Abstract
Break-induced replication (BIR) is a nonreciprocal recombination-dependent replication process that is an effective mechanism to repair a broken chromosome. We review key roles played by BIR in maintaining genome integrity, including restarting DNA replication at broken replication forks and maintaining telomeres in the absence of telomerase. Previous studies suggested that gene targeting does not occur by simple crossings-over between ends of the linearized transforming fragment and the target chromosome, but involves extensive new DNA synthesis resembling BIR. We examined gene targeting in Saccharomyces cerevisiae where only one end of the transformed DNA has homology to chromosomal sequences. Linearized, centromere-containing plasmid DNA with the 5′ end of the LEU2 gene at one end was transformed into a strain in which the 5′ end of LEU2 was replaced by ADE1, preventing simple homologous gene replacement to become Leu2+. Ade1+ Leu2+ transformants were recovered in which the entire LEU2 gene and as much as 7 kb of additional sequences were found on the plasmid, joined by microhomologies characteristic of nonhomologous end-joining (NHEJ). In other experiments, cells were transformed with DNA fragments lacking an ARS and homologous to only 50 bp of ADE2 added to the ends of a URA3 gene. Autonomously replicating circles were recovered, containing URA3 and as much as 8 kb of ADE2-adjacent sequences, including a nearby ARS, copied from chromosomal DNA. Thus, the end of a linearized DNA fragment can initiate new DNA synthesis by BIR in which the newly synthesized DNA is displaced and subsequently forms circles by NHEJ.
During the past several years, some old ideas about how recombination occurs have received strong experimental support. Meselson and Weigle (1) first proposed that crossing-over could be explained by a break-copy mechanism in which one end of a double-strand break (DSB) could invade an intact linear template molecule and initiate new DNA synthesis that could proceed to the end of the chromosome template. In essence, a recombination event led to the establishment of a unidirectional replication fork. Skalka (2) provided a more molecular view of this idea (a replicator's view of recombination, as she called it) to explain phage λ recombination. Mosig (3, 4) made a similar proposal to account for late DNA replication in phage T4. Formosa and Alberts (5) provided a key in vitro experimental demonstration for the formation of a replication fork by recombination. More recent studies by George and Kreuzer (6) of DSB-induced recombination, controlled by phage T4 genes in Escherichia coli have supported the idea that recombination leads to extensive replication. Similarly, recent experiments by Motamedi et al. (7) and by Kuzminov and Stahl (8) with phage λ have provided strong evidence that a major pathway to generate crossing-over involves extensive replication during break-copy recombination.
These ideas were applied by Kogoma (9, 10) to explain origin-independent, recombination-dependent replication of the E. coli genome. Several recent experiments have strongly supported the idea that break-induced replication (BIR) is an important process in restarting broken replication forks (11–13).
Possible Mechanisms of BIR
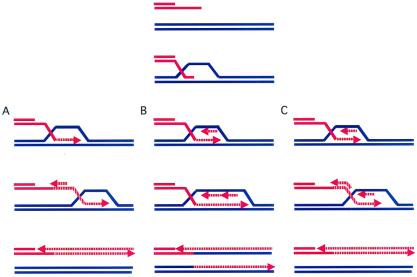
BIR events begin as one-ended recombination events, either because there is only one free DNA end or because only one of two ends of the DSB succeeds in strand invasion of a homologous sequence. One can imagine a number of ways in which the process can occur; there have not yet been any experimental observations to distinguish among them. In one scenario, strand invasion creates a D-loop that then migrates down the template (5); this process is analogous to the way RNA polymerase copies DNA, displacing a single strand of newly synthesized DNA (Fig. 1A). Subsequently, the single strand could be filled in—a process that might have different requirements than normal lagging-strand DNA synthesis—such that all of the newly synthesized DNA is associated with the initially broken end. Alternatively, the D loop could be transformed into a complete unidirectional replication fork that then migrates down the template chromosome (Fig. 1B). This process would result in two semiconservatively replicated molecules and a single Holliday junction that would have to be resolved. A third version imagines that the replication structure is acted on by branch migration enzymes, so that both newly synthesized leading and lagging strands are displaced and DNA synthesis is conservative (Fig. 1C). It is also not necessary that the replication structure reach the end of the template chromosome, because BIR might be terminated by an encounter with a converging replication fork.
Figure 1.
Alternative BIR mechanisms. A broken chromosome end will be resected by 5′ to 3′ exonucleases, allowing the 3′ end to interact with various recombination proteins to carry out strand invasion. (A) The 3′ end of the invading strand initiates DNA replication, leading to a migrating D-loop “bubble” as described by Formosa and Alberts (5). the displaced newly synthesized DNA strand can then be made double-stranded. (B) Strand invasion sets up a replication fork that will result in semiconservatively synthesized molecules. A Holliday junction will be resolved at some point. (C) Strand invasion sets up a replication fork in which branch migration enzymes displace both newly synthesized DNA strands as the replication structure migrates down the template.
BIR in Saccharomyces cerevisiae
Recombination in eukaryotes also appears to occur some of the time by recombination-dependent DNA replication processes. These experiments have been carried out mostly in S. cerevisiae. Esposito (14, 15) was the first to note examples of mitotic recombination in which there was a nonreciprocal recombination event that extended hundreds of kilobases down a chromosome arm. In this case, one daughter cell was identical to the parent diploid, in that it was still heterozygous for markers extending along the chromosome arm whereas the other cell was homozygous for all these alleles. Voelkel-Meiman and Roeder (16) saw similar events promoted by a mitotic “hot spot” and suggested that they could arise by extensive BIR. The idea that a broken chromosome end could acquire a new telomere by such a process was provided by Dunn et al. (17). They showed that a linearized plasmid with one end that lacked a telomere, but had homology to a subtelomeric Y′ region, could become stable by recombining with an intact, telomere-containing chromosome that also had an adjacent Y′ sequence. However, in these experiments, it was not possible to show whether the repair event was replicative or whether the transforming DNA acquired a new chromosome end at the expense of one of two sister chromosomes in the G2 phase of the cell cycle. At the same time, Walmsley et al. (18) also suggested that normal telomere maintenance could be achieved if one telomeric region used another as a template to extend the end by replication.
Bosco and Haber (19) extended this idea by studying the repair of a chromosomal DSB, created by the site-specific HO endonuclease, in which the centromere-distal side of the DSB had effectively no homology to any other site in the haploid genome, so that gene conversion repair could not occur. However, the centromere-proximal side of the DSB shared 70 bp homology with homologous sequences in the HMR locus, located 30 kb from the opposite end of the same chromosome, in the opposite orientation. The DSB was thus repaired by a recombination event leading to the formation of a 30-kb nonreciprocal translocation. This process required the Rad52p recombination protein. More recently, Malkova et al. (20) have shown that HO-induced BIR events also can be found in meiotic cells, especially when normal meiotic recombination is impaired.
