3.3 (0.6) | UTP-glucose-1-phosphate uridylyltransferase (Schizosaccharomyces pombe) | 6.00E-61 |
| Euro7/155 | ND | −1.2 (0.0) | |||
| 713/Euro7 | −2.31 | −2.75 | |||
| [17] AEST-11-E-06 | 713/155 | 3.11 | −1.4 (0.2) | Transcriptional regulator (Clostridium tetani E88) | 7.00E-22 |
| Euro7/155 | 2.91 | 2.4 (0.1) | |||
| 713/Euro7 | 1.96 | −3.36 | |||
| [18] AEST-11-F-02 | 713/155 | ND | 1.7 (0.4) | Ubiquitin/S27a fusion protein (Neurospora crassa) | 2.00E-81 |
| Euro7/155 | 1.93 | −1.1 (0.2) | |||
| 713/Euro7 | ND | 1.87 | |||
| [19] AEST-11-F-11 | 713/155 | −4.15 | −4.8 (0.6) | Chain A, α-1,2-mannosidase (Trichoderma reesei) | 6.00E-33 |
| Euro7/155 | ND | −1.4 (0.1) | |||
| 713/Euro7 | −2.55 | −3.43 | |||
| [20] AEST-12-F-11 | 713/155 | ND | 2.2 (0.3) | AP-1-like transcription factor (Neurospora crassa) | 4.00E-37 |
| Euro7/155 | 2.27 | 4.1 (0.9) | |||
| 713/Euro7 | ND | 0.54 | |||
| [21] AEST-12-G-04 | 713/155 | 4.85 | 6.0 (1.5) | GST (Schizosaccharomyces pombe) | 7.00E-39 |
| Euro7/155 | −3.11 | −4.5 (1.5) | |||
| 713/Euro7 | 4.31 | 27 | |||
| [22] AEST-14-H-02 | 713/155 | −3.00 | −2.5 (0.3) | Polyketide synthase (Mycobacterium leprae) | 1.00E-18 |
| Euro7/155 | ND | −2.8 (0.3) | |||
| 713/Euro7 | ND | 0.89 | /PICK> | ||
| [23] AEST-16-G-09 | 713/155 | ND | −1.9 (0.1) | Nuclear matrix protein (Homo sapiens) | 9.00E-03 |
| Euro7/155 | ND | 2.1 (0.3) | |||
| 713/Euro7 | −2.56 | −3.99 | |||
| [24] AEST-17-C-03 | 713/155 | −2.98 | −2.2 (0.3) | Aspartic protease precursor (Botryotinia fuckeliana) | 1.00E-31 |
| Euro7/155 | 2.15 | 2.2 (0.1) | |||
| 713/Euro7 | −3.33 | −4.85 | |||
| [25] AEST-17-H-07 | 713/155 | ND | 1.2 (0.2) | Probable 26S protease subunit (S. cerevisiae) | 2.00E-91 |
| Euro7/155 | ND | −1.4 (0.2) | |||
| 713/Euro7 | ND | 1.68 | |||
| [26] AEST-17-H-10 | 713/155 | 5.36 | 15.1 (5.0) | Isoamyl alcohol oxidase (Aspergillus oryzae) | 3.00E-42 |
| Euro7/155 | −2.02 | −3.9 (0.8) | |||
| 713/Euro7 | 3.27 | 58.89 | |||
| [27] AEST-22-A-11 | 713/155 | 3.76 | 4.6 (0.4) | Stomatin-like protein (Gibberella fujikuroi) | 2.00E-68 |
| Euro7/155 | 1.90 | 2.3 (0.3) | |||
| 713/Euro7 | 2.24 | 2.0 | |||
| [28] AEST-22-B-01 | 713/155 | ND | 1.9 (0.3) | Annexin XIV (Neurospora crassa) | 2.00E-34 |
| Euro7/155 | ND | −1.3 (0.2) | |||
| 713/Euro7 | ND | 2.47 | |||
| [29] AEST-22-D-09 | 713/155 | ND | 1.2 (0.2) | Ubiquitin-activating enzyme E1 (Candida albicans) | 1.00E-47 |
| Euro7/155 | ND | 1.0 (0.2) | |||
| 713/Euro7 | ND | 1.2 | |||
| [30] AEST-22-H-05 | 713/155 | ND | 4.5 (0.3) | Serine/threonine kinase (Rattus norvegicus) | 1.00E-04 |
| Euro7/155 | ND | 1.8 (0.5) | |||
| 713/Euro7 | 2.15 | 2.5 | |||
| [31] AEST-24-D-01 | 713/155 | 2.02 | 2.5 (0.4) | SnodProt1 precursor (Neurospora crassa) | 2.00E-32 |
| Euro7/155 | 2.92 | 2.9 (0.3) | |||
| 713/Euro7 | ND | 0.86 | |||
| [32] AEST-25-B-11 | 713/155 | −3.54 | −5.9 (1.3) | Nuclease P1 (Penicillium citrinum) | 1.00E-75 |
| Euro7/155 | ND | −1.1 (0.1) | |||
| 713/Euro7 | −3.18 | −5.36 | |||
| [33] AEST-26-B-03 | 713/155 | 3.94 | 8.2 (1.0) | Predicted protein (Neurospora crassa) | 1.00E-36 |
| Euro7/155 | −1.81 | 1.2 (0.4) | |||
| 713/Euro7 | 2.63 | 6.83 | |||
| [34] AEST-27-F-10 | 713/155 | −3.2 | −6.3 (0.4) | Pro1 (Neurospora crassa) | 4.00E-31 |
