Abstract
Axon degeneration is an evolutionarily conserved pathway that eliminates damaged or unneeded axons. Manipulation of this poorly understood pathway may allow treatment of a wide range of neurological disorders. In an RNAi-based screen performed in cultured mouse DRG neurons, we observed strong suppression of injury-induced axon degeneration upon knockdown of Sarm1 [SARM (sterile α-motif-containing and armadillo-motif containing protein)]. We find that a SARM-dependent degeneration program is engaged by disparate neuronal insults: SARM ablation blocks axon degeneration induced by axotomy or vincristine treatment, while SARM acts in parallel with a soma-derived caspase-dependent pathway following trophic withdrawal. SARM is a multidomain protein that associates with neuronal mitochondria. Deletion of the N-terminal mitochondrial localization sequence disrupts SARM mitochondrial localization in neurons but does not alter its ability to promote axon degeneration. In contrast, mutation of either the SAM (sterile α motif) or TIR (Toll-interleukin-1 receptor) domains abolishes the ability of SARM to promote axonal degeneration, while a SARM mutant containing only these domains elicits axon degeneration and nonapoptotic neuronal death even in the absence of injury. Protein–protein interaction studies demonstrate that the SAM domains are necessary and sufficient to mediate SARM–SARM binding. SARM mutants lacking a TIR domain bind full-length SARM and exhibit strong dominant-negative activity. These results indicate that SARM plays an integral role in the dismantling of injured axons and support a model in which SAM-mediated multimerization is necessary for TIR-dependent engagement of a downstream destruction pathway. These findings suggest that inhibitors of SAM and TIR interactions represent therapeutic candidates for blocking pathological axon loss and neuronal cell death.
Introduction
Axon degeneration is a specialized self-destructive program that mediates axon breakdown and clearance in development, injury, and disease (Coleman and Freeman, 2010; Wang et al., 2012). Like programmed cell death, axon degeneration plays beneficial roles during development and in response to damage, allowing developmental axon pruning (Luo and O'Leary, 2005) and promoting clearance of irreparably damaged axons following transection injury, a process termed “Wallerian” degeneration. Axonal degeneration is a prominent feature of diseases of the nervous system, including neurodegenerative diseases and neuropathies, that leads to dysfunctional neuronal connectivity (Saxena and Caroni, 2007). Identifying the molecular underpinnings of the axon degeneration program is therefore important for understanding nervous system development and may offer novel therapeutic options for neurological disorders.
Studies of mice bearing the axon-protective Wlds (Wallerian degeneration slow) mutation revealed that axon degeneration is a dynamically regulated destruction program (Coleman and Freeman, 2010). Wlds mice express a fusion protein containing Nmnat1, an enzyme in the NAD (nicotinamide adenine dinucleotide) biosynthetic pathway that is the axon-protective component (Coleman et al., 1998; Araki et al., 2004). While studies of Wlds mice have demonstrated the potential therapeutic benefit of suppressing axon degeneration in many neurological disorders, this gain-of-function mutation has not yielded direct insight into the genetic control of normal axon degeneration. Recent studies demonstrated that loss of dual leucine zipper kinase (DLK) slows axon degeneration in Drosophila and in mice, thus providing direct evidence for an axon destruction program (Miller et al., 2009). However, the limited duration of axon protection afforded by DLK ablation argues against its centrality in an axon degeneration pathway. Indeed, we and others have identified additional molecules whose loss or inhibition delays injury-induced axon degeneration (Barrientos et al., 2011; Gerdts et al., 2011; Wakatsuki et al., 2011; Bhattacharya et al., 2012; Babetto et al., 2013).
Here we report that SARM, a Toll-like receptor adaptor family member, is essential for injury-induced axon degeneration. SARM mutant mice exhibit dramatically delayed axon degeneration upon sciatic nerve transection. SARM contains a mitochondrial localization signal as well as SAM (sterile α motif) and TIR (Toll-interleukin-1 receptor) protein–protein interaction domains. Both fluorescently tagged human SARM and endogenous murine SARM associate with neuronal mitochondria, but a cytosolic SARM mutant effectively substitutes for wild-type SARM to promote injury-induced degeneration. Biochemical experiments demonstrate that SARM undergoes SAM-dependent multimerization and that SARM requires intact SAM domains to promote axon degeneration. Mutations within the TIR domain disrupt SARM function but do not disrupt SARM–SARM binding; moreover, the incorporation of TIR-less SARM molecules into SARM complexes renders them nonfunctional, indicating that SARM promotes axon degeneration as a complex that must contain multiple TIR domains to function. Finally, SARM containing the SAM and TIR domains but lacking the N-terminal 408 aa elicits axon degeneration and nonapoptotic neuronal death in the absence of injury, indicating the N terminus of SARM is a negative regulator of SARM-mediated degeneration. During these studies, Osterloh and colleagues (2012) independently reported the results of a Drosophila screen showing that SARM is important for injury-induced axon degeneration in Drosophila and mice.
