Abstract
Ribonucleotide reductase (RNR) is the key enzyme in the biosynthesis of deoxyribonucleotides. Alpha- and gammaherpesviruses express a functional enzyme, since they code for both the R1 and the R2 subunits. By contrast, betaherpesviruses contain an open reading frame (ORF) with homology to R1, but an ORF for R2 is absent, suggesting that they do not express a functional RNR. The M45 protein of murine cytomegalovirus (MCMV) exhibits the sequence features of a class Ia RNR R1 subunit but lacks certain amino acid residues believed to be critical for enzymatic function. It starts to be expressed independently upon the onset of viral DNA synthesis at 12 h after infection and accumulates at later times in the cytoplasm of the infected cells. Moreover, it is associated with the virion particle. To investigate direct involvement of the virally encoded R1 subunit in ribonucleotide reduction, recombinant M45 was tested in enzyme activity assays together with cellular R1 and R2. The results indicate that M45 neither is a functional equivalent of an R1 subunit nor affects the activity or the allosteric control of the mouse enzyme. To replicate in quiescent cells, MCMV induces the expression and activity of the cellular RNR. Mutant viruses in which the M45 gene has been inactivated are avirulent in immunodeficient SCID mice and fail to replicate in their target organs. These results suggest that M45 has evolved a new function that is indispensable for virus replication and pathogenesis in vivo.
Cytomegalovirus (CMV), a betaherpesvirus, is a widespread pathogen responsible for generally asymptomatic and persistent infections in healthy people. It may, however, cause severe disease in the absence of an effective immune response, as in immunologically immature and immunocompromised individuals (41). Strict species specificity has hampered investigation of human CMV (HCMV) in its natural host, and infection of mice with murine CMV (MCMV) has been extensively used as a model for studying the pathogenesis of HCMV infection (40, 52, 63). The two viruses are biologically similar in replication and pathogenesis (34, 50, 51), and their homologous genomes display similar genetic organizations and encode analogous gene products with similar functions (56). However, the functions of many viral gene products remain to be explored in order to determine the interactions of CMV with the host cell and its pathogenic mechanisms.
CMV replicates most efficiently in the absence of cellular DNA synthesis (23, 39). Moreover, it has evolved mechanisms to inhibit progression through the cell cycle and arrests cells with a G1 DNA content (5, 18, 47, 53, 60, 68). The viral DNA polymerase thus has competition-free access to the DNA precursors, and CMV replicates efficiently in vitro in quiescent cells (18, 45) and infects terminally differentiated cells in vivo (41, 51). However, since postmitotic cells do not replicate their genomes, the very low levels of deoxyribonucleotides (dNTP) and cell functions involved in DNA synthesis limit viral replication.
It has been demonstrated that CMV replication in quiescent fibroblasts depends on its ability to stimulate expression of the cellular enzymes involved in the biosynthesis of thymidylate, since they are not encoded by the viral genome (9, 27, 26, 28, 44, 46). However, it is still unclear how CMV expands the pools of all four dNTP in resting cells and provides a sufficient supply of DNA precursors to its polymerase. The key step in dNTP biosynthesis is carried out by the enzyme ribonucleotide reductase (RNR), which catalyzes the conversion of ribonucleoside diphosphates to the corresponding deoxyribonucleoside diphosphates. Both substrate specificity and overall activity are tightly controlled by binding of nucleoside triphosphate allosteric effectors. The expression of RNR is cell cycle regulated; it is very low or not detectable in resting cells and maximal in S-phase cells (3, 10, 20, 22, 57, 66).
RNR has been divided into three classes according to its mechanism for generation of the protein radical, metal cofactor requirement, and subunit composition (38). Mammalian cells, like most eukaryotic cells, contain a class Ia RNR. This form also exists in some prokaryotes, e.g., the well-studied nrdA/nrdB-encoded enzyme of Escherichia coli. Class Ia has an α2β2 form consisting of two homodimeric subunits, proteins R1 (α2) and R2 (β2). The R1 protein contains the active site and the binding sites for allosteric effectors. The R2 protein is a radical storage device containing an iron center-generated tyrosyl free radical.
The alpha- and gammaherpesviruses (2, 14, 16, 33, 43) express a functional RNR required for virus growth in nondividing cells and for viral pathogenesis and reactivation from latency in infected hosts (8, 17, 24, 25, 32, 37). The herpes simplex virus type 1 (HSV-1) RNR is the most extensively characterized. Like the mammalian and E. coli enzymes, it belongs to class Ia but completely lacks allosteric regulation (14).
Analysis of the protein-coding content of the HCMV and MCMV genomes has revealed the presence of an open reading frame (ORF), termed UL45 and M45, respectively (11, 56), which shows homology to the R1 subunit of other herpesvirus. However, like those of other betaherpesviruses, CMV genomes do not carry an ORF for the R2 subunit. It follows that these viruses are unlikely to express a functional RNR enzyme, and it is unclear how they can replicate in resting cells. The R1 homologs of CMV are still a biological enigma, because their expression, cellular localization, involvement in ribonucleotide reduction, and role in viral pathogenesis have not been described. All these points are now addressed in the present paper.
MATERIALS AND METHODS
Cells and culture conditions.
NIH 3T3 murine fibroblasts were grown as monolayers in Dulbecco's modified Eagle's medium (DMEM) (Gibco/BRL) supplemented with 10% calf serum (Gibco/BRL). Quiescent NIH 3T3 cells (arrested in G0/G1 phase) were obtained by culturing the subconfluent cultures for 48 h in DMEM plus 0.5% calf serum (low-serum medium). Flow cytometry at this time demonstrated more than 90% of cells arrested in G0/G1.
Virus preparation and infections.
MCMV (mouse salivary gland virus, strain Smith, ATCC VR.194) was purchased from the American Type Culture Collection (Manassas, Va.). Virus stocks were first produced in salivary glands of BALB/c mice and then propagated in vitro by infecting NIH 3T3 cells at a virus-to-cell ratio of 0.01. Cells were incubated in DMEM supplemented with 2% heat-inactivated calf serum, and virus was harvested, based on cytopathology, by sonication at about 10 days postinfection and was then clarified by centrifugation. Mock-infected fluid was prepared from NIH 3T3 cells by the procedure used to prepare MCMV. A virus stock solution containing approximately 107 to 108 PFU/ml (as determined by a plaque assay on the NIH 3T3 cell line) was used in all experiments. MCMV-GFP, bd-MCMV, MCMV-REV, IIIG2, and BamX have been described previously (6). MCMV-GFP (kindly provided by Martin Messerle) was generated by inserting the green fluorescent protein (GFP) gene into the ie2 region of the full-length MCMV bacterial artificial chromosome (BAC), pSM3fr, essentially as described previously (1); the BAC-derived MCMV (bd-MCMV) is a wild-type MCMV derived from pSM3fr (67); MCMV-REV is a revertant virus obtained by repairing the transposon insertion in the M45 mutant virus IID7 (6). These three viruses display a wild-type phenotype and were used as controls. IIIG2 is an M45 mutant virus carrying a transposon insertion at nucleotide position 62876 of the MCMV genome. BamX is a frameshift mutant of the M45 gene generated by replacing the transposon insertion in mutant IID7 (nucleotide position 62547) with an M45 sequence in which the BamHI restriction site was cut and filled in with Klenow polymerase, resulting in a 4-bp insertion (6). The frameshift mutation leads to a missense amino acid sequence after 188 out of 1,174 amino acids of the M45 protein. Thirty-three missense amino acids are added before a stop codon is encountered. A schematic map showing the transposon insertions and the BamX frameshift mutation is available as Supplemental Figure 2, published previously (6a).
FIG. 2.
(A) Time course of M45 expression during MCMV replication as determined by immunoblotting. Whole-cell lysates of mock-infected and MCMV-infected NIH 3T3 cells at various times after infection were separated by SDS-PAGE, transferred to a membrane, and probed with the anti-M45 antiserum or with the anti-actin monoclonal antibody. Lanes: 1, mock-infected cells; 2, 6 hpi; 3, 9 hpi; 4, 12 hpi; 5, 18 hpi; 6, 24 hpi; 7, 36 hpi; 8, 48 hpi. (B) Effect of PFA on M45 expression. Whole-cell extracts were prepared at 18 hpi (lanes 1 and 2), 24 hpi (lanes 3 and 4), or 48 hpi (lanes 5 and 6) from MCMV-infected NIH 3T3 cells treated with PFA (250 μg/ml) after virus adsorption (lanes 2, 4, and 6) or left untreated (lanes 1, 3, and 5). Protein expression was analyzed by immunoblotting with the anti-M45 antiserum or with the anti-actin monoclonal antibody. (C) Subcellular localization of M45 in MCMV-infected NIH 3T3 cells at 48 hpi, detected by immunofluorescence and confocal microscopy. Cells were incubated with the M45 antiserum and then with the secondary FITC-conjugated antibody. Nuclei were counterstained with propidium iodide. (D) Subcellular localization of transiently expressed M45 protein in NIH 3T3 cells transfected with the pcDNA3-45 vector at 24 h posttransfection. For immunofluorescence and confocal microscopy analysis, cells were incubated with the M45 antiserum and then with the secondary FITC-conjugated antibody. Nuclei were counterstained with propidium iodide. The merged pictures are shown.