| Euro7/155 | ND | −1.8 (0.2) | |||
| 713/Euro7 | −2.1 | −3.5 | |||
| [35] AEST-27-H-09 | 713/155 | ND | −1.5 (0.3) | Vivid PAS protein (Neurospora crassa) | 1.00E-38 |
| Euro7/155 | ND | 2.1 (0.2) | |||
| 713/Euro7 | −2.53 | −3.15 | |||
| [36] AEST-30-C-09 | 713/155 | −4.95 | −2.4 (0.3) | Mst12 (Magnaporthe grisea) | 6.00E-65 |
| Euro7/155 | ND | −1.47 (0.22) | |||
| 713/Euro7 | −1.97 | −1.63 | |||
| [37] AEST-30-G-04 | 713/155 | 2.94 | 4.5 (0.8) | Sir2-like (Saccharomyces cerevisiae) | 3.00E-27 |
| Euro7/155 | ND | −1.1 (0.2) | |||
| 713/Euro7 | 2.18 | 4.95 | |||
| [38] AEST-31-F-09 | 713/155 | 8.90 | 16.3 (2.0) | Fum12p (Gibberella moniliformis) | 5.00E-35 |
| Euro7/155 | 4.67 | 7.9 (0.5) | |||
| 713/Euro7 | ND | 2.06 | |||
| [39] AEST-32-A-06 | 713/155 | −5.44 | −15.5 (4.2) | MAS3 protein (Magnaporthe grisea) | 4.00E-11 |
| Euro7/155 | ND | −1.6 (0.4) | |||
| 713/Euro7 | −4.15 | −9.69 |
Real-time RT-PCR measurements of 39 clones. Measurements were made in triplicate for each clone by using two independent total RNA preparations. Clone IDs are indicated in column 1. Column 2 indicates the sources of RNA used for hybridization reactions and real-time RT-PCR measurements: 713/155, transcript accumulation in EP155/CHV1-EP713 relative to virus-free strain EP155; Euro7/155, transcript accumulation in EP155/CHV1-Euro7 relative to virus-free strain EP155; and 713/Euro7, transcript accumulation in EP155/CHV1-EP713 relative to EP155/CHV1-Euro7. Column 3 indicates the average change (n-fold) for each clone ID calculated from microarray experiments (six total hybridizations, from three sets of dye-swap experiments). Column 4 indicates the average change (n-fold) for each clone ID as measured by real-time RT-PCR, with the standard error of the mean in parentheses. Column 5 provides a brief biological process description from the BLAST hit provided in column 6. Underscored text indicates false-negative and boldface text indicates false-positive microarray predictions revealed by real-time RT-PCR analysis. Differential expression based on real-time RT-PCR measurements was defined as a change in transcript accumulation of twofold or greater. ND, no change detected by microarray analysis.
The discrepancy between microarray predictions and real-time RT-PCR measurements for AEST-02-A-07 in CHV1-EP713-infected EP155 prompted an examination of related AEST clones on the primary CHV1-EP713 and CHV1-Euro7 responsive gene lists. The gene SAHH (gi 28924052) is represented by four AEST clones on the C. parasitica microarray chip: AEST-22-B-11, AEST-40-D-10, AEST-01-H-09, and AEST-02-A-07. In this case, the gene was not included on the virus common list (Table 1) because none of the four AESTs were found on both the CHV1-EP713 and CHV1-Euro7 primary responsive lists; AEST-22-B-11, AEST-40-D-10, and AEST-01-H-09 were on the CHV1-EP713-responsive list and AEST-02-A-07 was on the CHV1-Euro7-responsive list. AEST-22-B-11, shown in parentheses, is the representative for gi number 28924052 on the CHV1-EP713 published list (1). The reader is referred to the microarray data management section of Materials and Methods for further explanation.