A direct demonstration of the replicative nature of BIR repair was provided by Morrow et al. (21), who showed that transforming yeast, with a DNA fragment with an origin of replication and a centromere with two oppositely oriented identical DNA segments at the ends, could result in the creation of an entirely new chromosome, in which both of the end segments had to recombine with the same unique homologous sequence on one chromosome and, both times, replicate all of the way to the chromosome end. Bosco and Haber (19) found similar results when they used HO endonuclease to lop off the end of a chromosome in a diploid, in which the DSB shared extensive homology only centromere-proximal to the DSB. They found that interchromosomal BIR could occur in G1 cells, producing two daughter cells with identical repair events. This result suggests that BIR can occur outside of normal S phase.
Two (or More) Pathways of Break-Induced Replication in S. cerevisiae
The genetic requirements of BIR have been determined by examining a diploid in which there is a single HO-induced DSB in the middle of the right arm of chromosome III. Normally, such a DSB would be repaired by “short patch” gene conversion, and indeed a rad52Δ diploid shows almost no repair of the broken chromosome; it is simply lost, creating a 2n-1 monosomic derivative. But, surprisingly, a rad51Δ strain eliminates gene conversions but still allows BIR to proceed (22). In colonies derived from single cells suffering a DSB, more than 80% of them give rise to at least a sector of cells within the colony that have retained the centromere and left arm of the broken chromosome by BIR, whereas the other cells in the colony had lost the broken chromosome. Signon et al. (23) have shown that a similar phenotype is found in rad54Δ, rad55Δ, and rad57Δ mutants, all of which eliminate gene conversions but allow BIR.
The idea that recombination-dependent initiation of repair DNA synthesis to the end of the chromosome could occur without the only known strand exchange protein, Rad51p, remains a great mystery. Recently, we have found that the sites where BIR is initiated in the absence of RAD51 are distinctly nonrandom. Virtually none of the repair events retain a marker on the broken chromosome that is 13 kb centromere-proximal to the DSB site. In fact, there appears to be a small, cis-acting DNA sequence (≤200 bp) located 34 kb proximal to the DSB site that is responsible for facilitating the majority of BIR events (24). We have speculated that this enhancer site permits the formation of a processive repair replication fork that is capable of traversing more than 150 kb to the end to the template chromosome. It is also possible that the small enhancer sequence acts similarly to the Chi sequence of E. coli, to “tame” exonucleolytic degradation of the chromosome and allow formation of an intermediate leading to BIR.
Because a DSB in the middle of a chromosome is so efficiently repaired by gene conversion in a RAD51 cell, it has not been possible to characterize a RAD51-dependent BIR process in the same diploid system. To examine this RAD51-dependent process, we have created a modified diploid in which the target chromosome is truncated such that there is only a very short segment of homology distal to the DSB that is too short to permit repair by gene conversion (M. Naylor, A. Malkova and J.E.H., unpublished results). In that strain, RAD51-mediated BIR is significantly more efficient than what is seen in the absence of RAD51. Moreover, the RAD51-dependent pathway does not require the distant facilitating sequence that promotes BIR in a rad51Δ diploid, and most of the repair events are initiated close to the site of the DSB.
Further genetic analysis of the RAD51-, RAD54-independent BIR pathway has revealed that it is largely dependent on another set of recombination genes: RAD50, MRE11, XRS2, RAD59, and TID1 (RDH54) (23). Double mutants, including rad51Δ rad50Δ, rad51Δ rad59Δ, and rad54Δ tid1Δ, fail to repair the DSB more than 90% to the time, leading to chromosome loss. However, 10% of the cells still give rise to sectors that appear to be BIR events by Southern blot and genetic analysis; thus none of these double mutants is as severely defective as a rad52Δ strain. This result is reminiscent of a study by Bai and Symington (25) of spontaneous heteroallelic recombination, in which a rad51 rad59 double mutant was still 3-fold less deficient in recombination than a rad52 strain. Perhaps there is still a third pathway to be discovered.
Yeast Telomere Maintenance in the Absence of Telomerase Appears To Employ BIR
In the absence of the telomerase enzyme, chromosome ends slowly shorten, in part because they fail to be replicated to the very end and perhaps because they are also resected by exonucleases. Depending on the initial length of the telomere, the rate of shortening, and the minimum size of a telomere repeat that is required to “cap” the chromosome end, cells can proliferate for many generations. In budding yeast, cells do not exhibit senescence for more than 50 generations, whereas in mice, the germline can be passed through 5 generations before clear evidence of genome instability is detected. In S. cerevisiae, Lundblad and Blackburn (26) first demonstrated that, among cells that cannot maintain chromosome ends by telomerase, there emerged survivors that had somehow managed to maintain telomere sequences at their ends. Survival depends on the RAD52 recombination gene. Many, but not all, of these survivors had also amplified subtelomeric Y′ elements that were originally present at some telomeres so that now virtually all ends had Y′ sequences. Yet curiously, the Y′ ends still carried telomere repeats. More recently, Teng and Zakian (27) showed that there were in fact two distinct types of survivors. Type I cells exhibited the amplification of Y′ sequences and telomere ends, whereas type II cells had managed to dramatically elongate the number of telomere repeats themselves, again by a RAD52-dependent recombination mechanism, without affecting subtelomeric Y′ distribution.
The demonstration of two distinct types of RAD52-dependent telomere maintenance in the absence of telomerase was consistent with the conclusions of Le et al. (28) that there were two genetically distinct pathways of recombination that could maintain telomeres. Deletion of the RAD51, RAD54, RAD55, and RAD57 genes caused an accelerated loss of viability, similar to rad52Δ cells, but there were still survivors. In contrast, deletions of RAD50, MRE11, and XRS2 caused a slower rate of senescence, and again there were survivors. However, a rad51Δ rad50Δ double mutant eliminated survivors, leading to the suggestion that there are two distinct RAD52-dependent recombination pathways. Teng et al. (29) showed that type I survivors are eliminated in a rad51Δ strain whereas Type II survivors are absent in a rad50Δ strain. Chen et al. (30) found that type II survivors also depend on RAD59. Recently, several labs have found that the Sgs1p, yeast's homologue of Bloom's and Werner's syndromes in humans, is also required for Type II events (31–33).
Thus, telomere maintenance in the absence of telomerase appears to obey the same genetic rules as for the repair of a single DSB created in a diploid: one pathway is RAD51 dependent and one is RAD50 dependent. There is one distinctive difference, however, and that is that Type II telomere recombination needs Sgs1p whereas this helicase had no apparent role in the analogous BIR event measured in the middle of a chromosome (23). The need for Sgs1p is one of several mysteries that surround recombinational lengthening of telomeres. First, both processes appear to involve recombination between the irregular TG1–3 telomere repeats, either between a degrading telomere end and similar sequences found at the junction by Y′ elements and other subtelomeric regions or between the terminal telomeric sequences themselves. Consequently in one yeast strain that lacks TG1–3 sequences centromere-proximal to Y′ elements, Type I events are virtually absent (33). It is also possible that recombination takes place between a telomere end and an autonomously replicating circular Y′ element with TG1–3 sequences connecting the ends. Recent observations in mammalian cells show that telomere sequences can form intrachromosomal loops, in which the telomere end has invaded more proximal telomere sequences (T-loops; ref. 34). This finding has raised the possibility that type II survivors involve intrachromosomal recombination. Such an invasion could create a rolling circle that could give rise to long telomere repeats.
