Abstract
Human immunodeficiency virus type 1 (HIV-1) drug resistance results from the accumulation of mutations in the viral genes targeted by the drugs. These genetic changes, however, are commonly detected and monitored by techniques that only take into account the dominant population of plasma virus. Because HIV-1-infected patients harbor a complex and diverse mixture of virus populations, the mechanisms underlying the emergence and the evolution of resistance are not fully elucidated. Using techniques that allow the quantification of resistance mutations in minority virus species, we have monitored the evolution of resistance in plasma virus populations from patients failing protease inhibitor treatment. Minority populations with distinct resistance genotypes were detected in all patients throughout the evolution of resistance. The emergence of new dominant genotypes followed two possible mechanisms: (i) emergence of a new mutation in a currently dominant genotype and (ii) emergence of a new genotype derived from a minority virus species. In most cases, these population changes were associated with an increase in resistance at the expense of a reduction in replication capacity. Our findings provide a preliminary indication that minority viral species, which evolve independently of the majority virus population, can eventually become dominant populations, thereby serving as a reservoir of diversity and possibly accelerating the development of drug resistance.
For many patients infected with human immunodeficiency virus type 1 (HIV-1), the administration of combinations of antiretroviral drugs, often referred to as highly active antiretroviral therapy (HAART), succeeds in suppressing detectable virus replication and reversing the evolution of the disease (41). In some patients, however, the suppressive effect of HAART is incomplete, which usually leads to selection of viral variants with resistance to antiretroviral agents. Resistance is mediated by mutations that emerge in the viral enzymes targeted by the drugs (8, 24, 42). For most antiretroviral drugs, a single resistance mutation does not result in maximal resistance. Thus, resistance is not an all-or-nothing phenomenon and generally increases over time (4, 35, 50). For example, the development of resistance to protease inhibitors is usually a gradual process, and the development of high levels of resistance usually requires the accumulation of multiple mutations in the enzyme itself (9, 10, 35), as well as, in certain cases, mutations at sites where Gag precursors are cleaved by this enzyme (11, 18, 31, 57). Protease enzymes expressing strong resistance to protease inhibitors often contain eight or more resistance-associated mutations, but many months of failing treatment must precede the appearance of such viral variants.
Several areas of uncertainty persist concerning the evolution of resistant viruses in patients failing treatment with protease inhibitors. First, it is commonly thought that resistance progresses by sequential steps. In this model, the acquisition of a new mutation by a virus results in an increase in selective advantage that allows the new viral species to outgrow its parental counterpart and to become the predominant species. This process will repeat itself for each addition of a new mutation, ultimately resulting in the predominance of a multiply mutated, highly resistant variant (32, 35) and to the disappearance of the less-resistant parental and intermediate species. This model, however, does not take into account the complexity of HIV populations in infected patients. It is now recognized that plasma virus is composed of a mixture of numerous quasispecies (8). In patients undergoing treatment and harboring resistant viral strains, the dominant quasispecies is accompanied by minority populations expressing distinct resistance genotypes (8, 14, 21, 34, 46). Because these viral species can evolve independently, an initially minority population, having followed a distinct evolutionary pathway, could emerge as the dominant population, either because it had evolved higher resistance or because a change in drug pressure gave that population a growth advantage over the prior majority population. This scenario, however, has never been documented.
A second area of uncertainty concerns the relative importance of drug resistance and viral replicative capacity in driving viral evolution. Although resistance mutations confer a decisive advantage to these variants over their parental wild-type counterparts for replication in the presence of antiretroviral drugs, there is accumulating evidence that resistant HIV strains are less fit than wild-type virus for replication in the absence of drug and that, overall, the replicative capacity of many drug-resistant HIV variants is reduced (1, 2, 4, 7, 13, 19, 21, 22, 37, 39). Resistance mutations in the protease are thought to modify the size and shape of the substrate-binding domain of the enzyme, thereby reducing the affinity of inhibitors for the enzyme (30, 39, 44, 47). These changes also impair the enzymatic cleavage of the natural substrates of the protease in Gag and Gag-Pol, accounting for the reduced replicative capacity of these viruses (12, 33, 39, 49, 55). It is also recognized that the major impact of certain mutations appearing under drug pressure is to restore, at least in part, impairments in viral replicative capacity resulting from the development of resistance (6, 11, 18, 31, 32, 43, 57). Some studies support the model that early mutations predominantly influence viral resistance, despite a loss in viral replicative capacity (15, 31, 32, 57), and that replicative capacity is later restored, at least partially, by compensatory mutations (32, 38). Other studies have found, however, that “compensatory” mutations were also instrumental in increasing resistance (38) and that the loss of replicative capacity generally increases with increasing numbers of mutations and increasing levels of resistance (4, 55) (A. Faye, E. Race, V. Obry, M. H. Prévot, V. Joly, S. Matheron, F. Damond, E. Dam, S. Paulous, and F. Clavel, Abstr. Third Int. Workshop Drug Resist. Treatment Strategies, abstr. 129, 1999), suggesting that improved resistance often remains the predominant determinant driving viral selection.
In the present study, we evaluated the evolution of resistance in patients failing treatment with HAART regimens including a protease inhibitor by using techniques that permitted the characterization of both the majority viral population as well as minority species expressing alternative resistance genotypes. Our goal was to evaluate the potential role of minority populations in the evolution of resistance and to characterize the contribution of changes in resistance and replicative capacity to the selection of variants in the course of viral evolution.
MATERIALS AND METHODS
Patient population.
Plasma samples from HIV-1-infected patients meeting the following criteria were evaluated in the present study: (i) a patient age of >18 years; (ii) the patient had begun treatment with a protease inhibitor for the first time and had been on continuous treatment with protease inhibitors over the time of evaluation; (iii) the patient subsequently experienced virologic failure, as defined by a rebound in plasma HIV-1 RNA after a period during which it had become undetectable or by the absence of such a period at any time after protease inhibitor treatment; (iv) an initial standard protease genotype, performed by direct sequencing of bulk PCR amplification products from plasma HIV-1 RNA after the onset of virologic failure, revealed the absence of resistance mutations V82A and/or L90M; (v) a similar genotype performed at a later time demonstrated the subsequent appearance of the L90M mutation. All patients were monitored at Hôpital Bichat-Claude Bernard, Paris, France. The patients (six men and one woman) had an average age of 33 ± 7 years. Six of the patients had previously been treated with nucleoside analogues. The mean viral load in plasma prior to treatment with protease inhibitors was 5.34 ± 0.33 log10 copies/ml, and the mean CD4+-T-cell count was 28 ± 15 cells/μl.
Quantification of minority viral populations in plasma samples.
All plasma samples with a detectable viral load obtained during the interval between the initiation of treatment with protease inhibitors and the detection of L90M resistance mutation on standard genotyping were evaluated. The techniques used to quantify viral populations expressing the V82A and L90M mutations have previously been described (21). Briefly, 1 ml of plasma was centrifuged (23,500 × g, 1 h, 4°C), and RNA was extracted from the pellet (QIAamp Viral RNA Mini Kit; Qiagen, Valencia, Calif.). Reverse transcription-PCR (RT-PCR) was performed with oligonucleotides ProtF1 and ProtR1 (21). The reaction products were diluted, and real-time PCR was performed in parallel reactions with primers and probes permitting either the nonselective amplification of all viral sequences or the preferential amplification of sequences containing the V82A or L90M resistance mutations. The percentage of mutated sequences was then calculated as follows: percent mutated sequences = (the quantity of mutated sequences in the sample/the quantity of total sequences in the sample) × 100. As previously described, this approach could detect minority populations representing 0.05% total sequences (L90M) and 1% of total sequences (V82A), respectively (21).
Evaluation of genotypes expressed by dominant and minority populations.