DISCUSSION
The recent development of a C. parasitica cDNA microarray platform (1) provides new opportunities to address a number of longstanding basic questions regarding hypovirus-mediated alterations of fungal phenotype and virulence. Allen et al. (1) used microarray analysis to confirm that hypovirus infection results in a stable alteration in transcript accumulation for a significant portion of the host genome (3, 17) and to expand the list of hypovirus-responsive C. parasitica genes from less than 20 to nearly 300. As illustrated here, microarray analysis also allows high-resolution comparisons of the effect of different hypovirus isolates on C. parasitica gene expression.
There is considerable evidence that hypoviruses are major contributors to the diversity in virulence levels and phenotypic traits observed for C. parasitica field isolates (6, 11-14, 22, 26). The constellation of altered traits that can accompany hypovirus-mediated attenuation of C. parasitica virulence is thought to significantly influence the effectiveness of hypovirulent strains as biological control agents for chestnut blight (9). For example, the severely reduced levels of asexual sporulation and loss of female fertility observed for many hypovirus-infected strains limit hypovirus transmission through and persistence within a C. parasitica population (22). The ability to monitor global changes in transcriptional responses to different hypoviruses provides unprecedented opportunities to gain insights into molecular mechanisms underlying differences in hypovirus-mediated symptom expression and virulence attenuation.
The high level of sequence similarity for CHV1-EP713 and CHV1-Euro7 coupled with the quite different phenotypic changes caused by the two viruses suggested several possible outcomes for microarray analysis. These ranged from (i) altered expression of a similar set of genes by both viruses but with a much greater magnitude of change by the severe virus to (ii) each virus causing alterations in transcript accumulation for an entirely distinct set of host genes. As indicated in Fig. 1, 3, and 4, the observed outcome fell between these two extremes. The number of host genes differentially expressed as a result of CHV1-Euro7 infection equaled approximately half the number that were responsive to CHV1-EP713 infection (166 versus 295, respectively). Of these, 80 genes were found to be responsive to both viruses.
Considered from another perspective, of the ca. 2,200 C. parasitica genes represented on the microarray chip, a total of 301 (13.6%) were scored as responsive to one virus or the other, and only 80 (3.6%) were responsive to both viruses, with 73 of those responding in the same direction. Extrapolations based on the estimate that the EST clones contained on the microarray represent ca. 22% of the predicted C. parasitica gene coding capacity (1) suggest that a total of only 363 host genes are altered in transcript accumulation as a general response to hypovirus infection. One could imagine that these virus-common responsive genes might include genes that are (i) altered in expression as part of a cellular defense response, (ii) involved in the control of hypovirus copy number, or (iii) altered in expression levels in order for viral RNA replication to proceed.
Differences were also evident in the magnitudes by which transcript accumulation changed in response to infection by the mild and severe hypovirus isolates. CHV1-Euro7 infection generally caused smaller changes in transcript accumulation than were observed for CHV1-EP713 infection. This difference in magnitude is clearly illustrated for the virus-common responsive genes in Fig. 4 and was supported by real-time RT-PCR analysis for a selected number of genes (Table 3). Additional experimental evidence for this general trend was generated by comparative hybridizations with cDNAs derived from RNA isolated from CHV1-EP713- and CHV1-Euro7-infected strain EP155. The expression change profile for the EP155/CHV1-EP713 versus EP155/CHV1-Euro7 hybridization was clearly more similar to that of EP155/CHV1-EP713 versus EP155 than to the profile derived from the EP155/CHV1-Euro7 versus EP155 hybridization (Fig. 3).
