Abstract
Genome-wide association studies are identifying novel Alzheimer's disease (AD) risk factors. Elucidating the mechanism underlying these polymorphisms is critical to the validation process and, by identifying rate-limiting steps in AD risk, may yield novel therapeutic targets. Here, we elucidate the mechanism of action of the AD-associated polymorphism rs3865444 in the promoter of CD33, a member of the sialic acid-binding Ig-superfamily of lectins (SIGLECs). Immunostaining established that CD33 is expressed in microglia in human brain. Consistent with this finding, CD33 mRNA expression correlated well with expression of the microglial genes CD11b and AIF-1 and was modestly increased with AD status and the rs3865444C AD-risk allele. Analysis of CD33 isoforms identified a common isoform lacking exon 2 (D2-CD33). The proportion of CD33 expressed as D2-CD33 correlated robustly with rs3865444 genotype. Because rs3865444 is in the CD33 promoter region, we sought the functional polymorphism by sequencing CD33 from the promoter through exon 4. We identified a single polymorphism that is coinherited with rs3865444, i.e., rs12459419 in exon 2. Minigene RNA splicing studies in BV2 microglial cells established that rs12459419 is a functional single nucleotide polymorphism (SNP) that modulates exon 2 splicing efficiency. Thus, our primary findings are that CD33 is a microglial mRNA and that rs3865444 is a proxy SNP for rs12459419 that modulates CD33 exon 2 splicing. Exon 2 encodes the CD33 IgV domain that typically mediates sialic acid binding in SIGLEC family members. In summary, these results suggest a novel model wherein SNP-modulated RNA splicing modulates CD33 function and, thereby, AD risk.
Introduction
Recent genome-wide association studies (GWASs) have identified a set of single nucleotide polymorphisms (SNPs) that are associated with Alzheimer's disease (AD) risk (Harold et al., 2009; Lambert et al., 2009; Jun et al., 2010; Hollingworth et al., 2011; Naj et al., 2011). Elucidating the mechanism of action of these SNPs may identify novel AD pathways. Indeed, genetic variation that modulates disease risk could be considered to biologically define rate-limiting steps in AD pathways and, as such, lead to robust new pharmacologic targets. Hence, an SNP with modest biological actions may reduce AD risk modestly, whereas a drug that acts strongly at the same target may have a large effect on AD risk reduction.
One of the AD-associated SNPs, rs3865444(Hollingworth et al., 2011; Morgan, 2011; Naj et al., 2011), is in the proximal promoter of CD33. CD33 is a member of the sialic acid-binding Ig-like lectin (SIGLEC) family of receptors, and the gene comprises seven coding exons. Exon 2 encodes the canonical IgV domain, exon 4 encodes the IgC structural domain, and exons 6 and 7 encode cytosolic immunotyrosine inhibitory motifs (ITIMs) (Varki and Angata, 2006; Perez-Oliva et al., 2011). The IgV domain likely mediates sialic acid binding because this domain has robust homology with the IgV domain of other SIGLECs, including Arg-122 that is critical for sialic acid binding (Varki and Angata, 2006). The binding of sialic acid activates CD33, leading to monocyte inhibition through the ITIM domains (Linnartz and Neumann, 2013). Here, we sought to identify the mechanism whereby rs3865444 modulates CD33 to alter AD risk. We report that CD33 is expressed in microglia. CD33 expression is modestly increased in AD and decreased with the minor, AD-protective rs3865444A allele. This allele is also associated with a robust increase in the proportion of CD33 lacking exon 2 (D2-CD33). rs12459419 likely mediates this association between rs3865444 and splicing because rs12459419 is coinherited with rs3865444 and directly modulates exon 2 splicing efficiency.
Materials and Methods
De-identified human brain specimens were provided by the University of Kentucky AD Center Neuropathology Core. Samples were from 30 women (14 non-AD and 16 AD) and 25 men (13 non-AD and 12 AD). AD and non-AD designations were by consensus conference, with disease being defined based on dementia and AD neuropathology, i.e., neuritic plaques and neurofibrillary tangles (Nelson et al., 2009). RNA and DNA were prepared from these samples as described previously (Zou et al., 2008; Ling et al., 2012).
CD33 immunostaining.
Paraffin-embedded tissue sections were cut at 5 μm, followed by antigen retrieval in citrate buffer, pH 6.0, using a pressure cooker (3 min). Sections were immersed in 5% goat serum in Tris-buffered saline, followed by an overnight incubation in anti-CD33 (clone PWS44; 1:100 dilution; Leica). After thorough rinsing in Tris-buffered saline, sections were incubated in biotinylated secondary antibody for 1 h, rinsed, incubated in ABC reagent (Vector Laboratories) for 1 h, developed in Nova Red chromagen (Vector Laboratories), and counterstained with Mayer's hematoxylin. For immunofluorescence, sections were treated similarly. Sections were then labeled with CD33 antibody (1:100), IBA-1 (rabbit polyclonal, 1:1000; Wako Chemicals), and/or GFAP ( rabbit polyclonal, 1:10,000; Dako), rinsed, and detected with labeled secondary antibodies (anti-rabbit DyLight 488-labeled anti-rabbit, 1:500; Jackson Laboratories; or Texas Red labeled anti-mouse, 1:500; Jackson Laboratories). Autofluorescence eliminator reagent (Millipore Bioscience Research Reagents) was used to block autofluorescence.
Identification of CD33 splice variants in human brain.
