Abstract
Reptilian reovirus is one of a limited number of nonenveloped viruses that are capable of inducing cell-cell fusion. A small, hydrophobic, basic, 125-amino-acid fusion protein encoded by the first open reading frame of a bicistronic viral mRNA is responsible for this fusion activity. Sequence comparisons to previously characterized reovirus fusion proteins indicated that p14 represents a new member of the fusion-associated small transmembrane (FAST) protein family. Topological analysis revealed that p14 is a representative of a minor subset of integral membrane proteins, the type III proteins Nexoplasmic/Ccytoplasmic (Nexo/Ccyt), that lack a cleavable signal sequence and use an internal reverse signal-anchor sequence to direct membrane insertion and protein topology. This topology results in the unexpected, cotranslational translocation of the essential myristylated N-terminal domain of p14 across the cell membrane. The topology and structural motifs present in this novel reovirus membrane fusion protein further accentuate the diversity and unusual properties of the FAST protein family and clearly indicate that the FAST proteins represent a third distinct class of viral membrane fusion proteins.
Biological membrane fusion is an essential cellular process mediated by specific fusion proteins (22, 57, 61). Extensive analysis of a number of enveloped virus fusion proteins has contributed to a model of protein-mediated membrane fusion. Enveloped virus fusion proteins are complex, multimeric, type I Nexoplasmic/Ccytoplasmic (Nexo/Ccyt) integral membrane proteins that facilitate virus entry into cells by mediating fusion between the viral envelope and the target cell membrane. Two distinct classes of enveloped virus fusion proteins have been identified: the class I fusion proteins exemplified by influenza virus and human immunodeficiency virus proteins and the class II proteins of the alpha- and flaviviruses (25, 47, 56, 57). For both classes, triggered conformational changes and/or multimer reorganization of their complex ectodomains are essential aspects of the fusion reaction (25, 56). This transition from a metastable to a low-energy form is believed to provide the energy to overcome the thermodynamic barriers that inhibit spontaneous membrane mergers (30, 56). However, the necessity and/or precise role of structural remodeling as a thermodynamic mediator of the fusion reaction remains unresolved (3, 13, 14, 37).
Since nonenveloped viruses lack a lipid bilayer, virus entry is not dependent on membrane fusion. As a result, nonenveloped viruses do not encode membrane fusion proteins. The rare exceptions to this generalization are the fusogenic reoviruses, an unusual group of syncytium-inducing nonenveloped viruses with segmented double-stranded RNA genomes (9, 32). Unlike those of enveloped viruses, the reovirus fusion proteins are not components of the virus particle and therefore are not involved in virus entry (7, 45). The reovirus fusion-associated small transmembrane (FAST) proteins are the only known examples of nonstructural viral proteins that induce cell-cell fusion in a manner that is not directly related to either entry or exit of the virus (10). The sole purpose of the FAST proteins appears to be the formation of polykaryons after the expression of the proteins inside virus-infected cells, a process that leads to rapid dissemination of the infection (10).
Two distinct reovirus membrane fusion proteins have been identified. The avian reoviruses (ARV) and Nelson Bay reovirus encode homologous 95- to 98-amino-acid fusion proteins, termed p10 (45). Baboon reovirus (BRV) encodes a unique 140-amino-acid p15 fusion protein which possesses no sequence similarity to p10 and a different arrangement of structural and functional motifs (7). Both p10 and p15 are integral membrane proteins that are modified by fatty acids: palmitate residues are added to an internal, membrane-proximal dicysteine motif in p10, while the N-terminal glycine residue in p15 is myristylated (7, 46). Both proteins also contain a positively charged region that is C-terminal to the transmembrane domain that may influence both protein topology in the membrane and the fusion event itself (7, 45). The ARV p10 protein localizes to the plasma membrane in an Nexo/Ccyt topology, placing a very small N-terminal domain (approximately 40 residues) external to the lipid bilayer (45). In the case of p15, the protein topology in the membrane has not been determined, although the presence of the N-terminal myristate moiety and two potential transmembrane domains suggests that p15 may assume an Ncyt bitopic or polytopic topology (7). The nonstructural nature and restricted role of the FAST proteins in the virus replication cycle may account for their small size and possible evolution toward the minimal protein determinants required to promote fusion of biological membranes.
The small size of the reovirus FAST proteins makes it difficult to envision how extensive conformational changes could play a role in either regulating the exposure of a buried fusion peptide or providing sufficient energy to overcome the thermodynamic barriers that maintain membrane structure. Consequently, the reovirus FAST proteins are unlikely to adhere to the current paradigm of protein-mediated membrane fusion that has emerged from studies of the enveloped virus fusion proteins (22, 47, 61). It seems likely that an analysis of each individual member of the FAST protein family, followed by a comparison of the motifs required for their fusion function, will contribute to an improved mechanistic understanding of these rudimentary membrane fusion machines.
We recently characterized a python reovirus, the prototype of a new species of fusogenic reovirus, reptilian reovirus (RRV) (1, 12). We now show that a p14 protein encoded by the first open reading frame (ORF) of a bicistronic mRNA represents a third distinct member of the reovirus FAST protein family with its own signature arrangement of structural motifs. Biochemical analysis revealed that p14 is a surface-localized, type III integral membrane protein (i.e., it utilizes an internal reverse signal-anchor sequence to direct an Nexo/Ccyt membrane topology). This topology results in the cotranslational translocation of a small, myristylated ectodomain across the lipid bilayer. Although the precise role of the external myristylated N terminus of p14 in the membrane fusion reaction is undetermined, this discovery adds a new element to be considered in models of protein-mediated membrane fusion.
MATERIALS AND METHODS
Cells and virus.
RRV was isolated from a python (Python regius) (1) and obtained from W. Ahne (University of Munich, Munich, Germany). Vero cells and quail QM5 cells (11) were maintained at 37°C in a 5% CO2 atmosphere in medium 199 with Earle's salts containing 100 U of penicillin and streptomycin per ml and 5% or 10% heat-inactivated fetal bovine serum, respectively.
Plasmids, cloning, and sequencing.
The procedure for cDNA synthesis and cloning of the RRV genome segments is described in detail elsewhere (9). The full-length S1 genome segment and individual ORFs were amplified by PCR and subcloned into the pcDNA3 mammalian expression vector (Invitrogen). The pcDNA3-p14 clone was used as a template to generate a number of tagged and/or substituted constructs. Two hemagglutinin (HA) epitope tags were added in tandem (separated by a glycine linker) to either the C terminus of p14 (p14-2HAC) or the N-terminal domain (p14-2HAN) after amino acid 7 (in order to avoid alteration of the myristylation consensus sequence). Site-specific substitutions in p14 were created by using a QuickChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's specifications. All subclones were confirmed by cycle sequencing (USB) according to the manufacturer's instructions before use in transient transfection and in vitro transcription-translation assays.
Anti-p14 antiserum production.
A recombinant baculovirus expressing the p14 fusion protein under the control of the polyhedrin promoter was created by using the Bac-To-Bac baculovirus cloning and expression system (Life Technologies). SF21 cells were grown in suspension cultures, and p14 was purified from the infected cell pellets by detergent disruption, affinity chromatography using TALON metal affinity resin (Clontech), and ion-exchange chromatography using HiTrap SP HP ion-exchange columns (Amersham Pharmacia Biotech). The purified p14 was used to generate polyclonal antiserum in rabbits, with Freund's complete adjuvant used for the primary injection and Freund's incomplete adjuvant used for five subsequent booster injections at 6-week intervals (500 μg of p14 per injection).
Cell staining, syncytial indexing, and antibody inhibition.