Extracellular virions were partially purified from tissue culture medium by two rounds of centrifugation through a 15% sucrose cushion in a Kontron TST 55.5 rotor (2 h, 26,000 rpm, 4°C). The partially purified virions, resuspended in TN (50 mM Tris [pH 7.4], 100 mM NaCl), were layered onto 9-ml potassium tartrate-glycerol gradients formed in TN and then centrifuged at 40,000 rpm for 15 min at 4°C in a Beckman SW41 rotor with slow acceleration and braking. Virions were visualized with incandescent light and were removed from the gradients.
For protein level determinations, NIH 3T3 cells were infected with MCMV at a multiplicity of infection (MOI) of 5 PFU/cell unless otherwise stated. Mock-infected control cultures were exposed to an equal volume of mock-infecting fluid. Virus adsorptions were carried out for 1 h at 37°C, and 0 h postinfection (0 hpi) was defined as the time immediately following this period. When quiescent cells were used, the low-serum medium removed from the cells before infection was returned to the plates to avoid any stimulation due to addition of fresh serum growth factors. Inactivation of virus by UV light was performed as described previously (44).
Preparation of protein extracts and immunoblotting.
Whole-cell extracts were prepared by resuspending pelleted cells in lysis buffer containing 125 mM Tris-Cl (pH 6.8), 3% sodium dodecyl sulfate (SDS), 20 mM dithiotreitol, 1 mM phenylmethylsulfonyl fluoride (PMSF), 4 μg of leupeptin/ml, 4 μg of aprotinin/ml, and 1 μg of pepstatin/ml. After a brief sonication, soluble proteins were collected by centrifugation at 15,000 × g. Supernatants were quantified for protein concentration with a Dc protein assay kit (Bio-Rad Laboratories) and were stored at −70° in 10% glycerol.
For immunoblotting, proteins were separated by SDS-polyacrylamide gel electrophoresis (PAGE) and transferred to Immobilon-P membranes (Millipore). Filters were then blocked with 5% nonfat dry milk in 10 mM Tris-Cl (pH 7.5)-100 mM NaCl-0.1% Tween 20 and were immunostained with anti-M45 polyclonal antibodies, the anti-R1 monoclonal antibody AD203, anti-R2 polyclonal antibodies (45), an anti-MCMV M44 monoclonal antibody (kindly provided by L. C. Loh, University of Saskatchewan), or an anti-actin mouse monoclonal antibody (Boehringer Mannheim). Immunocomplexes were then detected by means of sheep-anti mouse immunoglobulin (Ig) or goat anti-rabbit Ig antibodies, both conjugated to horseradish peroxidase (Amersham), and were visualized by using enhanced chemiluminescence (Super Signal; Pierce) according to the manufacturer's instructions.
Immunoprecipitation.
Whole-cell extracts were prepared by solubilization in lysis buffer (150 mM NaCl, 50 mM Tris-HCl [pH 8], 1% NP-40, 1 mM phenylmethylsulfonyl fluoride, 4 μg of leupeptin/ml, 4 μg of aprotinin/ml, and 1 μg of pepstatin/ml) and shaking for 20 min at 4°C. After centrifugation at 15,000 × g, the supernatants were incubated for 1 h at 4°C on a rocking platform with either anti-M45 antibodies or preimmune serum previously conjugated with CNBr-activated Sepharose 4 Fast Flow (Amersham Pharmacia) according to the manufacturer's instructions. The immunocomplexes were then centrifuged (700 × g) and washed three times with lysis buffer. The immunoprecipitated proteins were eluted with 0.1 M glycine buffer (pH 11.5), recovered as supernatants after centrifugation, resuspended in SDS gel sample buffer, separated by SDS-PAGE, and subjected to immunoblotting.
Immunofluorescence microscopy.
Cells grown on coverslips were infected with MCMV at an MOI of 0.5. At 48 hpi cells were washed with phosphate-buffered saline (PBS), fixed with 1% paraformaldehyde for 20 min at room temperature, and washed again with PBS. They were subsequently permeabilized with 0.2% Triton X-100 in PBS for 20 min at 4°C, washed with PBS-1% bovine serum albumin (BSA), and incubated with the anti-M45 antibody (diluted 250-fold) in a solution containing PBS-1% BSA and 0.2% Triton X-100 for 1 h at room temperature. After a wash with PBS-1% BSA and 0.05% Tween 20, cells were incubated with fluorescein isothiocyanate (FITC)-conjugated goat anti-rabbit antibodies in PBS-1% BSA and 0.2% Triton X-100 for 1 h. Coverslips were washed with a solution containing PBS-1% BSA and 0.05% Tween 20, and the nuclei were counterstained with propidium iodide. After an additional wash, the coverslips were mounted in 90% glycerol. Immunofluorescence microscopy was performed on an Olympus IX70 inverted confocal laser scanning microscope equipped with a krypton-argon ion laser (excitation wavelength, 488 nm; emission wavelength, 568 nm). Images derived from both channels (fluorescein and propidium iodide) were recorded simultaneously at identical apertures. The fluorescein-derived image was assessed with a green color, and the propidium iodide-derived image was assessed with a red color.
Plasmid constructs.
The coding sequence of the M45 gene was amplified by PCR from total DNA of NIH 3T3 cells infected with MCMV for 48 h. The two 5′ oligonucleotide primers were 5′-TCATCAACAACATATGGATCGCCAGCCCAAAGTC-3′ and 5′-TCATCAACAACATATGCACCATCATCATCATCATGATCGCCAGCCCAAAGTC-3′; the latter contained a sequence encoding a six-histidine tag immediately dowstream from the ATG. The 3′ oligonucleotide primer was 5′-GCTGCTCGAGGGATCCCTTTCAGCGATAATTCACGGA-3′. The PCR product without the histidine tag was cloned into the PCR cloning vector pGEM-T Easy (Promega), sequenced, and then subcloned into the mammalian expression vector pcDNA3 (Invitrogen). The PCR product containing the histidine tag was subcloned into the E. coli expression vector pET30a(+) (Novagen) and then sequenced. A 772-bp fragment encoding a C-terminal region of M45 (from nucleotide 2563 to nucleotide 3403) was excised with the restriction endonuclease FspI from the pGEM T Easy-M45 vector and subcloned in frame in the SmaI site of the E. coli expression vector pGEX-4T3 (Novagen) in order to express the M45 fragment as a fusion protein with glutathione S-transferase (GST). To generate the catalytically inactive mouse R1 C429A protein, the 2.8-kb R1 cDNA fragment contained in the pET3a vector (15) was subjected to site-directed mutagenesis with the QuikChange XL kit (Stratagene) and oligonucleotide primers 5′-CCATCAAATGCAGCAACCTGGCTACAGAAATAGTAGAGTACACC-3′ and 5′-GGTGTACTCTACTATTTCTGTAGCCAGGTTGCTGCATTTGATGG-3′. Underlining represents mutated nucleotides. The point mutations converting cysteine 429 into an alanine were confirmed by DNA sequence analysis. Transient transfections were performed as described previously (45).
Expression and purification of the recombinant proteins and immunizations.
Plasmid pET30a(+) containing the six-His-tagged full-length M45 sequence was transfected into Rosetta(DE3)pLysS bacteria (Novagen). An overnight culture of bacteria grown in Luria-Bertani broth plus kanamycin and chloramphenicol was used to infect a 1-liter culture of TerrificBroth containing the same antibiotics, and the culture was grown at 30°C to an A600 of 1.0. Then the temperature of the incubator was decreased to 15°C, and when the temperature of the liquid had reached 20°C, isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 0.05 mM. After overnight incubation at 15°C, the bacteria were harvested by centrifugation, washed, and lysed by freeze-thawing, and the extract was clarified by centrifugation in a Beckman 70 Ti rotor for 45 min at 45,000 rpm and 4°C. The protein extract was incubated with nickel-agarose in 50 mM Tris-Cl (pH 7.6)-0.3 m NaCl-10 mM imidazole for 1 h at 4°C and extensively washed with the same buffer. Adsorbed protein was eluted in 50 mM Tris-Cl (pH 7.6)-0.2 M imidazole and then immediately equilibrated with 50 mM Tris-Cl (pH 7.6)-0.1 M KCl on a Sephadex G-25 column. An aliquot of this material was further purified by chromatography on a Superdex 200 column (Amersham Biosciences) in 50 mM Tris-Cl (pH 7.6)-0.1 M KCl. The mouse R1 C429A protein was expressed and purified as described previously (15).