For three patients, the genotype of individual molecular clones derived from plasma HIV-1 RNA that did or did not express the L90M resistance mutation was determined at several time points in the evolution of resistance to protease inhibitors. To do this, a previously described approach was used that minimizes in vitro recombination (21). Briefly, 10-fold dilutions (10−2 to 10−6) of the original RT-PCR product were prepared, and 10 μl of each dilution was amplified with the primers F4 and R1 (21) by using the following reaction conditions: 1× buffer, 3 mM MgCl2, 800 nM concentrations of each deoxynucleoside triphosphate, 200 nM concentrations of each primer, and 2 U of AmpliTaq Gold Taq polymerase. The cycling parameters were 95°C for 10 min, followed by 15 cycles at 95°C for 30 s, 50°C for 30 s, and 72°C for 3 min each time, with a final step at 72°C for 10 min. After electrophoresis on agarose gels, ethidium bromide-stained products were examined, and the most-dilute initial sample that contained a visible band was used for cloning. The PCR products were cloned into pCR4-TOPO (Invitrogen, Carlsbad, Calif.). To identify clones containing the L90M mutation, individual colonies were transferred to 200 μl of Luria-Bertani medium, and after overnight incubation at 37°C, cultures were resuspended and diluted 1:100 with water, and 10-μl aliquots were evaluated by real-time PCR as described above. Clones with or without the L90M mutation were easily distinguished by comparing the number of cycles required to reach threshold fluorescence (Ct) obtained for reactions performed by using the nonselective and sequence-selective PCR conditions (mutant clones, ΔCt < 2 cycles; wild-type clones, ΔCt > 10 cycles). Plasmid DNA from representative clones expressing or not the L90M mutation was purified (Qiaprep Spin Miniprep kit [Qiagen]), and the inserts were sequenced. Sequencing confirmed the results of the screening by real-time PCR in all cases. The strong correlation between the percentage of sequences containing the L90M mutation detected in the initial quantification of plasma viruses and the percentage of clones containing this mutation obtained from the same sample (r2 = 0.98 by linear regression, P < 0.0001, n = 17) indicated that the clones were representative of the initial population of plasma viruses.
Sequence analysis of the protease gene.
Nucleotide sequences were aligned by using the CLUSTAL W program (53). Phylogenetic analysis was performed by using the DNAPARS (DNA parsimony) program in the PHYLogeny Inference Package (PHYLIP, version 3.5). Trees were rooted by using the genotype obtained prior to the initiation of treatment with protease inhibitors and were drawn by using TreeView (version 1.6.1) (40). Phylogenetic analysis indicated that some resistance mutations occurred on two or more occasions. To test the significance of this conclusion, phylogenetic distances were estimated by the neighbor-joining method by using the PAUP* 4.0 (phylogenetic analysis using parsimony and other methods) program (51). Independent analyses were performed without weighting and after weighting, and the resistance mutation was considered such that all genotypes with the mutation were constrained to be a monophyletic group. The constrained tree and the best estimated tree without constraints were compared by using the Kishino-Hasegawa test (28). If the null hypothesis was rejected (P < 0.05), a double occurrence of the mutation was considered more likely.
Construction of recombinant viruses.
DNA purified from pCR4-TOPO plasmids containing the protease sequence of interest was amplified by using the following oligonucleotides: Fx48 (5′-GTCTAGACAAGGAACTGTATCCTTTAGCTTCCCTCAGATCACTCTTTG) and R1c (5′-GGTACAGTATCGATAGGACTAATGGG). The resulting fragment was digested with XbaI and ClaI restriction endonucleases and cloned into pNL-4.3XCS (32) which had been previously digested with the same restriction enzymes. After transfection of Escherichia coli, colonies were expanded, and plasmid DNA was purified (Nucleobond AX; Machery-Nagel, Düren, Germany). All constructs were verified by sequencing. For each protease sequence evaluated here, multiple clones had been sequenced (8 to 16 independent clones derived from the patient's plasma). In each case, the recombinant viruses expressed the strictly consensus sequence.
Evaluation of viral resistance and replicative capacity.
The test used to measure viral resistance and replicative capacity has previously been described in detail (32, 56). Briefly, HeLa cells were cultivated in 25-cm3 flasks in Dulbecco Modified Eagle medium containing 10% fetal calf serum, 100 U of penicillin G/ml, and 100 μg of streptomycin/ml until 90% confluent and then transfected with 3 μg of HIV proviral plasmid DNA by using Lipofectamine (Qiagen). At 18 h after transfection, HeLa cells were treated with trypsin and resuspended in 20 ml of complete medium, and 150-μl aliquots were seeded into 96-well culture plates. Ten serial threefold dilutions of protease inhibitors (saquinavir or nelfinavir) were prepared in complete medium, and 50-μl aliquots of each dilution were added to quadruplicate wells (final concentrations of 0.017 to 3,000 nM). Additional wells received medium alone. After 24 h, the amount of HIV-1 p24 in the supernatant of cells cultured with medium alone was determined by enzyme-linked immunosorbent assay (NEN Life Science, Boston, Mass.). Based on this value, the amount of culture medium containing 2 ng of p24 antigen was determined, and this volume of medium from each well was used to infect P4 (HeLa-CD4 LTR-LacZ) cells (3), and the infectivity of the virions produced by HeLa cells was determined by using a previously described single-cycle colorimetric assay based on the cleavage of the substrate (chlorophenol red-β-d-galactopyranoside) by β-galactosidase expressed by P4 cells after induction by Tat. We have previously found in unpublished experiments that values obtained for replicative capacity were equivalent when the assessment of virus production was based on total reverse transcriptase activity, total RNA (as determined with branched DNA) and p24 antigen.
The quadruplicate optical density readings for each drug concentration were fitted to a sigmoidal dose-response curve with variable slope after the lower asymptote was fixed to the background of the assay. The 50% inhibitory concentration (IC50) and the IC90 were determined from the dose-response curve. The replicative capacity in the absence of drug was determined directly from the optical density readings obtained for P4 cells infected with virions produced by HeLa cells in the absence of drug. These values were not significantly different from the upper asymptote of the dose-response curve (data not shown). Wild-type virus (pNL-4.3XCS) and all mutants from the same patient were evaluated in parallel in all experiments. Each panel of viruses was studied in four to six independent experiments.
Statistical analysis.
Results are expressed as the mean ± the standard deviation unless otherwise indicated. Comparisons between groups were performed by analysis of variance. Post-test comparisons (only performed if P < 0.05) were made by using the Bonferroni test. Comparisons between the IC50s, IC90s, and replicative capacities of the viral mutants at successive steps in viral evolution were performed by using Student's paired t test. A P value of <0.05 was considered significant.
RESULTS
Kinetics of appearance of the V82A and L90M resistance mutations.
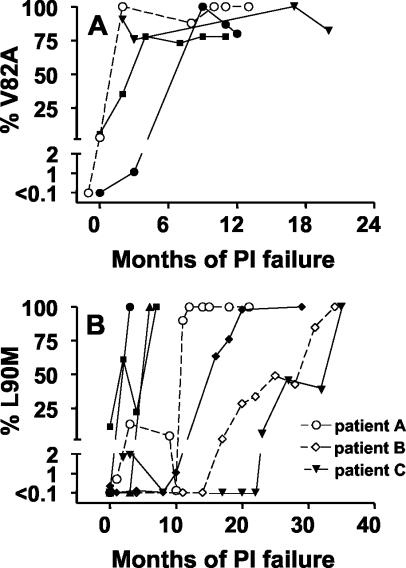
To characterize the kinetics of appearance of viruses expressing the protease resistance mutations V82A and L90M, we identified seven patients who had begun treatment with a protease inhibitor for the first time and for whom standard genotypic resistance testing performed after treatment failure ultimately revealed the presence of one or both of these mutations. All samples of plasma available for these patients since the beginning of treatment failure were then evaluated to determine the proportion of plasma viruses expressing V82A and/or L90M mutations. The V82A mutation, identified in four of the seven patients, was detectable within 3 months after treatment failure in all of these four patients and rapidly became fixed in the majority resistant viral population (Fig. 1A). The kinetics of appearance of the L90M mutation, however, ultimately identified in all seven patients, was more variable (Fig. 1B). In three individuals, the mutation was detected shortly after treatment failure and became fixed in the majority population of resistant viruses for the duration of the follow-up. Minority populations expressing L90M were also detected early in the development of resistance in three other patients, but these populations subsequently decreased in abundance or became undetectable. Thus, in the remaining four patients, the ultimate emergence of a majority species expressing L90M occurred later (8 to 34 months after the onset of treatment failure) and, in three patients, occurred in a progressive manner requiring several months. The protease inhibitor received at the onset of treatment was not predictive of the profile of appearance of L90M (data not shown).