Screening for CD33 splice variants was performed on a pool of four cDNA samples representing AD and non-AD individuals and varying rs3865444 genotypes. Nested PCR was used to amplify CD33 exons 1–7A and exons 1–7B in separate reactions. An initial 20 cycles of PCR (Platinum Taq; Invitrogen) was performed by using forward primer 5′-CTCAGACATGCCGCTGCT-3′ (Hernandez-Caselles et al., 2006) corresponding to exon 1 and reverse primers 5′-TTCAATGGCCATCATCTCCT-3′ and 5′-CATCCCATGAAAGTTGAGGG-3′, corresponding to exons 7A and 7B, respectively. The PCR product was then diluted 1:25 and subjected to 30 cycles of amplification using forward primer 5′-TACTGCTGCCCCTGCTGT-3′ from exon 1 and 5′-TGGCCATCATCTCCTGATCT-3′ and 5′-AATGCAGCTCCTCATCCATC-3′ corresponding to exons 7A and 7B, respectively. PCR consisted of an initial 3-min 94° incubation, followed by cycles of denaturation at 94° for 15 s, annealing at 60° for 15 s, and extension at 72° for 2 min (Veriti 96-Well Thermal Cycler; Invitrogen). PCR was conducted using 1 μm the indicated primers (1 μm) and ∼0.1 μg of cDNA template. After PCR, samples were incubated at 72° for 30 min and then cloned into pcDNA2.1 according to the instructions of the manufacturer (TA-Cloning Kit; Invitrogen). Twenty-five random clones were picked from each group amplification reaction (exons 1–7A, exons 1–7B) and sequenced by using M13 forward and reverse primers. In addition to common D2-CD33, one to two clones had both exons 7A and 7B, skipped other exons, or retained intron 1.
Quantitation of CD33 expression.
Quantitative PCR (qPCR) was used to quantify expression of total CD33 (forward, 5′-TGTTCCACAGAACCCAACAA-3′; reverse, 5′-GGCTGTAACACCAGCTCCTC-3′) primers corresponding to sequences within exons 4 and 5, respectively), as well as D2-CD33 (forward, 5′-CCCTGCTGTGGGCAGACTTG-3′; reverse, 5′- GCACCGAGGAGTGAGTAGTCC-3′) primers corresponding to sequences at the exon 1–3 junction and exon 3, respectively). The specificity of the 1 to 3 junctional primer was confirmed by testing on CD33 sequence fragments containing or lacking exon 2, as well as post-qPCR melting curve analysis and gel electrophoresis of PCR products. PCR was conducted using an initial 2 min incubation at 95°, followed by cycles of 10 s at 95°, 20 s at 60°, and 20 s at 72°. The 20 μl reactions contained 1 μm each primer, 1× PerfeCTa SYBR Green Super Mix (Quanta Biosciences), and 20 ng of cDNA. Experimental samples were amplified in parallel with serially diluted standards that were generated by PCR of cDNA using the indicated primers, followed by purification and quantitation by UV absorbance. Results from samples were compared relative to the standard curve to calculate copy number in each sample. Real-time assays were performed twice, and the average copy number was used for additional data analyses. To evaluate the correlation between CD33 and that of microglial mRNAs, we also quantified CD11b and AIF-1 expression. The copy number for each mRNA was then normalized to the geometric mean of reference genes RPL32 and EIF4H, previously quantified in this sample set (Zou et al., 2008; Ling et al., 2012). Because CD33 expression correlated with CD11b and AIF-1 (data not shown), we compared total CD33 expression to the geometric mean of CD11b and AIF-1 expression when analyzing CD33 expression relative to AD status and rs3865444 genotype, correcting for the microglial content of brain tissue samples from which the cDNA was originally prepared. Expression of D2-CD33 was analyzed relative to total CD33 expression. To screen for CD33 SNPs that may be tightly linked to rs3865444 and directly modulate exon 2 splicing, seven individuals homozygous for the major or minor allele of rs3865444 were sequenced from 400 bp 5′ to the transcription start site through exon 4. Nested PCR was conducted according to the directions of the manufacturer (Phusion High-Fidelity DNA Polymerase; New England Biolabs); 20 PCR cycles were conducted with forward primer 5′-CTGTGCCCGAGCTGTCTTAT-3′ and reverse primer 5′-AGGCTCCTTCCTACCTGAGC-3′. PCR products were then diluted 1:25 and used in a second round of PCR (25 cycles) using the forward primer 5′-GCTGCCACCTTCACTTTACC-3′ with reverse primer 5′-TTGTTGGGTTCTGTGGAACA-3′. The first round of PCR used ∼80 ng of genomic DNA in a 20 μl reaction with both PCRs conducted using 3% DMSO. This effort identified four SNPs in this region: (1) rs3865444, the AD-associated promoter SNP; (2) rs2459141 which is 142 bp upstream of the transcription start site; (3) rs12459419 at the fourth base of exon 2; and (4) rs2455069 at the 168th base of exon 2. Only rs12459419 was in perfect linkage with rs3865444 in these samples. Samples were subsequently genotyped for rs12459419 by using a TaqMan approach (Invitrogen); rs3865444 genotypes were determined by Nco1 restriction fragment-length polymorphism because a TaqMan assay is not available.