The fusion of the C-terminal portion of M45 to GST was produced in E. coli BL21(DE3) (Novagen) containing plasmid pGEX-4T3-M45. Bacteria were grown at 37°C until they reached an optical density at 600 nm (OD600) of 0.6 and were then induced with 1 mM IPTG for 2 h. The cells were pelleted, resuspended in lysis buffer (50 mM Tris-HCl [pH 8], 2 mM EDTA, 1 mM dithiothreitol [DTT], 1% Triton X-100, 500 μg of lysozyme/ml, 1 μg of pepstatin/ml, 2 μg of leupeptin/ml) at 30°C for 15 min, and briefly sonicated. Analysis of the soluble and insoluble fractions by SDS-PAGE followed by Coomassie blue staining revealed that more than 90% of the recombinant protein was present in the insoluble fraction (inclusion bodies). Inclusion bodies were washed twice with lysis buffer supplemented with 2 M urea and then solubilized in lysis buffer supplemented with 6 M urea. The denatured recombinant proteins were refolded by dialysis against folding buffer (0.1% NP-40, 0.2 mM EDTA, 1 mM DTT, 200 mM PMSF, 0.5 M NaCl, 0.1 M Tris-HCl [pH 7.5]) containing decreasing concentrations of urea from 5 to 0.5 M. The last dialysis was performed against folding buffer without urea. The refolded protein was concentrated through a centrifugal filter device (Amicon), analyzed by SDS-PAGE followed by Coomassie blue staining, and then used as an antigen for rabbit immunization in order to generate specific antisera. Rabbits were injected intramuscularly with 200 μg of protein five times and were bled after 3 months.
dNTP pool measurements.
Cell cultures were extracted with ice-cold trichloroacetic acid. The extracted nucleotides were separated directly by high-performance liquid chromatography (ribonucleoside triphosphates) or first run through a boronate affinity column (deoxyribonucleoside triphosphates) as described previously (35). Levels of nucleotide pools are expressed as percentages of the total nucleoside triphosphate pool (CTP + UTP + ATP + GTP + dCTP + dTTP + dATP + dGTP) to minimize variations due to small differences in cell number in the samples.
RNR assay.
The ability of the purified M45 protein to catalyze the reduction of [3H]CDP to [3H]dCDP at 37°C in the presence of recombinant mouse R2 protein, or to stimulate the reduction catalyzed by recombinant mouse R1 and R2 proteins or recombinant mouse R1 C429A mutant protein and R2 protein, was assayed as described elsewhere (21). In addition to the standard ATP-stimulated reduction, a putative effect of the M45 protein on the allosteric inhibition by dATP was assayed by the addition of 20 to 400 μM dATP to the assay mixture.
In vivo growth of MCMV mutants.
CB17 SCID mice (eight mice per group), maintained under pathogen-free conditions in our animal facility, were infected intraperitoneally with 106 PFU of either an M45 mutant virus or a control (wild-type) virus and were observed daily for survival. At 24 days postinfection, spleens, livers, lungs, kidneys, and salivary glands were aseptically harvested from two mice per group, homogenized, and titrated in duplicate on NIH 3T3 cells by plaque assay. Values given were calculated per gram of organ.
RESULTS
Generation and reactivity of an M45 antiserum.
In order to obtain protein for the generation of a specific antiserum, a fragment of the M45 ORF encoding the hydrophilic C-terminal domain was cloned into the prokaryotic expression vector pGEX-4T3 and expressed as a GST fusion protein in E. coli. The purified protein was then used to immunize rabbits, and the resulting serum was tested for its specificity by Western blot analysis. As shown in Fig. 1A, a strong signal corresponding to a protein of 150 to 160 kDa was observed in lysates from MCMV-infected NIH 3T3 cells at 48 hpi (lane 2), from cells transiently transfected with the M45 expression vector pcDNA3-M45 (lane 4), and from a lysate of IPTG-induced E. coli cells transformed with the expression vector pET30-M45 (lane 5). By contrast, no signal could be observed with lysates from mock-infected cells (lane 1) or from cells transfected with the empty pcDNA3 vector (lane 3) or on the same blot incubated with preimmune serum (data not shown). Immunoprecipitation followed by anti-M45 immunoblotting of lysates from MCMV-infected cells revealed M45 (150 to 160 kDa) and a second major polypeptide of about 116 kDa (Fig. 1B, lane 2) whose molecular weight corresponded to that of the faint band observed in Fig. 1A, lane 2. The same bands could not be detected when preimmune serum was used (Fig. 1B, lane 3). To determine whether the lower band corresponds to a proteolytic fragment of M45, the immunoprecipitates from infected cells were blotted and the Coomassie-stained bands were subjected to N-terminal sequencing. The N-terminal sequence of the 116-kDa polypeptide was AATMPPP, which corresponds to cleavage between tyrosine 277 and alanine 278 of M45. Thus, the lower band represents a polypeptide generated by specific M45 proteolysis. Altogether, these results demonstrate that a specific antiserum was generated for further analysis of M45 expression.
FIG. 1.
Identification of the MCMV M45 protein by the specific antiserum. (A) Immunoblot analysis of native or recombinant M45 expression in MCMV-infected NIH 3T3 cells, in cells transfected with an M45 expression vector, or in E. coli. Proteins of whole-cell lysates were separated by SDS-PAGE (5 to 15% acrylamide), transferred to a membrane, and probed with the M45 antiserum. Lanes: 1, extracts from mock-infected NIH 3T3 cells; 2, extracts from NIH 3T3 cells 48 h after infection with MCMV; 3, extracts from NIH 3T3 cells transiently transfected with the control vector pcDNA3; 4, extracts from NIH 3T3 cells transiently transfected with the M45 expression vector pcDNA3-M45; 5, extracts from IPTG-induced E. coli containing the M45 expression vector pET30-M45. (B) Immunoprecipitation of M45 by the specific antiserum. M45 was immunoprecipitated from cell extracts of MCMV-infected NIH 3T3 cells prepared at 48 hpi and was analyzed by immunoblotting with the M45 antiserum. Lanes: 1, cell extract from infected cells; 2, cell extract immunoprecipitated with the M45 antiserum; 3, cell extract immunoprecipitated with preimmune serum. Sizes of the molecular mass markers are shown on the left of each panel. Asterisk indicates the Ig heavy chains recognized by the secondary antibody.
Time course of M45 protein expression.
To investigate the kinetics of M45 expression during viral replication, cell lysates were harvested at various time points after MCMV infection and examined by immunoblotting with the M45 antiserum. A specific band at approximately 150 kDa was first detected at 12 hpi and accumulated throughout the course of infection (Fig. 2A). This finding is consistent with the kinetics of M45 mRNA accumulation, which have been reported previously (45).
Treatment with phosphonoformic acid (PFA), a specific inhibitor of viral DNA polymerase, did not affect M45 expression at 18 and 24 hpi but reduced its expression at 48 hpi (Fig. 2B). Therefore, M45 expression is not dependent on the onset of viral DNA synthesis.
Subcellular localization of M45.
Subcellular localization of M45 was determined by indirect immunofluorescence and confocal microscopy of both infected and transiently transfected cells. NIH 3T3 cells infected for 48 h with MCMV at an MOI of 0.1 PFU/cell were incubated with the M45-specific antiserum and then stained with fluorescein-conjugated anti-rabbit antibodies. Nuclei were counterstained with propidium iodide. As shown in Fig. 2C, M45 was quite dispersed throughout the cytoplasm, though a more intense signal was associated with granular formations. By contrast, no specific nuclear signal could be detected. A similar pattern of cytoplasmic staining was observed in cells transiently transfected with the M45 expression vector pcDNA3-M45, indicating that no other viral protein is required for M45 localization (Fig. 2D). No signal could be detected with mock-infected cells or when infected cells were stained with preimmune serum (data not shown).