Cluster plates containing subconfluent monolayers of Vero or QM5 cells were transfected with expression plasmids by use of Lipofectamine (Life Technologies) according to the manufacturer's instructions. Cell monolayers were fixed with methanol and stained at various times posttransfection with Wright-Giemsa stain. Alternatively, fixed cell monolayers were immunostained as follows: cells were preblocked with whole goat immunoglobulin G (IgG) (1:1,000) in Hank's buffered saline solution (HBSS) for 30 min at room temperature, and then a primary antibody (1:800 rabbit polyclonal anti-p14 antibody) was adsorbed to cells for 45 to 60 min at room temperature. After primary antibody binding, cell monolayers were washed six times in HBSS, and a secondary antibody (1:800 alkaline phosphatase-conjugated goat anti-rabbit antibody [Jackson Immunochemicals] in blocking buffer) was adsorbed to cells and washed as described above. After the addition of the substrate BCIP/NBT (5-bromo-4-chloro-3-indolylphosphate/nitroblue tetrazolium) to allow color development, cells were visualized under a Nikon Diaphot inverted microscope at a magnification of ×200. Image-Pro Plus software (v. 4.0) was used to capture images of stained cells. The relative ability of various p14 mutants to mediate syncytium formation was quantified by a syncytial index assay. The numbers of syncytial foci and syncytial nuclei present in five random fields of view were determined by microscopic examination of Giemsa-stained transfected cell monolayers at ×100 magnification. Results were reported as the means ± standard errors from three separate experiments. For antibody inhibition studies, twofold serial dilutions of complement-fixed polyclonal anti-p14 antiserum or normal rabbit serum were added to the medium of p14-transfected cells at 4 h posttransfection. At 14 to 18 h posttransfection, the cell monolayers were fixed with methanol, stained with Wright-Giemsa stain, and examined for the presence of multinucleated syncytia.
In vitro transcription and translation.
The full-length S1 genome segment, p14 ORF, σC ORF, and p14 constructs containing site-specific substitutions were transcribed and translated in vitro by use of nuclease-treated rabbit reticulocyte lysates (Promega). Translation products were characterized by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) using 15% acrylamide gels, and proteins were visualized by fluorography as previously described (7).
Radiolabeled cell lysates, immunoprecipitation, and membrane fractionation.
Transfected cell monolayers were radiolabeled at 12 to 18 h posttransfection for 1 h with 50 μCi of [3H]leucine/ml or for 3 h with 20 μCi of [3H]myristic acid (Amersham Pharmacia Biotech)/ml. Cells were lysed with RIPA buffer, nuclei were removed by centrifugation, and the cleared cell lysates were immunoprecipitated with 1:100 dilutions of rabbit polyclonal anti-p14 antiserum, normal rabbit serum, mouse monoclonal (IgG2b) anti-HA antiserum, or a mouse IgG2b isotype control, as previously described (7). The membrane and soluble fractions from radiolabeled, transfected cells were obtained by vesiculating cells with a 30-gauge needle followed by ultracentrifugation at 100,000 × g for 1 h as previously described (45). For the removal of peripherally associated membrane proteins, the membrane pellet was treated with either high salt (500 mM NaCl) or high pH (100 mM Na2CO3, pH 11.4) (15) on ice for 30 min, followed by a second centrifugation at 100,000 × g for 1 h to recover integral membrane proteins. All soluble and membrane fractions were then immunoprecipitated and analyzed by SDS-PAGE as described above.
Fluorescent cell staining.
Cells were seeded in culture plates containing coverslips and transfected 20 to 24 h after seeding as described above. At 6 h posttransfection, cell monolayers were washed twice with HBSS and either fixed with ice-cold methanol for staining of intracellular p14 or surface stained. Permeabilized cells were preblocked with whole goat or rat IgG (1:1,000) in HBSS for 30 min at room temperature, and then a primary antibody (1:800 rabbit polyclonal anti-p14 or 1:200 mouse monoclonal [IgG2b] anti-HA in blocking buffer) was adsorbed to cells for 45 to 60 min at room temperature. After primary antibody binding, cell monolayers were washed six times in HBSS, and a secondary antibody (1:400 fluorescein isothiocyanate [FITC]-conjugated goat anti-rabbit or 1:200 FITC-conjugated rat anti-mouse [Jackson Immunochemicals] in blocking buffer) was adsorbed to cells and washed as described above. Nonpermeabilized cells were stained as described above, with the following exceptions: antibody dilutions were 1:200 for rabbit anti-p14, 1:100 for mouse anti-HA, 1:100 for FITC-conjugated goat anti-rabbit, and 1:50 for FITC-conjugated rat anti-mouse (all in blocking buffer), and all staining was performed at 4°C. After primary antibody addition and washing, cells were fixed with ice-cold methanol and washed before the addition of secondary antibody. Stained cells on coverslips were mounted on glass slides by using fluorescent mounting medium (Dako), and cells were visualized and photographed under a Zeiss LSM510 scanning argon laser confocal microscope, using a ×100 objective.
RESULTS
The first ORF of the bicistronic S1 genome segment of RRV is responsible for cell-cell fusion.
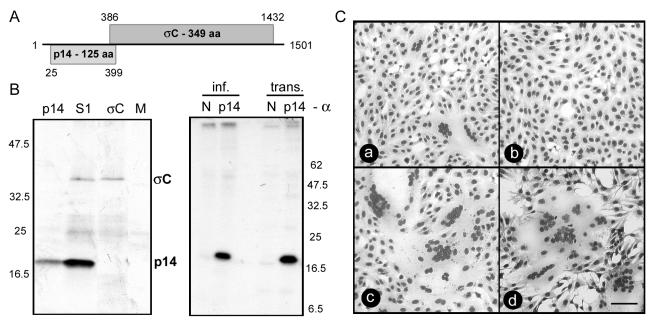
Sequence analysis identified two sequential, partially overlapping ORFs in the S1 genome segment of RRV (Fig. 1A). In vitro transcription and translation confirmed that the S1 genome segment is functionally bicistronic and encodes two protein products, namely a 35-kDa homolog of the ARV cell attachment protein σC and a 14-kDa RRV-specific gene product (Fig. 1B). Transfection analysis with the full-length S1 genome segment indicated that either or both of these S1 gene products induce cell-cell fusion (Fig. 1C). Subcloning and expression revealed that the 125-amino-acid p14 protein encoded by the first ORF, when expressed by itself in transfected cells, induced extensive multinucleated syncytium formation in both transfected Vero epithelial cells and QM5 quail cell fibroblasts (Fig. 1C). Immunoprecipitation confirmed the expression of p14 in both transfected and virus-infected cells (Fig. 1B). Gapped alignments of p14 with the other FAST proteins of ARV, Nelson Bay reovirus, and BRV revealed no significant sequence similarity (percent amino acid identities of <15%) (data not shown), indicating that the RRV p14 protein is a new member of the FAST protein family.
FIG. 1.
The first ORF on the RRV bicistronic S1 genome segment encodes a p14 fusion protein. (A) The names of the predicted gene products and the numbers of residues are shown within shaded rectangles representing the ORFs present in the RRV S1 genome segment. Numbers indicate the first and last nucleotides for each ORF (minus the termination codon). (B) Translation products generated by rabbit reticulocytes primed with in vitro-transcribed mRNAs representing the S1 genome segment, the p14 ORF, the σC ORF, and no exogenous mRNA (M) were detected by radiolabeling and SDS-PAGE (left panel). The locations of the p14 and σC translation products are indicated, along with the relative migration of molecular weight markers. Virus-infected (inf.) or p14-transfected (trans.) Vero cells were radiolabeled and immunoprecipitated with a polyclonal anti-p14 serum (p14) or normal rabbit serum (N) (right panel). The relative migration of molecular weight markers is indicated. (C) Cells were transfected with pcDNA3 expressing the bicistronic S1 genome segment (a), the σC ORF (b), or the p14 ORF (c and d). Panels a to c show transfected Vero cells that were Giemsa stained at 18 h posttransfection; panel d shows transfected QM5 cells that were Giemsa stained at 8 h posttransfection. Syncytial foci are clearly visible in panels a, c, and d. Scale bar = 100 μm.
