Abstract
Contemporary views of tumorigenesis regard its inception as a convergence of genetic mutation and developmental context. Glioma is the most common and deadly malignancy in the CNS; therefore, understanding how regulators of glial development contribute to its formation remains a key question. Previously we identified nuclear factor I-A (NFIA) as a key regulator of developmental gliogenesis, while miR-223 has been shown to repress NFIA expression in other systems. Using this relationship as a starting point, we found that miR-223 can suppress glial precursor proliferation via repression of NFIA during chick spinal cord development. This relationship is conserved in glioma, as miR-223 and NFIA expression is negatively correlated in human glioma tumors, and the miR-223/NFIA axis suppresses tumorigenesis in a human glioma cell line. Subsequent analysis of NFIA function revealed that it directly represses p21 and is required for tumorigenesis in a mouse neural stem cell model of glioma. These studies represent the first characterization of miR-223/NFIA axis function in glioma and demonstrate that it is a conserved proliferative mechanism across CNS development and tumorigenesis.
Introduction
The majority of CNS malignancies are glial in composition and origin, with glioma representing the most prevalent and deadly CNS tumor (Maher et al., 2001). A cardinal feature of glioma is a heterogeneous tumor mass that is largely comprised of pathological glial cells and their precursors, making the molecular and cellular processes that regulate gliogenesis potential therapeutic targets (Maher et al., 2001; Stiles and Rowitch, 2008). Indeed, the neural stem cell (NSC)/glial differentiative axis has previously been linked to glioma formation, with key transcriptional regulators of developmental gliogenesis having been implicated in tumorigenesis, including Olig2 and STAT3 (Ligon et al., 2007; de la Iglesia et al., 2008). While these studies indicate common transcriptional requirements for glial development and glioma tumorigenesis, how developmental gliogenesis contributes to tumorigenesis remains a poorly understood process that has important implications for the treatment of this largely fatal disease.
Previous studies identified nuclear factor I-A (NFIA) as a key transcription factor in the specification of glial identity and differentiation of astrocytes during spinal cord development (das Neves et al., 1999; Shu et al., 2003; Deneen et al., 2006; Namihira et al., 2009). Mechanistic studies into NFIA function during development indicate that it collaborates with Sox9 to regulate genetic programs associated with glial precursor metabolism and migration, two key developmental processes implicated in tumorigenesis (Kang et al., 2012). Previously, we also found that NFIA is overexpressed in human astrocytomas and glioblastoma multiforme (GBM; Song et al., 2010). Despite this knowledge and the extensive characterization of its function during glial development, its role in glioma tumorigenesis remains uncharacterized.
MicroRNAs (miRNAs) have emerged as critical regulatory elements that control gene expression in development and tumorigenesis (Dalmay and Edwards, 2006). In the hematopoietic system, miR-223 plays a key role in both development and tumorigenesis. In myeloid progenitors and myeloid leukemia, miR-223 suppresses cell proliferation; however, reduced expression is associated with enhanced survival, implying a more nuanced role for miR-223 in the myeloid lineage (Fazi et al., 2007; Stamatopoulos et al., 2009; Sun et al., 2010). In GBM, in silico analysis and bioinformatics has linked miR-223 expression to the proneural subtype, though its expression or function in glioma has not been validated in human tumors or models (Genovese et al., 2012).
Studies in the hematopoietic system indicate that miR-223 regulates expression of NFIA, though this relationship has not been characterized in the developing CNS and glioma tumorigenesis (Fazi et al., 2005). Given that both NFIA and miR-223 appear to be expressed in GBM, we reasoned an analogous regulatory relationship that exists during hematopoiesis also exists in the developing CNS and glioma tumorigenesis. Here we found that miR-223 suppresses glial precursor proliferation via repression of NFIA in the developing CNS. This relationship is conserved in human glioma, as miR-223 and NFIA expression is negatively correlated in human GBM, and the miR-223/NFIA axis suppresses glioma tumorigenesis. These studies represent the first characterization of miR-223/NFIA axis function in the CNS and reveal that it is a conserved proliferative mechanism across CNS development and tumorigenesis.
Materials and Methods
Chick experiments.
Expression constructs were cloned into the RCAS(B) (Morgan and Fekete, 1996), pCIG, or pEZX vectors and injected into the chick spinal cord at stage HH13–15 [approximately embryonic day 2 (E2)]. Harvested embryos were fixed in 4% PFA for 2–4 h, depending on stage. Electroporation was performed with a BTX Electro Square Porator (Momose et al., 1999). In situ hybridization (ISH) and immunocytochemistry (ICC) analysis was performed as described previously (Kang et al., 2012).
Antibodies.
Antibodies used for immunohistochemistry (IHC), chromatin immunoprecipitation (ChIP), and Western blotting in these studies were α-β-tubulin (Covance), α-BrdU (Abcam), α-caspase (R & D Systems), α-epidermal growth factor receptor (α-EGFR; DAKO), α-Gapdh (Millipore Bioscience Research Reagents), α-Ki67 (DAKO; Novocastra), α-Nestin (BD Biosciences), α-NeuN (Millipore), α-NFIA (Abcam), and α-SV40 (Santa Cruz Biotechnology).
Rat cortical culture.
Rat embryonic E14.5 cortices were dissected, dissociated, then plated at a density of 2 × 104 cells/cm2 on plates coated with 15 μg/ml polyornithine (Sigma) and 1 μg/ml human fibronectin (Biochemical Technologies). Cells were grown in DMEM/F-12 supplemented with N2 and B27 (Invitrogen) and 20 ng/ml bFGF (R & D Systems), as described by Gabay et al. (2003). Cells were expanded for 4 d followed by transfection with Lipofectamine 2000. BrdU (10 μm) was added to media and incubated for 4–6 h, followed by immunofluorescent BrdU detection.
Analysis of NFIA and miR-223 in human GBM.