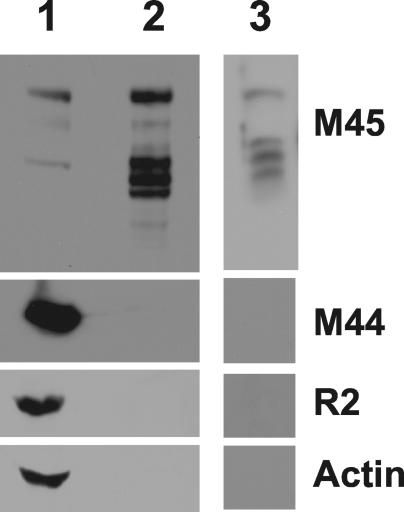
Virion association of M45.
Proteins from herpesviruses abundantly expressed at late times are often incorporated into virus particles during assembly; therefore, we looked for M45 in the MCMV particles. Extracellular virions were purified from tissue culture supernatants by two rounds of centrifugation through a 15% sucrose cushion or via density gradient centrifugation and were then subjected to Western blot analysis. An extract from infected NIH 3T3 cells at 48 hpi was used in parallel as a positive control (Fig. 3, lane 1). In the protein extracts of purified virions, the M45 antiserum detected a protein of approximately 150 kDa as well as lower-molecular-mass products, probably the result of proteolysis (Fig. 3, lanes 2 and 3). To assess the purity of the virion preparation, we performed immunoblot analyses with monoclonal antibodies against the nonstructural viral protein M44 and against the cellular proteins actin and the R2 subunit of RNR, which are constitutively expressed or highly induced by infection, respectively. M44, actin, and R2 were detected in the cell lysate (Fig. 3, lane 1), but not in the virion preparation (Fig. 3, lanes 2 and 3). This observation ruled out the possibility that the M45 signal in virions is due to contamination of the virion preparation with cellular material and demonstrated that M45 is associated with the virion particle.
FIG. 3.
Detection of M45 in purified MCMV particles by immunoblotting. Proteins from a whole-cell lysate of MCMV-infected NIH 3T3 cells at 48 hpi (lane 1) or from virus particles purified by two rounds of centrifugation through a 15% sucrose cushion (lane 2) or via density gradient centrifugation (lane 3) were separated by SDS-PAGE, blotted onto a membrane, and probed with antibodies against the viral proteins M45 and M44 and the cellular proteins R2 and actin.
M45 is not a functional RNR R1 subunit.
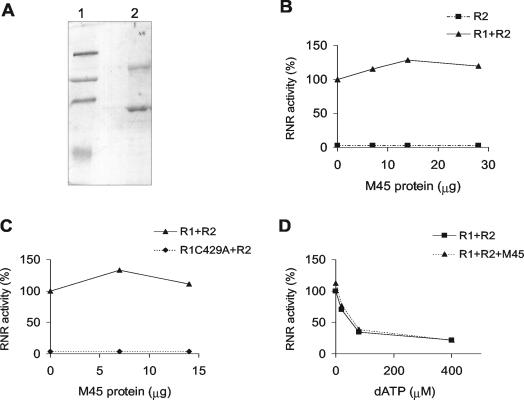
Since M45 displays the sequence features of a class Ia RNR R1 subunit, we looked to see whether it forms an enzymatically active RNR complex in vitro together with the cellular R2 subunit. Recombinant His-tagged M45 protein was purified by chromatography on a Ni-agarose column followed by chromatography on a Superdex 200 column. A 150- to 160-kDa protein band was clearly seen in the Ni-agarose column eluate, and its identity as M45 was confirmed in Western blots by using the M45 antiserum (data not shown). However, many other, smaller polypeptides were present in the eluate where the major bands react with the antibody; these most probably represent proteolytically degraded M45. On the Superdex column, the material eluting as a symmetrical peak with the void volume contains about 50% full-length M45 and 50% degraded protein (Fig. 4A). Since proteins with a molecular mass of around 600 kDa elute in this position, M45 is most probably eluting as an oligomeric complex.
FIG. 4.
RNR assays. (A) SDS-PAGE analyses of purified recombinant M45. Lane 1, molecular mass markers at 203, 120, 90, and 51 kDa; lane 2, recombinant His-tagged M45 purified by chromatography on a nickel-agarose column followed by chromatography on a Superdex 200 column. (B) Catalytic activity of recombinant M45 assayed in the presence of an excess of mouse R2 protein (10 μg) before and after the addition of a constant amount of mouse R1 protein (2 μg). The increasing amounts of purified recombinant M45 are 0, 7, 14, and 28 μg. (C) Catalytic activity of recombinant M45 assayed in the presence of an excess of mouse R2 (10 μg) together with a constant amount of mouse R1 (2 μg) or of the catalytically inactive R1 C429A protein (9 μg). The increasing amounts of purified recombinant M45 are 0, 7, and 14 μg. (D) Allosteric inhibition of ATP-stimulated CDP reduction by dATP. Catalytic activity of the mouse complex R1-R2 alone or together with recombinant M45 protein was assayed in the presence of 0, 20, 80, or 400 μM dATP.
In the RNR assay, our recombinant M45 alone or together with an excess of mouse R2 never showed any significant reduction of CDP in repeated assays and with different protein preparations. By contrast, there was a small but reproducible stimulation of the activity of mouse R1 assayed in the presence of an excess of R2 when M45 was added (Fig. 4B). This does not appear to be a general protein effect, since addition of truncated M45 protein or BSA resulted in no stimulation (data not shown). We tested the hypothesis that, in analogy with yeast Rnr3p (19), heterodimerization of R1 and M45 facilitates the recruitment of M45 to the holoenzyme. Therefore, we observed RNR activity when M45 was added to an assay system consisting of catalytically inactive mouse R1 C429A protein together with an excess of R2. Again, no CDP reduction was observed (Fig. 4C). Lastly, we determined whether addition of M45 to an assay system containing mouse R1 and R2 changed the allosteric regulation of the mouse RNR, and especially whether M45 made the activity more resistant to dATP. However, addition of M45 had no effect on dATP inhibition (Fig. 4D). This confirms an earlier observation that M45 does not bind to dATP-Sepharose, in contrast to R1. In conclusion, our recombinant M45 protein displays no RNR activity.
MCMV induces dNTP pool expansion through ribonucleotide reduction in resting cells.
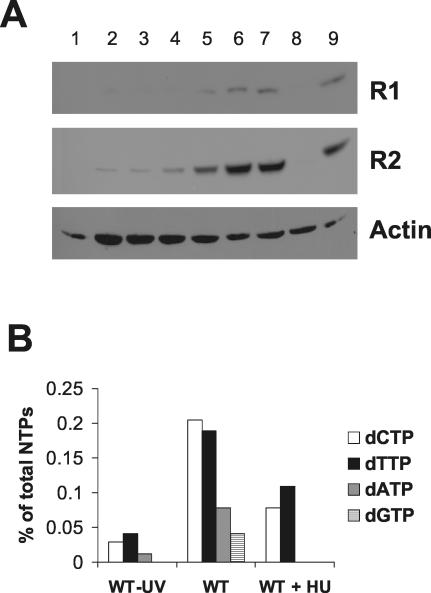
The finding that MCMV does not express a functional RNR R1 subunit prompted us to investigate whether it relies on the cellular enzyme to obtain dNTP in resting cells. To address this point, we first measured the steady-state levels of the cellular RNR subunits during the course of viral infection in serum-starved NIH 3T3 cells. Cell extracts were prepared at different times postinfection and analyzed by immunoblotting using anti-R1 monoclonal antibodies and anti-R2 polyclonal antibodies (Fig. 5A). As expected, R1 and R2 were not expressed in uninfected cells and were induced by serum stimulation. The expression of R2 was upregulated in the infected cells as early as 6 hpi, and its level increased significantly as the infection progressed. R1 was also induced by the infection, though to a lesser extent. The absence of any signal when cells were infected with UV-inactivated viruses indicates that neither potential serum contamination of viral preparations nor binding and entry of the inactivated virus particles were responsible for R1 and R2 induction.
FIG. 5.
Upregulation of cellular RNR expression and activity by MCMV. (A) Cellular R1 and R2 levels during MCMV infection. NIH 3T3 cells were growth-arrested in 0.5% calf serum and then either infected with active or UV-irradiated MCMV (MOI, 5 PFU/cell) or mock-infected. Whole-cell extracts were prepared at various times after infection, separated by SDS-PAGE, and analyzed by immunoblotting with the anti-R1, anti-R2, and anti-actin antibodies. A sample from quiescent cells stimulated with 10% calf serum for 24 h was also included. Lanes: 1, mock infection; 2, 6 hpi; 3, 12 hpi; 4, 24 hpi; 5, 36 hpi; 6, 48 hpi; 7, 60 hpi; 8, UV-inactivated MCMV 48 hpi; 9, noninfected cells grown in the presence of 10% serum. (B) Effect of MCMV infection on dNTP pool sizes in resting cells. NIH 3T3 cells were growth arrested in 0.5% calf serum and then infected with active or UV-inactivated MCMV (MOI, 5 PFU/cell). After virus adsorption, cells were either treated with 0.5 mM HU or left untreated. The nucleotides were extracted at 48 hpi, and their levels were measured by high-performance liquid chromatography. Levels of nucleotide pools are expressed as percentages of the total nucleoside triphosphate pool (CTP + UTP + ATP + GTP + dCTP + dTTP + dATP + dGTP) to minimize variations due to small differences in cell number in the samples.