Telomere maintenance in the absence of telomerase has also been observed in other fungi, notably Kluyveromyces lactis. McEachern and Blackburn (35) have shown a similar RAD52 dependence on the process, and recent studies have also suggested that telomere amplifications could come from recombination between the end and autonomous replicating circular telomere-containing DNA. Rolling circle replication could generate an elongated telomere at one end, and gene conversions (i.e., BIR) could then spread this sequence to other telomeres (M. McEachern, personal communication). McEachern's lab has also shown that a subtelomeric marker, initially present at a single telomere, can efficiently spread to most or all other telomeres in the cell when it lacks telomerase (M. McEachern, personal communication).
Mammalian Telomere Maintenance in the Absence of Telomerase: The Alternative Lengthening of Telomeres (ALT) Pathway
Although many immortalized human cells, including most tumor cells, exhibit a reactivation of the telomerase enzyme that is usually inactivated after birth, a subset of cell lines and tumor cells can maintain telomeres in the absence of such reactivation (36). These cells are said to have engaged an ALT pathway of telomere maintenance, and it is tempting to speculate that it proceeds by BIR. As yet, there are no genetic data to show whether this pathway depends on the expected cast of recombination proteins, but a recent paper by Reddel's lab (37) provides strong evidence that recombination is involved. An “ALT” cell line was transformed with a selectable marker inserted directly into telomere sequences; subsequently, this marker was found to proliferate to other chromosome ends during the growth of the cells. Such events were apparently less frequent in immortalized cells in which telomerase had been reactivated. Whether this outcome represents a nonreciprocal, proliferative (BIR-like) event has not been established. It is possible that a reciprocal ectopic recombination event among different telomeres occurs in G2, so that one daughter cell receives two copies of the marked sequence, but its sister cell would have none.
BIR and Its Relationship to Other Homologous Recombination Events
BIR begins when one end of a DSB invades a template and sets up a replication fork. It now seems that the initial events of BIR in S. cerevisiae may not be different from what occurs during gene conversion. Holmes and Haber (38) analyzed gene conversion of the MAT locus after an HO-induced DSB and showed that it required the same DNA polymerases and many of the same accessory factors that are required for normal DNA replication, including DNA polymerase α and for its associated primase. Assuming that the first newly synthesized strand in DNA repair would be initiated by the 3′ end of an invading DNA strand in a D-loop created by strand invasion, and not by an RNA primer, the need for Polα and primase would appear to reflect a role for lagging-strand DNA synthesis. Hence, gene conversion, like BIR, may involve both leading and lagging-strand synthesis, initiated at one end of the DSB, as shown in Fig. 1. In gene conversion, the repair replication fork may be captured by the second end of the DSB, thus terminating the repair event as a patch of new DNA synthesis rather than BIR continuing to the end of the chromosome. One fundamentally important (and unanswered) question is why BIR does not result more often when the two ends of a DSB are both homologous to the template. That is, why, having established a replication fork, does it not just proceed without engaging the second end?
Most likely the repair replication fork differs in significant ways from the origin-initiated replication fork. One difference may be that BIR does not use the putative helicase composed of six Mcm proteins, as does origin-initiated DNA replication (39, 40). Several studies of gene conversion suggest that the replication process during DSB repair is much less processive, much more prone to dissociation than normal replication, and probably less efficient. For example, Pâques et al. (41) showed that the efficiency of gap repair decreased 4-fold as the length of DNA that had to be gap-repaired was increased from a few base pairs to 10 kb. This study also demonstrated that there was a high level of dissociation of DNA polymerase (or at least its newly synthesized product) from its template during gene conversion. A DSB could be repaired even though it was necessary to copy sequences from two different chromosomal templates. For this event to occur, there must have been two dissociations of newly synthesized DNA from the two templates, to be annealed at the site of the DSB. The idea that DNA replication is likely to dissociate from its template will be discussed more below.
One key question about new DNA synthesis during DSB repair is whether the outcomes produced semiconservatively replicated donor and recipient molecules, as would be predicted by the DSB repair model of Szostak and colleagues (17) or whether the dissociated strands were both inherited into the recipient, repaired molecule, as is predicted by some synthesis-dependent strand invasion models [reviewed by Pâques and Haber (42)]. Genetic support for conservative inheritance of the newly synthesized DNA has been obtained from studies of gene conversion in which repair of the DSB involves the copying across an array of repeated sequences. In several studies in S. cerevisiae, using substrates carrying repeats of 375 bp or minisatellites of 36 bp or microsatellites of CTG sequences, it was found that there were frequent expansions and contractions in repeat number, and nearly all of these rearrangements were found in the recipient (41, 43–45).
Very strong physical evidence in favor of this kind of mechanism has been supplied by the elegant experiments of Arcangioli (46), studying mating-type gene switching in Schizosaccharomyces pombe. In fact, mat1 gene switching in S. pombe provides the only well-documented example in which a “programmed” single-strand nick or interruption in DNA produces a DSB, by the passage of a replication fork during S phase (46, 47). The newly generated DSB then appears to carry out gene conversion by a recombination process with a silent donor locus (mat2 or mat3). Arcangioli used heavy isotopes to label the newly synthesized DNA appearing at the mat1 locus and found that both newly synthesized DNA strands at the recipient locus. This is the first such demonstration in any gene conversion system in eukaryotes. It should now be possible to use this approach to learn whether BIR will also result in a conservative synthesis pattern, with both newly synthesized DNA strands annealed together.
Gene Targeting May also Involve BIR-Like Events
Another way to study the nature of DNA synthesis during repair is to study gene targeting of linearized DNA fragments. In contrast to gap repair, where the two ends of the DSB are oriented inward (ends-in), in gene targeting the two ends are oriented outward (ends-out). Conventionally, the integration of a fragment, to replace the original chromosomal sequences with sequences on the fragment, is represented as a pair of simple crossings-over at either end of the fragment (48, 49); certainly this seems to be the simplest way to explain the formation of very large (ca. 100 kb) deletions that can be created by transforming a linear fragment containing a selectable marker flanked by sequences that are homologous to two very distant sites on a chromosome arm (48). But several recent experiments in S. cerevisiae suggest that this kind of outcome is, at best, one of several alternative ways the ends may be processed (21, 50, 51). Many events may begin with two independent strand invasions, each setting up an outwardly moving replication fork (21).
In S. cerevisiae, gene targeting is an efficient process in which nearly all linearized DNA, with at least several hundred base pairs of homology on either side of a selectable marker, almost always integrates at the homologous chromosomal locus. Nonhomologous integration events can be studied in budding yeast if the transforming DNA is not homologous to the genome or if there is very limited homology (52, 53). Many of these integrations appear to occur at topoisomerase I sites (54). Capture of nonhomologous DNA also occurs efficiently at HO-generated DSB sites (55–58).
In mammalian cells, accurate gene targeting is less efficient, and linearized DNA frequently integrates at sites with which the fragment shares no homology. Moreover, when integration occurs at the desired locus, a significant fraction of the events appear to have integrated homologously whereas the opposite end has integrated nonhomologously (59–64).