Allen et al. (1) previously discussed the potential relevance of a number of CHV1-EP713-responsive genes with high database sequence matches to hypovirus-mediated symptom expression and virus replication. Interestingly, several of these genes also appeared in lists of hypovirus-responsive genes generated in the present study. Transcripts for the glutathione S-transferase (GST) homologue AEST-12-G-04 and the HSP70 homologue AEST-10-H-10 were found to be constitutively increased (6- to 13-fold and 2.7- to 2.8-fold, respectively) in CHV1-EP713-infected mycelia (1). GSTs are a superfamily of isoenzymes responsible for detoxifying the cellular environment by removing reactive oxygen through conjugation of thiol reduced glutathione to various harmful ligands, including plant phenols and aflatoxins (29). Microarray profiling studies (using Affymetrix GeneChips) conducted in Arabidopsis thaliana leaves independently infected with five different RNA viruses (31) identified four different GST homologues that are each induced in the presence of all five viruses. Cellular HSP70 proteins have been reported to be recruited during infection by a number of viruses to facilitate virion assembly or genome replication (7, 15, 30). Since the increased transcript accumulation of GST and HSP70 homologues was not accompanied by altered transcript accumulation for many additional heat shock and classical stress response genes represented on the C. parasitica EST microarray chip, Allen et al. (1) advanced the possibility that these homologues belong to a subset of stress-related genes induced by hypovirus infection to facilitate viral functions. As indicated in Table 1, the HSP70 homologue was increased in transcript accumulation after infection by both viruses. However, the GST homologue was downregulated by 4.5-fold after CHV1-Euro7 infection (Table 3 and Fig. 3); one of just seven genes that were found to be regulated in opposite direction by CHV1-EP713 and CHV1-Euro7. The observation that GST transcript accumulation is strongly increased in EP155/CHV1-EP713 and strongly decreased in EP155/CHV1-Euro7 justifies an examination of relative effects of the mild and severe hypovirus isolates on the cellular redox state and possible ramifications for differences in gene expression. The upregulation of HSP70 by both viruses adds some additional support for the speculation that HSP70 may play a role in virus replication.
S-Adenosyl-l-methionine synthetase (SAMS; AEST-08-F-10 and S-adenosyl-l-homocysteine hydrolase (SAHH; represented by AEST-22-B-11 on the CHV1-EP713-responsive list and by AEST-02-A-07 on the CHV1-Euro7-responsive list, see legend to Table 3) are responsible for generation of the primary methyl donor S-adenosyl-l-methionine (SAMe) and removal of S-adenosyl-l-homocysteine (SAH), the by-product of transmethylation reactions, respectively. Because of the central role played by SAMe in cellular metabolism, Allen et al. (1) considered the possibility that the constitutive increase in transcript accumulation for genes encoding both SAMS and SAHH in response to CHV1-EP713 infection could have significant metabolic or physiologic consequences for the host. The association between abnormal intracellular levels of SAMe and altered genome stability (18-20, 27), coupled with the reported influence of intracellular SAH levels on senescence and cell growth (21, 32), also prompted the suggestion that hypovirus infection may provide a useful model for examining the consequences of chronic RNA virus infection on host genome stability. These considerations are reinforced by the observation that homologues of both of these enzymes are upregulated by both the mild and severe isolates but to a greater extent by CHV1-EP713 (Tables 1 and 3).
Allen et al. (1) made the interesting observation that only 3 of 26 genes on the EST chip that fell under the molecular function category “transcription regulation/transcription factors” in Dawe et al. (8) were responsive to CHV1-EP713; each was reduced by at least threefold. These included the homologues of Mst12 (AEST-30-C-09) from Magnaporthe grisea, shown to be important for regulating infectious hyphae growth (24); Pro1 (AEST-27-F-10), involved in controlling sexual sporulation in several filamentous fungi (23); and HoxX (AEST-05-C-02), part of a bacterial two-component regulatory system (10). HoxX appears on the virus-common list (Table 1), and its downregulation by both viruses was confirmed by real-time RT-PCR (Table 3). Pro1 and Mst12 homologues were present only on the CHV1-EP713-responsive list (Table 3). However, a slight reduction in transcript accumulation (below the twofold cutoff) was observed for these genes by real-time RT-PCR measurements after CHV1-Euro7 infection. Finally, the homologue of the general transcription factor AP1 (AEST-12-F-11) was found by microarray analysis to be upregulated by CHV1-Euro7. This result was confirmed by real-time RT-PCR (Table 3), which also revealed a modest level of upregulation in CHV1-EP713-infected mycelia. This collection of hypovirus-regulated putative transcription factors represent prime candidates for future functional studies related to hypovirus-mediated alterations of host gene expression.