This result led us to investigate whether the virus-induced upregulation of the cellular RNR subunits results in increased ribonucleotide reduction.
Therefore, we studied the effect of hydroxyurea (HU), a specific RNR inhibitor, on the dNTP pools of quiescent MCMV-infected cells at 48 hpi. As in an earlier study (45), we observed that all four dNTP pools expanded after MCMV infection compared to a sample from cells infected with a UV-inactivated virus (Fig. 5B). When the pool measurements were performed on infected cells grown in the presence of 0.5 mM HU, a significant drop in the level of the pyrimidine pools was observed, while the purines totally disappeared, indicating that MCMV obtains dNTP through ribonucleotide reduction. We also measured the virus yield in the supernatants from the cell cultures used for the dNTP pool assay. The yield in untreated cells was 2.3 × 104 PFU/ml, whereas no infectious virus was found in HU-treated cells, in accordance with our previous observations (45). Taken together, these findings demonstrate that MCMV replication in resting cells is dependent on host ribonucleotide reduction induced by the virus itself.
In vivo growth of M45 MCMV mutants.
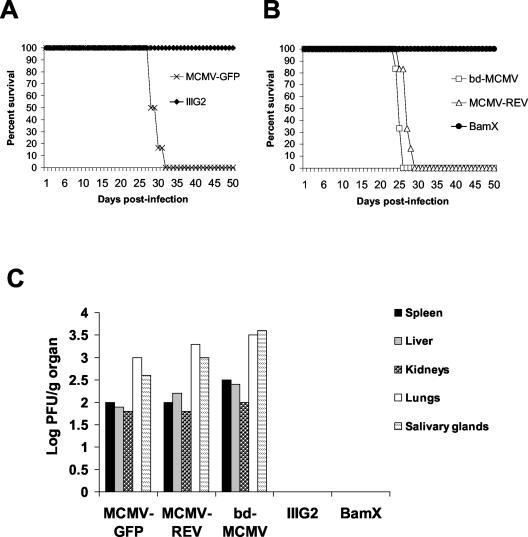
To determine whether M45 is required for viral replication in its natural host, the growth of the M45-mutant viruses IIIG2 and BamX was compared with that of a wild-type-like MCMV expressing GFP (MCMV-GFP), a BAC-derived wild-type MCMV (bd-MCMV), and a revertant MCMV in immunodeficient CB17 SCID mice, which lack functional T and B lymphocytes. SCID mice are sensitive to extremely low levels of MCMV replication and succumb within 4 weeks postinfection. Infection with the wild-type and revertant viruses resulted in 100% mortality within 5 weeks postinoculation. In contrast, infection with the same dose of IIIG2 or BamX produced no mortality. Figure 6A shows results for mice infected with one of the GFP-expressing viruses MCMV-GFP and IIIG2, whereas Fig. 6B shows results for mice infected with one of the viruses without GFP, i.e., bd-MCMV, MCMV-REV, and BamX. Because the M45 mutants did not cause lethal infection over the period of observation (up to 60 days), we determined virus titers in target organs by plaque assay 24 days after infection, which corresponds roughly to the time of death for the bd-MCMV-infected mice. As shown in Fig. 6C, infection with wild-type and revertant viruses yielded virus titers in the spleen, liver, kidneys, lungs, and salivary glands, indicating efficient viral spread and replication. In contrast, titers of IIIG2 and BamX were undetectable in all target organs. Hence, M45 mutant viruses are defective with respect to replication in their natural host.
FIG. 6.
(A) Survival curves of SCID mice infected with a wild-type virus (MCMV-GFP) or with an M45 mutant virus (IIIG2) expressing GFP. (B) Survival curves of SCID mice infected with wild-type (bd-MCMV), revertant (MCMV-REV), or M45 mutant (BamX) viruses without GFP. SCID mice were injected intraperitoneally with 106 PFU of virus and were monitored daily for survival. (C) Accumulation of infectious viruses in the spleen, liver, kidneys, lungs, and salivary glands. Two SCID mice for each group were infected as described above. The animals were killed 24 days after infection, and total levels of virus in homogenates of target organs were determined by plaque assays on NIH 3T3 cells. Each data point is the average for two animals.
DISCUSSION
This work addresses the long-standing question of whether the RNR R1 homolog encoded by CMV is a functional enzyme subunit. We carried out our study on the MCMV M45 protein because we were also interested in assessing its role in viral pathogenesis in vivo. The M45 reading frame has a length of 3,522 bp and encodes a 1,174-amino-acid protein with an expected molecular mass of 125 kDa. Since no data on gene expression from the M45 gene were available, we generated a specific polyclonal antiserum to study its product. M45 is detected as a polypeptide of 150 to 160 kDa in lysates from MCMV-infected cells and cells transfected with an M45 expression plasmid. The same result was obtained when recombinant M45 expressed in E. coli was analyzed. The discrepancy between the molecular mass estimated by SDS-PAGE and that expected from the sequence is most probably caused by an anomalous behavior of M45 in SDS-PAGE and not by posttranslational modifications, since these do not occur in E. coli. Immunofluorescence and Western blot analysis revealed that M45 is a cytoplasmic protein abundantly expressed at early and late times in infection irrespective of the onset of viral DNA synthesis. Moreover, we found that it is associated with purified viral particles, such as the R1 subunit of HSV-2 (61).
The idea that CMV encodes an RNR R1 subunit is based on the following arguments. Blast searches of the SwissProt protein database (http://www.ncbi.nlm.nih.gov:80/BLAST/) performed with the M45 amino acid sequence identify the C-terminal region of M45 as a homolog of a class Ia RNR R1 subunit. The closest matches occur with other herpesvirus R1 subunits (e.g., 27% amino acid identity to the HCMV R1 subunit, 24% to the human herpesvirus 6 [HHV-6] R1 subunit, and 22% to the HSV-1 and HSV-2 R1 subunits). Interestingly, like the HSV-1 and HSV-2 subunits, M45 possesses an N-terminal extension that makes it the longest herpesvirus R1 subunit. Moreover, an in silico structural and functional analysis of the HCMV genome yielded complete structural identification of the UL45 protein, a homolog of M45, as a RNR R1 subunit (54). Finally, both the UL45 and the M45 gene are positionally conserved with the gene for the RNR R1 subunit in other herpesviruses. However, the CMV genomes lack an apparent homolog of the RNR R2 subunit, which is positionally replaced by a gene encoding the processivity factor of the viral DNA polymerase (UL44 for HCMV and M44 for MCMV). Since M45 exhibits sequence features of a class Ia R1 subunit, it is expected to complex to an R2 subunit to form an active enzyme; it is highly improbable that it forms a single-subunit RNR like the class II enzymes expressed by Eubacteria and Archaebacteria. Therefore, we raised the hypothesis that it might usurp the place of the cellular R1 to form a hybrid version of the enzyme together with the cellular R2 subunit, whose expression is upregulated by MCMV infection (45; this paper). However, the results obtained when recombinant M45 was tested in the RNR assays indicate that it is not a functional equivalent of a class Ia R1 subunit, since it has no activity together with R2. Moreover, it has no activity alone, excluding the remote possibility that it behaves like a homomeric RNR. Similar results were obtained when native M45 was immunopurified from MCMV-infected cells and assayed for activity (data not shown). This finding rules out the possibility that M45 requires some kind of modifications carried out by the infected cells for its activity and that the His tag in the recombinant protein could affect its function. It is noteworthy that another betaherpesvirus R1 homolog, the U28 protein encoded by HHV-7, has no activity in an RNR assay (65).
The small but reproducible stimulation of the enzyme activity observed when M45 was added to an assay mixture containing R1 and R2 was reminiscent of the cross talk between the two R1 subunits encoded by Saccharomyces cerevisiae called Rnr1 and Rnr3. It has been proposed that heterodimerization of Rnr3 with Rnr1 facilitates the recruitment of Rnr3 to the holoenzyme and results in a synergism that is more evident when Rnr3 is allowed to form a complex with a catalytically inactive form of Rnr1 (19). However, when we assessed whether this model applies to M45, we found no M45 activity in an assay containing a catalytically inactive form of R1 and an excess of R2, indicating that M45 does not behave like the yeast Rnr3 subunit.