FIG. 1.
Quantification of the proportion of plasma virions expressing the V82A and L90M resistance mutations. At the indicated times after virologic treatment failure with protease inhibitor (PI), the proportion of cDNA sequences expressing the V82A (A) and L90M (B) resistance mutations was quantified by using sequence-selective real-time PCR as described in Materials and Methods. Each symbol represents the results for a different patient. The V82A mutation was detected by genotyping for only four of the seven patients evaluated. The symbols corresponding to patients A to C, whose cases were explored in greater detail by the analysis of clones, are indicated in panel B.
Role of minority populations in the evolution of resistance.
It was apparent from these initial results that viral populations expressing the L90M mutation could be present for many months before this mutation was present in the majority of circulating viruses. To determine whether the early minority populations expressing L90M participated in the ultimate emergence of the majority viral species expressing this mutation, the evolution of resistance in three patients was evaluated in greater detail. Viral RNA was extracted from plasma samples obtained at various times during the evolution of resistance, the protease region was amplified by RT-PCR, and the products were cloned. For each time point, 50 to 150 clones were screened by sequence-selective real-time PCR to identify clones containing sequences with or without the L90M resistance mutation, and representative clones from the two populations were sequenced. This information is summarized in Tables 1 to 3. Phylogenetic trees are also shown (Fig. 2).
TABLE 1.
Evolution of resistance genotypes for patient Aa
| Sequence no. | Parameter | Result at various times after protease inhibitor failure
|
||||
|---|---|---|---|---|---|---|
| 0 mo | 3 mo | 9 mo | 10 mo | 11 mo | ||
| Genotype (% total population)b | ||||||
| Baseline 63P-77I | 100 | 8 | 2 | |||
| 1 | 93L | 100 | 5 | |||
| 2 | 82A | 38 | ||||
| 3 | 82A 90M | <1 | 13 | |||
| 4 | 36I 82A | 15 | ||||
| 5 | 82A 93L | 30 | 29 | |||
| 6 | 90M 93L | <1 | ||||
| 7 | 36I 82A 93L | 8 | ||||
| 8 | 54V 71V 82A 93L | 58d | 5 | |||
| 9 | 54V 71V 82A 90M 93L | 44d | ||||
| 10 | 36I 54V 71V 82A 90M 93L | 44d | ||||
| Total no. of clones sequenced (with L90M/without L90M) | 2/13 | 4/6 | 0/3 | 4/5 | ||
| % Total clones with L90M | 1.1 | 13 | 0 | 88 | ||
| Treatmentc | d4T, ddI, RTV | d4T, 3TC, IDV | d4T, IDV, DLV | d4T, IDV, DLV | ||
Predominant genotype values are shown in boldface.
The percent total population with the indicated genotype was calculated as the (percentage of total clones without L90M × the number of sequences with genotype)/the total clones without L90M sequenced (for genotypes without L90M) and as the (percentage of total clones with L90M × the number of sequences with genotype)/the total clones with L90M sequenced (for genotypes with L90M).
Abbreviations: d4T, stavudine; ddI, didanosine; RTV, ritonavir; 3TC, lamivudine; IDV, indinavir; DLV, delavirdine; AZT, zidovudine; NFV, nelfinavir; EFV, efavirenz; SQV, saquinavir.
Origin from the prior dominant viral species.
TABLE 3.
Evolution of resistance genotypes for patient Ca
| Sequence no. | Parameter | Result at various times after protease inhibitor failure
|
|||||
|---|---|---|---|---|---|---|---|
| 0 or 1 mo | 3 mo | 23 mo | 24 mo | 34 mo | 37 mo | ||
| Genotype (% total population)b | |||||||
| Baseline (20R-36I-63P) | 100 | ||||||
| 1 | 82A | 98 | |||||
| 2 | 82A 90M | 2 | |||||
| 3 | 54V 71V 82A | 13 | |||||
| 4 | 46L 54V 71V 82A | 75d | 13 | ||||
| 5 | 53L 54V 71V 82A | 13 | |||||
| 6 | 54V 71V 82A 90M | 1 | |||||
| 7 | 53L 54V 71V 82A 90M | 14 | |||||
| 8 | 54V 71V 82A 93L | 25 | 38e | 9 | 6 | ||
| 9 | 53L 54V 71V 82A 93L | 13 | |||||
| 10 | 54V 71V 82A 84V 93L | 64d | 6 | ||||
| 11 | 54V 71V 82A 90M 93L | 9 | 9 | 74e | |||
| 12 | 46L 54V 71V 82A 90M 93L | 4 | |||||
| 13 | 54V 71V 82A 84V 90M 93L | 14 | |||||
| Total no. of clones sequenced (with L90M/without L90M) | 0/14 | 1/3 | 0/4 | 8/7 | 7/8 | 6/6 | |
| % Total clones with L90M | 0 | 2 | 0 | 10 | 27 | 88 | |
| Treatmentc | d4T, 3TC, RTV | d4T, 3TC, NFV | EFV, ddI, SQV/RTV | EFV, ddI, SQV/RTV | EFV, d4T-ddI, NFV | ||
FIG.2.
Evolution of the protease gene in patients failing retroviral treatment. At the indicated times after treatment failure, RNA was extracted from plasma, and the viral protease gene was amplified by RT-PCR and cloned. Clones with or without the L90M mutation were identified and sequenced. On the top of each panel, the viral load and the percentage of total sequences expressing the L90M mutation are shown. Below, the phylogenetic trees obtained by DNAPARS are shown, along with the corresponding protease genotypes. The results for patients A, B, and C are shown in the corresponding panels.
For patient A, resistance mutations in the majority population accumulated in the following order: I93L → V82A → I54V + A71V → L90M. In each case, the resistance mutations were added sequentially to the genotype of the previously existing majority population. Several other features are noteworthy. First, minority viral populations with distinct genotypes were present at most times studied. Second, the L90M mutation was independently selected on more than one occasion. The initial minority populations with this mutation, present at 3 and 9 months after treatment failure, had only the L90M and I93L mutations or the V82A and L90M mutations. The majority population expressing the L90M mutation that emerged at 11 months had a distinct origin. In this case, the mutation was added to a preexisting genotype that contained the I54V, A71V, V82A, and I93L mutations. Indeed, the maximum-likelihood score of phylogenetic trees in which a single origin of L90M was imposed was significantly lower than that of trees with two or three independent origins of L90M (P < 0.001 [Kishino-Hasegawa test]). Similar arguments indicated that the M36I mutation had been selected on at least two occasions (data not shown). After the incorporation of delavirdine into the previously failing regimen, a transient 2.5-log10 fall in viral load occurred at month 10 (Fig. 2A). At this time point, only viruses expressing a phenotype very close to that present before the initiation of therapy were detected, suggesting that a subpopulation of virus not subjected to drug pressure had continued to replicate in this individual. Viral species expressing genotype I54V A71V V82A I93L reemerged quickly (month 11) and then went on to add L90M and subsequently M36I.