Glioblastoma (GBM) and normal tissue array (US Biomax GL806a) was subjected to in situ hybridization and immunohistochemistry. GBM samples from these tissue arrays were not classified into subtypes. Exiqon miRCURY LNA-miR-223 or control probe was used according to manufacturer's directions. Immunohistochemistry for NFIA and Ki-67 was performed using DAB and ABC reagent kits (Vector Laboratories). For statistical analyses, using the NFIA and miR-223 correlation as a specific example, Log2 transformation was applied to expression data across 24 tumor samples, and the profiles for miR-223 and NFIA had Pearson correlation coefficients (PCCs) of −0.39, and Spearman's rank correlations (rho) of −0.41, both indicating significant negative correlations (p = 0.03 for PCC and p = 0.02 for Spearman's rho). With NFIA serving as observation, least squares regression gave a linear model with slope −0.905 and an R2 value of 0.155. The quadratic function fitted by least squares regression showed an improved R2 value (0.188) and clear trends of negative correlation between log2 scale profiles of miR-223 and NFIA. This same analysis was performed in the correlation of miR-223/Ki-67 and NFIA/Ki67. All above analysis were done using the statistics toolbox of Matlab 7.1 (R14).
Generation of cell lines and in vitro proliferation assays.
Lentiviruses containing miR-223 or scrambled control were generated, and the human U87 glioma cell line was infected. Shortly after infection, cell lines were subjected to puromycin selection to generate stably expressing populations. For NFIA rescue experiments, the U87-miR-223 cell line was infected with a lentivirus containing NFIA and dsRed, and was enriched using FACS analysis. Once generated, cell lines were stored or used for up to six passages. Thereafter, new cells were thawed and resubjected to puromycin selection for subsequent experiments. For proliferation assays, cells were plated and counted every day over the course of 6 d. For the cell cycle analysis, U87 cell lines were harvested and fixed with 70% ethanol for 2 h. After several 1× PBS washes, cells were resuspended in a propidium iodide/Triton X-100 staining solution with RNaseA for 30 min at 37°C. Cells were collected and resuspended in FACS buffer for analyses on FACS Aria. Cell cycle distributions were analyzed using FlowJO software. For soft agar assays, 5 × 104 cells were plated in 10% FBS/Iscove's media mixed with 0.7% noble agar, and growth was monitored over the course of 2 weeks, as described by Deneen et al. (2003).
Tumor studies.
Mouse injections were performed as described previously, using female severe combined immune deficiency (SCID) mice (Ahmed et al., 2010). Mouse brains were injected with 2.5 × 105 cells of the respective U87 cell lines that also coexpress luciferase. Tumor growth was monitored over 2 weeks by bioluminescent imaging of the luciferase. After 2 weeks mice were killed for analyses.
Mice were injected with BrdU (200 μg/g body weight), and brains were harvested 16 h later. Brains were fixed overnight in 4% paraformaldehyde and sectioned for immunofluorescence or hemotoxylin and eosin staining.
For tumor-sphere experiments, neurospheres were infected with retroviruses containing SV40 LargeT and EGFR-vIII (Deneen and Denny, 2001; Bachoo et al., 2002). Stable cell lines were generated via puromycin selection and expression confirmed via immunohistochemistry. Subsequently, 1 × 106 cells of each cell line were injected subcutaneously into female SCID mice and visually monitored for tumor formation. Upon formation of a 1.5 cm tumor, the mice were killed and the tumor harvested. All animal studies were performed in accordance with institutional Institutional Animal Care and Use Committee policies and with prior approval.
Quantitative reverse transcriptase PCR and luciferase assays.
Total RNA was isolated from cell lines using an RNeasy mini isolation kit (Qiagen). Superscript III (Invitrogen) was used for reverse transcription of 1 μg of total RNA samples. Quantitative reverse transcriptase PCR (qRT-PCR) was performed using PerfeCta SYBR Green Fast Mix (Quanta Biosciences) and a LightCycler 480 (Roche). Primer sequences used are given in Table 1.
Table 1.
Primers used for qRT-PCR
| Primer | Sequence 5′–3′ |
|---|---|
| Ms p21-F | CGATATCCAGACATTCAGAGCCACA |
| Ms p21-R | CGAAGAGACAACGGCACACTTTGCT |
| Ms NFIA-F | TAATCCAGGGCTCTGTGTCC |
| Ms NFIA-R | TGGCTGGGACTTTCAGATTG |
| Ms cyclophilin-F | GTCTCCTTCGAGCTGTTTGC |
| Ms cyclophilin-R | GATGCCAGGACCTGTATGCT |
| hup21-F | TGGAGACTCTCAGGGTCGAAA |
| hup21-R | GGCGTTTGGAGTGGTAGAAATC |
For the luciferase assays, U87 and HEK293 cell lines cells were transfected with pGL3-reporter constructs and a pEZX or CMV-β-galactosidase vector using Superfect transfection reagent (Qiagen). The p21-luciferase and NFIA 3′UTR-luciferase reporters were described previously (Fazi et al., 2005; Ligon et al., 2007). Cells were harvested 48 h later and lysates analyzed for luciferase activity. β-galactosidase was used to normalize for transfection efficiency.
ChIP assays.
U87 cell lines were harvested and processed for ChIP assay. Harvested cells were fixed with 1% formaldehyde for 10 min. Cross-linked chromatin was then sheared by sonication and cleared by centrifugation. The samples were precleared with protein G beads and immunoprecipitated using NFIA antibody or control IgG (Santa Cruz Biotechnology). Immunoprecipitated complexes were isolated, the cross-links reversed, and proteins digested with proteinase K. The DNA was purified, and PCR was performed using region-specific primers. Primer sequences used are given in Table 2.
Table 2.
Primers used for ChIP assays
| Primer | Sequence 5′–3′ |
|---|---|
| Hu p21 219-7F | TCCTGCAGCACGCGAGGTT |
| Hu p21 219-7R | GACTTCGGCAGCTGCTCACA |
| Hu p21 4018-3732F | TCTGTGAAAACATGCCCAGCGGC |
| Hu p21 4018-3272R | GACACGGTCCCCTGTTTCAA |
Neurosphere culture.