Structural motifs in p14.
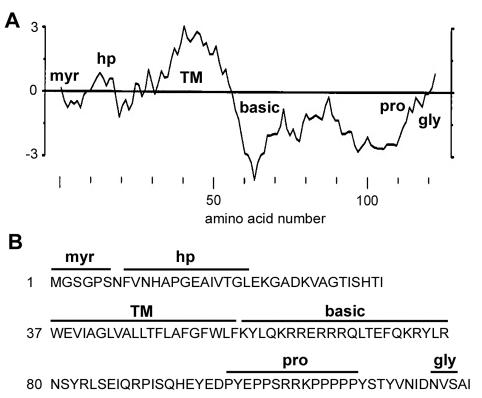
The RRV p14 protein contains several predicted structural motifs (Fig. 2). A hydropathy plot of p14 and sequence analysis identified a predicted transmembrane (TM) domain, suggesting that p14 resides as an integral membrane protein. Sequence analysis also showed that p14 lacks a cleavable N-terminal signal sequence (29), suggesting that the membrane-spanning domain may function as an internal signal anchor (21). The only other region in p14 with any hydrophobic character occurs in the N-terminal domain (Fig. 2A), a region we termed the hydrophobic patch. The N-terminal domain also contains a consensus sequence for N-terminal myristylation (MGXXXS/T/A) (52). The C-terminal domain is comprised of two different regions, namely a highly basic, membrane-proximal region (10 of the 22 residues immediately following the TM domain are basic) and a C-terminal proline-rich region (eight prolines between residues 99 and 112) that includes a stretch of five consecutive prolines. The C-terminal domain also contains a consensus sequence for N-linked glycosylation (NXS/T) at Asn121. The functional significance of these sequence-predicted structural motifs was further investigated.
FIG. 2.
Sequence-predicted structural motifs in p14. (A) Hydropathy profile of the p14 protein averaged over a window of 11 residues. Hydrophobic residues are above the horizontal line. The locations of the myristylation consensus sequence (myr), the hydrophobic patch (hp), the TM, the polybasic region (basic), the polyproline region (pro), and the N-linked glycosylation consensus sequence (gly) are indicated. (B) Predicted amino acid sequence of p14. The locations of the structural motifs described for panel A are indicated above the sequence.
p14 assumes an Nexo/Ccyt surface membrane topology.
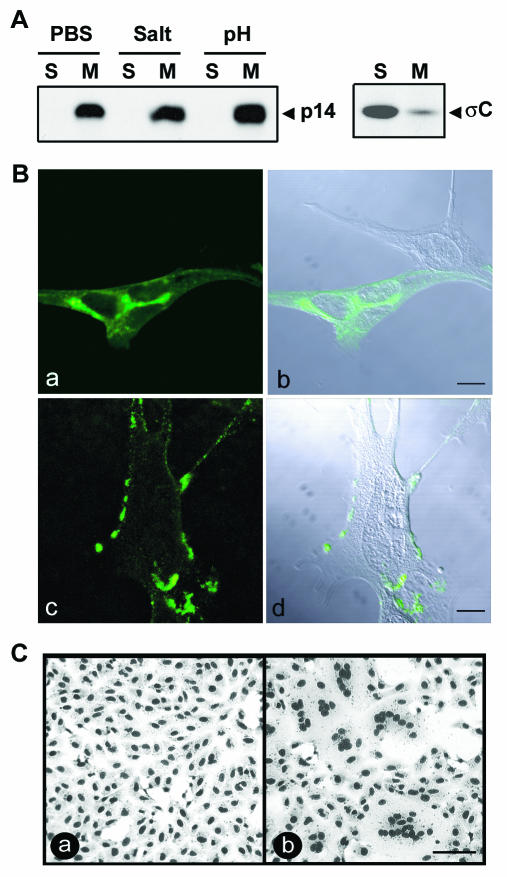
Analysis of the soluble and membrane fractions from transfected QM5 cells indicated that p14 localizes exclusively to the membrane pellet (Fig. 3A), suggesting that p14 is cotranslationally inserted into cellular membranes. Treatment of the membrane pellet with either high salt or high pH to extract peripheral membrane proteins (15) did not alter the p14 distribution (Fig. 3A), indicating that the predicted p14 transmembrane domain is functional and that p14 exists exclusively as an integral membrane-spanning protein. Consistent with the membrane localization of p14, immunostaining of permeabilized cells revealed a reticular staining pattern, with concentrations of p14 in the perinuclear region and numerous punctate foci throughout the cytoplasm radiating out to the plasma membrane. Similar staining of nonpermeabilized cells showed a patchy ring fluorescence at the surfaces of cells (Fig. 3B). Furthermore, a polyclonal anti-p14 antiserum inhibited syncytium formation (Fig. 3C). These results suggest that p14 localizes to the endoplasmic reticulum (ER)-Golgi pathway and that at least a portion of p14 traffics to the cell surface, where it is directly involved in promoting the membrane fusion reaction.
FIG. 3.
p14 is a surface-localized integral membrane protein. (A) The membrane pellet from radiolabeled, p14-transfected QM5 cells was isolated by centrifugation. Membrane pellets were treated with phosphate-buffered saline (PBS) or stripped with either high salt or high pH to remove peripheral proteins, and the integral membrane (M) or soluble (S) fractions were isolated by centrifugation. The presence of p14 in each fraction was detected by immunoprecipitation using an anti-p14 polyclonal antiserum. A similar analysis was performed with transfected cells expressing the soluble ARV σC protein (right panel) as a control for the membrane isolation protocol. (B) p14-transfected QM5 cells were immunostained at 6 h posttransfection with an anti-p14 polyclonal antibody, and antibody distribution was detected by immunofluorescence microscopy using a FITC-conjugated secondary antibody (a and c). The corresponding differential interference microscopy (DIC) images overlaid with the fluorescent images are also shown (b and d). Cells were permeabilized by methanol fixation prior to antibody staining to reveal intracellular p14 distribution (a and b) or were stained live to reveal cell surface expression of p14 (c and d). Scale bar = 10 μm. (C) Transfected Vero cells expressing p14 were treated with anti-p14 polyclonal antiserum (a) or normal rabbit serum (b). Cells were fixed with methanol at 14 h posttransfection and Giemsa stained to reveal the inhibition of syncytium formation by the anti-p14 antiserum. Scale bar = 100 μm.
For examination of the p14 membrane topology, constructs were created that contained two HA epitope tags either added to the C terminus of p14 or inserted between residues seven and eight within the N-terminal domain. The addition of the double-epitope tag to the C terminus (p14-2HAC) slowed, but did not inhibit, the extent of cell-cell fusion, while insertion of the epitope tag in the N-terminal domain (p14-2HAN) abolished polykaryon formation (Fig. 4A). Cells transfected with p14-2HAN or p14-2HAC were immunostained with an anti-HA monoclonal antibody, either after fixation and permeabilization to reveal intracellular fluorescence or with live cells to detect the surface-expressed ectodomain. In permeabilized cells, both constructs revealed the characteristic reticular staining pattern of authentic p14 (Fig. 4B). Positive surface staining was only obtained with the p14-2HAN construct, indicating that p14 assumes an Nexo/Ccyt surface topology in the plasma membrane. Furthermore, the potential N-linked glycosylation site near the C terminus of p14 was nonfunctional (Fig. 5), as evidenced by the lack of a gel mobility shift due to glycosylation when p14 was expressed in transfected cells in the absence or presence of the glycosylation inhibitor tunicamycin. Therefore, we infer that p14 assumes an exclusive Nexo/Ccyt topology.