The development of resistance in patient B began with the addition of M46I and V82T mutations. Subsequently, two populations coexisted in this individual for at least 6 months: one that used the ACC codon for V82T (Table 2, genotypes 2 and 3) and a second, initially minority, population that expressed I93L and used the ACT codon for V82T (Table 2, genotypes 4 to 6). Starting at 39 months after failure, only viruses from this second lineage were detectable. The majority species was first represented by viruses that expressed L24I but not L90M. Four months later, viruses that expressed L90M but not L24I had become the dominant genotype. It is noteworthy that this later strain had been present as a minority species for 10 months before its emergence as the dominant strain at 43 months after failure. Only a single occurrence of the L90M mutation was detected in this patient, a finding consistent with the observation that minority species expressing this mutation were not detected during the first 19 months of treatment failure (Fig. 2B).
TABLE 2.
Evolution of resistance genotypes for patient Ba
| Sequence no. | Parameter | Result at various times after protease inhibitor failure
|
|||||
|---|---|---|---|---|---|---|---|
| 0 mo | 19 mo | 27 mo | 33 mo | 39 mo | 43 mo | ||
| Genotype (% total population)b | |||||||
| Baseline 63P-77I | 100 | ||||||
| 1 | 46I 82T | 100 | |||||
| 2 | 24I 46I 54V 71V 82T | 99d | 64 | ||||
| 3 | 46I 82T 90M 93L | 1 | |||||
| 4 | 46I 54V 71V 82T 90M 93L | 36 | 45 | 100e | |||
| 5 | 24I 46I 54V 71V 82T 93L | 55e | |||||
| Total no. of clones sequenced (with L90M/without L90M) | 1/5 | 4/6 | 3/12 | 9/0 | |||
| % Total clones with L90M | 1 | 36 | 45 | 100 | |||
| Treatmentc | AZT, 3TC, NFV | AZT, 3TC, NFV | AZT, 3TC, NFV | AZT, 3TC, NFV | AZT, 3TC, NFV | ||
The development of resistance in patient C began with the V82A mutation. A minority population expressing V82A and L90M was detected early but did not persist (Fig. 2C and Table 3). At 23 months after failure, I54V and A71V had been added, and two distinct lineages had developed. The predominant lineage at 23 months did not have I93L (Table 3, genotypes 4 to 8). Minority populations within this lineage were found a month later, at which point more clones were analyzed. These included variants expressing either the M46L, F53L, or L90M mutations. The second lineage, which had added I93L, was found to undergo considerable diversification at 34 months (Table 3, genotypes 9 to 13), whereas the patient was still receiving saquinavir. At that time point, the dominant viral population belonged to the I93L lineage and expressed I84V but not L90M. Three months later, after a switch to nelfinavir, a population with L90M but not I84V, which had been continuously present as a minority population for more than a year, became the dominant population. Thus, overall, L90M was selected on at least three occasions in this individual (P < 0.001 comparing the maximum-likelihood score for phylogenetic trees in which a single origin of L90M was imposed, with that of scores obtained for trees with three independent origins of L90M). Similar analysis suggested that both the M46L and F53L mutations had also been selected on two distinct occasions in the course of viral evolution in this patient (P < 0.001 [Kishino-Hasegawa test]).
Several features of the evolution of resistance were common to these patients. First, more than one viral population was detected at most times (3.3 ± 2.1 genotypes detected at each time point). Second, the minority populations contributed to the evolution of resistance. For the patients studied here, a preexisting dominant resistance genotype was replaced by a new dominant genotype on nine occasions. In five cases, the change in genotype was explained by the addition of new resistance mutations to the previous majority genotype. In four cases, however, the change resulted from the emergence of a genotype derived from a previously minority viral population. In two cases, these minority populations were demonstrated to have been present for 10 or more months prior to their emergence as the majority population.
Role of recombination in generating diversity.
A potential source for the diversification of genotypes is recombination, which is known to occur in vivo (36) and can also be produced as an artifact of PCRs (16, 27). Several findings indicated that artifactual recombination occurring in vitro did not account for the genetic diversity observed. First, control experiments, performed by using the techniques used here to generate clones, did not produce evidence for in vitro recombination (21). Second, in a number of cases, the identical genotype was observed in two or more independent samples obtained at several-month intervals. Third, several examples were observed where mutations were always mutually exclusive, including mutations present at the N- and C-terminal ends of the amplified fragment. For example, 28 clones from patient B had either the L24I or L90M mutation but never both mutations. Similarly, 31 clones from patient A, obtained at three different time points, had either the I54V mutation or a polymorphism at position 72, but no clone had both. These results would not be expected if recombination was occurring frequently during PCR.
We examined the possible role of in vivo recombination in circumstances in which the same mutation appeared to occur on two or more occasions in different contexts. In many cases, genetic analysis favored the de novo appearance of the same mutation in a new context over generation of the novel genotype by recombination. Recombination between sequences 5 or 8 and sequence 10 (patient A) was an unlikely explanation for the appearance of M36I in genotype 11 because sequences 10 and 11 had a silent polymorphism at codon 19 not found in sequences 5 or 8, the other genotypes expressing M36I. Similar arguments militated against recombination as an explanation for the appearance of L24I in genotype 6 from patient B, F53L in genotype 10 from patient C, and L90M in genotype 12 from patient C (data not shown). In other cases, no genetic evidence for or against recombination was present. In some of these cases, however, the time of appearance of the mutation did not favor recombination. For example, the appearance of L90M in genotype 7 from patient C could have occurred by recombination between genotypes 3 and 4, but genotype 7 was first detected at 24 months, whereas genotype 3 was only detected at 3 months.
Recombination was a likely explanation for only one genotype identified in the present study (genotype 4, patient B), since the recombination between genotypes 2 and 5, rather than the appearance of I54Vand A71V on two distinct occasions appears more probable. It is noteworthy, however, that genotype 5 was not detected in this patient until 6 months after genotype 4, although it may have been present as a low prevalence minority population at earlier times.
Role of resistance and replicative capacity in the selection of new majority populations.
For two patients, we evaluated the role of changes in drug resistance and viral replicative capacity in the selection of new majority populations. To do so, the protease corresponding to that present in the last three majority viral populations identified in these patients was cloned into pNL4-3, viruses were obtained by transfection of HeLa cells, and the drug resistances and replicative capacities of these recombinant viruses were compared between themselves and those of the wild-type virus. For patient B, who had received previous treatment with ritonavir and indinavir, the viral evolution occurred during a time when the only protease inhibitor received was nelfinavir. The majority population at month 27 (genotype L24I M46I I54V A71V V82T) had a large increase in resistance but reduced replicative capacity compared to that of wild-type virus (Fig. 3 and 4). The subsequent majority population (month 39) added the I93L mutation. This resulted in viruses displaying a further increase in nelfinavir resistance, especially at higher drug concentrations, but a further significant fall in replicative capacity in the absence of drug. At month 43, a viral population without the L24I mutation but that had added L90M, became the majority population. Nelfinavir resistance for recombinant viruses with this protease was not significantly different from that of its predecessor. This new majority species did, however, have a significant increase in replicative capacity compared to that of its predecessor. RT resistance genotypes covering the time span evaluated here were as follows: for month 33, M41M/L A62A/V D67N T69T/A K70R M184V T215F, and for month 43, M41L D67N K70R M184V T215F K219Q.
FIG. 3.