Mouse E13.5 cortex was dissected, dissociated, and plated on ultra-low attachment plates in media containing DMEM/F-12, B27 and N12 supplements (Invitrogen), 10 ng/ml EGF, and 10 ng/ml bFGF. Neurospheres formed in 4–6 d. Neurospheres used in this study were passaged no more than three times. For BrdU incorporation assays, BrdU (10 μm) was added to the media for 4–6 h followed by immunofluorescent detection of BrdU. For differentiation experiments, neurospheres were plated on coverslips coated with 15 μg/ml of polyornithine (Sigma) and 1 μg/ml human fibronectin (Biochemical Technologies) and allowed to differentiate for 7 d. Differentiation media consisted of DMEM/F-12 and B27 and N2 supplements without growth factors.
Results
miR-223 suppress glial precursor proliferation through repression of NFIA
To examine a potential relationship between miR-223 and NFIA, we characterized the extent of their coexpression during CNS development by combining ISH for miR-223 with ICC for NFIA (see Materials and Methods). Our analysis revealed that miR-223 and NFIA demonstrate generally nonoverlapping patterns of expression in the developing cortex in both the mouse and chick (Fig. 1A–F). In germinal centers, NFIA is expressed in the ventricular zone (VZ), which is consistent with previous studies (Plachez et al., 2008), while miR-223 is expressed in the intermediate zone (IZ; Fig. 1A,D). In upper cortical regions, including the cortical plate, NFIA and miR-223 also demonstrate nonoverlapping patterns of expression (Fig. 1B–B″,E–E″). The mutually exclusive expression dynamics in the developing CNS, coupled with our observation that the miR-223 target sequence is conserved across human, mouse, and chick (Fig. 1G), suggests that miR-223 suppresses NFIA expression. To test this possibility, we performed ectopic overexpression of miR-223 in the embryonic chick spinal cord and assessed its effect on NFIA expression. Our analysis revealed that overexpression of miR-223 resulted in decreased NFIA expression at E6 (Fig. 2B), while the scrambled control had no effect (Fig. 2I). Further analysis of the miR-223 overexpression phenotypes revealed decreased expression of the glial precursor marker GLAST without a concomitant increase in neurogenesis (Fig. 2C,D). These observations suggest that miR-223 overexpression influences glial precursor proliferation. To assess cell proliferation, we analyzed proliferating cell nuclear antigen (PCNA) expression and found that it was decreased in the presence of miR-223 (Fig. 2E,V). In addition, we performed BrdU labeling on chick embryos at E6 and found decreased incorporation in the presence of miR-223,without any effect on cell death or survival. (Fig. 2F,W,X). To confirm these observations in an in vitro setting, we performed analogous experiments in E14.5 rat cortical progenitor cultures and found a 50% decrease in BrdU incorporation in the presence of miR-223, compared to the scrambled control (Fig. 2G,N,Z). Importantly, miR-223 overexpression did not alter the glial constituency of these cultures, further suggesting that it influences proliferation (Fig. 2Y). Together, our chick and glial culture studies indicate overexpression of miR-223 can suppress glial precursor proliferation during CNS development.
Figure 1.
miR-223 and NFIA expression patterns in the developing CNS. A–C, Expression of miR-223 and NFIA at E16.5 in the embryonic mouse cortex. D–F, Expression of miR-223 and NFIA at E8 in the embryonic chick cortex. A–A″ and D–D″ demonstrate expression in germinal centers of mouse and chick cortex. B–C and E–F demonstrate expression in the cortical plate (CP). C and F are NFIA antibody controls, where the miR-223 ISH was treated with only secondary antibody as a control for the specificity of NFIA staining. The lack of staining with this control demonstrates the specificity of the staining in B and E. G, Cross-species alignment of the NFIA 3′UTR demonstrating that the miR-223 target site is highly conserved across human, mouse, and chick.
Figure 2.
miR-223 can suppress glial precursor proliferation through NFIA. A–F, miR-223 overexpression in chick spinal cord. H–M, Scrambled control. O–T, mir-223+NFIA. Arrowheads in B–F denote proliferative phenotypes on the electroporated (Ep) sides of miR-223 embryos. G, N, and U show the same manipulations performed in E14.5 rat cortical culture. Quantification in V–X is derived from at least three sections from four independently electroporated chick embryos. In Y and Z, the values are derived from two independent experiments performed in triplicate. Error bars represent SEM. *p < 0.001 (scrambled vs miR-223); **p < 0.01 (in rescue studies, i.e., miR-223 vs miR-223+NFIA). Con, Control, or nonelectroporated, side of the embryo.
Given the key role of NFIA in the specification of gliogenesis, we examined whether the decrease in glial precursor proliferation by miR-223 is caused by its suppression of NFIA. To this end, we coelectroporated miR-223 and an HA-tagged version of NFIA that does not contain the 3′UTR region targeted by miR-223 (Fig. 1G). Analysis at E6 revealed that coexpression of NFIA with miR-223 rescued expression of PCNA and GLAST and restored BrdU incorporation levels to those comparable to the scrambled control, both in the chick and in vitro (Fig. 2O–Z). Collectively, our data indicate that the suppression of glial precursor proliferation by miR-223 is mediated by its repression of NFIA expression.
miR-223/NFIA axis regulates glioma cell proliferation
Given that many developmental mechanisms are reused during tumorigenesis, it stands to reason that the processes regulating glial precursor proliferation during CNS development may also contribute to the generation of glial tumors. Therefore, we next examined whether the miR-223/NFIA regulatory axis contributes to the generation of glioma. As a first step in establishing this relationship, we examined the correlation of miR-223 and NFIA expression in human GBM tissue arrays using ISH and IHC, respectively (Fig. 3A–F). GBM is the most prevalent and deadly form of glioma, and is therefore a suitable model to examine this relationship. For each tumor, the number of miR-223- or NFIA-expressing cells found in adjacent patient tumor sections was quantified. These values were correlated across 24 patient samples and subjected to Spearman's rank correlation (rho), where we found that the expression of miR-223 and NFIA is negatively correlated across this set of GBMs (Fig. 3J). Given our observation that the miR-223/NFIA axis regulates glial precursor proliferation during development, we next examined whether their expression in GBM is correlated with the proliferative marker Ki-67. Similar to the above studies, we used IHC to quantify the number of Ki-67-expressing cells in tumor array sections adjacent to those used in the above analysis. Our Spearman's rank correlation (rho) analysis found that Ki-67 is positively correlated with NFIA (Fig. 3K), while miR-223 is negatively correlated (Fig. 3L). Together, these data suggest that the expression dynamics of the miR-223/NFIA axis is linked to proliferating populations within GBM.