Materials and Methods
Primary neuronal culture.
Dorsal root ganglia (DRGs) from embryonic day (E) 12.5 mice were dissected in DMEM and dissociated in 0.25% trypsin at 37°, then washed in Neurobasal media (Invitrogen) containing 2% B27 supplement (Invitrogen). Complete DRG media comprised Neurobasal media supplemented with l-glutamine (Invitrogen), 2% B27 (Invitrogen), 50 ng/ml nerve growth factor (Harlan Laboratories), 1 μm 5-fluoro-2′-deoxyuridine (Sigma-Aldrich), and 1 μm uridine (Sigma-Aldrich). Axon insult experiments were performed after 7 d growth in vitro. For cortical neuron cultures, cortices were isolated from 12.5 to 14.5 d embryos in DMEM, dissociated in 0.25% trypsin at 37°, then washed in Neurobasal media containing 2% B27 supplement. Cell suspensions containing 105 neurons were applied to poly-d-lysine/laminin-coated coverslips in 24-well plates and imaging experiments were performed at 5 d in vitro.
Lentiviral transduction.
Lentiviral shRNA constructs in the pLKO.1 vector (Moffat et al., 2006) targeting murine Sarm1 (target sequences: 5′-CTGGTTTCTTACTCTACGAAT-3′; 5′-CTTCTAAGACTCACAGATGAA-3′; 5′-CAGCCAGAGAAATGCTACATT-3′; listed in order as presented in Fig. 1A), Myd88 (5′-GCGACTGATTCCTATTAAATA-3′), and GFP (5′-TGCCCGACAACCACTACCTGA-3′) were obtained from the Washington University RNAi Core. BclXl (Vohra et al., 2010), Cre (Dasgupta and Milbrandt, 2007), and mitoDSRed (Misko et al., 2012) constructs have been previously described. All other mammalian expression constructs were derived from the FCIV lentiviral vector (Sasaki et al., 2009b), which contains a ubiquitin promoter and Venus marker driven by an internal ribosomal entry sequence (IRES). Human SARM1 and MYD88 cDNA (Open Biosystems) were PCR amplified and subcloned into FCIV using the homologous recombination-based InFusion system (Clontech). A Flag epitope was added to the C terminus of SARM (SARM-Flag) to facilitate affinity purification. Venus-tagged SARM fusion proteins were created by restriction enzyme-based removal of the IRES sequence of FCIV and insertion of SARM cDNA in frame with Venus, leaving a three-residue Ala-Thr-Thr peptide linker between the SARM C terminus and Venus. SARM deletion mutants lacking residues 2–27 (ΔN27), 2–408 (SAM-TIR), 2–560 (TIR), 409–560 (ΔSAM), 409–724 (ΔSAMΔTIR), 2–408/561–724 (SAM), and 561–724 (ΔTIR) were similarly subcloned into FCIV using the InFusion system. SARM mutant constructs bearing eight-residue alanine replacements (8×Ala) at residues 410–417, 445–452, 481–488, 517–524, 625–632, 661–668, 697–704, and 715–721 were generated by the megaprime PCR method (Kammann et al., 1989). Successful insertion of all clones was verified by sequencing. Double-floxed inverse orf (DIO) constructs (DIO-SAM-TIR-Venus and DIO-SAM-TIR[697-704A]-Venus) were derived by subcloning the loxP/lox2722 flanked insert from pAAV-Ef1a-DIO enhanced yellow fluorescent protein (Sohal et al., 2009; acquired from AddGene) into our FCIV vector and then subcloning SAM-TIR-Venus in an inverted orientation between the NheI and BstXI sites.
Figure 1.