Replicative capacities of recombinant viruses containing protease sequences present at different times in the evolution of resistance. DNA fragments corresponding to the protease present in the predominant viral species at different times in the evolution of resistance for patient B (top panel) and patient C (bottom panel) were cloned into pNL-4.3XCS. Viruses produced after transfection of HeLa cells were recovered and used to infect in quadruplicate P4 cells (2 ng of p24/well) cultured in 96-well plates, and the induction of β-galactosidase was determined by a colorimetric assay 36 h later. The results for individual experiments, expressed as the percentage of wild-type virus from the same experiment, are shown by open symbols. Solid triangles represent the mean ± the standard error of the mean for all determinations. Viruses for patient B (top) had the following resistance genotypes: 1, L24I M46I I54V A71V V82T (dominant at months 27 and 33); 2, L24I M46I I54V A71V V82T I93L (month 39); and 3, M46I I54V A71V V82T L90M I93L (month 43). Virus genotypes for patient C (bottom) were as follows: 1, I54V A71V V82A I93L (month 24); 2, I54V A71V V82A I84V I93L (month 34); and 3, I54V A71V V82A L90M I93L (month 37). In two cases, polymorphisms were detected in the virions expressing the following resistance genotypes: patient B, 43 months, 55K/R, and patient C, 34 months, 37D/E. Only the most abundant population was studied here (55K and 37D, respectively). WT, wild type.
FIG. 4.
Resistance to protease inhibitors of recombinant viruses containing protease sequences present at different times in the evolution of resistance. Recombinant viruses were produced as described in the Fig. 3 legend and used to transfect HeLa cells. At 18 h after transfection, HeLa cells were treated with trypsin, and aliquots were seeded into 96-well plates containing serial threefold dilutions of saquinavir (SQV) or nelfinavir (NFV). After 24 h, aliquots of supernatant corresponding to 2 ng of p24 (based on the amount of p24 in the supernatant of HeLa cells cultured with medium alone) were used to infect in quadruplicate P4 cells cultured in 96-well plates, and the induction of β-galactosidase was determined by a colorimetric assay 36 h later. The results for each virus were fitted to a sigmoid curve with variable slope, and the IC50 and IC90 were determined. The results are presented as the mean ± the standard error of the mean of the fold increase in IC50 (A and B) or IC90 (C and D) compared to that of wild-type virus (patient B, n = 6 experiments; patient C, n = 5 experiments for saquinavir and n = 4 experiments for nelfinavir). The viruses from patient B (A and B) and patient C (C and D) are as described in the Fig. 3 legend. The mean IC50 and IC90 values ± the standard error of the mean for wild-type virus were, respectively, 10.9 ± 5.6 and 110.3 ± 70.1 ng/ml for nelfinavir and 1.7 ± 1.51 and 8.1 ± 3.9 ng/ml for saquinavir.
For patient C, viral evolution accompanied several changes in protease inhibitor treatment. The majority population at month 24 after treatment failure, a time when the patient had been receiving saquinavir for 1 month (genotype I54V A71V V82A I93L) had a large increase in resistance to that drug but a loss in replicative capacity compared to that of wild-type virus (Fig. 3 and 4). At month 34, the majority population had added the I84V mutation, which resulted in a further improvement in resistance to saquinavir at the expense of a significant fall in replicative capacity. At month 37, treatment had been changed from saquinavir to nelfinavir. This change was accompanied by the emergence of a new majority population, expressing the L90M, but not the I84V mutation. This population displayed a considerably greater resistance to nelfinavir than did its predecessor (Fig. 5) and also had a small improvement in replicative capacity in the absence of drug. Despite its better resistance to nelfinavir, the drug the patient was receiving, the resistance of this virus to saquinavir was lower than that of its predecessor (Fig. 5). For patient C, a minority viral population expressing the F53L mutation was also tested (Table 3, genotype 8). The replicative capacity for this virus in the absence of drug was very low, precluding the measurement of drug resistance (data not shown). At month 28, the RT genotype was M41L L74V V108V/I V118I Y181C G190S/A L210W T215Y. The genotype at month 39 was identical, except that the subpopulations of virions expressing V108I and G190A were no longer detectable.
FIG. 5.
Dose-response curves for the inhibition of viral replication of recombinant viruses from patient C by saquinavir (SQV) and nelfinavir (NFV). The effect of serial dilutions of saquinavir and nelfinavir on the replication of wild-type virus (WT [gray triangles]), virus 2 (○), and virus 3 (•) was evaluated as described in the Fig. 4 legend. The data for each virus from a single experiment were fitted to sigmoid curves with variable slopes. At each of the indicated drug concentrations, the replication, expressed as a percentage of that observed for wild-type virus in the absence of drug, was determined. The values from five (saquinavir) or four (nelfinavir) independent experiments were used to generate the sigmoid curves shown in the figure.
Overall, for the six evolutionary steps evaluated, an improvement in protease resistance was observed on five occasions. In four cases the higher drug resistance was associated with a moderate, although significant, loss in replicative capacity. On only one occasion was an improvement in viral replicative capacity observed without an improvement in resistance. In the one case examined, a change in treatment resulted in the rapid selection of a previously existing minority population with better resistance to the newly introduced protease inhibitor.
DISCUSSION
HIV-1 has the capacity to generate genetically diverse progeny at a high rate because of the highly mutagenic and recombinogenic nature of RT (23, 29, 36, 45, 48) and because of the rapid turnover of infected cells in vivo (8, 20, 25). The concurrent selection of the best-fit variants is thought to account for the rapid evolution of HIV-1 sequences in the face of pressure from the immune system and for the selection of drug-resistant variants in the course of incompletely suppressive antiretroviral therapy (8, 17). Our results, which reveal the importance of minority viral species appearing in the course of treatment failure, further underscore the ability of HIV-1 to generate and select best-fit variants. Like the dominant viral species present at any time in the evolution of resistance, these minority populations can continue to evolve, can subsequently emerge as dominant populations, and can thus serve as a reservoir of diversity that may accelerate the development of drug resistance.
Several findings suggest that the resistance mutations retained by the predominant plasma viral population represent only a fraction of all resistance mutations occurring in an infected individual. First, we identified several cases in which the same mutation occurred on more than one occasion in different contexts. Second, we found that a number of mutations that were absent from the majority virus population were expressed in minority species. The prevalence of such minority genotypes was probably underestimated by our study due to the limited number of clones sequenced. Indeed, we found a direct correlation between the number of clones sequenced in each sample and the number of distinct genotypes observed (P < 0.001; data not shown). Thus, the occurrence of a resistance mutation is not a rare event, and once established, these mutations are not necessarily tenaciously retained in the dominant genotype.
There could be several explanations for the coexistence and coevolution of virus populations expressing different genotypes in a single infected individual. In theory, the large diversity of HIV quasispecies that exists before antiretroviral treatment should make it possible for selection to be initiated in distinct founders. These distinct founder species could then follow related but slightly different evolutionary pathways, a possibility supported by our observation that, in the same patient, the same mutations could emerge independently in distinct backgrounds. Alternatively, minority species could have diverged from a single founder. Indeed, despite the generally high level of diversity of resistance genotypes observed in the present study, we did find clear evidence for evolutionary bottlenecks. In particular, for the three patients studied in detail, evolution was characterized by the appearance of I54V and A71V (in the context of V82A/T). Subsequent to the addition of this pair of mutations, considerable diversification rapidly occurred, but all of these new variants appeared to be derived from a single founder species.