Figure 3.
miR-223 and NFIA expression is negatively correlated in GBM. A–C, Representative in situ hybridization analysis of miR-223 in human brain and GBM. D–F, Representative immunohistochemistry analysis of NFIA in human brain and GBM. G–L, Representative immunohistochemistry analysis of Ki-67 in human brain and GBM. J–L, Spearman's rank correlation plot; linear regression results in log2 scale indicating that miR-223 and NFIA expression is negatively correlated (J), NFIA and Ki-67 are positively correlated (K), and miR-223 and Ki-67 are negatively correlated (L; for details of statistical analysis, see Materials and Methods). Arrows in J–L denote where the representative GBM images in A–I lie on the correlation plot.
That the expression of miR-223 and NFIA in GBM is congruent with their developmental patterns of expression suggests that their functional relationship may also be conserved. Therefore, we next investigated whether the miR-223/NFIA axis also regulates glioma proliferation by overexpressing miR-223 or scrambled control in the human U87 glioma cell line. Here we infected U87 cells with lentivirus containing miR-223 or scrambled control and generated stably expressing cell lines via drug selection. Upon the generation of a stable cell line, we confirmed expression of miR-223 via qRT-PCR and found that it both repressed activity of the NFIA 3′UTR reporter and suppressed NFIA protein expression, indicating that NFIA is a direct target of miR-223 in U87 glioma cells (Fig. 4A,B). Next we examined cell proliferation and found that the miR-223 overexpressing cell line grew significantly slower than the scrambled control and demonstrated an increase in the number of cells in G1 phase of the cell cycle (Fig. 4C,D). To further assess the transforming properties of these tumor cell lines, we measured anchorage-independent growth by performing a soft agar assay. Consistent with our previous findings, the miR-223-expressing cell line demonstrated reduced growth in soft agar compared to the scrambled control (Fig. 4E–G). Together, these data indicate that miR-223 suppresses the proliferation of a glioma cell line.
Figure 4.
miR-223 suppress glioma cell line proliferation through NFIA. A, Reporter assay in U87 cell lines indicating that miR-223 represses activation of the 3′UTR of NFIA. B, Western blot analysis indicating repression of NFIA in U87 cells overexpressing miR-223. C, Growth curve of U87 cells overexpressing miR-223 or scrambled control over the course of 6 d. Analysis was performed three times, in triplicate, and the plot is representative of these studies. D, Cell cycle analysis of U87 cells overexpressing miR-223, scrambled control, and miR-223+NFIA. Studies were performed three times, in triplicate; graph is representative of these studies. E–G, Soft agar assay of the same U87 cell lines described in D. These studies were performed three times, in duplicate. Error bars in A, C, and D represent SD.
Next, we examined whether suppression of glioma cell proliferation by miR-223 is mediated by repression of NFIA expression. Here we generated lentivirus-containing NFIA and the dsRed reporter (or empty vector control), infected the existing the miR-223-expressing cell line with each virus, and generated stable cell lines via FACS isolation. Analysis of cell cycle and soft agar growth revealed that NFIA overexpression rescues the growth phenotypes manifest in the miR-223-expressing cell line (Fig. 4D–G). Collectively, our data indicate that, analogous to CNS development, suppression of glioma cell proliferation by miR-223 is mediated through its repression of NFIA.
miR-223/NFIA axis suppresses tumorigenesis and regulates p21 expression
The preceding data indicate that the miR-223/NFIA regulatory axis plays a crucial role in the proliferation of glial precursors during CNS development and in human glioma cell lines. Next we sought to determine whether miR-223 overexpression in glioma cell lines impacts tumorigenesis. To this end we injected U87 cells expressing miR-223, scrambled control, or miR-223+NFIA into the mouse brain and found that the cell lines overexpressing miR-223 demonstrated decreased tumor formation compared to the scrambled control (Fig. 5A,B,J). This reduction in tumor formation is correlated with decreased expression of Ki-67 (Fig. 5D,E,K) and BrdU incorporation (Fig. 5G,H,L), linking the defects in cell proliferation to the impaired tumorigenesis. Furthermore, the effects of miR-223 on tumorigenesis and cell proliferation are rescued by overexpression of NFIA, indicating that repression of NFIA by miR-223 mediates these phenotypes (Fig. 5C,F,I–L).
Figure 5.
miR-223/NFIA axis regulates tumorigenesis and represses p21. A–I, Analysis of tumors generated from U87 cells expressing scrambled control (A, D, G), miR-223 (B, E, H), or miR-223+NFIA (C, F, I). Experiments were performed three times total mice for each cell line (scrambled, n = 11; miR-223, n = 11; miR-223+NFIA, n = 8). Quantification of bioluminescent imaging in J is from a set of eight from each group. Values in J represent relative increases in bioluminescence within a given group over time by comparing the bioluminescence reading at Day 1 with that at Day 14. Quantification in K and L is derived from five tumors, four adjacent sections per tumor, from each cell line. M, qRT-PCR analysis of NFIA and p21 expression in U87 cells expressing miR-223 or scrambled control. N, Reporter assays using p21-luciferase combined with overexpression of miR-223, scrambled control, or miR-223+NFIA. O, Representative map of NFIA binding sites in human p21 promoter. P, ChIP assays on U87 cells using antibodies to NFIA. Arrowheads in O denote primers used for ChIP PCR. Error bars represent SD. *p < 0.001 (scrambled vs miR-223); **p < 0.001 (rescue studies, i.e., miR-223 vs miR-223+NFIA).