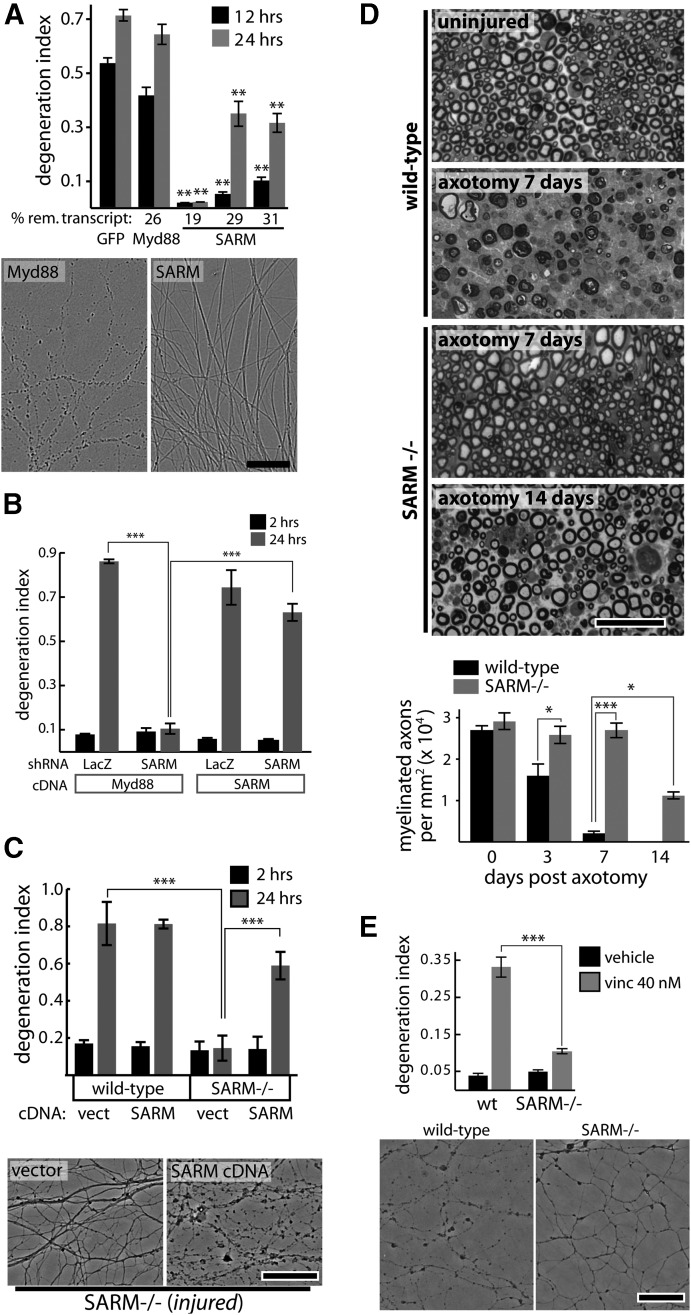
SARM is required for injury-induced axon degeneration. A, shRNAs targeting GFP (control) or Myd88 do not suppress axotomy-induced axon degeneration while SARM knockdown with three independent shRNA vectors protects axons; protection is correlated with knockdown efficiency measured by qRT-PCR (percentage remaining transcript shown below bar graph). **p < 0.01; comparisons made to shRNA sequence targeting GFP at each time point. Images of axons treated with Myd88 or SARM shRNA at 24 h postaxotomy are shown. B, Axotomy-induced axon degeneration is blocked by SARM shRNA but not control (LacZ) shRNA. Expression of human SARM cDNA does not cause direct axon degeneration (see 2 h postaxotomy) but restores normal injury-induced axon degeneration. C, Axons of wild-type DRG neurons undergo degeneration by 24 h postaxotomy while SARM−/− neurons do not. Expression of human SARM cDNA restores normal injury-induced degeneration. D, Toluidine blue-stained sciatic nerve cross sections distal to nerve transection show complete axon degeneration (loss of normal myelin profiles) by 7 d postinjury in wild-type animals while SARM−/− axons show no degeneration at 7 d and only partial degeneration by 14 d postinjury. E, Treatment with vincristine (40 nm) for 24 h causes axon fragmentation in wild-type DRG neurons but not SARM−/− neurons. Representative phase-contrast images are shown. *p < 0.05; **p < 0.01; ***p < 0.001; error bars show standard error of the mean (SEM), scale bars, 50 μm.
Lentiviral particles were produced by cotransfection of lentiviral expression vectors (pLKO.1 and FCIV) with lentiviral packaging plasmids into HEK293T cells as described previously (Sasaki et al., 2009b). Lentiviral particles were concentrated 10-fold from packaging cell media using LentiX Concentrator (Clontech), resuspended in PBS, and stored at −80°C. Concentrated lentiviral stocks were added to neuronal culture media at 50–200-fold final dilution. For axotomy experiments using shRNA vectors (Fig. 1A,B), BclXl expression lentivirus was added to cultures to avoid apoptosis of DRG neurons that is induced by some shRNA vectors (Gerdts et al., 2011).