Several mechanisms could also account for the remarkable persistence of some genotypes as minority populations during resistance evolution. Usually, the minority species found in our patients had as many or occasionally more resistance mutations than the current dominant genotype and persisted for many months as minority species. Since their replicative capacity in the presence of drug was probably close to that of the dominant species, it is most likely that these different populations were produced at sites where the selective pressure exerted by antiretroviral drugs was comparable. In some cases, however, the minority populations corresponded to strains with fewer resistance mutations and with lower resistance than the dominant species. Possibly, these populations represented vestiges of previously dominant populations, representing earlier stages of HIV-1 evolution toward resistance, and that were destined to disappear. Alternatively, these plasma virions could originate from tissue compartments in which the selective pressure by antiretroviral drugs is lower than in other compartments. The presence of compartments harboring viruses whose resistance genotype is distinct from that of plasma virus has been described (G. Tirado, G. R. Jove, and Y. Yamamura, Abstr. 11th Int. HIV Drug Resist. Workshop: Basic Principles Clin. Implications, abstr. 79, 2002; J. Ghosn, J. P. Viard, M. De Almeida, I. Aaron, C. Goujard, D. Salmon, C. Katlama, R. Tubiana, M. Leruez-Ville, C. Rouzioux, and M. L. Chaix, Abstr. 11th Int. HIV Drug Resist. Workshop: Basic Principles Clin. Implications, abstr. 54, 2002). As an extreme example of this phenomenon, we found evidence for the persistent low-level replication of essentially wild-type virus, presumably in a privileged compartment, in an individual (patient A) who had been on continuous treatment for 10 months and whose dominant viral population had evolved 4 resistance-associated mutations. Such persistent replication of wild-type virus in treated individuals has been described by others (26, 54; D. Irlbeck, M. J. Stanhope, C. Douady, H. M. Amrine, T. Melby, D. R. McClernon, and E. R. Lanier, Abstr. 11th Int. HIV Drug Resist. Workshop: Basic Principles Clin. Implications, abstr. 66, 2002).
An important finding in our study was the observation that minority species with genotypes distinct from the prevailing majority population played an important role in the subsequent evolution of resistance. We identified several occasions in which resistance evolved by the selection of an additional resistance mutation to the previously existing predominant resistance genotype. In almost as many cases, however, the appearance of a new dominant resistance genotype resulted from the emergence of a viral species derived from a previously minority viral population. Thus, the existence of multiple viral populations with distinct resistance genotypes appears to increase the avenues available for subsequent evolution of resistance and therefore is likely to accelerate the process. The recruitment of these populations could occur either by direct selection or after recombination. Although the recruitment of the same resistance mutation in different contexts was observed, the analysis of accompanying polymorphisms suggested that recombination, occurring either in vivo or in vitro, was an unlikely explanation for several of these events.
The heterogeneity of resistance genotypes has important implications for the treatment of patients experiencing drug resistance. Even if the resistance profile of the currently dominant genotype is known with precision and serves as the basis for the choice of a new therapeutic regimen, the possibility remains that minority species may exist against which the treatment will prove less effective. This problem is clearly demonstrated by the evolution of resistance seen in patient C after a change in treatment from saquinavir to nelfinavir. Prior to the change in treatment, the predominant viral population expressed the resistance genotype I54V A71V V82A I84V I93L, a genotype that resulted in significantly higher resistance to saquinavir than nelfinavir. However, a viral population expressing the resistance genotype I54V A71V V82A L90M I93L had also been present in this patient in modest proportions for at least 10 months, and this genotype produced significantly higher resistance to nelfinavir. After the change to nelfinavir, this preexisting population rapidly emerged as the dominant species, after only a transient and incomplete response to the new regimen.
Having identified the changes in viral populations occurring during the evolution of viral resistance, it was possible to directly compare the accompanying changes in drug resistance and replicative capacity. These analyses were conducted with recombinant viruses carrying only protease sequences from patient-derived plasma virus in an otherwise identical genetic background (46, 49). Thus, the effects of mutations in gag cleavage sites or reverse transcriptase mutations on viral fitness were not evaluated (5, 11, 18, 31, 49, 52, 57) and could have participated in driving viral evolution. However, for the two patients studied here, RT resistance genotypes showed little evolution (no new mutations for patient C and the addition of one thymidine analog mutation to four preexisting thymidine analog mutations for patient B). Thus, despite these caveats, our data suggest that protease resistance, as measured by changes in IC90 values, was a major driving force for the evolution of virus populations. Indeed, drug resistance improved in five of the six evolutionary steps studied and, in the remaining case resistance, already quite high, did not decrease. Our findings are less compatible with a model in which strong resistance develops first and is followed by the appearance of compensatory changes that improve defects in replicative capacity. In most of the population changes analyzed, the improvement in resistance was usually associated with a reduction in replicative capacity. The emergence of new majority populations with improved replicative capacity was observed on two occasions, as described by others (32, 38), but in neither case did this occur at the expense of a decrease in resistance. Interestingly, the replicative capacity of recombinant viruses representing majority species was never severely impaired (≥62% of wild-type levels). In contrast, a marked impairment was observed in the single recombinant virus studied that contained a genotype that was only observed in a minority population. Overall, during changes in majority populations, the losses in replicative capacity appeared to be easily outbalanced by even small gains in resistance. This was illustrated in patient C, in whom the predominant genotype observed while the patient was receiving saquinavir had significantly lower replicative capacity than the coexisting minority population (64% versus 75% that of wild-type virus), suggesting that the relatively small twofold increase in the IC90 of saquinavir was sufficient to drive selection, despite the negative effects of the new genotype on replicative capacity.
In summary, the present study underscores the considerable ability of HIV-1 to generate resistance mutations, resulting in the coexistence of numerous viral quasispecies with distinct resistance genotypes throughout the evolution of resistance. These minority populations can continue to evolve and ultimately emerge as dominant viral populations. Thus, they appeared to play an active role in the development of resistance in the patients studied here and may accelerate the process. Further studies are needed to confirm the role of minority populations in the evolution of resistance to other antiretroviral agents. The existence of minority populations may also create important obstacles for the accurate assessment of viral resistance, further complicating the choice of alternate regimens for patients experiencing treatment failure with antiretroviral drugs. The development of strategies for the evaluation and treatment of patients with drug resistance that adequately take into consideration this additional level of complexity might produce clinical benefit.
Acknowledgments
We gratefully acknowledge the help of Françoise Brun-Vezinet and Diane Descamps in performing these studies.
This study was supported in part by a grant from the Agence Nationale de Recherches sur le SIDA (ANRS) and a grant from the European Union (grant no. QLK2_CT_2001_02360).