We next sought to investigate the molecular mechanism by which the miR-223/NFIA axis regulates cell proliferation. NFI genes have been implicated in the regulation of p21, a key inhibitor of cell cycle and proliferation, whose dysregulation is associated with several malignancies, including glioma (Ouellet et al., 2006). Given this potential link and our observation that miR-223 results in decreased NFIA expression and cell proliferation, we reasoned that the miR-223/NFIA axis regulates cell proliferation by modulating p21 expression. To test this hypothesis we performed qRT-PCR in U87 cells overexpressing miR-223 or the scrambled control and found that p21 expression is increased in the presence of miR-223 and correlated with reduced NFIA expression (Fig. 5M). Next we performed reporter assays with the p21 promoter and found that miR-223 results in increased p21 promoter activity compared to scrambled control, while NFIA blocks miR-223-induced activity and represses p21 activity on its own (Fig. 5N). These data suggest that increased p21 expression in the presence of miR-223 is the result of derepression due to reduced NFIA expression. That the p21 promoter contains several NFIA binding sites (Fig. 5O) led us to hypothesize that it represses p21 expression by associating with the p21 promoter. To this end, we performed ChIP assays on U87 cells and found that NFIA associates with the p21 promoter, suggesting that it directly regulates p21 expression (Fig. 5P). Together, these data indicate that the miR-223/NFIA axis plays a central role in glioma tumorigenesis by regulating key mechanisms that control cell cycle and proliferation.
NFIA regulates NSC proliferation and tumorigenesis
The foregoing epistasis and functional studies, together with our previous findings, suggest that in glioma NFIA operates at the interface of cell proliferation, fate determination, and tumorigenesis. Therefore, to examine NFIA function in a model that couples these facets of developmental and tumor biology, we used NSCs from E13.5 wild-type, NFIA+/−, and NFIA−/− embryonic cortex (das Neves et al., 1999). Consistent with a role for NFIA in proliferation, we found that neurosphere formation and BrdU incorporation were impaired in the absence of NFIA (Fig. 6A,E,H,I). In parallel, we examined p21 expression and found increased levels of p21 in the absence of NFIA (Fig. 6B). Next we examined the differentiative potential of these NSC populations, where, upon exposure to differentiation conditions, we found that NFIA−/− NSCs demonstrated an increase in the number of neurons and concomitant decrease in the number of nestin-expressing cells, compared to wild-type and NFIA+/− NSC populations (Fig. 6C–I). Collectively, these data indicate that loss of NFIA suppresses NSC proliferation and produces a more differentiated phenotype.
Figure 6.
NFIA regulates NSC proliferation and tumorigenesis. A, Analysis of neurosphere formation in wild-type, NFIA+/−, and NFIA−/− NSCs. Dissociated NSCs were grown in suspension, and the number of neurospheres generated was quantified over the course of 6 d. B, qRT-PCR analysis of NFIA and p21 expression in NSCs. For both A and B, experiments were performed in duplicate three times, and graphs are representative. C–I, Analysis of NFIA+/− and NFIA−/− NSCs differentiated derivates (C, D, F, G) and proliferative properties (E, H). Quantification in I is derived from three independent experiments performed in duplicate; error bars represent SD and *p < 0.001. J, K, IHC demonstrating ectopic expression of SV40 and EGFR-vIII in NSC populations. N, Kaplan–Meier survival curve for the NFIA+/−;SV40/EGFR-vIII and NFIA−/−;SV40/EGFR-vIII cell lines. Experiments were performed twice on a total of eight mice per group. L, M, Analysis on NFIA+/−;SV40/EGFR-vIII tumors demonstrating SV40 and EGFR-vIII expression (L) and tumor pathology that resembles GBM (M).
That loss of NFIA alters the proliferative capacity and developmental potential of NSCs raises the question of whether the tumorigenecity of these populations is similarly impacted. To determine whether NFIA is required for tumorigenesis in an NSC model, we infected NFIA+/− and NFIA−/− NSCs with retroviruses containing EGFR-vIII and SV40-LargeT; stable lines were generated via drug selection. After confirming ectopic expression of both transgenes (Fig. 6J,K), each cell line was injected subcutaneously into SCID mice, and tumorigenesis was monitored. As indicated in Figure 6N, all mice injected with NFIA+/− NSCs formed tumors that pathologically resembled GBM (Fig. 6M), while the NFIA−/− NSCs did not form any tumors. These data indicate that NFIA expression is required for tumorigenesis in NSCs transformed by EGFR-vIII/SV40-LargeT, thus linking its role in NSC proliferation and differentiation to cellular transformation by these oncogenic mechanisms.
Discussion
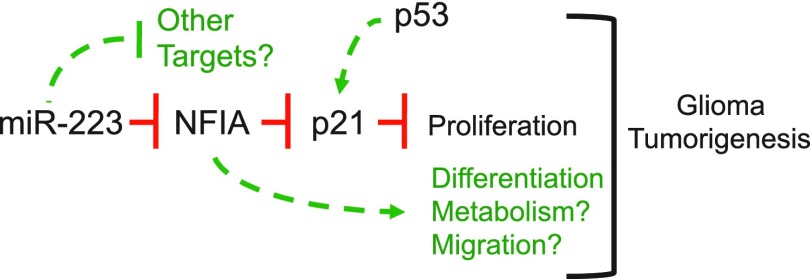
In this study, we demonstrate that the miR-223/NFIA axis regulates the proliferation of glial precursors and glioma cells through regulation of p21. During CNS development, we found that miR-223 suppresses glial precursor proliferation through repression of NFIA expression. Expression of NFIA and miR-223 in human GBM is inversely correlated, and functional studies revealed that the miR-223/NFIA axis similarly regulates the proliferation and tumorigenicity of glioma cell lines (Fig. 7). Finally, we demonstrate that NFIA promotes the proliferation of NSC populations and is required for tumorigenesis in a NSC model of glioma. Collectively, we demonstrate that the miR-223/NFIA axis is a proliferative mechanism that is conserved across development and tumorigenesis in the CNS.
Figure 7.