There is also an unexpected class of transformants in which the transforming DNA must have initially encountered the homologous target and initiated recombination, but eventually the fragment, with its newly appended DNA sequences copied from the donor locus, integrated at a distant location. Scheerer and Adair (65) used a truncated portion of the APRT gene to correct an aprt mutation. A significant fraction of Aprt+ transformants arose after the transforming fragment was extended by replicating the remaining part of the mutant aprt gene to produce a wild-type APRT fragment, which then integrated at a nonhomologous site. This appears to be the result of a one-ended BIR-like event in which the newly synthesized DNA must dissociate from its template.
BIR Events Coupled to Nonhomologous Recombination in Higher Eukaryotic Cells
Recently Richardson and Jasin (66) reported an example of an apparently one-ended homologous recombination event during the repair of an I-SceI-induced DSB on a chromosome. The system was designed such that there were truncated but overlapping segments of a gene, one of which contained an I-SceI cleavage site in the region of shared homology. Recombinants containing an intact gene were recovered; but, surprisingly, almost all of them had not resulted from a simple crossover event that would create an intact gene. Instead, recombination appears to have been initiated by one end of the DSB, after which newly synthesized DNA copied from the template was reinserted into the original broken chromosome by nonhomologous end-joining (NHEJ). This event resembles those obtained by transformation by Scheerer and Adair (65), discussed above.
An apparently similar process has been reported in tobacco plant cells by Puchta (67) studying I-SceI-induced recombination between two nonfunctional gene segments in ectopic locations. Here, too, the majority of events were those where homologous sequences were apparently added only from one of the two ends of the DSB, so that the second junction was apparently created by NHEJ.
It should be noted that the genetic requirements for the end-joining of broken chromosome ends may not be the same as for the nontargeted integration of transfected DNA. Liang et al. (68) have shown that NHEJ repair of an I-SceI-directed DSB is the same as for VDJ recombination; that is, there is a great reduction in the recovery of end-joinings in the absence of the Ku80p. However, there was little effect of the Ku80 mutation on the recovery of random Neo-containing DNA transformed into cells. Whether the nonhomologous events that appear to terminate BIR events in the cases discussed above would be Ku80p dependent is not known.
BIR Events During Transformation in Budding Yeast
In this paper we report experiments in budding yeast designed to explore homology-driven events that require a nonhomologous recombination event for their completion. We find that many of these events occur by a BIR-like mechanism in which there is extensive DNA synthesis but in which the newly synthesized DNA is dissociated from the template. These “hit-and-run” events strongly resemble events described in higher eukaryotes and provide an opportunity to study their mechanism in a simple eukaryote.
Materials and Methods
Strains and Plasmids.
Strain WYL395 (MATa his3–11,14, ura3∷HIS3 ade1 (ADE1)∷leu2) contains a 1.7-kb insertion of ADE1 that replaces the XhoI-Asp718 segment, including the upstream region and 5′ end of the LEU2 gene. Plasmid pWYL137 is a derivative of pBR322-based plasmid YCp50 containing URA3 and CEN4, into which the XhoI-SalI LEU2 fragment was inserted at the SalI site, such that the 3′ end of LEU2 was oriented toward the EcoRI site of pBR322. Subsequently, EcoRI-digested DNA was re-ligated to remove the 3′ end of LEU2. Plasmid pRS315 is an LEU2-containing centromeric plasmid (69). Strain YEK105 (hoΔ hmlΔ∷ADE1 mataΔ hmrΔ∷ADE1 ade1 lys5 trp1∷hisG ura3Δ∷LEU2) is a derivative of YFP17 (41) deleted for the URA3 gene from nucleotides 115,911 to 116,812 on chromosome V. Transformation was carried out by using short homologous segments of ADE2 appended by PCR amplification to a URA3 gene corresponding to the nucleotides 115,918 to 116,810 so that there is no homology with the original URA3 locus.
Transformation Conditions.
Transformation of plasmids and PCR fragments was carried out according to the procedure of Gietz et al. (70). PCR amplification of LEU2 and of the ade2Δ∷URA3 targeting fragment was carried out according to the manufacturers' instructions. Primer sequences are available on request. The construction of short regions of ADE2 homology next to URA3 was carried out as described by Wach et al. (52). Cells were grown at 30°C.
DNA Analysis.
Southern blots of restriction endonuclease-digested DNA and of separated chromosomes was carried out as described (19). Chromosome-separating gel electrophoresis was performed with a Bio-Rad CHEF-DRII system, according to the manufacturer's instructions.
Results
Template-Extended Nonhomologous Recombination.
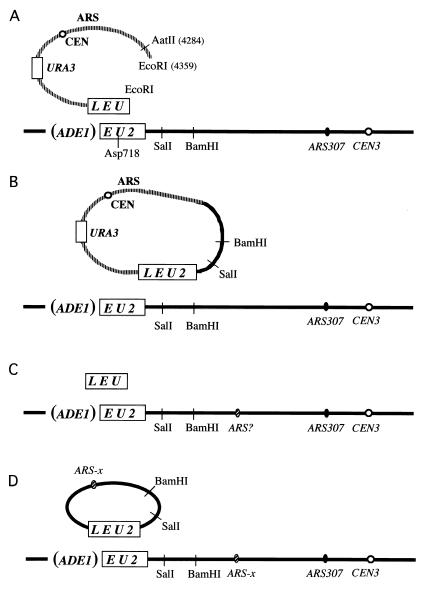
Strain WYL395 lacks the 5′ end of the LEU2 gene, replaced by an ADE1 gene, hereafter designated as (ADE1)EU2 (Fig. 2A). This strain was transformed with an EcoRI-digested linearized fragment of centromeric, replicating plasmid pWL37, containing a deletion of the 3′ end of the coding region of LEU2 truncated at the EcoRI site (hereafter designated LEU). The plasmid and chromosomal segments of leu2 share 390 bp of homology. In order for the cells to become Leu2+, an intact LEU2 gene must be created by a recombination event between the plasmid and chromosomal leu2 sequences. A simple crossover between a recircularized plasmid and the (ADE1)EU2 locus on the chromosome could produce an LEU2 gene, but this is unlikely, because plasmid integration would introduce a second centromere into chromosome III—a lethal event.
Figure 2.
BIR-dependent formation of LEU2 recombinants. (A) EcoRI-digested plasmid pWYL37 has homology only at one end to sites in the yeast genome. (B) The “LEU” segment at one end of the DSB may initiate new DNA synthesis, but the completion of the event requires that the newly synthesized DNA is displaced from the template and must rejoin to the other end of the DSB by a nonhomologous end-joining event. (C) A <1-kb “LEU” DNA fragment was transformed into the same strain shown in A. This fragment has no ARS sequence and cannot replicate autonomously. (D) Hit-and-run transformants containing a circular derivative of chromosome III sequences were recovered. A putative origin of replication, designated ARS-x, is apparently responsible for the ability of these all-yeast circles to replicate.