Lentiviral transduction efficiency and expression level of Venus-tagged SARM constructs were monitored by high-content microscopy and automated image analysis. DRG neurons expressing each construct were fixed and stained with DAPI. Images of Venus and DAPI fluorescence were acquired from four wells per virus using an Operetta high-content imaging system (PerkinElmer). Following image background subtraction, average Venus pixel intensity per cell was quantified within regions drawn around >100 DAPI-stained nuclei. Box plots showing the medians, upper and lower quartiles, and upper and lower 95% boundaries of these intensity distributions are shown in Figure 4C. For all viruses tested, viral transduction efficiency was >95%. To verify that all constructs were adequately expressed for functional analysis shown in Figure 4C, median fluorescence intensity of each construct was measured and found to be above that of a low-titer positive control (SARM-V low) that effectively restored axon degeneration competence in SARM−/− neurons.
Figure 4.
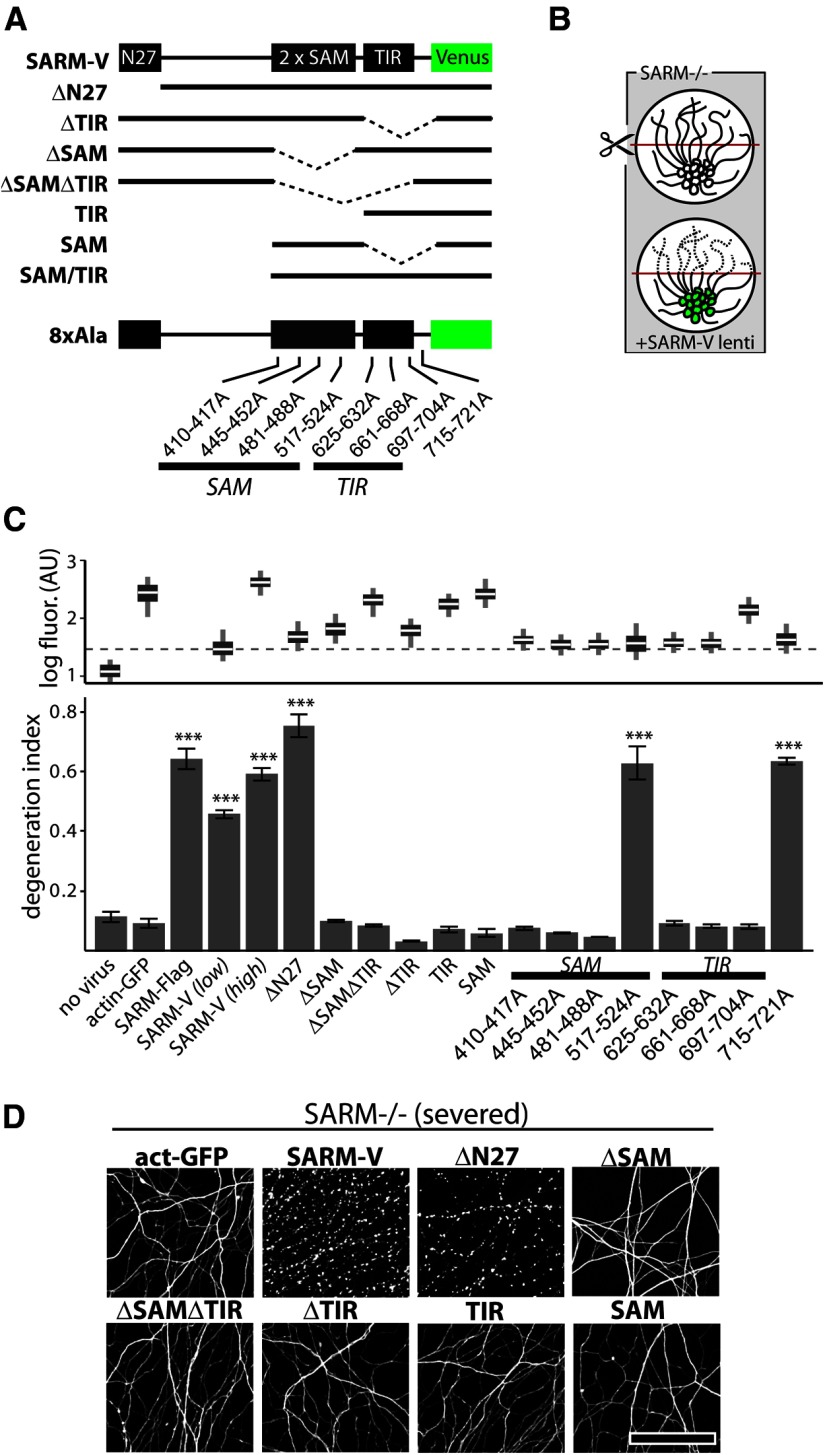
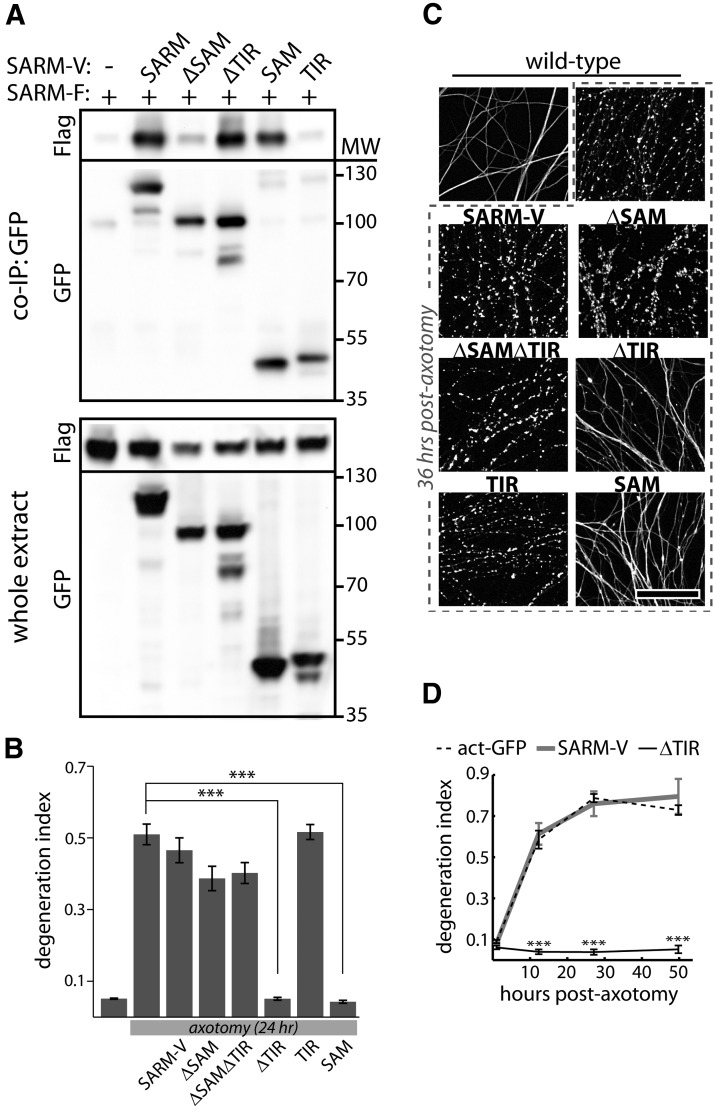
SAM and TIR domains of SARM are required to mediate injury-induced axon degeneration. A, Diagram of Venus-tagged full-length SARM (SARM-V) and various mutants analyzed. Dotted lines indicate deleted regions. N27, Residues 1–27. Eight separate alanine replacement mutants (8×Ala) contain