CD33 minigenes containing exon 1 through exon 4 and differential for rs12459419C/T were generated by PCR and cloned into pcDNA3.1 (Invitrogen). Sequencing confirmed that the inserts differed only at rs12459419C/T. BV2 microglial cells were maintained in DMEM/F-12 (Invitrogen) supplemented with a final concentration of 10% fetal bovine serum, 50 U/ml penicillin, and 50 μg/ml streptomycin. Cells were seeded in six-well plates (2 × 105 cells per well) and allowed to grow for 24 h before transfection with 1 μg of allele-specific CD33 minigene vector in 6 μl of Lipofectamine 2000 reagent (Invitrogen) and 94 μl of Opti-MEM, per the recommendations of the manufacturer. Eighteen hours after transfection, poly(A+) RNA was prepared and reverse transcribed by using random hexamers per the directions of the manufacturer (SuperScript III; Invitrogen). Three transfections were performed in duplicate for each allele. CD33 and D2-CD33 expression from the minigene was quantified by qPCR using primers corresponding to exon 3–4 junction and 3′ vector-derived sequence (5′CAGCTCAACGTCACCTATGTTC and 5′CGTAGAATCGAGACCGAGGA) and 5′ vector and the exon 1–3 junction (5′TGCTTACTGGCTTATCGAAATTA and 5′TGTGGGTCAAGTCTGCCC), respectively. A one-tailed t test was used to analyze the results.
Results
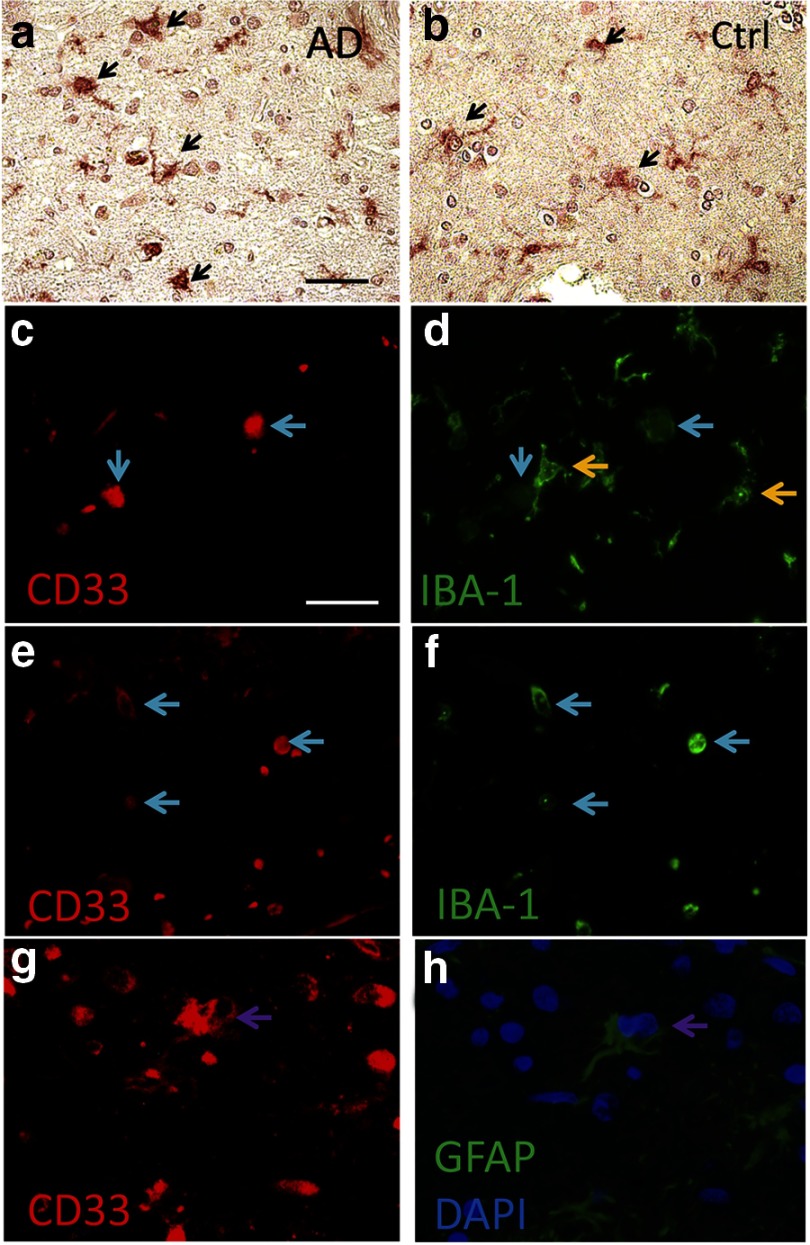
To elucidate the actions of rs3865444 and CD33 in AD, we began by performing immunocytochemistry to localize CD33 expression in human brain. CD33 expression was observed in microglia as discerned by morphology (Fig. 1A,B). Microglial localization was confirmed by double labeling brain sections for CD33 and IBA-1, a microglial protein, or GFAP, an astrocytic protein (Fig. 1C–F). Overall, the predominant microglial localization is consistent with the hypothesized role of CD33 as a sialic acid receptor that inhibits monocyte-lineage cell activation (Lajaunias et al., 2005).
Figure 1.