Unlike the cellular enzymes, the herpesvirus RNRs completely lack allosteric regulation as well as most of the residues involved in effector binding in the E. coli and mammalian enzymes at both the activity and specificity sites (12, 42), indicating that the function is unnecessary or perhaps detrimental for viral replication. These considerations, along with the observation that M45 does not bind to the inhibitory effector dATP, led us to test the hypothesis that M45 may functionally interact with the cellular enzyme by affecting its allosteric control. However, our results demonstrated that M45 does not make the activity of the cellular enzyme resistant to dATP inhibition.
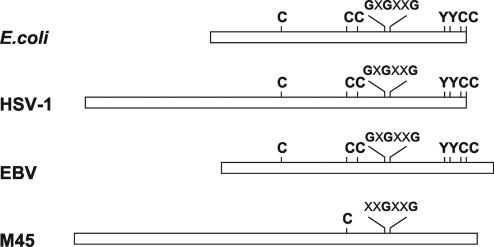
Finally, along with the lack of a functional interaction with host RNR, we could not find any physical interactions between M45 and cellular R1 and R2 in the immunoprecipitates from infected cell extracts (data not shown). Taken together, the results from the enzymatic assays indicate that during viral coevolution with the host, M45 has lost its direct involvement in ribonucleotide reduction and has become catalytically inactive. Such loss of function is further highlighted by the amino acid sequence comparison between M45 and the R1 subunit of E. coli, which represents the prototype of class Ia, and the R1 proteins of HSV-1 and Epstein-Barr virus (EBV), chosen as representatives of alpha- and gammaherpesviruses. As depicted in Fig. 7, sequence alignment analysis with the ClustalW program (http://www.ebi.ac.uk/clustalw/index.html) revealed that the residues shown to have a catalytic role in the E. coli R1 subunit (38) are highly conserved in the HSV-1 and EBV R1 proteins. These include E. coli cysteines 225, 439, 462, 754, and 759 and tyrosines 730 and 731. By contrast, only cysteine 814 (corresponding to cysteine 439 in E. coli) is conserved in the M45 sequence. Moreover, the proposed GxGxxG nucleotide-binding site (E. coli residues 514 to 519) is only partially conserved in the M45 sequence (xxGxxG at residues 890 to 895). When we performed sequence alignment analysis with the R1 proteins of all the betaherpesviruses for which sequencing information is available, we found that the lack of important catalytic residues is a general feature of the subfamily. These findings are consistent with the view that betaherpesviruses differ from the other two subfamilies in the strategy they have evolved to satisfy their need for DNA precursors. The alpha- and gammaherpesvirus genomes encode a functional RNR, thymidine kinase (TK), and dUTPase, which are essential enzymes for nucleotide anabolism and presumably are expressed to provide dNTP for viral DNA synthesis in resting cells where the cellular enzymes are not expressed (58). Although we cannot exclude the possibility that another viral protein is required for M45 RNR activity, betaherpesviruses seem to have abandoned the strategy of supplying enzymes involved in the biosynthesis of DNA precursors, since the genes for a TK and the RNR R2 subunit are absent and the genes for the dUTPase and the RNR R1 subunit are mutated and encode catalytically inactive proteins (48, 49, 65; this work). It was previously reported that CMV has evolved the ability to induce the expression of the cellular enzymes involved in the biosynthesis of thymidylate, such as folylpolyglutamate synthase (9), dihydrofolate reductase (44, 46), thymidylate synthase (26, 28), and dCMP deaminase (27), in order to replicate in resting cells. Consistent with the view that CMV relies on cellular enzymes for dNTP biosynthesis is the finding that both MCMV (this paper) and HCMV (62) induce expression of the R1 and R2 subunits of the cellular RNR. Moreover, we have demonstrated that increased expression of these subunits results in expansion of all four dNTP pools in resting fibroblasts. The finding that treatment of infected cells with HU leads to a specific drop in both purine and pyrimidine pools confirmed that virus-induced pool expansion is a consequence of ribonucleotide reduction and not of the triggering of a salvage pathway. Importantly, inhibition of MCMV replication by HU revealed that the virus-induced host RNR activity is essential for a productive infection in resting cells. We propose that emergence of this peculiar replicative strategy during the course of CMV evolution has allowed many genes involved in nucleotide metabolism to be selectively eliminated from the viral genome or to mutate and gain new functions.
FIG. 7.
Line diagram representing the amino acid sequence alignment of M45 with the R1 proteins of E. coli, HSV-1, and EBV. The proposed nucleotide-binding site (GxGxxG) and the five cysteines and two tyrosines that have a direct catalytic role, as discussed in the text, are highly conserved in the E. coli, HSV-1, and EBV R1 proteins. By contrast, most of these residues are absent in the M45 sequence.
Several considerations suggest that M45 is not a vestigial gene encoding a useless function. First, since its reading frame has remained open, it can be presumed to encode a functional protein. Second, its abundant expression at late times and its association with the viral particle may point to its involvement in virus maturation or a function immediately after virus entry, when it is delivered to the cytoplasm of newly infected cells. This question is the subject of current investigation. Third, previous results have shown that MCMV mutants carrying an inactivated M45 gene do not replicate in two different endothelial cell lines and have a greatly reduced ability to grow on macrophages, whereas they grow almost normally on fibroblasts (6). M45 was shown to protect cells from premature apoptosis induced by the virus (6, 7). This antiapoptotic activity is seen in different cell types but is most prominent in endothelial cells, where it is required for virus replication and spread. Apparently, endothelial cells are particularly sensitive to virus-induced apoptosis. Surprisingly, the HCMV UL45 protein seems to be dispensable for HCMV growth in human umbilical endothelial cells (29). Because macrophages and endothelial cells play key roles in CMV dissemination (4, 13, 36, 55, 59, 64), viral gene products which regulate growth in these cell types should significantly affect MCMV pathogenesis in vivo. This has, for instance, been shown for MCMV mutants with deletions in the m139-to-m141 region, which are growth deficient in macrophages and attenuated in mice (30, 31). To directly assess the role of M45 in viral pathogenesis, we investigated the virulence of two M45 mutant viruses (a transposon insertion mutant and a frameshift mutant) in SCID mice. We found that these viruses are avirulent in SCID mice, since they do not kill the animals, and their replication is undetectable in target organs. This failure to replicate in SCID mice is highly significant, as these animals are extremely susceptible to MCMV infection, and warrants further investigation to define the mechanism underlying this highly attenuated phenotype. In summary, the arguments outlined above support the view that M45 has abandoned its function as a RNR subunit and acquired a new function, probably completely unrelated to RNR activity, that is essential for cellular tropism and viral pathogenesis in vivo. Protecting cells from virus-induced apoptosis is clearly a new function of M45, and it makes sense that MCMV incorporates such a protein into the virion for immediate availability in the infected cell. Since herpesvirus proteins are often multifunctional, it is possible that M45 has acquired an additional function, which adds to the observed phenotype. Further studies are required to assess whether this loss and gain of function apply to other R1 homologs of the Betaherpesvirinae subfamily, and to understand their newly acquired functions.
Acknowledgments
We thank Margareta Thelander for excellent technical assistance and Peter Reichard for manuscript revision.
This work was supported by grants from the Associazione Italiana per la Ricerca sul Cancro, from Ricerca Sanitaria Finalizzata (Regione Piemonte), and from Program 40% (MIUR). U.K. and W.B. were supported by the Deutsche Forschungsgemeinschaft (SFB 455 and SFB 479, respectively).
Footnotes
This work is dedicated to the memory of Giorgio Cavallo.