REFERENCES
- 1.Archer, R. H., C. Dykes, P. Gerondelis, A. Lloyd, P. Fay, R. C. Reichman, R. A. Bambara, and L. M. Demeter. 2000. Mutants of human immunodeficiency virus type 1 (HIV-1) reverse transcriptase resistant to nonnucleoside reverse transcriptase inhibitors demonstrate altered rates of RNase H cleavage that correlate with HIV-1 replication fitness in cell culture. J. Virol. 74:8390-8401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Back, N. K., and B. Berkhout. 1997. Limiting deoxynucleoside triphosphate concentrations emphasize the processivity defect of lamivudine-resistant variants of human immunodeficiency virus type 1 reverse transcriptase. Antimicrob. Agents Chemother. 41:2484-2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barbosa, P., P. Charneau, N. Dumey, and F. Clavel. 1994. Kinetic analysis of HIV-1 early replicative steps in a coculture system. AIDS Res. Hum. Retrovir. 10:53-59. [DOI] [PubMed] [Google Scholar]
- 4.Barbour, J. D., T. Wrin, R. M. Grant, J. N. Martin, M. R. Segal, C. J. Petropoulos, and S. G. Deeks. 2002. Evolution of phenotypic drug susceptibility and viral replication capacity during long-term virologic failure of protease inhibitor therapy in human immunodeficiency virus-infected adults. J. Virol. 76:11104-11112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bleiber, G., M. Munoz, A. Ciuffi, P. Meylan, and A. Telenti. 2001. Individual contributions of mutant protease and reverse transcriptase to viral infectivity, replication, and protein maturation of antiretroviral drug-resistant human immunodeficiency virus type 1. J. Virol. 75:3291-3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borman, A. M., S. Paulous, and F. Clavel. 1996. Resistance of human immunodeficiency virus type 1 to protease inhibitors: selection of resistance mutations in the presence and absence of the drug. J. Gen. Virol. 77:419-426. [DOI] [PubMed] [Google Scholar]
- 7.Caliendo, A. M., A. Savara, D. An, K. DeVore, J. C. Kaplan, and R. T. D'Aquila. 1996. Effects of zidovudine-selected human immunodeficiency virus type 1 reverse transcriptase amino acid substitutions on processive DNA synthesis and viral replication. J. Virol. 70:2146-2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coffin, J. M. 1995. HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science 267:483-489. [DOI] [PubMed] [Google Scholar]
- 9.Condra, J. H., D. J. Holder, W. A. Schleif, O. M. Blahy, R. M. Danovich, L. J. Gabryelski, D. J. Graham, D. Laird, J. C. Quintero, A. Rhodes, H. L. Robbins, E. Roth, M. Shivaprakash, T. Yang, J. A. Chodakewitz, P. J. Deutsch, R. Y. Leavitt, F. E. Massari, J. W. Mellors, K. E. Squires, R. T. Steigbigel, H. Teppler, and E. A. Emini. 1996. Genetic correlates of in vivo viral resistance to indinavir, a human immunodeficiency virus type 1 protease inhibitor. J. Virol. 70:8270-8276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Condra, J. H., W. A. Schleif, O. M. Blahy, L. J. Gabryelski, D. J. Graham, J. C. Quintero, A. Rhodes, H. L. Robbins, E. Roth, M. Shivaprakash, et al. 1995. In vivo emergence of HIV-1 variants resistant to multiple protease inhibitors. Nature 374:569-571. [DOI] [PubMed] [Google Scholar]
- 11.Cote, H. C., Z. L. Brumme, and P. R. Harrigan. 2001. Human immunodeficiency virus type 1 protease cleavage site mutations associated with protease inhibitor cross-resistance selected by indinavir, ritonavir, and/or saquinavir. J. Virol. 75:589-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Croteau, G., L. Doyon, D. Thibeault, G. McKercher, L. Pilote, and D. Lamarre. 1997. Impaired fitness of human immunodeficiency virus type 1 variants with high-level resistance to protease inhibitors. J. Virol. 71:1089-1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dauber, D. S., R. Ziermann, N. Parkin, D. J. Maly, S. Mahrus, J. L. Harris, J. A. Ellman, C. Petropoulos, and C. S. Craik. 2002. Altered substrate specificity of drug-resistant human immunodeficiency virus type 1 protease. J. Virol. 76:1359-1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dybul, M., M. Daucher, M. A. Jensen, C. W. Hallahan, T. W. Chun, M. Belson, B. Hidalgo, D. C. Nickle, C. Yoder, J. A. Metcalf, R. T. Davey, L. Ehler, D. Kress-Rock, E. Nies-Kraske, S. Liu, J. I. Mullins, and A. S. Fauci. 2003. Genetic characterization of rebounding human immunodeficiency virus type 1 in plasma during multiple interruptions of highly active antiretroviral therapy. J. Virol. 77:3229-3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eastman, P. S., J. Mittler, R. Kelso, C. Gee, E. Boyer, J. Kolberg, M. Urdea, J. M. Leonard, D. W. Norbeck, H. Mo, and M. Markowitz. 1998. Genotypic changes in human immunodeficiency virus type 1 associated with loss of suppression of plasma viral RNA levels in subjects treated with ritonavir (Norvir) monotherapy. J. Virol. 72:5154-5164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fang, G., G. Zhu, H. Burger, J. S. Keithly, and B. Weiser. 1998. Minimizing DNA recombination during long RT-PCR. J. Virol. Methods 76:139-148. [DOI] [PubMed] [Google Scholar]
- 17.Gaschen, B., J. Taylor, K. Yusim, B. Foley, F. Gao, D. Lang, V. Novitsky, B. Haynes, B. H. Hahn, T. Bhattacharya, and B. Korber. 2002. Diversity considerations in HIV-1 vaccine selection. Science 296:2354-2360. [DOI] [PubMed] [Google Scholar]
- 18.Gatanaga, H., Y. Suzuki, H. Tsang, K. Yoshimura, M. F. Kavlick, K. Nagashima, R. J. Gorelick, S. Mardy, C. Tang, M. F. Summers, and H. Mitsuya. 2002. Amino acid substitutions in Gag protein at non-cleavage sites are indispensable for the development of a high multitude of HIV-1 resistance against protease inhibitors. J. Biol. Chem. 277:5952-5961. [DOI] [PubMed] [Google Scholar]
- 19.Goudsmit, J., A. de Ronde, E. de Rooij, and R. de Boer. 1997. Broad spectrum of in vivo fitness of human immunodeficiency virus type 1 subpopulations differing at reverse transcriptase codons 41 and 215. J. Virol. 71:4479-4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haase, A. T. 1999. Population biology of HIV-1 infection: viral and CD4+ T-cell demographics and dynamics in lymphatic tissues. Annu. Rev. Immunol. 17:625-656. [DOI] [PubMed] [Google Scholar]
- 21.Hance, A. J., V. Lemiale, J. Izopet, D. Lecossier, V. Joly, P. Massip, F. Mammano, D. Descamps, F. Brun-Vezinet, and F. Clavel. 2001. Changes in human immunodeficiency virus type 1 populations after treatment interruption in patients failing antiretroviral therapy. J. Virol. 75:6410-6417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harrigan, P. R., S. Bloor, and B. A. Larder. 1998. Relative replicative fitness of zidovudine-resistant human immunodeficiency virus type 1 isolates in vitro. J. Virol. 72:3773-3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harris, R. S., K. N. Bishop, A. M. Sheehy, H. M. Craig, S. K. Petersen-Mahrt, I. N. Watt, M. S. Neuberger, and M. H. Malim. 2003. DNA deamination mediates innate immunity to retroviral infection. Cell 113:803-809. [DOI] [PubMed] [Google Scholar]
- 24.Hirsch, M. S., F. Brun-Vezinet, B. Clotet, B. Conway, D. R. Kuritzkes, R. T. D'Aquila, L. M. Demeter, S. M. Hammer, V. A. Johnson, C. Loveday, J. W. Mellors, D. M. Jacobsen, and D. D. Richman. 2003. Antiretroviral drug resistance testing in adults infected with human immunodeficiency virus type 1: 2003 recommendations of an International AIDS Society-USA Panel. Clin. Infect. Dis. 37:113-128. [DOI] [PubMed] [Google Scholar]
- 25.Ho, D. D., A. U. Neumann, A. S. Perelson, W. Chen, J. M. Leonard, and M. Markowitz. 1995. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature 373:123-126. [DOI] [PubMed] [Google Scholar]
- 26.Imamichi, H., K. A. Crandall, V. Natarajan, M. K. Jiang, R. L. Dewar, S. Berg, A. Gaddam, M. Bosche, J. A. Metcalf, R. T. Davey, Jr., and H. C. Lane. 2001. Human immunodeficiency virus type 1 quasi species that rebound after discontinuation of highly active antiretroviral therapy are similar to the viral quasi species present before initiation of therapy. J. Infect. Dis. 183:36-50. [DOI] [PubMed] [Google Scholar]