Compared with transformation with uncut plasmid, transformation with EcoRI-cleaved pWL37 gave rise to Ura3+ transformants ≈20% of the time, consistent with other studies showing efficient ligation of linearized plasmids with cohesive ends. We examined five Leu2− plasmids and confirmed in each case that they had restored the EcoRI site. Among these Ura3+ plasmids, Leu2+ transformants were found at a frequency of 2.1 ± 0.4 × 10′4. Leu2+ Ura3+ plasmids were recovered at a similar frequency (3.3 × 10−4) when pWL37 was cleaved with both EcoRI and AatII (Fig. 1), so that the DNA ends were not perfectly matched. The small increase may reflect the fact that the ends of the plasmid cannot be efficiently re-ligated, providing more opportunity for recombination to be initiated with a chromosomal site.
We identified three classes of Leu2+ Ura3+ transformants. The great majority (53/61, or 87%) contained an autonomously replicating plasmid carrying both URA3 and LEU2 (Class I). These cells could readily lose the URA3 marker and hence become 5-fluoroorotic acid (5-FOA)-resistant (71); when they lost URA3, they simultaneously became Leu2−, while remaining Ade1+. Class II cells (6/61 events) were, like Class I, Ade1+ but were 5-FOA sensitive. Subsequent analysis showed that 5-FOA-sensitive (Ura3+) cells could nevertheless lose both the LEU2 and pBR322 sequences (data not shown). Consequently, these transformants may have resulted from two events, one of which was the formation of an LEU2-containing plasmid, as in Class I, but where there was also a gene conversion of the chromosomal ura3–52 locus to URA3. There were also two Class III cells that were Leu2+ Ura3+ but Ade1−. These transformants apparently created an LEU2 gene by integrating a centromere-less part of pWYL137 at the LEU2 locus, with loss of at least part of the ADE1 gene. These two events were not characterized further.
Homologous Recombination-Associated Nonhomologous Recombination.
Eighteen independent Leu2+ Ura3+ Ade1+ transformants of Class I were first analyzed by Southern blots of DNA digested with several restriction enzymes and probed either with the 5′ end of LEU2 that is absent from the chromosomal copy or with pBR322 sequences, or with other LEU2 segments. The original parent plasmid pWL37 contains a single Asp-718 restriction site, located 390 bp 5′ of the EcoRI site (Fig. 2A). Each of the LEU2 transformants carried a plasmid that yielded an Asp-718 restriction fragment larger than the original linearized fragment in plasmid pWL37. This result is expected if the plasmid now contains a complete LEU2 ORF, which extends 454 bp beyond the EcoRI site. Cleavage with SalI and BamHI was used to determine roughly how much DNA 3′ of the EcoRI site each of these LEU2 plasmids had acquired. Approximately 94% (17/18) of the LEU2 genes extended more than 477 bp beyond the end of the LEU2 ORF, and included the SalI site (Fig. 2). Ten of 18 plasmids (56%) extended as far as a BamHI site, 3260 bp distal to the end of the LEU2 gene.
To examine the extent of new DNA synthesis beyond the end of the LEU2 gene and to determine how these sequences had been joined to the pBR322 sequences on the plasmid, we transformed 7 plasmids into E. coli, selecting for ampicillin resistance, and then sequenced the junction between pBR322 and chromosome III sequences. In each case, we could identify a precise junction between the LEU2-adjacent sequences and pBR322. The exact junctions are described in Table 1. There are a number of different junctions, although one particular junction was recovered in four of the seven independent isolates. We did not find any special features surrounding this junction. Between 15 and 46 bp of pBR322 had been lost. Each junction contained 1 to 3 bp of homology shared between the end of the pBR322 sequence and the extended LEU2 sequence. In some cases there are several additional possible base pairs that could form between the two ends, separated from the junction by one or two mismatched bases. These junctions are very similar to nonhomologous end-joinings seen in the repair of chromosomal DSBs induced by the HO endonuclease or by dicentric chromosome breakage (72) and to those arising during end-joining in mammalian cells (63).
Table 1.
DNA sequence analysis of junctions formed between pBR322 and the end of BIR-extended DNA adjacent to LEU2
| Clone | Sequence | Nucleotide at junction | |
|---|---|---|---|
| G423 | pBR322 | ACATTAACCTAT | 4313 (−46) |
| G466 | Junction | ACATTATCCGGT | |
| G574 | Chromosome III | TTACTATCCGGT | 92912 (+1328) |
| G578 | pBR322 | GAGGCCCTTTCG | 4347 (−12) |
| Junction | GAGGCCAAGTCT | ||
| Chromosome III | AAAGACAAGTCT | 95241 (+3657) | |
| G575 | pBR322 | AGGCCCTTTCGT | 4348 (−11) |
| Junction | AGGCCAAGTCTC | ||
| Chromosome III | AAGACAAGTCTC | 95571 (+3987) | |
| G576 | pBR322 | CACGAGGCCCTT | 4344 (−15) |
| Junction | CACGAGTAGCAT | ||
| Chromosome III | TAGGAGTAGCAT | 96632 (+5048) |
Nonhomologous junctions formed by replication-extension of the “LEU” segment of LEU2 and pBR322, illustrated in Fig. 2B. Initially, the ends of EcoRI-linearized pWL37 corresponded to position 4359 in the pBR322 sequence and the EcoRI site of the cloned “LEU” region, corresponding to nucleotide 91584 in the chromosome III sequence (78). The number of nucleotides lost (indicated by a minus sign) or gained (plus sign) from the two EcoRI ends is also shown.
Taken together, these results argue that the LEU2 gene present on the plasmid was created by a homologous strand invasion event that initiated new DNA synthesis, followed by the dissociation of the newly synthesized DNA and its subsequent ligation, by NHEJ, to the opposite, resected end of the transformed plasmid. We refer to these events as “hit-and-run” transformations.
Interestingly, in no case did the extension of sequences extend more than about 5 kb from the EcoRI site where strand invasion and the initiation of replication occurred, even though LEU2 is located more than 20 kb from the centromere so that additional sequences theoretically could have been replicated before end-joining occurred. It is unlikely that this constraint reflects any significant limitation on plasmid size in yeast, because we have created, by transformation and gap-repair, plasmids of more than 40 kb (73).
Hit-and-Run Transformation with Linearized DNA Lacking Replication Origin Sequences.
In the system described above, the great majority of hit-and-run transformations created a circular, autonomously replicating plasmid. In that system, the 5′ end of the truncated LEU2 gene (“LEU”) shared no homology with the chromosome and was protected from any degradation by more than 4 kb of plasmid sequences. To determine whether we could recover Leu2+ transformants with small LEU fragments, where one end was close to the 5′ promoter region, we transformed a PCR-amplified fragment 898 bp long, where the 5′ end was 272 bp upstream of the LEU2 ORF (Fig. 2C). The frequency of obtaining Leu2+ transformants was ≈6%, as efficient as transformation of an equivalent molar amount of intact circular pRS315 plasmid DNA.