CD33 immunohistochemistry in human brain. CD33-immunopositive cell profiles (arrows) show morphology consistent with microglia in both AD and non-AD samples (a, b). Immunofluorescence was used to help distinguish the CD33-immunopositive cell types. c/d, e/f, and g/h show the same microscope fields. Sections were immunostained for CD33 and counterstained for IBA-1 (a microglial/macrophage lineage marker) or GFAP (an astrocyte lineage marker), as indicated. Microglia with rounded morphologies tended to colocalize with both CD33 and IBA-1 (blue arrows in a–d), with more ramified IBA-1-immunopositive microglia staining less positively for CD33 (yellow arrows); these results suggest that CD33 expression is increased in ameboid microglia. In addition to apparent double labeling in microglial cells, there are areas in which GFAP-positive label colocalizes with CD33 immunopositivity, indicating either astrocytic expression of CD33 or the engulfment of CD33-positive cells by astrocytes (note the two DAPI-labeled nuclei present in the CD33-positive/GFAP-positive area indicated by arrows in g, h). Sections are from superior/middle temporal gyri of individuals with AD pathology (a, c–h) or without (b) AD pathology. Ctrl, Control. Scale bar: a, b, 50 μm; c–h, 20 μm.
CD33 is encoded by seven exons, including the alternatively spliced exons 7A and 7B (Brinkman-Van der Linden et al., 2003). To identify common CD33 isoforms in human brain, we performed PCR amplification on human brain cDNA using primers corresponding to exon 1 and exon 7A or 7B. Sequencing 50 random clones revealed frequent isoforms lacking the 381 bp exon 2 (D2-CD33). Because the codon reading frame is maintained in the absence of exon 2, D2-CD33 encodes a protein that is identical to CD33 but lacks the IgV domain that mediates sialic acid binding in SIGLEC family members (Perez-Oliva et al., 2011; for review, see Varki and Angata, 2006). Both D2-CD33 and CD33 are found on the cell surface of transfected cells (Perez-Oliva et al., 2011).
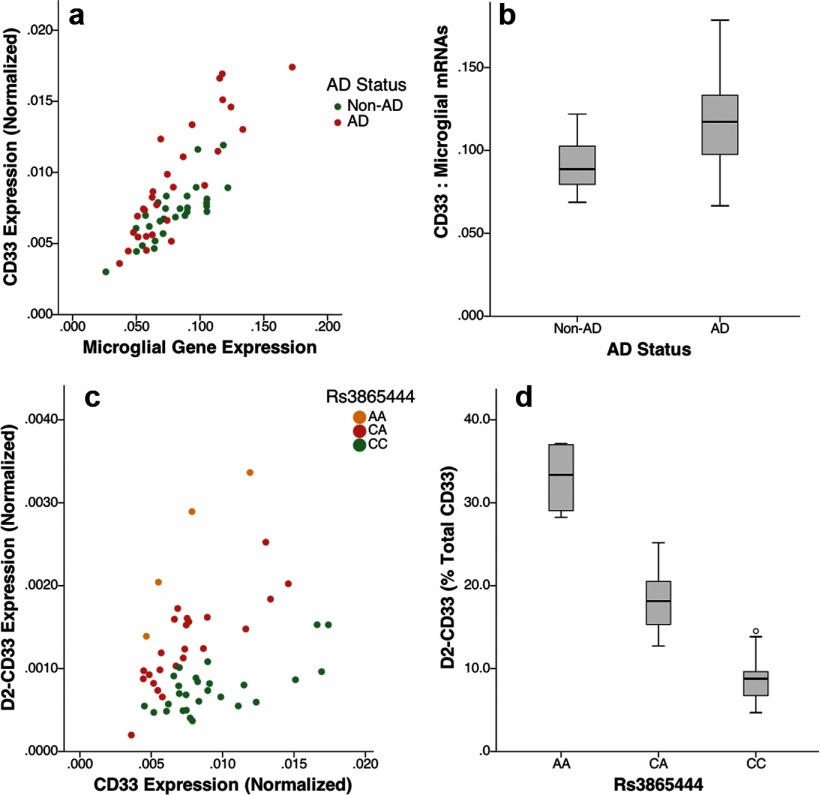
We initially hypothesized that rs3865444 modulates CD33 expression because this SNP is 372 bp upstream of the CD33 transcription start site. To evaluate this possibility, CD33 expression was assessed by performing qPCR with primers corresponding to sequence within exons 4 and 5. Because immunostaining suggested that CD33 localized primarily to microglia, we analyzed CD33 expression relative to the geometric mean of two microglial mRNAs, CD11b and AIF-1 (IBA-1), as well as AD status and rs3865444 genotype. CD33 expression correlated strongly with microglial mRNA expression, was increased in AD, and was modestly decreased with the AD-protective rs3865444A allele (Fig. 2A,B); regression analysis showed a highly significant model (adjusted r2 = 0.76) wherein CD33 expression correlated significantly with microglial gene expression (p = 3.3 × 10−16, standardized β coefficient of 0.78), AD status (p = 5.9 × 10−6, standardized β coefficient of 0.29), and rs3865444 genotype (p = 0.012, standardized β coefficient of −0.17). These results are consistent with CD33 expression in microglia, increased CD33 expression in AD, and, more modestly, decreased CD33 expression with the protective rs3865444A allele.
Figure 2.
CD33 isoform expression relative to AD status. CD33 expression correlated well with microglial gene expression (a, presented as geometric mean of CD11b and AIF-1, r2=0.64), as well as each microglial mRNA individually (data not shown). An association between CD33 and AD status was visualized by considering the ratio of CD33 to the geometric mean of the microglial reference genes (b). D2-CD33 correlated well with CD33 expression (c; r2 = 0.88, 0.67, and 0.51 for the AA, CA, and CC genotypes, respectively). The percentage of CD33 expressed as D2-CD33 was strongly associated with rs3865444 genotype (d, p = 1.2 × 10−13).