The microarray analyses described here have resulted in the initial assignment of >20% of the C. parasitica-expressed genes to one of the following categories for the experimental conditions used in the present study: (i) nonresponsive to hypovirus infection, (ii) upregulated by both mild and severe hypoviruses, and (iii) downregulated by both mild and severe hypoviruses, and (iv) regulated in opposite directions by the two viruses. As indicated in Table 1 and online supplementary lists, the genes assigned an “unknown” biological function form the largest category in both the virus-specific and virus-common responsive gene lists reported in the present study. Consequently, a comprehensive understanding of the significance of the transcriptional responses to hypovirus infection will require additional functional studies of the differentially expressed genes identified through microarray profiling. However, it is anticipated that further refinement of the virus-specific and virus-common lists through the addition of differential expression data for other experimental conditions and hypoviruses, e.g., chimeric viruses, will reveal trends and patterns that will drive the direction of future mechanistic studies.
Acknowledgments
This study was supported in part by Public Health Service grant GM55981 to D.L.N.
REFERENCES
- 1.Allen, T. D., A. L. Dawe, and D. L. Nuss. 2003. Use of cDNA microarrays to monitor transcriptional responses of the chestnut blight fungus Cryphonetria parasitica to infection by virulence-attenuating hypoviruses. Eukaryot. Cell 2:1253-1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anagnostakis, S. L. 1982. Biological control of chestnut blight. Science 215:466-471. [DOI] [PubMed] [Google Scholar]
- 3.Chen, B., S. Gao, G. H. Choi, and D. L. Nuss. 1996. Extensive alteration of fungal gene transcript accumulation and elevation of G-protein-regulated cAMP levels by a virulence-attenuating hypovirus. Proc. Natl. Acad. Sci. USA 93:7996-8000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, B., and D. L. Nuss. 1999. Infectious cDNA clone of hypovirus CHV1-Euro7: a comparative virology approach to investigate virus-mediated hypovirulence of the chestnut blight fungus Cryphonectria parasitica. J. Virol. 73:985-992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choi, G. H., and D. L. Nuss. 1992. Hypovirulence of chestnut blight fungus conferred by an infectious viral cDNA. Science 257:800-803. [DOI] [PubMed] [Google Scholar]
- 6.Chung, P., P. J. Bedker, and B. I. Hillman. 1994. Diversity of Cryphonectria parasitica hypovirulence-associated double-stranded RNAs within a chestnut population in New Jersey. Phytopathology 84:984-990. [Google Scholar]
- 7.Cripe, T. P., S. E. Delos, P. A. Estes, and R. L. Garcea. 1995. In vivo and in vitro association of hsc70 with polyomavirus capsid proteins. J. Virol. 69:7807-7813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dawe, A. L., V. C. McMains, M. Panglao, S. Kasahara, B. Chen, and D. L. Nuss. 2003. An ordered collection of expressed sequences from Cryphonectria parasitica and evidence of genomic microsynteny with Neurospora crassa and Magnaporthe grisea. Microbiology 149:2373-2384. [DOI] [PubMed] [Google Scholar]
- 9.Dawe, A. L., and D. L. Nuss. 2001. Hypoviruses and chestnut blight: exploiting viruses to understand and modulate fungal pathogenesis. Annu. Rev. Genet. 35:1-29. [DOI] [PubMed] [Google Scholar]
- 10.Durmowicz, M. C., and R. J. Maier. 1997. Roles of HoxX and HoxA in biosynthesis of hydrogenase in Bradyrhizobium japonicum. J. Bacteriol. 179:3676-3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elliston, J. E. 1978. Pathogenicity and sporulation in normal and diseased strains of Endothia parasitica in American chestnut. p. 95-100. In W. L. MacDonald, F. C. Cech, J. Luchok, and C. Smith (ed.), Proceedings of the American chestnut symposium. West Virginia University Press, Morgantown.