Model of miR-223/NFIA axis function in tumorigenesis. miR-223 repression of NFIA expression results in derepression of p21, which influences the proliferation of precursor populations and glioma cells in the CNS. p53 is included as a permissive factor, required for p21 induction and a key mediator of the NFIA/p21 transcriptional interplay. That deletion NFIA resulted in a complete loss of tumorigenesis in NSC populations transformed by SV40/EGFR-vIII strongly suggests that it influences other pathways that contribute to tumorigenesis. This possibility is noted by a dashed arrow. Likewise, it is possible that miR-223 also influences the expression of other targets that impact tumorigenesis, both negatively and positively, which may explain why the effects of deletion of NFIA are more profound than miR-223 overexpression.
Our studies, for the first time, implicate miR-223 as a key regulator of glial precursor and glioma proliferation. Previous in silico analysis correlated miR-223 expression with the proneural subtype of GBM, though expression was never validated in primary samples (Genovese et al., 2012). While our characterization of miR-223 does not make this distinction, we found heterogeneous expression across GBM samples, suggesting that its expression may indeed be restricted to a specific GBM subtype. Additionally, previous functional analysis suggests that miR-223 does not affect tumorigenesis in a p53−/−;PTEN−/− mouse NSC model of GBM (Genovese et al., 2012). This is in contrast to our results in human U87 glioma cells and is likely due to an inability to induce p21 expression in the p53−/−;PTEN−/− NSC model, thereby attenuating the effects of miR-223/NFIA regulatory axis in this context. That U87 cells contain intact p53 (Gomez-Manzano et al., 1997), together with the p53−/−;PTEN−/− NSC results, suggest that suppression of glioma growth by miR-223 requires intact p53. Put together, it appears that p53 functions as a permissive factor in the transcriptional interplay between NFIA and p21 (Fig. 7), where modulation of NFIA expression influences the repression of p21, but not its activation. Therefore, in cases where NFIA expression is reduced, the concordant upregulation of p21, and subsequent suppression of proliferation, would require the presence of p53, a well-established inducer of p21 expression. Indeed, repression of glial precursor proliferation by miR-223 during development occurs in the presence of p53, further supporting this notion.
Previous studies have implicated miR-223 in tumorigenesis, though its role varies: in gastric cancers, its expression is correlated with poor prognosis, whereas in acute myeloid leukemia (AML) it suppresses cell proliferation (Fazi et al., 2007; Sun et al., 2010). The basis for these differences remains undefined; however, in AML, miR-223 suppresses cell growth through repression of E2Fs (Pulikkan et al., 2010). We found that miR-223 similarly suppresses glioma cell growth, though it appears to operate through repression of NFIA and subsequent upregulation of p21 (Fig. 7). It will be important to determine whether these represent tissue-specific mechanisms of growth suppression by miR-223 or are conserved across tissues and tumor types. That NFIA is also expressed in the myeloid lineage suggests that it is possible that the miR-223/NFIA/p21 axis is conserved in AML (Fazi et al., 2005). Alternatively, one explanation for the potentially disparate mechanisms of miR-223 action across tumors is that the impact of a given microRNA on cell physiology is largely dependent upon the repertoire of available targets. Therefore, it is possible that in the presence of NFIA (and p53), miR-223 suppresses cell proliferation, and in the absence of NFIA (or p53) it does not. Further analysis correlating miR-223 function and the expression of its targets across tumor types, and normal counterparts, will reveal new insights into the diversity of cellular processes it impacts.
While the expression patterns of miR-223 and NFIA in GBM are negatively correlated (Fig. 3), there are tumors that contain high miR-223 and NFIA expression and likely contain overlapping patterns of expression. One explanation for such a phenomenon is that other regulatory mechanisms may override miR-223 inhibition of NFIA in this context. For example, NFIA is regulated by Sox9 and Notch signaling (Kang et al., 2012; Namihira et al., 2009); therefore, if Sox9 expression or Notch activation is elevated, the amount of NFIA transcript is also likely to be elevated, which could titrate away the effects of the miR-223 inhibition of NFIA. The converse is also true; other miR-223 targets may be elevated in a subset of GBMs, serving as an miR-223 “sink,” allowing for its effect on NFIA expression to minimized. Given the heterogeneous and genetically aberrant nature of GBM, these scenarios are likely and further illustrate the importance in defining the molecular and cellular signatures of these tumors.
The basis for our glioma studies is that miR-223 represses NFIA expression in the developing CNS and that this relationship regulates glial precursor proliferation. The relationship between miR-223 and NFIA was initially described during hematopoiesis, where repression of NFIA by miR-223 promoted granulocyte differentiation (Fazi et al., 2005). While the effects of miR-223 in the developing CNS appear to be restricted to glial precursor proliferation, we previously found that inhibition of NFIA blocks gliogenesis and promotes neuronal differentiation (Deneen et al., 2006). Analysis of NFIA−/− NSCs corroborates these findings and, together with its role in granulocyte development, suggests that NFIA functions as a suppressor of differentiation in these lineages. The suppression of differentiation is generally balanced by a concomitant promotion of cellular proliferation, suggesting that NFIA itself promotes cell proliferation. This is supported by our observations that it rescues cell proliferation defects in the presence of miR-223 in both chick spinal cord and in U87 glioma cell lines. Moreover, we found that neurosphere production and BrdU incorporation are significantly decreased in NFIA−/− NSCs, and that NFIA directly represses expression of p21, indicating that it is required for NSC proliferation as well. These findings are in contrast to previous studies in the developing hippocampus that imply NFIA suppresses precursor proliferation at late embryonic stages. While these studies did not assess NSC properties from these regions in vitro, it is likely that such differences reflect region or stage-specific functions for NFIA (Piper et al., 2010).