Because D2-CD33 was a common splice variant, we next quantified D2-CD33 by performing qPCR with primers corresponding to the exon 1–3 junction and exon 3. D2-CD33 expression was compared with total CD33 expression; isoform-specific standard curves were analyzed in parallel with samples, allowing absolute quantitation of each isoform. D2-CD33 expression corresponded well with CD33 expression and strikingly well with rs3865444 genotype (Fig. 2C). When D2-CD33 expression was considered as a percentage of total CD33 expression, the association with rs3865444 genotype was readily apparent (Fig. 2D); this impression was confirmed by linear regression analyses that found an overall highly significant model (adjusted r2 = 0.75) with D2-CD33 strongly associated with rs3865444 genotype (p = 1.01 × 10−13, standardized β coefficient of 0.81), as well as CD33 expression (p = 2.3 × 10−10, standardized β coefficient of 0.58), and modestly decreased with AD status (p = 0.013, standardized β coefficient of −0.19). Hence, the proportion of CD33 expressed as D2-CD33 showed a dose-dependent relationship with rs3865444 allele; the percentage of CD33 expressed as D2-CD33 increased by 10.7 ± 0.8% per copy of the AD-protective rs3865444A allele. Future work will include confirmation of these mRNA changes at the protein level.
Because rs3865444 resides in the CD33 promoter, this SNP is not likely to directly modulate exon 2 splicing. We hypothesized that rs3865444 is coinherited with an SNP near or within exon 2 that modulates exon 2 splicing efficiency. To investigate this hypothesis, we sequenced CD33 from 400 bp 5′ of the transcription start site through exon 4 in four rs3865444C/C and three rs3865444A/A individuals. We found four SNPs in these samples: (1) rs3865444; (2) rs2459141 (142 bp upstream of the transcription start site); (3) rs12459419 (the fourth base of exon 2); and (4) rs2455069 (the 168th base of exon 2). Among these variations, only rs12459419 was coinherited with rs3865444 in all seven individuals, i.e., rs3865444AA individuals were also rs12459419TT, whereas rs3865444CC individuals were rs12459419CC. Subsequent rs12459419 genotyping of the samples depicted in Figure 2 found that rs12459419 major and minor alleles were perfectly coinherited with rs3865444 major and minor alleles. Thus, the rs12459419C/T alleles substitute perfectly for the rs3865444C/A alleles in Figure 2.
To evaluate whether rs12459419 is a functional polymorphism and directly modulates exon 2 splicing efficiency, we generated CD33 minigenes for each rs12459419 allele. These minigenes included exon 1 through exon 4, along with intervening introns. The only difference in minigene sequences was the rs12459419 alleles. These minigenes were transfected into BV2 microglial cells, and expression of minigene-derived D2-CD33 and CD33 was quantified. This analysis found that D2-CD33 as a percentage of total CD33 increased approximately threefold between cells transfected with rs12459419C (3.4 ± 1.4, mean ± SE; n = 3) versus rs12459419T minigenes (10.3 ± 2.3, mean ± SE; n = 3, p = 0.034). These findings confirm that rs12459419 is a functional polymorphism and are consistent with the human brain results. Although the mechanism underlying rs12459419 actions is beyond the scope of this study, in silico analysis of RNA binding proteins (Smith et al., 2006) for the RNA sequence containing rs12459419, GGG(C/U)CUG, predicts that the splicing factor SRSF2 binds when rs12459419C is present but not when rs124594149U is present (scores of 4.1 and 1.4, respectively, threshold score for binding of 2.4). SRSF2 is widely expressed, including immune cells (Visconte et al., 2012). In summary, the most likely interpretation of these overall results is that rs12459419 mediates the rs3865444 association with D2-CD33 expression in brain because rs12459419 is highly coinherited with rs3865444, is the only SNP in exons 1–4 that is coinherited with rs3865444, resides within exon 2, and has a plausible mechanism to modulate splicing.
Discussion
This report provides new insights into a previously described AD risk allele, rs3865444, and CD33. The primary findings are that CD33 expression is primarily microglial, increased in AD, and that the AD-protective rs3865444A allele acts via its proxy, rs12459419T, to strongly increase the proportion of CD33 expressed as D2-CD33. Our results support a hypothetical model wherein splicing regulates the ability of CD33 to inhibit microglial activation. Furthermore, the paradigm that we used, considering quantitative gene expression as a function of splice variants and of cell-type variation among brain samples, may be generally applicable. These experiments identify a novel mechanism for the protective allele of CD33 that opens opportunities for CD33 pharmacologic modulation in AD.