FIG. 4.
p14 assumes an Nexo/Ccyt surface membrane topology. (A) Vero cells were transfected with authentic p14 (a), p14-2HAC (b), p14-2HAN (c), or the pcDNA3 vector alone (d). Cells were immunostained at 18 h postinfection with an anti-p14 antiserum and alkaline phosphatase-conjugated secondary antibody to reveal antigen-positive foci. p14-2HAC displayed reduced fusion kinetics, while p14-2HAN was fusion negative. Scale bar = 100 μm. (B) Vero cells transfected with p14-2HAN (a and c) or p14-2HAC (b and d) were immunostained at 8 h posttransfection with an anti-HA monoclonal antibody and FITC-conjugated secondary antibody. For the detection of intracellular p14, cells were fixed with methanol before staining (a and b); for surface-localized p14 (c and d), staining was performed on live cells. Scale bar = 10 μm.
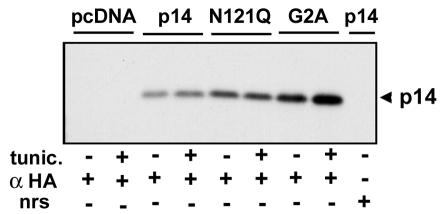
FIG. 5.
p14 is not N-glycosylated. 2HAC-tagged p14-, p14-N121Q (removes C-proximal glycosylation signal)-, p14-G2A (removes N-terminal myristylation signal)-, and pcDNA3-transfected Vero cells in the presence or absence of the N-linked glycosylation inhibitor tunicamycin (tunic.) were radiolabeled at 20 h posttransfection and immunoprecipitated with anti-HA monoclonal antibody (α HA) or normal rabbit serum (nrs). The relative gel mobilities of the precipitated p14 proteins were determined by SDS-PAGE and fluorography. The presence of a single species of p14 in all samples is indicative that the C-proximal glycosylation signal present in p14 is not functional.
The C terminus of p14 is dispensable for fusion activity.
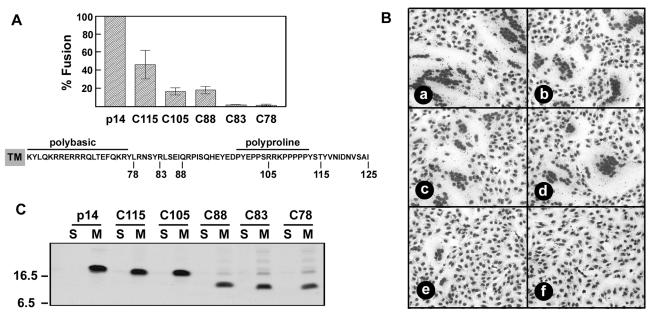
Proline-rich regions can form a type II polyproline helix and are often involved in protein-protein interactions (36). For an examination of the influence of the unusual C-proximal polyproline motif on p14 fusion activity, five p14 C-terminal deletion mutants (named for their lengths, in amino acids) were assessed by a quantitative fusion assay based on the average number of syncytial nuclei per field. Deletion of the C-terminal 10 to 20 amino acids of p14, including the five consecutive proline residues and the potential polyproline helix (residues 108 to 112), reduced the rate of p14-induced syncytium formation, as evidenced by a decreased syncytial index at early times posttransfection (Fig. 6A). However, the overall extent of fusion remained unimpaired, as both p14-C105 and p14-C115 mediated the formation of large multinucleated syncytia (Fig. 6B) that eventually encompassed the entire cell monolayer. The p14-C88 deletion, which removed all of the proline residues from the C-terminal domain, also displayed an impaired rate, but not extent, of fusion (Fig. 6). Therefore, the C-terminal 37 amino acids of p14, representing the proline-rich region and potential polyproline helix motif, are dispensable for the mechanism of p14-mediated fusion.
FIG. 6.
C-terminal 37 amino acids of p14 are dispensable for fusion. (A) Vero cells transfected with authentic p14 or with various p14 C-terminal deletion constructs, named according to the number of the last C-terminal residue, were Giemsa stained at 16 h posttransfection. The average number of nuclei present in syncytia was determined by microscopic examination of five random fields, and results are expressed as percentages of wild-type fusion ± standard errors. The sequence of p14 from the TM to the C terminus is shown, and the locations of the C-terminal truncations, the polybasic region, and the polyproline region are indicated. (B) Vero cells were transfected with p14 (a), C115 (b), C105 (c), C88 (d), C83 (e), or C78 (f) and Giemsa stained at 24 h posttransfection. The C88, C105, and C115 truncations induced extensive syncytium formation, in spite of the reduced kinetics indicated in panel A. The C83 truncation induced a limited number of small syncytia that failed to progress, while the C78 truncation was devoid of syncytium-inducing activity. (C) QM5 cells were transfected with p14 or the C-terminal deletion mutants (C115, C105, C88, C83, and C78), and the distribution of the p14 constructs in the soluble (S) versus the membrane (M) fractions was determined by radiolabeling and immunoprecipitation as described in the legend for Fig. 3A. The relative gel mobilities of molecular weight standards are indicated on the left.
Conversely, the p14-C78 construct was devoid of fusion activity, while the p14-C83 construct retained minimal fusion activity, initiating the formation of small syncytia that failed to progress in size over time (Fig. 6). The different fusion activities of the C-terminal truncations did not correlate with their relative expression-detection levels or efficiencies of membrane insertion; all of the truncated p14 constructs localized exclusively to the membrane fraction (Fig. 6C). Immunoprecipitation results did indicate a reduced detection of the C88, C83, and C78 constructs in the membrane fraction, possibly due to decreased protein expression or stability and/or the loss of a C-proximal epitope recognized by the polyclonal antiserum (Fig. 6C). However, the relative fusion activity of these constructs did not correlate with their detection in the membrane fraction. Therefore, a minimum size or structure of the p14 C-terminal domain most likely influences p14 fusion activity by altering events downstream of membrane insertion.
N-terminal myristylation of p14 is essential for fusion activity.
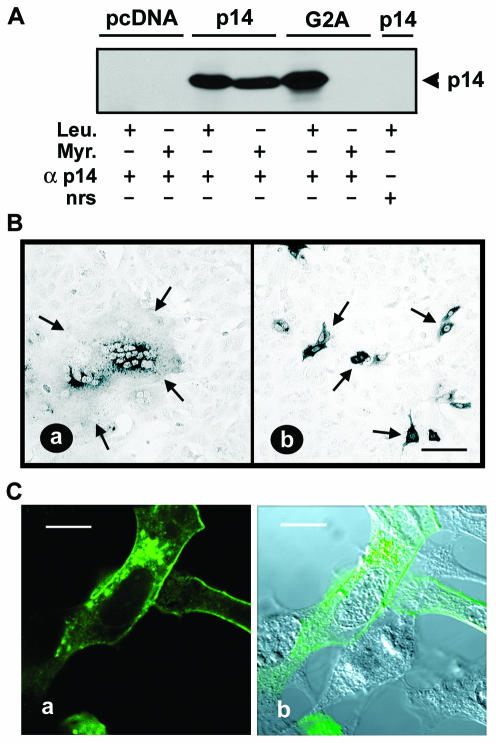
The myristate moiety of myristylated proteins is almost always associated with the cytoplasmic leaflet of lipid bilayers (38). In view of the predicted Nexo/Ccyt topology of p14, we anticipated that the myristylation consensus sequence would be nonfunctional and therefore irrelevant for the p14 function. This was not the case. Radiolabeling indicated the incorporation of [3H]myristic acid into authentic p14, but not into a construct containing a G2A substitution that removed the myristylation consensus sequence (Fig. 7A). The loss of labeling of the G2A construct by myristic acid confirmed that the labeling of authentic p14 reflected the incorporation of [3H]myristic acid and not the metabolic redistribution of the radiolabel, and it implied that p14 is a myristylated integral membrane protein. In addition, the myristate moiety appears to be an essential component of p14, since the p14-G2A construct did not mediate cell-cell fusion (Fig. 7B). The altered fusion activity of the myristylation-negative construct did not reflect altered protein expression (Fig. 7A) or p14 subcellular localization, as determined by immunofluorescent staining (Fig. 7C). Therefore, the N-terminal myristylation consensus sequence of p14 is both functional and essential for p14-induced syncytium formation.