Among more than 500 Leu2+ transformants, none became Ade1−. Thus, none of the transformants appeared to have arisen by a process in which the 3′ end of the fragment recombined homologously whereas the 5′ end, which has no homology to the chromosome, integrated by a nonhomologous insertion. The nature of the event creating Leu2+ Ade1+ transformants was examined first by using chromosome-separating gels to determine into which chromosome the LEU2 gene had integrated. Surprisingly, in 12 of 12 cases, the LEU2-hybridizing DNA did not comigrate with any of yeast's 16 chromosomes (data not shown). This result suggested that the LEU2 gene was present in an autonomously replicating episome. This conclusion was supported by the fact that, in each case, nonselective growth of the Leu2+ colonies on YEPD plates, followed by subcloning, resulted in the appearance of Leu2− colonies, at frequencies ranging from 15 to 32% (data not shown). Thus, the LEU2 gene was present in a moderately stable, replicating unit.
The sizes of these autonomously replicating elements was determined by using BamHI and SalI restriction endonucleases that cut rarely in the chromosomal region containing LEU2. The sizes of the unstable DNA ranged from 8.5 kb to <15 kb (data not shown). Surprisingly, at least five of nine independent LEU2 circles that were analyzed did not extend to the first known ARS sequence (ARS307) that lies 16.15 kb centromere-proximal to the end of LEU2 at location 108,578 bp from the left telomere of chromosome III. This result suggests that, at least in our strain, there is a novel ARS (ARS-x) sequence located within 9 kb of the 3′ end of LEU2. The precise position of this putative ARS-x has not been determined.
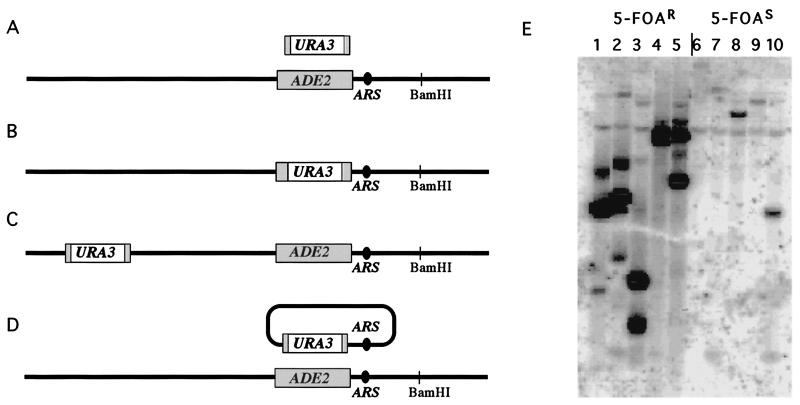
To show that transformation could create autonomously replicating circles containing a known ARS, we also did a gene-targeting experiment using ADE2 sequences, where an ARS lies only a few base pairs upstream of the translation start site (within positions 566190 to 566789 on chromosome XV; refs. 74 and 75). In this experiment, we attached 50 bp of ADE2 sequences (positions 564740–564790 and 565740–565790) to either side of a URA3 gene by PCR techniques (52) in a strain completely deleted for the endogenous URA3 locus. With so little homology, only 48.6% (100/204) were Ade− Ura+, expected for accurate replacements of the ADE2 locus by gene targeting. Of the remaining 51.4% (Ade+ Ura+) colonies, 91/104 were nonhomologous integrations of URA3 at some other chromosome location, as described (53, 76). These strains were sensitive to 5-FOA and displayed a single new URA3-homologous band on a Southern blot. However, ≈13/104 (6.3%) of the Ade+ Ura+ colonies proved to be 5-FOA-resistant because they harbored an autonomously replicating plasmid and could easily give rise to Ura− (5-FOAR) cells. Examples are shown in Fig. 3E, where it can be seen that these colonies have a much greater intensity of hybridization to the URA3 gene than is seen with the nontargeted integrants.
Figure 3.
(A) Gene targeting at the ADE2 locus using an URA3 gene carrying only 50 bp of ADE2 sequences on either end. (B) An ARS sequence lies just upstream of the ADE2 gene, which is transcribed right to left. Three types of transformants were recovered. About half had replaced ADE2 with URA3 sequences. (C) Another group had inserted the URA3 nonhomologously at different sites in the genome. (D) A third group had formed unstable, apparently circular autonomously replicating sequences in which the ADE2-adjacent ARS had been copied onto the DNA of the transforming fragment, presumably by a BIR-like event. (E) Examples of BamHI-digested DNA from 5-FOA-resistant (5-FOAR) colonies harboring an unstable replicating URA3 gene (lanes 1 to 5), which is present in high copy number when probed with URA3 sequences. There is no BamHI site within 5000 bp upstream and 2200 bp downstream of the ADE2 gene. Supercoiled, nicked circular and linearized forms are evident. In lanes 6–10 are 5-FOA-sensitive colonies where the URA3 gene had integrated at unknown locations, present at single copy.
PCR amplification and DNA sequencing of sequences adjacent to URA3 in two of the autonomously replicating episomes confirmed that they had acquired ADE2-adjacent DNA sequences at both ends of the plasmid, including the adjacent ARS, which is more than 1000 bp from the end of the targeting oligonucleotide (data not shown). These results show that both ends of the linearized fragment, with only 50 bp of homology at each end, had engaged in BIR, which then culminated in the formation of an unstable circular molecule containing the ADE2-adjacent ARS (Fig. 3D). The restriction enzyme BamHI does not cleave within 5 kb upstream of ADE2 or 2.2 kb downstream and does not cleave any of the five replicating ADE2 circles shown in Fig. 3E. Note that the sizes of the BamHI digests of these independently isolated transformants are different in every case. This result suggests that the amount of newly synthesized DNA in the circular molecules was different in each case. These events are similar to those described by Morrow et al. (21), except that, in these cases, recombination-dependent replication did not extend to the chromosome end but rather resulted in circular, autonomously replicating molecules.
Discussion
In this paper, we present a model system for studying one-ended homologous recombination processes that must terminate by a nonhomologous recombination event. This system produced results analogous to the recombination events studied by Richardson and Jasin (66) in mammalian cells and Puchta (67) in tobacco, where recombination between two truncated and overlapping gene segments produces an intact gene, but without any crossing-over. For this outcome to occur, the newly synthesized DNA must be displaced from its template so that it can then participate in an illegitimate recombination event. This dissociation of DNA during BIR appears to be one of the distinctive differences between repair DNA synthesis and normal DNA replication.
One surprising result from our work was the frequent recovery of autonomously replicating transformants obtained when the transforming fragment did not contain an ARS sequence. We have shown that, with as little as 50 bp of homologous sequence at the end of a selectable marker that shares no homology to the genome, BIR events can occur to acquire an ARS sequence more than 1 kb away. In at least some cases, both short ends of the targeting vector engaged in hit-and-run behavior, culminating in the production of a circular, replicating product.
BIR can occur when only one end of a DSB shares homology with a template. This situation arises at degraded telomeres and may also arise at stalled replication forks. At least in S. cerevisiae, there seem to be two or more RAD52-dependent processes that can carry out this type of repair (23). BIR could also occur if, for some reason, one end of a DSB is more efficient in initiating strand invasion. This possibility may explain the apparent one-ended recombination events that were seen in the kinetic analysis of recombination intermediates initiated by HO cleavage of a plasmid containing inverted homologous DNA segments (77). Both ends of the DSB shared homology with the template, but one of the two ends produced what appeared to be a crossover product an hour earlier than the second crossover was detected. We think this intermediate was likely to have arisen by BIR.