REFERENCES
- 1.Angulo, A., P. Ghazal, and M. Messerle. 2000. The major immediate-early gene ie3 of mouse cytomegalovirus is essential for viral growth. J. Virol. 74:11129-11136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Averett, D. R., C. Lubbers, G. B. Elion, and T. Spector. 1983. Ribonucleotide reductase induced by herpes simplex type 1 virus. Characterization of a distinct enzyme. J. Biol. Chem. 258:9831-9838. [PubMed] [Google Scholar]
- 3.Bjorklund, S., S. Skog, B. Tribukait, and L. Thelander. 1990. S-phase-specific expression of mammalian ribonucleotide reductase R1 and R2 subunit mRNAs. Biochemistry 29:5452-5458. [DOI] [PubMed] [Google Scholar]
- 4.Brautigam, A. R., F. J. Dutko, L. B. Olding, and M. B. Oldstone. 1979. Pathogenesis of murine cytomegalovirus infection: the macrophage as a permissive cell for cytomegalovirus infection, replication and latency. J. Gen. Virol. 44:349-359. [DOI] [PubMed] [Google Scholar]
- 5.Bresnahan, W. A., I. Boldogh, E. A. Thompson, and T. Albrecht. 1996. Human cytomegalovirus inhibits cellular DNA synthesis and arrests productively infected cells in late G1. Virology 224:150-160. [DOI] [PubMed] [Google Scholar]
- 6.Brune, W., C. Menard, J. Heesemann, and U. H. Koszinowski. 2001. A ribonucleotide reductase homolog of cytomegalovirus and endothelial cell tropism. Science 291:303-305. [DOI] [PubMed] [Google Scholar]
- 6a.Brune, W., C. Menard, J. Heesemann, and U. H. Koszinowski. 12 January 2001, posting date. A ribonucleotide reductase homolog of cytomegalovirus and endothelial cell tropism. Supplementary material. Science 291:303-305. [Online.] http://www.sciencemag.org/cgi/content/full/291/5502/303/DC1. [DOI] [PubMed]
- 7.Brune, W., M. Nevels, and T. Shenk. 2003. Murine cytomegalovirus m41 open reading frame encodes a Golgi-localized antiapoptotic protein. J. Virol. 77:11633-11643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cameron, J. M., I. McDougall, H. S. Marsden, V. G. Preston, D. M. Ryan, and J. H. Subak-Sharpe. 1988. Ribonucleotide reductase encoded by herpes simplex virus is a determinant of the pathogenicity of the virus in mice and a valid antiviral target. J. Gen. Virol. 69:2607-2612. [DOI] [PubMed] [Google Scholar]
- 9.Cavallo, R., D. Lembo, G. Gribaudo, and S. Landolfo. 2001. Murine cytomegalovirus infection induces cellular folylpolyglutamate synthetase activity in quiescent cells. Intervirology 44:224-226. [DOI] [PubMed] [Google Scholar]
- 10.Chabes, A., and L. Thelander. 2000. Controlled protein degradation regulates ribonucleotide reductase activity in proliferating mammalian cells during the normal cell cycle and in response to DNA damage and replication blocks. J. Biol. Chem. 275:17747-17753. [DOI] [PubMed] [Google Scholar]
- 11.Chee, M. S., A. T. Bankier, S. Beck, R. Bohni, C. M. Brown, R. Cerny, T. Horsnell, C. A. Hutchison, T. Kouzarides, J. A. Martignetti, et al. 1990. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr. Top. Microbiol. Immunol. 154:125-169. [DOI] [PubMed] [Google Scholar]
- 12.Cohen, G. H. 1972. Ribonucleotide reductase activity of synchronized KB cells infected with herpes simplex virus. J. Virol. 9:408-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Collins, T. M., M. R. Quirk, and M. C. Jordan. 1994. Biphasic viremia and viral gene expression in leukocytes during acute cytomegalovirus infection of mice. J. Virol. 68:6305-6311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Conner, J., H. Marsden, and J. B. Clements. 1994. Ribonucleotide reductase of herpes viruses. Rev. Med. Virol. 4:25-34. [Google Scholar]
- 15.Davis, R., M. Thelander, G. J. Mann, G. Behravan, F. Soucy, P. Beaulieu, P. Lavallee, A. Graslund, and L. Thelander. 1994. Purification, characterization, and localization of subunit interaction area of recombinant mouse ribonucleotide reductase R1 subunit. J. Biol. Chem. 269:23171-23176. [PubMed] [Google Scholar]
- 16.Davison, A. J., and J. E. Scott. 1986. The complete DNA sequence of varicella-zoster virus. J. Gen. Virol. 67:1759-1816. [DOI] [PubMed] [Google Scholar]
- 17.de Wind, N., A. Berns, A. Gielkens, and T. Kimman. 1993. Ribonucleotide reductase-deficient mutants of pseudorabies virus are avirulent for pigs and induce partial protective immunity. J. Gen. Virol. 74:351-359. [DOI] [PubMed] [Google Scholar]
- 18.Dittmer, D., and E. S. Mocarski. 1997. Human cytomegalovirus infection inhibits G1/S transition. J. Virol. 71:1629-1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Domkin, V., L. Thelander, and A. Chabes. 2002. Yeast DNA damage-inducible Rnr3 has a very low catalytic activity strongly stimulated after the formation of a cross-talking Rnr1/Rnr3 complex. J. Biol. Chem. 277:18574-18578. [DOI] [PubMed] [Google Scholar]
- 20.Engstrom, Y., S. Eriksson, I. Jildevik, S. Skog, L. Thelander, and B. Tribukait. 1985. Cell cycle-dependent expression of mammalian ribonucleotide reductase. Differential regulation of the two subunits. J. Biol. Chem. 260:9114-9116. [PubMed] [Google Scholar]
- 21.Engstrom, Y., S. Eriksson, L. Thelander, and M. Akerman. 1979. Ribonucleotide reductase from calf thymus. Purification and properties. Biochemistry 18:2941-2948. [DOI] [PubMed] [Google Scholar]
- 22.Eriksson, S., A. Graslund, S. Skog, L. Thelander, and B. Tribukait. 1984. Cell cycle-dependent regulation of mammalian ribonucleotide reductase. The S phase-correlated increase in subunit M2 is regulated by de novo protein synthesis. J. Biol. Chem. 259:11695-11700. [PubMed] [Google Scholar]
- 23.Fortunato, E. A., A. K. McElroy, I. Sanchez, and D. H. Spector. 2000. Exploitation of cellular signaling and regulatory pathways by human cytomegalovirus. Trends Microbiol. 8:111-119. [DOI] [PubMed] [Google Scholar]
- 24.Goldstein, D. J., and S. K. Weller. 1988. Factor(s) present in herpes simplex virus type 1-infected cells can compensate for the loss of the large subunit of the viral ribonucleotide reductase: characterization of an ICP6 deletion mutant. Virology 166:41-51. [DOI] [PubMed] [Google Scholar]
- 25.Goldstein, D. J., and S. K. Weller. 1988. Herpes simplex virus type 1-induced ribonucleotide reductase activity is dispensable for virus growth and DNA synthesis: isolation and characterization of an ICP6 lacZ insertion mutant. J. Virol. 62:196-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gribaudo, G., L. Riera, D. Lembo, M. De Andrea, M. Gariglio, T. L. Rudge, L. F. Johnson, and S. Landolfo. 2000. Murine cytomegalovirus stimulates cellular thymidylate synthase gene expression in quiescent cells and requires the enzyme for replication. J. Virol. 74:4979-4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gribaudo, G., L. Riera, P. Caposio, F. Maley, and S. Landolfo. 2003. Human cytomegalovirus requires cellular deoxycytidylate deaminase for replication in quiescent cells. J. Gen. Virol. 84:1437-1441. [DOI] [PubMed] [Google Scholar]
- 28.Gribaudo, G., L. Riera, T. L. Rudge, P. Caposio, L. F. Johnson, and S. Landolfo. 2002. Human cytomegalovirus infection induces cellular thymidylate synthase gene expression in quiescent fibroblasts. J. Gen. Virol. 83:2983-2993. [DOI] [PubMed] [Google Scholar]
- 29.Hahn, G., H. Khan, F. Baldanti, U. H. Koszinowski, M. G. Revello, and G. Gerna. 2002. The human cytomegalovirus ribonucleotide reductase homolog UL45 is dispensable for growth in endothelial cells, as determined by a BAC-cloned clinical isolate of human cytomegalovirus with preserved wild-type characteristics. J. Virol. 76:9551-9555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hanson, L. K., J. S. Slater, Z. Karabekian, H. W. Virgin IV, C. A. Biron, M. C. Ruzek, N. van Rooijen, R. P. Ciavarra, R. M. Stenberg, and A. E. Campbell. 1999. Replication of murine cytomegalovirus in differentiated macrophages as a determinant of viral pathogenesis. J. Virol. 73:5970-5980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hanson, L. K., J. S. Slater, Z. Karabekian, G. Ciocco-Schmitt, and A. E. Campbell. 2001. Products of US22 genes M140 and M141 confer efficient replication of murine cytomegalovirus in macrophages and spleen. J. Virol. 75:6292-6302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heineman, T. C., and J. I. Cohen. 1994. Deletion of the varicella-zoster virus large subunit of ribonucleotide reductase impairs growth of virus in vitro. J. Virol. 68:3317-3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Henry, B. E., R. Glaser, J. Hewetson, and D. J. O'Callaghan. 1978. Expression of altered ribonucleotide reductase activity associated with the replication of the Epstein-Barr virus. Virology 89:262-271. [DOI] [PubMed] [Google Scholar]
- 34.Ho, M. 1991. Cytomegalovirus: biology and infection, 2nd ed. Plenum Publishing Corp., New York, N.Y.