eight Ala residues in place of the indicated SARM residues. B, Genetic rescue experiment schematic: SARM−/− axons do not degenerate upon injury whereas axons of SARM−/− neurons infected with SARM lentivirus are competent to undergo injury-induced degeneration. C, Bottom, Axons of SARM−/− DRG neurons expressing actin-GFP (control) do not degenerate by 36 h postaxotomy; normal axon degeneration is restored by expression of Flag-tagged SARM (SARM-Flag), SARM-V, and ΔN27 constructs but not ΔSAM, ΔSAMΔTIR, ΔTIR, TIR, or SAM constructs. Three of four 8×Ala mutants disrupting the SAM domains and three of three disrupting the TIR domain abolished axon degeneration function of SARM while a C-terminal 8×Ala replacement outside the TIR domain did not block function. These results are also summarized in Tables 1 and 2. Top, Box plot depicts log fluorescence intensity measured from >100 cells per group (see Materials and Methods); wide boxes denote lower and upper quartiles, white lines denote median values, and whiskers extend to upper and lower 95th percentiles. Median fluorescence of all mutants exceeds that of a functional low-virus control (SARM-V low; dashed line). ***p < 0.001; one-way ANOVA with Tukey–Kramer post-test. D, Representative α-tubulin-stained axon images for deletion mutants shown in B. Error bars = SEM, scale bars, 50 μm.
In vitro axon injury and quantification of fragmentation.