FIG. 7.
N-terminal myristylation of p14 is essential for cell fusion. (A) 2HAC-tagged p14-, p14-G2A-, and pcDNA3-transfected Vero cells were labeled with either [3H]myristic acid (Myr.) or [3H]leucine (Leu.) and immunoprecipitated with anti-p14 polyclonal antiserum (α p14) or normal rabbit serum (nrs). Precipitates were fractionated by SDS-PAGE and radiolabeled p14 was detected by fluorography. (B) Vero cells transfected with either 2HAC-tagged p14 (a) or p14-G2A (b) were fixed with methanol at 18 h posttransfection and immunostained with anti-p14 polyclonal antiserum and an alkaline phosphatase-conjugated secondary antibody. Arrows indicate the limits of an antigen-positive syncytial focus (a) or individual antigen-positive cells expressing the fusion-negative p14-G2A construct (b). Scale bar = 100 μm. (C) QM5 cells transfected with p14-G2A were permeabilized with methanol and immunostained at 9 h posttransfection with anti-p14 polyclonal antiserum and FITC-conjugated secondary antibody. Fluorescence microscopy (a) revealed punctate, perinuclear intracellular staining of p14-G2A and a ring surface fluorescence characteristic of authentic p14. The corresponding DIC image overlaid with the fluorescent image is also shown (b). Scale bar = 10 μm.
p14 translocates its myristylated N terminus across the membrane.
The unexpected placement of a myristylated N-terminal domain external to the plasma membrane prompted the need for further confirmation of the p14 topology. An N-linked glycosylation site was engineered into the N-terminal domain of p14 (V9T substitution), and N-glycosylation mapping was used to examine the topology of the p14-V9T construct. In vitro translation of authentic p14 in the presence or absence of canine microsomal membranes (CMM) produced polypeptides with identical gel mobilities (Fig. 8A), confirming the sequence-predicted absence of a cleavable N-terminal signal peptide. Conversely, in vitro translation of p14-V9T in the presence of CMM resulted in two protein species, one with a retarded gel mobility relative to authentic p14 or to p14-V9T translated in the absence of CMM (Fig. 8A). The mobility shift of approximately 3 kDa approximated the expected shift per core glycan (2.5 kDa) (6) and was in accord with the mobility shift of a known glycosylated control protein (Fig. 8A). The nonmyristylated p14-G2A construct in the presence of CMM demonstrated a minor population with the same gel mobility as the glycosylated species of p14-V9T (Fig. 8A), suggesting that a small percentage of p14-G2A molecules may adopt the inverse Ncyt/Cexo topology, leading to the glycosylation of Asn121. Therefore, myristylation may be a minor contributor for determining an exclusive Nexo/Ccyt topology for p14, at least in vitro.
FIG. 8.
p14 translocates its myristylated N terminus across the membrane. (A) 2HAC-tagged p14, p14-G2A, or p14-V9T were in vitro transcribed and translated in the presence or absence of canine microsomal membranes (CMM). The relative gel mobilities of the radiolabeled authentic (p14) and glycosylated (p14*) species of p14 are indicated on the right, and molecular weight markers are shown on the left. The control lane (Con.) indicates the glycosylated and nonglycosylated species generated by the translation of yeast α factor mRNA. (B) Vero cells were transfected with 2HAC-tagged p14 or p14-V9T or were mock transfected (M) with pcDNA3 vector. Transfected cells were labeled with [3H]myristic acid (Myr.) or [3H]leucine (Leu.) in the presence or absence of tunicamycin (tunic.) and were immunoprecipitated with anti-p14 antiserum (α p14) or normal rabbit serum (nrs). The relative gel mobilities of the radiolabeled authentic (p14) and glycosylated (p14*) species of p14 are indicated on the right, and molecular weight markers are shown on the left.
Analysis of p14-V9T in transfected Vero cells also detected two different species of p14-V9T, with a loss of the slower migrating form after the treatment of cells with tunicamycin, confirming that this polypeptide represented glycosylated p14 (Fig. 8B). To exclude the possibility that there were two populations of p14, one with the myristylated N-terminal domain inside the cell and the other nonmyristylated form in the Nexo/Ccyt topology, p14-V9T-transfected cells were labeled with [3H]myristic acid. The N-terminal domain of p14-V9T was both N-glycosylated and myristylated, as evidenced by the ability of [3H]myristic acid to label both forms of p14-V9T (Fig. 8B). These results conclusively establish that the myristylated N-terminal domain of p14 is translocated into the ER lumen, where we believe it interacts with the luminal leaflet of the membrane, and after trafficking to the plasma membrane, resides in the exoplasmic leaflet outside the cell.
An additional observation noted during in vivo analysis was the loss of syncytium-inducing ability imparted by the V9T substitution (data not shown). The loss of fusion activity was not the result of steric hindrance from the addition of a large carbohydrate moiety, since the fusion ability of p14-V9T was not restored by the inhibition of N-linked glycosylation by tunicamycin. Transport of the p14-V9T construct was also not affected, as a population of p14-V9T molecules was clearly visualized accumulating at the plasma membrane by immunofluorescence staining of permeabilized cells (data not shown). Therefore, a minor alteration near the N terminus of the p14 ectodomain hydrophobic patch affects the membrane fusion activity of the protein, independent of any effects on p14 membrane insertion, protein topology, or cell surface localization. The small p10 ectodomain contains a similar hydrophobic patch that may function in an analogous manner as the fusion peptide motifs found in enveloped virus fusion proteins (45). However, the relatively low overall hydrophobicity of these FAST protein hydrophobic patches and the likelihood that these potential fusion peptide motifs are not sequestered within a complex tertiary structure distinguish these motifs from the typical fusion peptides of enveloped viruses (47, 57).
DISCUSSION
Structural motifs in p14, the newest member of the diverse FAST protein family.
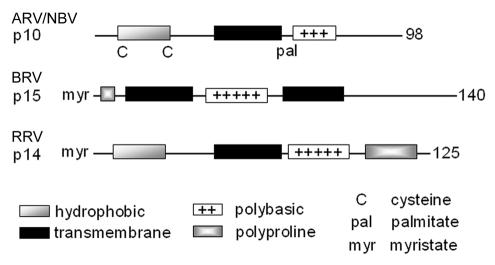
The present study indicates that the RRV p14 protein is a novel reovirus FAST protein and the newest member of a diverse family of atypical viral membrane fusion proteins. While p14 shares no significant sequence similarity to previously identified FAST proteins (7, 45), some structural commonality between the FAST proteins does exist (Fig. 7). All of the FAST proteins are small, basic, acylated integral membrane proteins. However, the nature of the acylation, the arrangement of the hydrophobic motifs, and the presence or absence of essential cysteine residues or proline-rich regions (Fig. 9) lead to each FAST protein containing its own signature arrangement of structural motifs. Moreover, key structural features found in enveloped viral fusion proteins, such as coiled-coil domains, typical fusion peptide motifs, and large, complex, multimeric ectodomains, are absent from the FAST proteins. Clearly, the reovirus FAST proteins represent a third class of viral membrane fusion proteins that are distinct from the class I and class II enveloped virus fusion proteins (25, 47). The unique structural features of the FAST proteins are likely to have important implications for the mechanism of FAST-mediated membrane fusion.