In gene targeting, BIR can apparently occur from both ends of a transforming fragment and can in fact create an entire new chromosome, beginning with a fragment of a few kilobases (21). Here, we show that gene targeting can also abort, yielding a quasi-stable, autonomously replicating circular product dependent both on BIR and on NHEJ. The frequency with which such an event occurs most likely depends on the proximity of the targeting sequences to an ARS sequence that will support DNA replication in subsequent generations. It will be interesting to see whether we can force a linearized fragment to integrate more often by homologous recombination at one end and nonhomolo-gous insertion at the other if we chose a chromosomal target that is distant from any sequence that will function as an ARS.
Acknowledgments
Maureen Barlow provided excellent technical assistance. Titia de Lange and members of the Haber lab offered valuable comments and suggestions. Work on BIR in this laboratory has been funded by National Institutes of Health Grant GM20056 and by Department of Energy Grant 99ER62729.
Abbreviations
- DSB
double-strand break
- BIR
break-induced replication
- NHEJ
nonhomologous end-joining
- 5-FOA
5-fluoroorotic acid
Footnotes
This paper results from the National Academy of Sciences colloquium, “Links Between Recombination and Replication: Vital Roles of Recombination,” held November 10–12, 2000, in Irvine, CA.
References
- 1.Meselson M, Weigle J. Proc Natl Acad Sci USA. 1961;47:857–868. doi: 10.1073/pnas.47.6.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Skalka A. In: Mechanisms in Recombination. Grell R F, editor. New York: Plenum; 1974. pp. 421–432. [Google Scholar]
- 3.Mosig G, Werner R. Proc Natl Acad Sci USA. 1969;64:747–754. doi: 10.1073/pnas.64.2.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mosig G. Annu Rev Genet. 1998;32:379–413. doi: 10.1146/annurev.genet.32.1.379. [DOI] [PubMed] [Google Scholar]
- 5.Formosa T, Alberts B M. Cell. 1986;47:793–806. doi: 10.1016/0092-8674(86)90522-2. [DOI] [PubMed] [Google Scholar]
- 6.George J W, Kreuzer K N. Genetics. 1996;143:1507–1520. doi: 10.1093/genetics/143.4.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Motamedi M R, Szigety S K, Rosenberg S M. Genes Dev. 1999;13:2889–2903. doi: 10.1101/gad.13.21.2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuzminov A, Stahl F W. Genes Dev. 1999;13:345–356. doi: 10.1101/gad.13.3.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kogoma T. Cell. 1996;85:625–627. doi: 10.1016/s0092-8674(00)81229-5. [DOI] [PubMed] [Google Scholar]
- 10.Kogoma T. Microbiol Mol Biol Rev. 1997;61:212–238. doi: 10.1128/mmbr.61.2.212-238.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuzminov A. Mol Microbiol. 1995;16:373–384. doi: 10.1111/j.1365-2958.1995.tb02403.x. [DOI] [PubMed] [Google Scholar]
- 12.Michel B. Trends Biochem Sci. 2000;25:173–178. doi: 10.1016/s0968-0004(00)01560-7. [DOI] [PubMed] [Google Scholar]
- 13.Seigneur M, Bidnenko V, Ehrlich S D, Michel B. Cell. 1998;95:419–430. doi: 10.1016/s0092-8674(00)81772-9. [DOI] [PubMed] [Google Scholar]
- 14.Esposito M S. Proc Natl Acad Sci USA. 1978;75:4436–4440. doi: 10.1073/pnas.75.9.4436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Golin J E, Esposito M S. Genetics. 1984;107:355–365. doi: 10.1093/genetics/107.3.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Voelkel-Meiman K, Roeder G S. Genetics. 1990;126:851–867. doi: 10.1093/genetics/126.4.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dunn B, Szauter P, Pardue M L, Szostak J W. Cell. 1984;39:191–201. doi: 10.1016/0092-8674(84)90205-8. [DOI] [PubMed] [Google Scholar]
- 18.Walmsley R W, Chan C S, Tye B K, Petes T D. Nature (London) 1984;310:157–160. doi: 10.1038/310157a0. [DOI] [PubMed] [Google Scholar]
- 19.Bosco G, Haber J E. Genetics. 1998;150:1037–1047. doi: 10.1093/genetics/150.3.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Malkova A, Klein F, Leung W-Y, Haber J E. Proc Natl Acad Sci USA. 2000;97:14500–14505. doi: 10.1073/pnas.97.26.14500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morrow D M, Connelly C, Hieter P. Genetics. 1997;147:371–382. doi: 10.1093/genetics/147.2.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Malkova A, Ivanov E L, Haber J E. Proc Natl Acad Sci USA. 1996;93:7131–7136. doi: 10.1073/pnas.93.14.7131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Signon L, Malkova A, Naylor M, Haber J E. Mol Cell Biol. 2001;21:2048–2056. doi: 10.1128/MCB.21.6.2048-2056.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Malkova A, Signon L, Schaefer C B, Naylor M, Theis J F, Newlon C S, Haber J E. Genes Dev. 2001;15:1055–1160. doi: 10.1101/gad.875901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bai Y, Symington L S. Genes Dev. 1996;10:2025–2037. doi: 10.1101/gad.10.16.2025. [DOI] [PubMed] [Google Scholar]
- 26.Lundblad V, Blackburn E H. Cell. 1993;73:347–360. doi: 10.1016/0092-8674(93)90234-h. [DOI] [PubMed] [Google Scholar]
- 27.Teng S C, Zakian V A. Mol Cell Biol. 1999;19:8083–8093. doi: 10.1128/mcb.19.12.8083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Le S, Moore J K, Haber J E, Greider C. Genetics. 1999;152:143–152. doi: 10.1093/genetics/152.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Teng S, Chang J, McCowan B, Zakian V A. Mol Cell. 2000;6:947–952. doi: 10.1016/s1097-2765(05)00094-8. [DOI] [PubMed] [Google Scholar]
- 30.Chen Q, Ijpma A, Greider C W. Mol Cell Biol. 2001;21:1819–1827. doi: 10.1128/MCB.21.5.1819-1827.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnson F B, Marciniak R A, McVey M, Stewart S A, Hahn W C, Guarente L. EMBO J. 2001;20:905–913. doi: 10.1093/emboj/20.4.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cohen H, Sinclair D A. Proc Natl Acad Sci USA. 2001;98:3174–3179. doi: 10.1073/pnas.061579598. . (First Published March 6, 2001; 10.1073/pnas.061579598) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang P, Pryde F E, Lester D, Maddison R L, Borts R H, Hickson I D, Louis E J. Curr Biol. 2001;11:125–129. doi: 10.1016/s0960-9822(01)00021-5. [DOI] [PubMed] [Google Scholar]