DRG neurons were cultured in 96-well microtiter plates with cell bodies sequestered to allow imaging of axons by automated microscopy (Gerdts et al., 2011). Axotomy was performed by manually severing DRG axons near the soma using a 3 mm wide flat blade. Six bright-field images per well of live axons were acquired at indicated time points post-insult using an Operetta high-content imaging system (PerkinElmer) with a 20× objective. Axon degeneration was quantified based on axon morphology and was reported as the “degeneration index” (“DI” ) using an ImageJ-based script we have previously described (Sasaki et al., 2009a). This metric ranges from 0 (perfectly intact) to 1 (perfectly fragmented) where values >0.5 correspond to extensive axon degeneration. Six images per well were measured as technical replicates, and 4–6 wells per condition were analyzed and used for statistical comparisons. To deprive DRG neurons of NGF, wells were rinsed twice with media lacking NGF before the addition of media containing anti-NGF antibody (Vohra et al., 2010). Axonal fragmentation was quantified as described above at 36 or 72 h postinsult. Vincristine (Araki et al., 2004) was added to DRG culture media at 40 nm. Immunostaining of DRG neurons was performed as described previously (Gerdts et al., 2011) using 1:2000 α-tubulin monoclonal antibody (Sigma-Aldrich). Photomontages shown in Figure 2D were created from eight individual fields using Adobe Photoshop.
Figure 2.
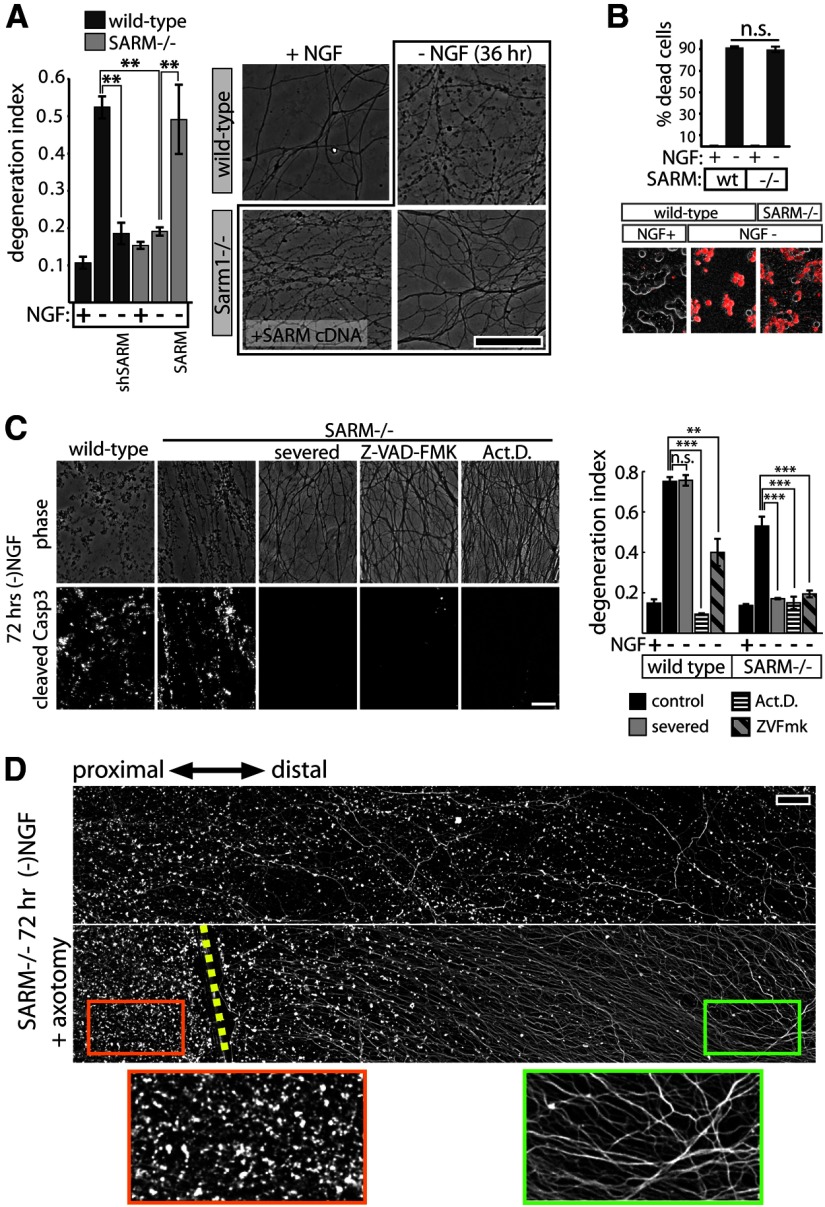
SARM plays a role in axon degeneration elicited by trophic factor withdrawal. A, NGF deprivation elicits axon degeneration in wild-type neurons that is suppressed by SARM knockdown (shSARM). SARM−/− neurons show suppressed axon degeneration that is restored by expression of SARM. Representative phase-contrast images are shown at the right. B, SARM ablation does not affect apoptosis of DRG neurons measured after 24 h of NGF deprivation using ethidium homodimer (EH) exclusion assay. Top, Quantification of EH-positive cells shows no difference in cell death between genotypes. Bottom, Representative images of EH positivity (red stain). C, Following 72 h trophic factor withdrawal, SARM−/− axons exhibit fragmentation and cleaved caspase-3 (Casp3) immunoreactivity comparable to wild-type axons. Axon degeneration and Casp3 are blocked by severing the axons, treating with caspase inhibitor Z-VAD-FMK (10 μg/ml), or the transcriptional inhibitor actinomycin D (Act.D.; 1 μg/ml). Axon degeneration quantification is shown in the bar graph (right). D, Fluorescence (α-tubulin stain) montages showing axotomy-induced axon protection in SARM−/− DRGs deprived of NGF for 72 h. Cut site is indicated by yellow dashed line. **p < 0.01; ***p < 0.001; error bars = SEM, scale bars, 50 μm.
Fluorescence microscopy.