FIG. 9.
Structural motifs in reovirus FAST proteins. The repertoires and arrangement of structural motifs present in the RRV p14, ARV p10, and BRV p15 FAST proteins are depicted in a linear fashion, drawn approximately to scale. Numbers refer to amino acid residues.
Topological analysis indicates that p14 is a type III (Nexo/Ccyt) membrane protein.
The absence of a cleavable signal peptide (Fig. 8), the fact that p14 resides exclusively as an integral membrane protein (Fig. 3), and the Nexo/Ccyt topology (Fig. 4, 5, and 8) imply that p14 utilizes an internal reverse signal-anchor to localize the small, approximately 40-residue N-terminal domain external to the membrane. All of these properties identified p14 as a type III (Nexo/Ccyt) membrane protein (17, 48). Unlike type I (Nexo/Ccyt) and type II (Cexo/Ncyt) proteins, type III proteins translocate N-terminal residues that are transiently exposed to the cytosol before the signal-anchor emerges from the ribosome (17). Topogenic signals favoring a type III membrane topology, all of which are present in p14, include (i) positively charged residues on the C-terminal side of the signal-anchor that promote cytosolic retention (54); (ii) a long signal-anchor sequence, as longer and more hydrophobic sequences favor N-terminal translocation (40, 55); and (iii) the small size of the translocated N-terminal domain (8). Other examples of type III membrane proteins include cytochrome P-450, CLN3, mouse synaptogamins I and II, and the influenza viral proteins M2 and NB (24, 26, 28, 42, 58). However, p14 is the first demonstrated example of a type III membrane fusion protein.
The type III character of p14 is shared by the p10 FAST protein of ARV, which also assumes an Nexo/Ccyt topology and contains topogenic signals typical of type III membrane proteins, including the absence of an N-terminal signal peptide, positive charges distributed on the C-terminal side of the signal-anchor, and a small N-terminal ectodomain (45). While the membrane topology of BRV p15 remains undefined, this protein also lacks any evidence of a cleavable signal peptide and contains a small, myristylated N-terminal domain and a basic region C-terminal to a predicted transmembrane anchor (7). However, p15 also contains a second possible membrane-spanning domain, leading to speculation that the protein may be a multispanning membrane protein (7). At this time, no compelling evidence exists to rule out the possibility that p15, like the other reoviral FAST proteins, may also be a type III (Nexo/Ccyt) membrane protein.
Cotranslational translocation of the p14 myristylated ectodomain.
Myristic acid mediates reversible protein interactions with lipid bilayers (38). The stable association of soluble, myristylated proteins with membranes requires additional signals, such as another lipid modification or an electrostatic interaction (33). The unique ability of myristic acid to reversibly associate with the cytosolic leaflet of the plasma membrane in response to the availability of a second signal regulates the activities of a number of cellular signaling proteins (35, 38, 41).
In addition to soluble proteins, a limited number of integral membrane proteins are N-terminally myristylated, such as the type II (Cexo/Ncyt) CLA-1 protein (CD36 family) (5). Some integral membrane proteins are also myristylated at internal lysine or cysteine residues (2, 18, 49, 50, 53). In the case of myristylated integral membrane proteins, stable membrane association derives from the TM domain and the myristic acid serves some undefined function other than protein anchoring in the membrane. The role of myristate in these integral membrane proteins is speculative, but it has been suggested to involve the regulation of enzymatic activity by the modulation of protein-protein or protein-lipid interactions or by an alteration of the ability of the protein to be subsequently modified.
Our discovery that the myristylated p14 N terminus is translocated across the membrane and resides outside the cell was surprising, but not without precedent. The large surface antigens of both human and duck hepatitis B viruses are polytopic proteins that adopt two functional membrane topologies, one of which externalizes the myristylated N-terminal pre-S1 domain of the protein on the virion surface, where it mediates effective entry of the virus into hepatocytes (4, 51). However, unlike the case for p14, which cotranslationally translocates a myristylated N terminus, interactions of the duck hepatitis B virus pre-S1 domain with cytosolic Hsc70 lead to posttranslational externalization of the myristylated N terminus (16, 27, 34, 51).
As far as we are aware, the hepatitis virus L protein is the only other reported example of an external myristylated N-terminal domain. However, other examples may exist. The vaccinia virus L1R protein is an N-terminally myristylated polytopic protein, and a topological analysis inferred that the myristylated N terminus of L1R resides in the ER lumen (43, 60). Similarly, CLN3 and the human integrin beta-6 chain assume Nexo/Ccyt membrane topologies, and each contains an N-terminal myristylation consensus sequence (28, 44). It remains to be determined whether these consensus sequences are functional and whether additional examples of myristylated ectodomains exist.
A myristylated ectodomain is essential for p14 fusion activity.
The type II topology (Cexo/Ncyt) of N-terminally myristylated cellular membrane proteins or the locations of the internally myristylated lysine or cysteine residues in these proteins position the myristic acid on the cytosolic side of the membrane, similar to the situation with myristylated cytosolic proteins. In contrast, the essential myristic moiety present in p14 must exert its influence on p14-induced membrane fusion via interactions with the external leaflet of the membrane bilayer. Although the role of the p14 myristic acid is unclear, several possible roles can be envisioned. Acylation of both viral and cellular proteins can target these proteins to detergent-resistant membrane microdomains within the plasma membrane (31, 39). Although the myristylation-negative p14-G2A still trafficked to the cell surface (Fig. 5), the loss of myristylation could influence p14 localization to membrane microdomains. A second possibility is based on the assumption that the myristic acid associates with the luminal leaflet of the ER membrane after translocation, in a manner analogous to the membrane association of signal peptides before their cleavage (19, 20). Signal peptide interactions with the luminal leaflet of the ER membrane can promote the correct folding of the nascent protein (59); a similar situation could contribute to folding of the p14 ectodomain into a fusion-competent conformation. An intriguing third possibility reflects the unique ability of myristic acid to reversibly associate with a lipid bilayer (38). During the close apposition of membranes that must occur prior to fusion, myristic acid may disassociate from the outer leaflet of the donor membrane and interact with the outer leaflet of the target membrane. Such interactions of myristic acid, from a sufficient number of p14 molecules, with the target and/or donor membranes could alter the lipid packing of one or both membranes to support the lipid rearrangements required for fusion to proceed. This hypothesis is supported by the greater membrane permeabilization exhibited by a myristylated hydrophobic peptide than by its nonacylated partner (23).
Our discovery of this newest FAST protein with its unusual membrane topology further underscores the diversity of class III viral membrane fusion proteins. A collection of membrane interaction motifs, including signal-anchors, hydrophobic patches, polybasic regions, and now an externalized fatty acid, have been assembled into these rudimentary membrane fusion proteins. Though the mechanism of FAST-mediated membrane fusion remains undetermined, it is evident that the FAST proteins do not have the size capability or structural features to promote membrane fusion by using large energy-releasing triggered conformational changes, as occurs with the class I and class II enveloped virus fusion proteins (47). Continued structure-function analysis of the FAST proteins should provide alternate insights into the minimal determinants of protein-mediated fusion of biological membranes.