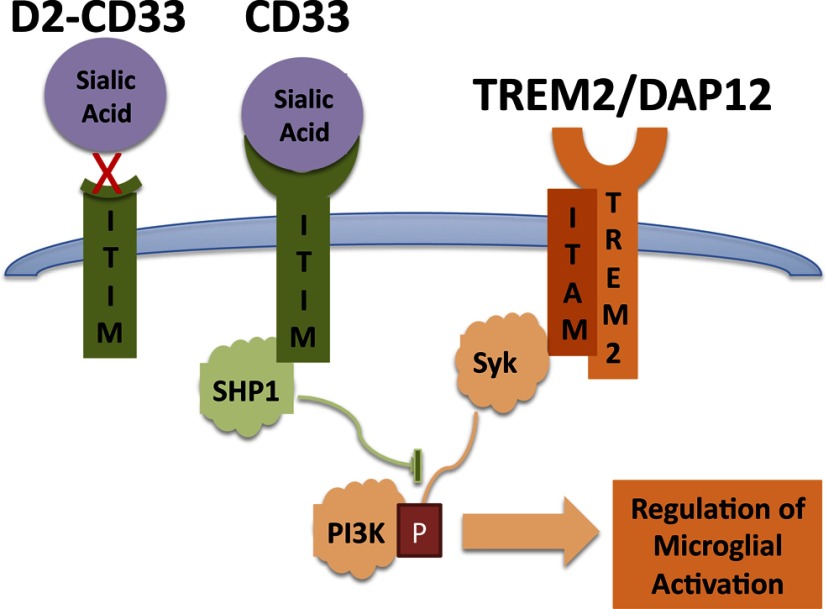
This study contributes to an emerging understanding that AD risk is modulated by genetic factors that influence microglial activation. Recently, increased AD risk was strongly associated with apparent inactivating mutations in the microglial activator TREM2 (Guerreiro et al., 2013; Jonsson et al., 2013); TREM2 acts via DAP12 to activate Syk-mediated tyrosine phosphorylation to promote microglial phagocytosis (Fig. 3; for review, see Linnartz et al., 2010; Neumann and Daly, 2013). In the current study, rs3865444A, which is associated with decreased AD risk (Hollingworth et al., 2011; Naj et al., 2011), is associated with lower overall CD33 expression as well as exclusion of CD33 exon 2. These findings are consistent with recent reports that CD33 expression is reduced with the rs3865444A allele (Griciuc et al., 2013). When quantitation of CD33 expression relies on an exon 2-encoded epitope, the loss of exon 2 could amplify apparent decreases in CD33 expression with the rs3865444A allele (Bradshaw et al., 2013). Sialic acids activate CD33 to stimulate SHP1/SHP2 tyrosine phosphatases, resulting in inhibition of phagocytosis (Fig. 3; for review, see Linnartz et al., 2010; Neumann and Daly, 2013). Because D2-CD33 lacks the IgV domain that is predicted to mediate sialic acid binding, D2-CD33 likely encodes a nonfunctional protein (for review, see Varki and Angata, 2006); consistent with this possibility, phagocytosis as measured by Aβ42 uptake is inhibited when BV2 cells are transfected with CD33 but not CD33 lacking the IgV domain (Griciuc et al., 2013). Hence, D2-CD33 likely represents a loss of CD33 function. Overall, these results provide a genetic basis to support a model wherein TREM2 and CD33 act in opposing directions to modulate tyrosine phosphorylation and, thereby, microglial activation and AD risk (Linnartz et al., 2010; Neumann and Daly, 2013). Alleles that inhibit TREM2 function increase AD risk, whereas alleles that inhibit CD33 function reduce AD risk.
Figure 3.
CD33 and TREM2 represent opposing forces in microglial activation. Sialic acid binding to CD33 results in activation of SHP1 phosphatase, which inhibits immune cell activation. The D2-CD33 isoform lacks the exon that encodes the apparent sialic acid binding domain and hence is proposed to be nonresponsive to sialic acid and inactive. In contrast, an unknown ligand binds TREM2 that signals through DAP12 to activate the tyrosine kinase Syk, resulting in microglial activation. Genetic evidence suggests that loss of CD33 function decreases AD risk, whereas loss of TREM2 function increases AD risk.
The approach used here to elucidate the actions of rs3865444 may be applicable to other polymorphisms identified using GWAS. Elucidating these molecular mechanisms has been challenging for several reasons. First, because GWASs evaluate a stochastic subset of polymorphisms, the polymorphisms identified from these studies are typically not functional but rather are coinherited with a functional polymorphism. If the coinheritance between the AD SNP and the functional SNP is modest or if there is more than one functional SNP, discerning an association between the AD SNP and functional changes is especially challenging. Second, genetic expression studies often use microarray data (Guerreiro et al., 2010; Allen et al., 2012) that are often unable to accurately quantify individual mRNA isoforms. Finally, human brain samples are heterogeneous with regard to cell types, and most expression studies do not control for variation in the proportion of cell type. This issue is especially germane considering the usually small size of research tissue samples and the 2–4 mm thickness of human gray matter. Hence, variation in cell type contributes to substantial experimental variation. Overall, the combination of qPCR assays specific to individual CD33 isoforms, controlling for microglial content in samples, and the fact that the functional polymorphism is tightly linked with the AD SNP contributed to the success of this study.
Although the effect of CD33 polymorphisms on AD risk is modest, elucidating the underlying mechanism may lead to AD treatments with high impact. GWASs identify natural genetic variants that alter disease risk. Elucidating the actions of these variants reveals “bottlenecks” in AD risk pathways, i.e., the polymorphisms biologically define rate-limiting steps in AD. From that perspective, an SNP with modest biologic actions may reduce AD risk only marginally, whereas a drug that acts strongly at the same target may have a large effect on AD risk. In this regard, although CD33 antibodies activate CD33 in the short-term, antibody-mediated downregulation of CD33 inhibits CD33 and promotes monocyte activation (Lajaunias et al., 2005). Moreover, humanized CD33 antibodies have been developed as therapies for acute myeloid leukemia because these cancers often express high levels of CD33 (Amadori and Stasi, 2008). Because such antibodies efficiently downregulate CD33 in vivo in the periphery (Scheinberg et al., 1991), we speculate that these CD33 antagonists may have merit with regards to AD.