- 34.Griffith J D, Comeau L, Rosenfield S, Stansel R M, Bianchi A, Moss H, de Lange T. Cell. 1999;97:503–514. doi: 10.1016/s0092-8674(00)80760-6. [DOI] [PubMed] [Google Scholar]
- 35.McEachern M J, Blackburn E H. Genes Dev. 1996;10:1822–1834. doi: 10.1101/gad.10.14.1822. [DOI] [PubMed] [Google Scholar]
- 36.Bryan T M, Englezou A, Dalla-Pozza L, Dunham M A, Reddel R R. Nat Med. 1997;3:1271–1274. doi: 10.1038/nm1197-1271. [DOI] [PubMed] [Google Scholar]
- 37.Dunham M A, Neumann A A, Fasching C L, Reddel R R. Nat Genet. 2000;26:447–450. doi: 10.1038/82586. [DOI] [PubMed] [Google Scholar]
- 38.Holmes A, Haber J E. Cell. 1999;96:415–424. doi: 10.1016/s0092-8674(00)80554-1. [DOI] [PubMed] [Google Scholar]
- 39.Aparicio O M, Weinstein D M, Bell S P. Cell. 1997;91:59–69. doi: 10.1016/s0092-8674(01)80009-x. [DOI] [PubMed] [Google Scholar]
- 40.Labib K, Tercero J A, Diffley J F. Science. 2000;288:1643–1647. doi: 10.1126/science.288.5471.1643. [DOI] [PubMed] [Google Scholar]
- 41.Pâques F, Leung W Y, Haber J E. Mol Cell Biol. 1998;18:2045–2054. doi: 10.1128/mcb.18.4.2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pâques F, Haber J E. Microbiol Mol Biol Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Richard G F, Goellner G M, McMurray C T, Haber J E. EMBO J. 2000;19:2381–2390. doi: 10.1093/emboj/19.10.2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Richard G F, Dujon B, Haber J E. Mol Gen Genet. 1999;261:871–882. doi: 10.1007/s004380050031. [DOI] [PubMed] [Google Scholar]
- 45.Pâques F, Richard G-F, Haber J E. Genetics. 2001;158:155–166. doi: 10.1093/genetics/158.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Arcangioli B, de Lahondes R. EMBO J. 2000;19:1389–1396. doi: 10.1093/emboj/19.6.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dalgaard J Z, Klar A J. Nature (London) 1999;400:181–184. doi: 10.1038/22139. [DOI] [PubMed] [Google Scholar]
- 48.Surosky R T, Tye B K. Proc Natl Acad Sci USA. 1985;82:2106–2110. doi: 10.1073/pnas.82.7.2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rothstein R J. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- 50.Leung W, Malkova A, Haber J E. Proc Natl Acad Sci USA. 1997;94:6851–6856. doi: 10.1073/pnas.94.13.6851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Negritto M T, Wu X, Kuo T, Chu S, Bailis A M. Mol Cell Biol. 1997;17:278–286. doi: 10.1128/mcb.17.1.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wach A, Brachat A, Pohlmann R, Philippsen P. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- 53.Schiestl R H, Dominska M, Petes T D. Mol Cell Biol. 1993;13:2697–2705. doi: 10.1128/mcb.13.5.2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhu J, Schiestl R H. Mol Cell Biol. 1996;16:1805–1812. doi: 10.1128/mcb.16.4.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Teng S C, Kim B, Gabriel A. Nature (London) 1996;383:641–644. doi: 10.1038/383641a0. [DOI] [PubMed] [Google Scholar]
- 56.Moore J K, Haber J E. Nature (London) 1996;383:644–646. doi: 10.1038/383644a0. [DOI] [PubMed] [Google Scholar]
- 57.Yu X, Gabriel A. Mol Cell. 1999;4:873–881. doi: 10.1016/s1097-2765(00)80397-4. [DOI] [PubMed] [Google Scholar]
- 58.Ricchetti M, Fairhead C, Dujon B. Nature (London) 1999;402:96–100. doi: 10.1038/47076. [DOI] [PubMed] [Google Scholar]
- 59.Doetschman T, Maeda N, Smithies O. Proc Natl Acad Sci USA. 1988;85:8583–8587. doi: 10.1073/pnas.85.22.8583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Berinstein N, Pennell N, Ottaway C A, Shulman M J. Mol Cell Biol. 1992;12:360–367. doi: 10.1128/mcb.12.1.360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Villemure J F, Belmaaza A, Chartrand P. Mol Gen Genet. 1997;256:533–538. doi: 10.1007/s004380050598. [DOI] [PubMed] [Google Scholar]
- 62.Richard M, Gusew N, Belmaaza A, Chartrand P. Somatic Cell Mol Genet. 1997;23:75–81. doi: 10.1007/BF02679957. [DOI] [PubMed] [Google Scholar]
- 63.Roth D B, Wilson J H. Mol Cell Biol. 1986;6:4295–4304. doi: 10.1128/mcb.6.12.4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jasin M, Elledge S J, Davis R W, Berg P. Genes Dev. 1990;4:157–166. doi: 10.1101/gad.4.2.157. [DOI] [PubMed] [Google Scholar]
- 65.Scheerer J B, Adair G M. Mol Cell Biol. 1994;14:6663–6673. doi: 10.1128/mcb.14.10.6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Richardson C, Jasin M. Mol Cell Biol. 2000;20:9068–9075. doi: 10.1128/mcb.20.23.9068-9075.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Puchta H. Genetics. 1999;152:1173–1181. doi: 10.1093/genetics/152.3.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liang F, Romanienko P J, Weaver D T, Jeggo P A, Jasin M. Proc Natl Acad Sci USA. 1996;93:8929–8933. doi: 10.1073/pnas.93.17.8929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sikorski R S, Hieter P. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gietz D, St. Jean A, Woods R A, Schiestl R H. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Boeke J D, Lacroute F, Fink G R. Mol Gen Genet. 1984;197:345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- 72.Kramer K M, Brock J A, Bloom K, Moore J K, Haber J E. Mol Cell Biol. 1994;14:1293–1301. doi: 10.1128/mcb.14.2.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wu X, Haber J E. Cell. 1996;87:277–285. doi: 10.1016/s0092-8674(00)81345-8. [DOI] [PubMed] [Google Scholar]
- 74.Stotz A, Linder P. Gene. 1990;95:91–98. doi: 10.1016/0378-1119(90)90418-q. [DOI] [PubMed] [Google Scholar]
- 75.Janatova I, Gourdon P, Meilhoc E, Klein R D, Masson J M. Curr Genet. 2000;37:298–303. doi: 10.1007/s002940050531. [DOI] [PubMed] [Google Scholar]
- 76.Schiestl R H, Petes T D. Proc Natl Acad Sci USA. 1991;88:7585–7589. doi: 10.1073/pnas.88.17.7585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rudin N, Sugarman E, Haber J E. Genetics. 1989;122:519–534. doi: 10.1093/genetics/122.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Oliver S G, van der Aart Q J, Agostoni-Carbone M L, Aigle M, Alberghina L, Alexandraki D, Antoine G, Anwar R, Ballesta J P, Benit P, et al. Nature (London) 1992;357:38–46. doi: 10.1038/357038a0. [DOI] [PubMed] [Google Scholar]