Acknowledgments
We thank Jingyun Shou for expert technical assistance and Ryan Liebscher for assistance with preliminary experiments characterizing p14 myristylation.
This research was supported by grants from the Canadian Institutes of Health Research (CIHR). R.D. is the recipient of a CIHR Investigator Award.
REFERENCES
- 1.Ahne, W., I. Thomsen, and J. Winton. 1987. Isolation of a reovirus from the snake, Python regius. Arch. Virol. 94:135-139. [DOI] [PubMed] [Google Scholar]
- 2.Armah, D. A., and K. Mensa-Wilmot. 1999. S-Myristoylation of a glycosylphosphatidylinositol-specific phospholipase C in Trypanosoma brucei. J. Biol. Chem. 274:5931-5938. [DOI] [PubMed] [Google Scholar]
- 3.Bentz, J., and A. Mittal. 2000. Deployment of membrane fusion protein domains during fusion. Cell Biol. Int. 24:819-838. [DOI] [PubMed] [Google Scholar]
- 4.Bruss, V., J. Jagelstein, E. Gerhardt, and P. R. Galle. 1996. Myristylation of the large surface protein is required for hepatitis B virus in vitro infectivity. Virology 218:396-399. [DOI] [PubMed] [Google Scholar]
- 5.Calvo, D., and M. A. Vega. 1993. Identification, primary structure, and distribution of CLA-1, a novel member of the CD36/LIMPII gene family. J. Biol. Chem. 268:18929-18935. [PubMed] [Google Scholar]
- 6.Daniels, R., B. Kurowski, A. E. Johnson, and D. N. Hebert. 2003. N-linked glycans direct the cotranslational folding pathway of influenza hemagglutinin. Mol. Cell 11:79-90. [DOI] [PubMed] [Google Scholar]
- 7.Dawe, S. J., and R. Duncan. 2002. The S4 genome segment of Baboon reovirus is bicistronic and encodes a novel fusion-associated small transmembrane protein. J. Virol. 76:2131-2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Denzer, A. J., C. E. Nabholz, and M. Spiess. 1995. Transmembrane orientation of signal-anchor proteins is affected by the folding state but not the size of the N-terminal domain. EMBO J. 14:6311-6317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duncan, R. 1999. Extensive sequence divergence and phylogenetic relationships between the fusogenic and nonfusogenic orthoreoviruses: a species proposal. Virology 260:316-328. [DOI] [PubMed] [Google Scholar]
- 10.Duncan, R., Z. Chen, S. Walsh, and S. Wu. 1996. Avian reovirus-induced syncytium formation is independent of infectious progeny virus production and enhances the rate, but is not essential, for virus-induced cytopathology and virus egress. Virology 224:453-464. [DOI] [PubMed] [Google Scholar]
- 11.Duncan, R., and K. Sullivan. 1998. Characterization of two avian reoviruses that exhibit strain-specific quantitative differences in their syncytium-inducing and pathogenic capabilities. Virology 250:263-272. [DOI] [PubMed] [Google Scholar]
- 12.Duncan, R., J. Corcoran, J. Shou, and D. Stoltz. 2004. Reptilian reovirus: a new fusogenic orthoreovirus species. Virology 319:131-140. [DOI] [PubMed]
- 13.Epand, R. M., and R. F. Epand. 2003. Irreversible unfolding of the neutral pH form of influenza hemagglutinin demonstrates that it is not in a metastable state. Biochemistry 42:5052-5057. [DOI] [PubMed] [Google Scholar]
- 14.Epand, R. M., and R. F. Epand. 2002. Thermal denaturation of influenza virus and its relationship to membrane fusion. Biochem. J. 365:841-848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fujiki, Y., A. L. Hubbard, S. Fowler, and P. B. Lazarow. 1982. Isolation of intracellular membranes by means of sodium carbonate treatment: application to endoplasmic reticulum. J. Cell Biol. 93:97-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gallina, A., and G. Milanesi. 1993. Trans-membrane translocation of a myristylated protein amino terminus. Biochem. Biophys. Res. Commun. 195:637-642. [DOI] [PubMed] [Google Scholar]
- 17.Goder, V., and M. Spiess. 2001. Topogenesis of membrane proteins: determinants and dynamics. FEBS Lett. 504:87-93. [DOI] [PubMed] [Google Scholar]
- 18.Hedo, J. A., E. Collier, and A. Watkinson. 1987. Myristyl and palmityl acylation of the insulin receptor. J. Biol. Chem. 262:954-957. [PubMed] [Google Scholar]
- 19.Hegde, R. S., and V. R. Lingappa. 1999. Regulation of protein biogenesis at the endoplasmic reticulum membrane. Trends Cell Biol. 9:132-137.10203789 [Google Scholar]
- 20.Heinrich, S. U., W. Mothes, J. Brunner, and T. A. Rapoport. 2000. The Sec61p complex mediates the integration of a membrane protein by allowing lipid partitioning of the transmembrane domain. Cell 102:233-244. [DOI] [PubMed] [Google Scholar]
- 21.High, S., and V. Laird. 1997. Membrane protein biosynthesis—all sewn up? Trends Cell Biol. 7:206-210. [DOI] [PubMed] [Google Scholar]
- 22.Jahn, R., T. Lang, and T. C. Südhof. 2003. Membrane fusion. Cell 112:519-533. [DOI] [PubMed] [Google Scholar]
- 23.Joseph, M., and R. Nagaraj. 1995. Interaction of peptide corresponding to fatty acylation sites in proteins with model membranes. J. Biol. Chem. 270:16749-16755. [DOI] [PubMed] [Google Scholar]
- 24.Kida, Y., M. Sakaguchi, M. Fukuda, K. Mikoshiba, and K. Mihara. 2000. Membrane topogenesis of a type I signal-anchor protein, mouse synaptotagmin II, on the endoplasmic reticulum. J. Cell Biol. 150:719-729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kielian, M. 2002. Structural surprises from the flaviviruses and alphaviruses. Mol. Cell 9:454-456. [DOI] [PubMed] [Google Scholar]
- 26.Lamb, R. A., S. L. Zebedee, and C. D. Richardson. 1985. Influenza virus M2 protein is an integral membrane protein expressed on the infected-cell surface. Cell 40:627-733. [DOI] [PubMed] [Google Scholar]
- 27.Loffler-Mary, H., M. Werr, and R. Prange. 1997. Sequence-specific repression of cotranslational translocation of the hepatitis B virus envelope proteins coincides with the binding of heat shock protein Hsc70. Virology 18:144-152. [DOI] [PubMed] [Google Scholar]
- 28.Mao, Q., B. J. Foster, H. Xia, and B. L. Davidson. 2003. Membrane topology of CLN3, the protein underlying Batten disease. FEBS Lett. 541:40-46. [DOI] [PubMed] [Google Scholar]
- 29.Martoglio, B., and B. Dobberstein. 1998. Signal sequences: more than just greasy peptides. Trends Cell Biol. 8:410-415. [DOI] [PubMed] [Google Scholar]
- 30.Melikyan, G. B., R. M. Markosyan, H. Hemmati, M. K. Delmedico, D. M. Lambert, and F. S. Cohen. 2000. Evidence that the transition of HIV-1 gp41 into a six-helix bundle, not the bundle configuration, induces membrane fusion. J. Cell Biol. 151:413-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Melkonian, K. A., A. G. Ostermeyer, J. Z. Chen, M. G. Roth, and D. A. Brown. 1999. Role of lipid modifications in targeting proteins to detergent-resistant membrane rafts. J. Biol. Chem. 274:3910-3917. [DOI] [PubMed] [Google Scholar]
- 32.Nibert, M. L., and L. A. Schiff. 2000. Reoviruses and their replication, p. 793-842. In D. M. Knipe and P. M. Howley (ed.), Fundamental virology, 3rd ed. Lippincott-Raven Press, Philadelphia, Pa.