In summary, CD33 expression is increased in AD and in individuals with the rs3865444C allele that is associated with increased AD risk. Moreover, rs3865444 is associated with CD33 exon 2 splicing efficiency through the exon 2 polymorphism rs12459419. For this reason, rs12459419 may be a more appropriate polymorphism for future AD association studies. Targeting CD33 and its role in microglial activation may yield AD-protective agents.
Footnotes
This work was made possible by National Institutes of Health Grants P01-AGO30128 (SE), P30-AG028383 (D.W.F., P.T.N.), and P20-GM103436 (D.W.F.) and the University of Kentucky Bucks for Brains program (M.M.). The University of Kentucky has filed a provisional patent for CD33 inhibition in Alzheimer's disease that includes M.M., J.F.S., I.P., and S.E.
References
- Allen M, Zou F, Chai HS, Younkin CS, Crook J, Pankratz VS, Carrasquillo MM, Rowley CN, Nair AA, Middha S, Maharjan S, Nguyen T, Ma L, Malphrus KG, Palusak R, Lincoln S, Bisceglio G, Georgescu C, Schultz D, Rakhshan F, et al. Novel late-onset Alzheimer disease loci variants associate with brain gene expression. Neurology. 2012;79:221–228. doi: 10.1212/WNL.0b013e3182605801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amadori S, Stasi R. Integration of monoclonal antibodies and immunoconjugates into the treatment of acute myeloid leukemia. Curr Opin Hematol. 2008;15:95–100. doi: 10.1097/MOH.0b013e3282f3de84. [DOI] [PubMed] [Google Scholar]
- Bradshaw EM, Chibnik LB, Keenan BT, Ottoboni L, Raj T, Tang A, Rosenkrantz LL, Imboywa S, Lee M, Von Korff A, Alzheimer Disease Neuroimaging I, Morris MC, Evans DA, Johnson K, Sperling RA, Schneider JA, Bennett DA, De Jager PL. CD33 Alzheimer's disease locus: altered monocyte function and amyloid biology. Nat Neurosci. 2013;16:848–850. doi: 10.1038/nn.3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkman-Van der Linden EC, Angata T, Reynolds SA, Powell LD, Hedrick SM, Varki A. CD33/Siglec-3 binding specificity, expression pattern, and consequences of gene deletion in mice. Mol Cell Biol. 2003;23:4199–4206. doi: 10.1128/MCB.23.12.4199-4206.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griciuc A, Serrano-Pozo A, Parrado AR, Lesinski AN, Asselin CN, Mullin K, Hooli B, Choi SH, Hyman BT, Tanzi RE. Alzheimer's disease risk gene CD33 inhibits microglial uptake of amyloid beta. Neuron. 2013;78:631–643. doi: 10.1016/j.neuron.2013.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerreiro R, Wojtas A, Bras J, Carrasquillo M, Rogaeva E, Majounie E, Cruchaga C, Sassi C, Kauwe JS, Younkin S, Hazrati L, Collinge J, Pocock J, Lashley T, Williams J, Lambert JC, Amouyel P, Goate A, Rademakers R, Morgan K, et al. TREM2 variants in Alzheimer's disease. N Engl J Med. 2013;368:117–127. doi: 10.1056/NEJMoa1211851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerreiro RJ, Beck J, Gibbs JR, Santana I, Rossor MN, Schott JM, Nalls MA, Ribeiro H, Santiago B, Fox NC, Oliveira C, Collinge J, Mead S, Singleton A, Hardy J. Genetic variability in CLU and its association with Alzheimer's disease. PLoS One. 2010;5:e9510. doi: 10.1371/journal.pone.0009510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, Hamshere ML, Pahwa JS, Moskvina V, Dowzell K, Williams A, Jones N, Thomas C, Stretton A, Morgan AR, Lovestone S, Powell J, Proitsi P, Lupton MK, Brayne C, Rubinsztein DC, et al. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer's disease. Nat Genet. 2009;41:1088–1093. doi: 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández-Caselles T, Martínez-Esparza M, Péerez-Oliva AB, Quintanilla-Cecconi AM, García-Alonso A, Alvarez-López DM, García-Peñarrubia P. A study of CD33 (SIGLEC-3) antigen expression and function on activated human T and NK cells: two isoforms of CD33 are generated by alternative splicing. J Leukoc Biol. 2006;79:46–58. doi: 10.1189/jlb.0205096. [DOI] [PubMed] [Google Scholar]
- Hollingworth P, Harold D, Sims R, Gerrish A, Lambert JC, Carrasquillo MM, Abraham R, Hamshere ML, Pahwa JS, Moskvina V, Dowzell K, Jones N, Stretton A, Thomas C, Richards A, Ivanov D, Widdowson C, Chapman J, Lovestone S, Powell J, et al. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer's disease. Nat Genet. 2011;43:429–435. doi: 10.1038/ng.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson T, Stefansson H, Steinberg S, Jonsdottir I, Jonsson PV, Snaedal J, Bjornsson S, Huttenlocher J, Levey AI, Lah JJ, Rujescu D, Hampel H, Giegling I, Andreassen OA, Engedal K, Ulstein I, Djurovic S, Ibrahim-Verbaas C, Hofman A, Ikram MA, et al. Variant of TREM2 associated with the risk of Alzheimer's disease. N Engl J Med. 2013;368:107–116. doi: 10.1056/NEJMoa1211103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun G, Naj AC, Beecham GW, Wang LS, Buros J, Gallins PJ, Buxbaum JD, Ertekin-Taner N, Fallin MD, Friedland R, Inzelberg R, Kramer P, Rogaeva E, St George-Hyslop P, St George-Hyslop P, Cantwell LB, Dombroski BA, Saykin AJ, Reiman EM, Bennett DA, et al. Meta-analysis confirms CR1, CLU, and PICALM as alzheimer disease risk loci and reveals interactions with APOE genotypes. Arch Neurol. 