That NFIA itself inhibits NSC proliferation led us to further investigate its role in glioma, where we found, for the first time, that it is required for tumorigenesis. These results, combined with our findings that it is expressed in GBM and various grades of astrocytoma, suggest that NFIA directly contributes to the pathology of human glioma (Song et al., 2010). Our mechanistic studies point to its repression of p21 as being a catalyst for its role in cell proliferation; however, given that none of the NFIA−/−;SV40;EGFR-vIII NSC populations formed tumors, it seems likely that NFIA also influences other aspects of cell physiology that contribute to oncogenesis (Fig. 7). Previously, we found that NFIA collaborates with Sox9 to regulate a set of genes that control key aspects of glial precursor development that have implications in oncogenesis, including metabolism and cell migration (Kang et al., 2012). Therefore, understanding how these developmental programs are reutilized during tumorigenesis may give more insight into how NFIA contributes to glioma formation. In addition to NFIA's relationship with Sox9, another set of relevant developmental relationships includes its interactions with other glial fate determinants implicated in glioma, including Olig2 and STAT3 (Ligon et al., 2007; de la Iglesia et al., 2008; Swartling et al., 2012). NFIA associates with Olig2, and both function to repress p21, suggesting that they have either a redundant or collaborative relationship in this context (Deneen et al., 2006). While there is no direct evidence that STAT3 and NFIA associate, they collaborate to regulate GFAP expression in differentiating astrocytes, suggesting that this relationship may serve to impair tumor proliferation, rather than promote it (Namihira et al., 2009). Indeed, the role of STAT3 in glioma is dependent upon genetic context; therefore, it is possible that the role of NFIA in glioma is similarly nuanced and linked to its relationships with other glial fate determinants. Understanding how these key relationships that regulate glial development are reutilized and contribute to glioma formation will be critical to our understanding of this disease and its eventual treatment.
Footnotes
This work was supported by grants from the Sontag Foundation (B.D.); the V Foundation for Cancer Research (B.D.); National Institutes of Health Grants R01 NS071153 (B.D.), 5-T32HL092332-08 (S.M.G.), and U54CA149169 (S.T.C.W.); a pilot project award from the Center for Pediatric Cancer Genomics and Therapeutics at Texas Children's Cancer Center (B.D.); and the T.T. and W.F. Chao Foundation (S.T.C.W.).
References
- Ahmed N, Salsman VS, Kew Y, Shaffer D, Powell S, Zhang YJ, Grossman RG, Heslop HE, Gottschalk S. HER2-specific T cells target primary glioblastoma stem cells and induce regression of autologous experimental tumors. Clin Cancer Res. 2010;16:474–485. doi: 10.1158/1078-0432.CCR-09-1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachoo RM, Maher EA, Ligon KL, Sharpless NE, Chan SS, You MJ, Tang Y, DeFrances J, Stover E, Weissleder R, Rowitch DH, Louis DN, DePinho RA. Epidermal growth factor receptor and Ink4a/Arf: convergent mechanisms governing terminal differentiation and transformation along the neural stem cell to astrocyte axis. Cancer Cell. 2002;1:269–277. doi: 10.1016/S1535-6108(02)00046-6. [DOI] [PubMed] [Google Scholar]
- Dalmay T, Edwards DR. MicroRNAs and the hallmarks of cancer. Oncogene. 2006;25:6170–6175. doi: 10.1038/sj.onc.1209911. [DOI] [PubMed] [Google Scholar]
- de la Iglesia N, Konopka G, Puram SV, Chan JA, Bachoo RM, You MJ, Levy DE, Depinho RA, Bonni A. Identification of a PTEN-regulated STAT3 brain tumor suppressor pathway. Genes Dev. 2008;22:449–462. doi: 10.1101/gad.1606508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- das Neves L, Duchala CS, Tolentino-Silva F, Haxhiu MA, Colmenares C, Macklin WB, Campbell CE, Butz KG, Gronostajski RM, Godinho F. Disruption of the murine nuclear factor I-A gene (Nfia) results in perinatal lethality, hydrocephalus, and agenesis of the corpus callosum. Proc Natl Acad Sci U S A. 1999;96:11946–11951. doi: 10.1073/pnas.96.21.11946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deneen B, Denny CT. Loss of p16 pathways stabilizes EWS/FLI1 expression and complements EWS/FLI1 mediated transformation. Oncogene. 2001;20:6731–6741. doi: 10.1038/sj.onc.1204875. [DOI] [PubMed] [Google Scholar]
- Deneen B, Welford SM, Ho T, Hernandez F, Kurland I, Denny CT. PIM3 proto-oncogene kinase is a common transcriptional target of divergent EWS/ETS oncoproteins. Mol Cell Biol. 2003;23:3897–3908. doi: 10.1128/MCB.23.11.3897-3908.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deneen B, Ho R, Lukaszewicz A, Hochstim CJ, Gronostajski RM, Anderson DJ. The transcription factor NFIA controls the onset of gliogenesis in the developing spinal cord. Neuron. 2006;52:953–968. doi: 10.1016/j.neuron.2006.11.019. [DOI] [PubMed] [Google Scholar]
- Fazi F, Rosa A, Fatica A, Gelmetti V, De Marchis ML, Nervi C, Bozzoni I. A minicircuitry comprised of microRNA-223 and transcription factors NFI-A and C/EBPalpha regulates human granulopoiesis. Cell. 2005;123:819–831. doi: 10.1016/j.cell.2005.09.023. [DOI] [PubMed] [Google Scholar]
- Fazi F, Racanicchi S, Zardo G, Starnes LM, Mancini M, Travaglini L, Diverio D, Ammatuna E, Cimino G, Lo-Coco F, Grignani F, Nervi C. Epigenetic silencing of the myelopoiesis regulator microRNA-223 by the AML1/ETO oncoprotein. Cancer Cell. 2007;12:457–466. doi: 10.1016/j.ccr.2007.09.020. [DOI] [PubMed] [Google Scholar]