- 33.Peitzsch, R. M., and S. McLaughlin. 1993. Binding of acylated peptides and fatty acids to phospholipid vesicles: pertinence to myristoylated proteins. Biochemistry 32:10436-10443. [DOI] [PubMed] [Google Scholar]
- 34.Prange, R., M. Werr, and H. Loffler-Mary. 1999. Chaperones involved in hepatitis B virus morphogenesis. Biol. Chem. 380:305-314. [DOI] [PubMed] [Google Scholar]
- 35.Randazzo, P. A., T. Terui, S. Sturch, H. M. Fales, A. G. Ferrige, and R. A. Kahn. 1995. The myristoylated amino terminus of ADP-ribosylation factor a is a phospholipid- and GTP-sensitive switch. J. Biol. Chem. 270:14809-14815. [DOI] [PubMed] [Google Scholar]
- 36.Reiersen, H., and A. R. Rees. 2001. The hunchback and its neighbors: proline as an environmental modulator. Trends Biochem. Sci. 26:679-684. [DOI] [PubMed] [Google Scholar]
- 37.Remeta, D. P., M. Krumbiegel, C. A. Minetti, A. Puri, A. Ginsburg, and R. Blumenthal. 2002. Acid-induced changes in thermal stability and fusion activity of influenza hemagglutinin. Biochemistry 41:2044-2054. [DOI] [PubMed] [Google Scholar]
- 38.Resh, M. D. 1999. Fatty acid acylation: new insights into membrane targeting of myristoylated and palmitoylated proteins. Biochim. Biophys. Acta 1451:1-16. [DOI] [PubMed] [Google Scholar]
- 39.Robbins, S. M., N. A. Quintrell, and J. M. Bishop. 1995. Myristoylation and differential palmitoylation of HCK protein-tyrosine kinases govern their attachment to membranes and association with caveolae. Mol. Cell. Biol. 15:3507-3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rösch, K., D. Naeher, V. Liard, V. Goder, and M. Spiess. 2000. The topogenic contribution of uncharged amino acids on signal sequence orientation in the endoplasmic reticulum. J. Biol. Chem. 275:14916-14922. [DOI] [PubMed] [Google Scholar]
- 41.Saouaf, S. J., A. Wolven, M. D. Resh, and J. B. Bolen. 1997. Palmitoylation of Src family tyrosine kinases regulates functional interaction with a B-cell substrate. Biochem. Biophys. Res. Commun. 234:325-329. [DOI] [PubMed] [Google Scholar]
- 42.Sato, T., M. Sakaguchi, K. Mihara, and T. Omura. 1990. The amino-terminal structures that determine topological orientation of cytochrome P-450 in microsomal membrane. EMBO J. 9:2391-2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Senkevich, T. G., C. L. White, E. V. Koonin, and B. Moss. 2000. A viral member of the ERV1/ALR protein family participates in a cytoplasmic pathway of disulfide bond formation. Proc. Natl. Acad. Sci. USA 97:12068-12073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sheppard, D., C. Rozzo, L. Starr, V. Quaranta, D. J. Erle, and R. Pytela. 1990. Complete amino acid sequence of a novel integrin beta subunit (beta 6) identified in epithelial cells using the polymerase chain reaction. J. Biol. Chem. 265:11502-11507. [PubMed] [Google Scholar]
- 45.Shmulevitz, M., and R. Duncan. 2000. A new class of fusion-associated small transmembrane (FAST) proteins encoded by the non-enveloped fusogenic reoviruses. EMBO J. 19:902-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shmulevtiz, M., J. Salsman, and R. Duncan. 2003. Palmitoylation, membrane-proximal basic residues, and transmembrane glycine residues in the reovirus p10 protein are essential for syncytium formation. J. Virol. 77:9769-9779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Skehel, J. J., and D. C. Wiley. 2000. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu. Rev. Biochem. 69:531-569. [DOI] [PubMed] [Google Scholar]
- 48.Speiss, M. 1995. Heads or tails—what determines the orientation of proteins in the membrane. FEBS Lett. 369:76-79. [DOI] [PubMed] [Google Scholar]
- 49.Stevenson, F. T., S. L. Bursten, C. Fanton, R. M. Locksley, and D. H. Lovett. 1993. The 31-kDa precursor of interleukin 1α is myristoylated on specific lysines within the 16 kDa N-terminal propiece. Proc. Natl. Acad. Sci. USA 90:7245-7249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stevenson, F. T., S. L. Bursten, R. M. Locksley, and D. H. Lovett. 1992. Myristyl acylation of the tumor necrosis factor alpha precursor on specific lysine residues. J. Exp. Med. 176:1053-1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Swameye, I., and H. Schaller. 1997. Dual topology of the large envelope protein of duck hepatitis B virus: determinants preventing pre-S translocation and glycosylation. J. Virol. 71:9434-9441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Towler, D. A., S. P. Adams, S. R. Eubanks, D. S. Towery, E. Jackson-Machelski, L. Glaser, and J. I. Gordon. 1988. Myristoyl CoA:protein N-myristoyltransferase activities from rat liver and yeast possess overlapping yet distinct peptide substrate specificities. J. Biol. Chem. 263:1784-1790. [PubMed] [Google Scholar]
- 53.Vassilev, A. O., N. Plesofsky-Vig, and R. Brambl. 1995. Cytochrome c oxidase in Neurospora crassa contains myristic acid covalently linked to subunit 1. Proc. Natl. Acad. Sci. USA 92:8680-8684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.von Heijne, G., and Y. Gavel. 1988. Topogenic signals in integral membrane proteins. Eur. J. Biochem. 174:671-678. [DOI] [PubMed] [Google Scholar]
- 55.Wahlberg, J. M., and M. Spiess. 1997. Multiple determinants direct the orientation of signal-anchor proteins: the topogenic role of the hydrophobic signal domain. J. Cell Biol. 137:555-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weissenhorn, W., A. Dessen, L. J. Calder, S. C. Harrison, J. J. Skehel, and D. C. Wiley. 1999. Structural basis for membrane fusion by enveloped viruses. Mol. Membr. Biol. 16:3-9. [DOI] [PubMed] [Google Scholar]
- 57.White, J. M. 1990. Viral and cellular membrane fusion proteins. Annu. Rev. Physiol. 52:675-697. [DOI] [PubMed] [Google Scholar]
- 58.Williams, M. A., and R. A. Lamb. 1986. Determination of the orientation of an integral membrane protein and sites of glycosylation by oligonucleotide-directed mutagenesis: influenza B virus NB glycoprotein lacks a cleavable signal sequence and has an extracellular NH2-terminal region. Mol. Cell. Biol. 6:4317-4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wilson, I. A., J. J. Skehel, and D. C. Wiley. 1981. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 Å resolution. Nature 289:366-373. [DOI] [PubMed] [Google Scholar]
- 60.Wolffe, E. J., S. Vijaya, and B. Moss. 1995. A myristylated membrane protein encoded by the vaccinia virus L1R open reading frame is the target of potent neutralizing monoclonal antibodies. Virology 211:53-63. [DOI] [PubMed] [Google Scholar]
- 61.Zimmerberg, J., S. S. Vogel, and L. V. Chernomordik. 1993. Mechanisms of membrane fusion. Annu. Rev. Biophys. Biomol. Struct. 22:433-466. [DOI] [PubMed] [Google Scholar]