2010;67:1473–1484. doi: 10.1001/archneurol.2010.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lajaunias F, Dayer JM, Chizzolini C. Constitutive repressor activity of CD33 on human monocytes requires sialic acid recognition and phosphoinositide 3-kinase-mediated intracellular signaling. Eur J Immunol. 2005;35:243–251. doi: 10.1002/eji.200425273. [DOI] [PubMed] [Google Scholar]
- Lambert JC, Heath S, Even G, Campion D, Sleegers K, Hiltunen M, Combarros O, Zelenika D, Bullido MJ, Tavernier B, Letenneur L, Bettens K, Berr C, Pasquier F, Fiévet N, Barberger-Gateau P, Engelborghs S, De Deyn P, Mateo I, Franck A, et al. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer's disease. Nat Genet. 2009;41:1094–1099. doi: 10.1038/ng.439. [DOI] [PubMed] [Google Scholar]
- Ling IF, Bhongsatiern J, Simpson JF, Fardo DW, Estus S. Genetics of clusterin isoform expression and Alzheimer's disease risk. PLoS One. 2012;7:e33923. doi: 10.1371/journal.pone.0033923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linnartz B, Neumann H. Microglial activatory (immunoreceptor tyrosine-based activation motif)- and inhibitory (immunoreceptor tyrosine-based inhibition motif)-signaling receptors for recognition of the neuronal glycocalyx. Glia. 2013;61:37–46. doi: 10.1002/glia.22359. [DOI] [PubMed] [Google Scholar]
- Linnartz B, Wang Y, Neumann H. Microglial immunoreceptor tyrosine-based activation and inhibition motif signaling in neuroinflammation. Int J Alzheimers Dis. 2010;2010.pii:587463. doi: 10.4061/2010/587463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan K. The three new pathways leading to Alzheimer's disease. Neuropathol Appl Neurobiol. 2011;37:353–357. doi: 10.1111/j.1365-2990.2011.01181.x. [DOI] [PubMed] [Google Scholar]
- Naj AC, Jun G, Beecham GW, Wang LS, Vardarajan BN, Buros J, Gallins PJ, Buxbaum JD, Jarvik GP, Crane PK, Larson EB, Bird TD, Boeve BF, Graff-Radford NR, De Jager PL, Evans D, Schneider JA, Carrasquillo MM, Ertekin-Taner N, Younkin SG, et al. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer's disease. Nat Genet. 2011;43:436–441. doi: 10.1038/ng.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson PT, Braak H, Markesbery WR. Neuropathology and cognitive impairment in Alzheimer disease: a complex but coherent relationship. J Neuropathol Exp Neurol. 2009;68:1–14. doi: 10.1097/NEN.0b013e3181919a48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann H, Daly MJ. Variant TREM2 as risk factor for Alzheimer's disease. N Engl J Med. 2013;368:182–184. doi: 10.1056/NEJMe1213157. [DOI] [PubMed] [Google Scholar]
- Pérez-Oliva AB, Martínez-Esparza M, Vicente-Fernández JJ, Corral-San Miguel R, García-Peñarrubia P, Hernández-Caselles T. Epitope mapping, expression and post-translational modifications of two isoforms of CD33 (CD33M and CD33m) on lymphoid and myeloid human cells. Glycobiology. 2011;21:757–770. doi: 10.1093/glycob/cwq220. [DOI] [PubMed] [Google Scholar]
- Scheinberg DA, Lovett D, Divgi CR, Graham MC, Berman E, Pentlow K, Feirt N, Finn RD, Clarkson BD, Gee TS, Larson SM, Oettgen HF, Old LJ. A phase I trial of monoclonal antibody M195 in acute myelogenous leukemia: specific bone marrow targeting and internalization of radionuclide. J Clin Oncol. 1991;9:478–490. doi: 10.1200/JCO.1991.9.3.478. [DOI] [PubMed] [Google Scholar]
- Smith PJ, Zhang C, Wang J, Chew SL, Zhang MQ, Krainer AR. An increased specificity score matrix for the prediction of SF2/ASF-specific exonic splicing enhancers. Hum Mol Genet. 2006;15:2490–2508. doi: 10.1093/hmg/ddl171. [DOI] [PubMed] [Google Scholar]
- Varki A, Angata T. Siglecs—the major subfamily of I-type lectins. Glycobiology. 2006;16:1R–27R. doi: 10.1093/glycob/cwj008. [DOI] [PubMed] [Google Scholar]
- Visconte V, Makishima H, Maciejewski JP, Tiu RV. Emerging roles of the spliceosomal machinery in myelodysplastic syndromes and other hematological disorders. Leukemia. 2012;26:2447–2454. doi: 10.1038/leu.2012.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou F, Gopalraj RK, Lok J, Zhu H, Ling IF, Simpson JF, Tucker HM, Kelly JF, Younkin SG, Dickson DW, Petersen RC, Graff-Radford NR, Bennett DA, Crook JE, Estus S. Sex-dependent association of a common low-density lipoprotein receptor polymorphism with RNA splicing efficiency in the brain and Alzheimer's disease. Hum Mol Genet. 2008;17:929–935. doi: 10.1093/hmg/ddm365. [DOI] [PMC free article] [PubMed] [Google Scholar]