- 35.Hofer, A., J. T. Ekanem, and L. Thelander. 1998. Allosteric regulation of Trypanosoma brucei ribonucleotide reductase studied in vitro and in vivo. J. Biol. Chem. 273:34098-34104. [DOI] [PubMed] [Google Scholar]
- 36.Ibanez, C. E., R. Schrier, P. Ghazal, C. Wiley, and J. A. Nelson. 1991. Human cytomegalovirus productively infects primary differentiated macrophages. J. Virol. 65:6581-6588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jacobson, J. G., D. A. Leib, D. J. Goldstein, C. L. Bogard, P. A. Schaffer, S. K. Weller, and D. M. Coen. 1989. A herpes simplex virus ribonucleotide reductase deletion mutant is defective for productive acute and reactivatable latent infections of mice and for replication in mouse cells. Virology 173:276-283. [DOI] [PubMed] [Google Scholar]
- 38.Jordan, A., and P. Reichard. 1998. Ribonucleotide reductases. Annu. Rev. Biochem. 67:71-98. [DOI] [PubMed] [Google Scholar]
- 39.Kalejta, R. F., and T. Shenk. 2002. Manipulation of the cell cycle by human cytomegalovirus. Front. Biosci. 7:d295-d306. [DOI] [PubMed] [Google Scholar]
- 40.Koszinowski, U. H., M. Del Val, and M. J. Reddehase. 1990. Cellular and molecular basis of the protective immune response to cytomegalovirus infection. Curr. Top. Microbiol. Immunol. 154:189-220. [DOI] [PubMed] [Google Scholar]
- 41.Landolfo, S., M. Gariglio, G. Gribaudo, and D. Lembo. 2003. The human cytomegalovirus. Pharmacol. Ther. 98:269-297. [DOI] [PubMed] [Google Scholar]
- 42.Langelier, Y., and G. Buttin. 1981. Characterization of ribonucleotide reductase induction in BHK-21/C13 Syrian hamster cell line upon infection by herpes simplex virus (HSV). J. Gen. Virol. 57:21-31. [DOI] [PubMed] [Google Scholar]
- 43.Lankinen, H., A. Graslund, and L. Thelander. 1982. Induction of a new ribonucleotide reductase after infection of mouse L cells with pseudorabies virus. J. Virol. 41:893-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lembo, D., A. Angeretti, M. Gariglio, and S. Landolfo. 1998. Murine cytomegalovirus induces expression and enzyme activity of dihydrofolate reductase in quiescent cells. J. Gen. Virol. 78:2803-2808. [DOI] [PubMed] [Google Scholar]
- 45.Lembo, D., G. Gribaudo, A. Hofer, L. Riera, M. Cornaglia, A. Mondo, A. Angeretti, M. Gariglio, L. Thelander, and S. Landolfo. 2000. Expression of an altered ribonucleotide reductase activity associated with the replication of murine cytomegalovirus in quiescent fibroblasts. J. Virol. 74:11557-11565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lembo, D., G. Gribaudo, R. Cavallo, L. Riera, A. Angeretti, L. Hertel, and S. Landolfo. 1999. Human cytomegalovirus stimulates cellular dihydrofolate reductase activity in quiescent cells. Intervirology 42:30-36. [DOI] [PubMed] [Google Scholar]
- 47.Lu, M., and T. Shenk. 1996. Human cytomegalovirus infection inhibits cell cycle progression at multiple points, including the transition from G1 to S. J. Virol. 70:8850-8857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McGeehan, J. E., N. W. Depledge, and D. J. McGeoch. 2001. Evolution of the dUTPase gene of mammalian and avian herpesviruses. Curr. Protein Peptide Sci. 2:325-333. [DOI] [PubMed] [Google Scholar]
- 49.McGeoch, D. J., and A. J. Davison. 1999. The molecular evolutionary history of herpesviruses, p. 441-465. In E. Domingo, R. Webster, and J. Holland (ed.), Origin and evolution of viruses. Academic Press, London, United Kingdom.
- 50.Mocarski, E. S. 1993. Cytomegalovirus biology and replication, p. 173-226. In B. Roizman, R. Whitley, and C. Lopez (ed.), The human herpesviruses. Raven Press, New York, N.Y.
- 51.Mocarski, E. S., and C. T. Courcelle. 2001. Cytomegaloviruses and their replication, p. 2629-2673. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology. Lippincott Williams and Wilkins, Philadelphia, Pa.
- 52.Mocarski, E. S., G. B. Abenes, W. C. Manning, L. C. Sambucetti, and J. M. Cherrington. 1990. Molecular genetic analysis of cytomegalovirus gene regulation in growth, persistence and latency. Curr. Top. Microbiol. Immunol. 154:47-74. [DOI] [PubMed] [Google Scholar]
- 53.Noris, E., C. Zannetti, A. Demurtas, J. Sinclair, M. De Andrea, M. Gariglio, and S. Landolfo. 2002. Cell cycle arrest by human cytomegalovirus 86-kDa IE2 protein resembles premature senescence. J. Virol. 76:12135-12148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Novotny, J., I. Rigoutsos, D. Coleman, and T. Shenk. 2001. In silico structural and functional analysis of the human cytomegalovirus (HHV5) genome. J. Mol. Biol. 310:1151-1166. [DOI] [PubMed] [Google Scholar]
- 55.Percivalle, E., M. G. Revello, L. Vago, F. Morini, and G. Gerna. 1993. Circulating endothelial giant cells permissive for human cytomegalovirus (HCMV) are detected in disseminated HCMV infections with organ involvement. J. Clin. Investig. 92:663-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rawlinson, W. D., H. E. Farrell, and B. G. Barrell. 1996. Analysis of the complete DNA sequence of murine cytomegalovirus. J. Virol. 70:8833-8849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Reichard, P. 1988. Interactions between deoxyribonucleotide and DNA synthesis. Annu. Rev. Biochem. 57:349-374. [DOI] [PubMed] [Google Scholar]
- 58.Roizman, B., and D. M. Knipe. 2001. Herpes simplex viruses and their replication, p. 2399-2459. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology. Lippincott Williams and Wilkins, Philadelphia, Pa.
- 59.Saltzman, R. L., M. R. Quirk, and M. C. Jordan. 1988. Disseminated cytomegalovirus infection. Molecular analysis of virus and leukocyte interactions in viremia. J. Clin. Investig. 81:75-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Salvant, B. S., E. A. Fortunato, and D. H. Spector. 1998. Cell cycle dysregulation by human cytomegalovirus: influence of the cell cycle phase at the time of infection and effects on cyclin transcription. J. Virol. 72:3729-3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Smith, C. C., and L. Aurelian. 1997. The large subunit of herpes simplex virus type 2 ribonucleotide reductase (ICP10) is associated with the virion tegument and has PK activity. Virology 234:235-242. [DOI] [PubMed] [Google Scholar]
- 62.Song, Y. J., and M. F. Stinski. 2002. Effect of the human cytomegalovirus IE86 protein on expression of E2F-responsive genes: a DNA microarray analysis. Proc. Natl. Acad. Sci. USA 99:2836-2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Staczek, J. 1990. Animal cytomegaloviruses. Microbiol. Rev. 54:247-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stoddart, C. A., R. D. Cardin, J. M. Boname, W. C. Manning, G. B. Abene, and E. S. Mocarski. 1994. Peripheral blood mononuclear phagocytes mediate dissemination of murine cytomegalovirus. J. Virol. 68:6243-6253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sun, Y., and J. Conner. 1999. The U28 ORF of human herpesvirus-7 does not encode a functional ribonucleotide reductase R1 subunit. J. Gen. Virol. 80:2713-2718. [DOI] [PubMed] [Google Scholar]
- 66.Thelander, L., and P. Reichard. 1979. Reduction of ribonucleotides. Annu. Rev. Biochem. 48:133-158. [DOI] [PubMed] [Google Scholar]
- 67.Wagner, M., S. Jonjic, U. H. Koszinowski, and M. Messerle. 1999. Systematic excision of vector sequences from the BAC-cloned herpesvirus genome during virus reconstitution. J. Virol. 73:7056-7060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wiebusch, L., and C. Hagemeier. 1999. Human cytomegalovirus 86-kilodalton IE2 protein blocks cell cycle progression in G1. J. Virol. 73:9274-9283. [DOI] [PMC free article] [PubMed] [Google Scholar]