Cortical and DRG neurons were grown on poly-d-lysine/laminin-coated coverglass and treated with mitochondria-targeted DsRed (Misko et al., 2012) and SARM-Venus expression lentiviruses at 2 d in vitro. After 4 d, coverglass was mounted on glass slides with DAPI-containing mounting media. Images were acquired using a Nikon Eclipse 80i microscope with Plan Apo 40× objective using MetaMorph image acquisition software. Intensity scaling and multichannel pseudocoloring were performed with ImageJ software (National Institutes of Health).
Immunofluorescence.
Wild-type and SARM−/− DRG neurons were isolated as described above and seeded in a 16-well chamber slide. At DIV 2, cells were infected with lentivirus expressing mitoDSred. At DIV 7, cells were fixed in 3.7% paraformaldehyde. Fixed cells were blocked and permeabilized in for 1 h in PBS plus 0.5% Triton X-100 and 5% BSA (PBS-T). Slides were incubated with anti-SARM antisera (1:300 in-house rabbit polyclonal) in blocking buffer overnight. Slides were thoroughly washed in PBS-T and incubated with Alexa488-conjugated anti-rabbit antibody. After thoroughly washing with PBS-T, slides were mounted and neurons were visualized with a Nikon D-Eclipse C1 confocal microscope using a 60× objective. Images are z-projections of confocal stacks. Wild-type and SARM−/− neurons were prepared simultaneously and images collected using identical settings. Anti-SARM rabbit polyclonal antisera were generated by Yenzym antibodies using synthetic peptides corresponding to amino acids 447–465 and 704–724 from human SARM. Antisera was purified by affinity chromatography using the peptides used as the immunogen.
Surgical procedures and histology.
Mice of either sex were anesthetized by intraperitoneal injection of 2-2-2 tribromoethanol at a dose of 500 mg/kg. The sciatic nerve was exposed at the hip and transected using small surgical scissors. The distal stump was deflected onto an adjacent muscle to prevent regeneration. Nerve lesions were produced on the right side and the contralateral nerve was left intact and served as the uninjured control. To analyze the nerve structure, the distal segments of the sciatic nerves (injured and control) were fixed with 3% glutaraldehyde, rinsed with 0.1 m phosphate buffer, pH 7.0, and incubated overnight at 4°C with 1% osmium tetroxide in phosphate buffer. Specimens were then dehydrated in graded ethanol and embedded in epoxy (Araldite 502). Nerve cross sections (0.4 μm) were stained with 1% toluidene blue and morphometric analysis was performed using light microscope (Axoskop 50, Zeiss) and morphometry software (Leco IA32, Leco Instruments), as previously described (Hunter et al., 2007). Three age-matched C57BL/6 and SARM−/− mice were analyzed at 3 and 7 d post-transection while contralateral uninjured nerves served as a control. Three additional SARM−/− mice were analyzed at 14 d post-transection.
Mitochondrial fractionation.