- 12.Elliston, J. E. 1985. Characteristics of dsRNA-free and dsRNA-containing strains of Endothia parasitica in relation to hypovirulence. Phytopathology 75:151-158. [Google Scholar]
- 13.Enebak, S. A., W. L. MacDonald, and B. I. Hillman. 1994. Effect of dsRNA associated with isolates of Cryphonectria parasitica from the central Appalachians and their relatedness to other dsRNAs from North America and Europe. Phytopathology 84:528-534. [Google Scholar]
- 14.Griffin, G. J. 1986. Chestnut blight and its control. Hort. Rev. 8:291-336. [Google Scholar]
- 15.Holstein, S. E., H. Ungewickell, and E. Ungewickell. 1996. Mechanism of clathrin basket dissociation: separate functions of protein domains of the DnaJ homologue auxilin. J. Cell Biol. 135:925-937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson, M. R., K. Wang, J. B. Smith, M. J. Heslin, and R. B. Diasio. 2000. Quantitation of dihydropyrimidine dehydrogenase expression by real-time reverse transcription polymerase chain reaction. Anal. Biochem. 278:175-184. [DOI] [PubMed] [Google Scholar]
- 17.Kang, H.-S., J.-W. Choi, S.-M. Park, B. Cha, M.-S. Yang, and D.-H. Kim. 2000. Ordered differential display from Cryphonectria parasitica. Plant Pathol. J. 16:142-146. [Google Scholar]
- 18.Kricker, M. C., J. W. Drake, and M. Radman. 1992. Duplication-targeted DNA methylation and mutagenesis in the evolution of eukaryotic chromosomes. Proc. Natl. Acad. Sci. USA 89:1075-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laird, P. W., L. Jackson-Grusby, A. Fazeli, S. L. Dickinson, W. E. Jung, E. Li, R. A. Weinberg, and R. Jaenisch. 1995. Suppression of intestinal neoplasia by DNA hypomethylation. Cell 81:197-205. [DOI] [PubMed] [Google Scholar]
- 20.Laird, P. W., and R. Jaenisch. 1994. DNA methylation and cancer. Hum. Mol. Genet. 3:1487-1495. [DOI] [PubMed] [Google Scholar]
- 21.Lo, S. C., L. Hamer, and J. E. Hamer. 2002. Molecular characterization of a cystathionine β-synthase gene, CBS1, in Magnaporthe grisea. Eukaryot. Cell 1:311-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.MacDonald, W. L., and D. W. Fulbright. 1991. Biological control of chestnut blight: use and limitation of transmissible hypovirulence. Plant Dis. 75:656-661. [Google Scholar]
- 23.Masloff, S., S. Jacobsen, S. Poggeler, and U. Kuck. 2002. Functional analysis of the C6 zinc finger gene pro1 involved in fungal sexual development. Fung. Genet. Biol. 36:107-116. [DOI] [PubMed] [Google Scholar]
- 24.Park, G., C. Xue, L. Zheng, S. Lam, and J. R. Xu. 2002. MST12 regulates infectious growth but not appressorium formation in the rice blast fungus Magnaporthe grisea. Mol. Plant-Microb. Interact. 15:183-192. [DOI] [PubMed] [Google Scholar]
- 25.Parsley, T. B., B. Chen, L. M. Geletka, and D. L. Nuss. 2002. Differential modulation of cellular signaling pathways by mild and severe hypovirus strains. Eukaryot. Cell 1:401-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peever, T. L., Y.-C. Liu, and M. G. Milgroom. 1997. Diversity of hypoviruses and other double-stranded RNAs in Cryphonectria parasitica in North America. Phytopathology 87:1026-1033. [DOI] [PubMed] [Google Scholar]
- 27.Selker, E. U. 1990. DNA methylation and chromatin structure: a view from below. Trends Biochem. Sci. 15:103-107. [DOI] [PubMed] [Google Scholar]
- 28.Shapira, R., G. H. Choi, and D. L. Nuss. 1991. Virus-like genetic organization and expression strategy for a double-stranded RNA genetic element associated with biological control of chestnut blight. EMBO J. 10:731-739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sheehan, D., G. Meade, V. M. Foley, and C. A. Dowd. 2001. Structure, function, and evolution of glutathione transferases: implications for classification of non-mammalian members of an ancient enzyme superfamily. Biochem. J. 360:1-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ungewickell, E., H. Ungewickell, S. E. Holstein, R. Lindner, K. Prasad, W. Barouch, B. Martin, L. E. Greene, and E. Eisenberg. 1995. Role of auxilin in uncoating clathrin-coated vesicles. Nature 378:632-635. [DOI] [PubMed] [Google Scholar]
- 31.Whitham, S. A., S. Quan, H. S. Chang, B. Cooper, B. Estes, T. Zhu, X. Wang, and Y. M. Hou. 2003. Diverse RNA viruses elicit the expression of common sets of genes in susceptible Arabidopsis thaliana plants. Plant J. 33:271-283. [DOI] [PubMed] [Google Scholar]
- 32.Xu, D., R. Neville, and T. Finkel. 2000. Homocysteine accelerates endothelial cell senescence. FEBS Lett. 470:20-24. [DOI] [PubMed] [Google Scholar]