- Gabay L, Lowell S, Rubin LL, Anderson DJ. Deregulation of dorsoventral pattering by FGF confers trilineage differentiation capacity on CNS stem cells in vitro. Neuron. 2003;40:485–499. doi: 10.1016/S0896-6273(03)00637-8. [DOI] [PubMed] [Google Scholar]
- Genovese G, Ergun A, Shukla SA, Campos B, Hanna J, Ghosh P, Quayle SN, Rai K, Colla S, Ying H, Wu CJ, Sarkar S, Xiao Y, Zhang J, Zhang H, Kwong L, Dunn K, Wiedemeyer WR, Brennan C, Zheng H, Rimm DL, Collins JJ, Chin L. microRNA regulatory network inference identifies miR-34a as a novel regulator of TGF-β signaling in glioblastoma. Cancer Discov. 2012;2:736–749. doi: 10.1158/2159-8290.CD-12-0111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Manzano C, Fueyo J, Kyritsis AP, McDonnell TJ, Steck PA, Levin VA, Yung WK. Characterization of p53 and p21 functional interactions in glioma cells en route to apoptosis. J Natl Cancer Inst. 1997;89:1036–1044. doi: 10.1093/jnci/89.14.1036. [DOI] [PubMed] [Google Scholar]
- Kang P, Lee HK, Glasgow S, Finley M, Donti T, Garber Z, Foster AE, Novitch BG, Gronostajski RM, Deneen B. Sox9 and NFIA coordinate a transcriptional regulatory cascade during the onset of gliogenesis. Neuron. 2012;74:79–94. doi: 10.1016/j.neuron.2012.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligon KL, Huillard E, Mehta S, Kesari S, Liu H, Alberta JA, Bachoo RM, Kane M, Louis DN, Depinho RA, Anderson DJ, Stiles CD, Rowitch DH. Olig2-regulated lineage-restricted pathway controls replication competence in neural stem cells and malignant glioma. Neuron. 2007;15:503–517. doi: 10.1016/j.neuron.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher EA, Furnari FB, Bachoo RM, Rowitch DH, Louis DN, Cavenee WK, DePinho RA. Malignant glioma: genetics and biology of a grave matter. Genes Dev. 2001;15:1311–1333. doi: 10.1101/gad.891601. [DOI] [PubMed] [Google Scholar]
- Momose T, Tonegawa A, Takeuchi J, Ogawa H, Umesono K, Yasuda K. Efficient targeting of gene expression in chick embryos by microelectroporation. Dev Growth Differ. 1999;41:335–344. doi: 10.1046/j.1440-169X.1999.413437.x. [DOI] [PubMed] [Google Scholar]
- Morgan BA, Fekete DM. Manipulating gene expression with replication-competent retroviruses. Methods Cell Biol. 1996;51:185–218. doi: 10.1016/S0091-679X(08)60629-9. [DOI] [PubMed] [Google Scholar]
- Namihira M, Kohyama J, Semi K, Sanosaka T, Deneen B, Taga T, Nakashima K. Committed neuronal precursors confer astrocytic potential on residual neural precursor cells. Dev Cell. 2009;16:245–255. doi: 10.1016/j.devcel.2008.12.014. [DOI] [PubMed] [Google Scholar]
- Ouellet SVF, Lessard M, Leclerc S, Drouin R, Guerin S. Transcriptional regulation of the cyclin dependent kinase inhibitor 1A (p21) gene by NFI in proliferating human cells. Nuc Acid Res. 2006;34:6472–6487. doi: 10.1093/nar/gkl861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper M, Barry G, Hawkins J, Mason S, Lindwall C, Little E, Sarkar A, Smith AG, Moldrich RX, Boyle GM, Tole S, Gronostajski RM, Bailey TL, Richards LJ. NFIA controls telencephalic progenitor cell differentiation through repression of the Notch effector Hes1. J Neurosci. 2010;30:9127–9139. doi: 10.1523/JNEUROSCI.6167-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plachez C, Lindwall C, Sunn N, Piper M, Moldrich RX, Campbell CE, Osinski JM, Gronostajski RM, Richards LJ. Nuclear factor I gene expression in the developing forebrain. J Comp Neurol. 2008;508:385–401. doi: 10.1002/cne.21645. [DOI] [PubMed] [Google Scholar]
- Pulikkan JA, Dengler V, Peramangalam PS, Peer Zada AA, Müller-Tidow C, Bohlander SK, Tenen DG, Behre G. Cell-cycle regulator E2F1 and microRNA-223 comprise an autoregulatory negative feedback loop in acute myeloid leukemia. Blood. 2010;115:1768–1778. doi: 10.1182/blood-2009-08-240101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu T, Butz KG, Plachez C, Gronostajski RM, Richards LJ. Abnormal development of forebrain midline glia and commissural projections in Nfia knock-out mice. J Neurosci. 2003;23:203–212. doi: 10.1523/JNEUROSCI.23-01-00203.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H, Gonzalez-Gomez I, Suh GS, Commins DL, Sposto R, Gilles FH, Deneen B, Erdreich-Epstein A. Nuclear factor IA is expressed in astrocytomas and is associated with improved survival. Neuro Oncol. 2010;12:122–132. doi: 10.1093/neuonc/nop044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatopoulos B, Meuleman N, Haibe-Kains B, Saussoy P, Van Den Neste E, Michaux L, Heimann P, Martiat P, Bron D, Lagneaux L. microRNA-29c and microRNA-223 down-regulation has in vivo significance in chronic lymphocytic leukemia and improves disease risk stratification. Blood. 2009;113:5237–5245. doi: 10.1182/blood-2008-11-189407. [DOI] [PubMed] [Google Scholar]
- Stiles CD, Rowitch DH. Glioma stem cells: a midterm exam. Neuron. 2008;58:832–846. doi: 10.1016/j.neuron.2008.05.031. [DOI] [PubMed] [Google Scholar]
- Sun W, Shen W, Yang S, Hu F, Li H, Zhu TH. miR-223 and miR-142 attenuate hematopoietic cell proliferation, and miR-223 positively regulates miR-142 through LMO2 isoforms and CEBP-beta. Cell Res. 2010;20:1158–1169. doi: 10.1038/cr.2010.134. [DOI] [PubMed] [Google Scholar]
- Swartling FJ, Savov V, Persson AI, Chen J, Hackett CS, Northcott PA, Grimmer MR, Lau J, Chesler L, Perry A, Phillips JJ, Taylor MD, Weiss WA. Distinct neural stem cell populations give rise to disparate brain tumors in response to N-MYC. Cancer Cell. 2012;21:601–613. doi: 10.1016/j.ccr.2012.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]