- 27.Judo, M. S., A. B. Wedel, and C. Wilson. 1998. Stimulation and suppression of PCR-mediated recombination. Nucleic Acids Res. 26:1819-1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kishino, H., and M. Hasegawa. 1989. Evaluation of the maximum likelihood estimate of the evolutionary tree topologies from DNA sequence data, and the branching order in hominoidea. J. Mol. Evol. 29:170-179. [DOI] [PubMed] [Google Scholar]
- 29.Lecossier, D., F. Bouchonnet, F. Clavel, and A. J. Hance. 2003. Hypermutation of HIV-1 DNA in the absence of the Vif protein. Science 300:1112.. [DOI] [PubMed] [Google Scholar]
- 30.Mahalingam, B., P. Boross, Y. F. Wang, J. M. Louis, C. C. Fischer, J. Tozser, R. W. Harrison, and I. T. Weber. 2002. Combining mutations in HIV-1 protease to understand mechanisms of resistance. Proteins 48:107-116. [DOI] [PubMed] [Google Scholar]
- 31.Mammano, F., C. Petit, and F. Clavel. 1998. Resistance-associated loss of viral fitness in human immunodeficiency virus type 1: phenotypic analysis of protease and gag coevolution in protease inhibitor-treated patients. J. Virol. 72:7632-7637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mammano, F., V. Trouplin, V. Zennou, and F. Clavel. 2000. Retracing the evolutionary pathways of human immunodeficiency virus type 1 resistance to protease inhibitors: virus fitness in the absence and in the presence of drug. J. Virol. 74:8524-8531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martinez-Picado, J., A. V. Savara, L. Sutton, and R. T. D'Aquila. 1999. Replicative fitness of protease inhibitor-resistant mutants of human immunodeficiency virus type 1. J. Virol. 73:3744-3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Metzner, K. J., S. Bonhoeffer, M. Fischer, R. Karanicolas, K. Allers, B. Joos, R. Weber, B. Hirschel, G. K. L., and H. F. Gunthard. 2003. Emergence of minor populations of human immunodeficiency virus type 1 carrying the M184V and L90M mutations in subjects undergoing structured treatment interruptions. J. Infect. Dis. 188:1433-1443. [DOI] [PubMed] [Google Scholar]
- 35.Molla, A., M. Korneyeva, Q. Gao, S. Vasavanonda, P. J. Schipper, H. M. Mo, M. Markowitz, T. Chernyavskiy, P. Niu, N. Lyons, A. Hsu, G. R. Granneman, D. D. Ho, C. A. Boucher, J. M. Leonard, D. W. Norbeck, and D. J. Kempf. 1996. Ordered accumulation of mutations in HIV protease confers resistance to ritonavir. Nat. Med. 2:760-766. [DOI] [PubMed] [Google Scholar]
- 36.Negroni, M., and H. Buc. 2001. Mechanisms of retroviral recombination. Annu. Rev. Genet. 35:275-302. [DOI] [PubMed] [Google Scholar]
- 37.Nijhuis, M., S. Deeks, and C. Boucher. 2001. Implications of antiretroviral resistance on viral fitness. Curr. Opin. Infect. Dis. 14:23-28. [DOI] [PubMed] [Google Scholar]
- 38.Nijhuis, M., R. Schuurman, D. de Jong, J. Erickson, E. Gustchina, J. Albert, P. Schipper, S. Gulnik, and C. A. Boucher. 1999. Increased fitness of drug resistant HIV-1 protease as a result of acquisition of compensatory mutations during suboptimal therapy. AIDS 13:2349-2359. [DOI] [PubMed] [Google Scholar]
- 39.Olsen, D. B., M. W. Stahlhut, C. A. Rutkowski, H. B. Schock, A. L. vanOlden, and L. C. Kuo. 1999. Non-active site changes elicit broad-based cross-resistance of the HIV-1 protease to inhibitors. J. Biol. Chem. 274:23699-23701. [DOI] [PubMed] [Google Scholar]
- 40.Page, R. D. 1996. TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 41.Palella, F. J., Jr., K. M. Delaney, A. C. Moorman, M. O. Loveless, J. Fuhrer, G. A. Satten, D. J. Aschman, S. D. Holmberg, et al. 1998. Declining morbidity and mortality among patients with advanced human immunodeficiency virus infection. N. Engl. J. Med. 338:853-860. [DOI] [PubMed] [Google Scholar]
- 42.Parikh, U., C. Calef, B. Larder, R. Schinazi, and J. W. Mellors. 2001. Mutations in retroviral genes associated with drug resistance, p. 191-277. In Carla Kuiken, Brian Foley, Beatrice Hahn, Preston Marx, Francine McCutchan, John Nellors, Steven Wolinsky, and Bette Korber (ed.), HIV sequence compendium 2001. NIAID, Los Alamos, N.Mex.
- 43.Peters, S., M. Munoz, S. Yerly, V. Sanchez-Merino, C. Lopez-Galindez, L. Perrin, B. Larder, D. Cmarko, S. Fakan, P. Meylan, and A. Telenti. 2001. Resistance to nucleoside analog reverse transcriptase inhibitors mediated by human immunodeficiency virus type 1 p6 protein. J. Virol. 75:9644-9653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Prabu-Jeyabalan, M., E. A. Nalivaika, N. M. King, and C. A. Schiffer. 2003. Viability of a drug-resistant human immunodeficiency virus type 1 protease variant: structural insights for better antiviral therapy. J. Virol. 77:1306-1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Preston, B. D., B. J. Poiesz, and L. A. Loeb. 1988. Fidelity of HIV-1 reverse transcriptase. Science 242:1168-1171. [DOI] [PubMed] [Google Scholar]
- 46.Resch, W., N. Parkin, E. L. Stuelke, T. Watkins, and R. Swanstrom. 2001. A multiple-site-specific heteroduplex tracking assay as a tool for the study of viral population dynamics. Proc. Natl. Acad. Sci. USA 98:176-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ridky, T. W., A. Kikonyogo, J. Leis, S. Gulnik, T. Copeland, J. Erickson, A. Wlodawer, I. Kurinov, R. W. Harrison, and I. T. Weber. 1998. Drug-resistant HIV-1 proteases identify enzyme residues important for substrate selection and catalytic rate. Biochemistry 37:13835-13845. [DOI] [PubMed] [Google Scholar]
- 48.Roberts, J. D., K. Bebenek, and T. A. Kunkel. 1988. The accuracy of reverse transcriptase from HIV-1. Science 242:1171-1173. [DOI] [PubMed] [Google Scholar]
- 49.Robinson, L. H., R. E. Myers, B. W. Snowden, M. Tisdale, and E. D. Blair. 2000. HIV type 1 protease cleavage site mutations and viral fitness: implications for drug susceptibility phenotyping assays. AIDS Res. Hum. Retrovir. 16:1149-1156. [DOI] [PubMed] [Google Scholar]
- 50.Ross, L., M. Johnson, R. DeMasi, Q. Liao, N. Graham, M. Shaefer, and M. St. Clair. 2000. Viral genetic heterogeneity in HIV-1-infected individuals is associated with increasing use of HAART and higher viremia. AIDS 14:813-819. [DOI] [PubMed] [Google Scholar]
- 51.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 52.Simon, V., N. Padte, D. Murray, J. Vanderhoeven, T. Wrin, N. Parkin, M. Di Mascio, and M. Markowitz. 2003. Infectivity and replication capacity of drug-resistant human immunodeficiency virus type 1 variants isolated during primary infection. J. Virol. 77:7736-7745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Van Rompay, K. K., T. B. Matthews, J. Higgins, D. R. Canfield, R. P. Tarara, M. A. Wainberg, R. F. Schinazi, N. C. Pedersen, and T. W. North. 2002. Virulence and reduced fitness of simian immunodeficiency virus with the M184V mutation in reverse transcriptase. J. Virol. 76:6083-6092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Watkins, T., W. Resch, D. Irlbeck, and R. Swanstrom. 2003. Selection of high-level resistance to human immunodeficiency virus type 1 protease inhibitors. Antimicrob. Agents Chemother. 47:759-769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zennou, V., F. Mammano, S. Paulous, D. Mathez, and F. Clavel. 1998. Loss of viral fitness associated with multiple Gag and Gag-Pol processing defects in human immunodeficiency virus type 1 variants selected for resistance to protease inhibitors in vivo. J. Virol. 72:3300-3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang, Y. M., H. Imamichi, T. Imamichi, H. C. Lane, J. Falloon, M. B. Vasudevachari, and N. P. Salzman. 1997. Drug resistance during indinavir therapy is caused by mutations in the protease gene and in its Gag substrate cleavage sites. J. Virol. 71:6662-6670. [DOI] [PMC free article] [PubMed] [Google Scholar]