Cortical neurons isolated from E12.5 mouse embryos were infected at DIV 1 with lentiviral SARM-V. At DIV 7, cells were lysed in mitochondrial isolation buffer (10 mm HEPES, pH7.4, 1 mm EDTA, 250 mm sucrose, 1 mm PMSF, protease inhibitor mixture from Roche) by dounce homogenization. Cell extracts were centrifuged at 500 × g for 5 min to remove cell debris. Nuclei were pelleted at 1500 × g for 5 min. The postnuclear fraction was spun at 10,000 × g for 10 min. The supernatant was saved (cytosolic fraction) and the crude mitochondrial fraction was washed once with mitochondrial isolation buffer and saved for analysis by SDS-PAGE and Western immunoblotting. For proteinase K treatment, rat cortical neurons isolated from E17 embryos were treated as described above (in lysis buffer lacking protease inhibitor). Cytosolic and mitochondrial fractions were incubated with 0.5 mg/ml proteinase K at 25°C for 30 min. The reaction was stopped with 10 mm PMSF. Anti-GFP was from Invitrogen, anti-Tom40 (D-2) was from Santa Cruz Biotechnology, anti-Opa1 was from BD Biosciences, and anti-Hsp90 (C45G5) was from Cell Signaling Technology.
Coimmunoprecipitation.
HEK293 cells were cotransfected with SARM-Flag and wild-type SARM-V (or the indicated mutant). Twenty-four hours post-transfection, cells were harvested in coimmunoprecipitation buffer (1 × PBS, 10 mm HEPES, pH 7.4, 0.1% Triton X, 1 mm EDTA, 1 mm PMSF, 1× protease inhibitor mixture from Roche), sonicated, and precleared of cell debris at 3000 rpm for 3 min. Precleared cell extracts were incubated with rabbit polyclonal anti-GFP antisera (Invitrogen) and protein-G beads (Roche). Beads were thoroughly washed in coimmunoprecipitation buffer and SARM-V complexes resuspended in sample buffer, then analyzed by SDS-PAGE and Western immunoblotting with anti-Flag (Anti-Flag M2, Sigma-Aldrich).
Quantitative RT-PCR.
Knockdown efficiency of shRNA constructs was tested using DRG neuronal cultures grown in 24-well plates at a seeding density of 104 cells per well. Cells were infected after 2 d in vitro. RNA was harvested after 7 d in vitro and purified by phenol-chloroform extraction using Ribozol reagent (Amresco). Transcript levels relative to GAPDH were determined by qPCR (standard curve method) using an ABI Prism 7900HT sequence detection instrument (Applied Biosystems) with two technical replicates per sample per primer set.
Cell viability measurement.
To quantify neuronal cell death, cells were stained by direct addition of 20 μm ethidium homodimer (Botium) to the cell media and three 10× fluorescent and bright-field fields per well were acquired using the Operetta imaging system. Ethidium homodimer-positive (dead) and ethidium homodimer-negative (live) neurons were manually counted in each field (50–100 neurons per field), and the fraction of dead cells per well was used in statistical comparisons. To quantify viability of HEK cells transfected with SARM constructs, 104 HEK293 cells were seeded in each well of a 96-well microtiter plate and grown in DMEM supplemented with 10% fetal bovine serum. Cells in each well were transfected with 50 ng of each plasmid DNA (6 wells per group) by addition of plasmid diluted in 10 μl of Opti-MEM (Invitrogen) and mixed with 0.15 μl of X-TremeGene 9 transfection reagent (Roche). Thirty-six hours post-transfection, cell media was replaced with warmed (37°) phenol-free DMEM containing 0.01 mg/ml resazurin sodium salt (Sigma-Aldrich). After 1 h incubation, fluorescence in each well was read on a POLARstar Optima plate reader (BMG Labtech) with 544 excitation and 590 emission settings. Fluorescence values were normalized by subtracting background fluorescence (the mean fluorescence measured in wells without cells but containing incubated DMEM/resazurin) and then dividing by the mean fluorescence of the vector control group.
Statistics.
Results presented from all in vitro experiments are representative of ≥3 independent experiments. All statistical comparisons were calculated using Matlab software (Mathworks). Unless otherwise noted, statistical comparisons shown in figures indicate unpaired t tests with Bonferroni multiple-comparison adjustment.
Results
SARM is required for injury-induced axon degeneration
To identify genes required for degeneration of injured axons, we developed a quantitative, image-based screening paradigm using primary mouse DRG neurons grown in 96-well microtiter plates and injured by axotomy (Gerdts et al., 2011). Axon fragmentation is morphologically scored and reported as the DI, which ranges from 0 (perfectly intact) to 1 (perfectly fragmented; Sasaki et al., 2009a). Using this system, we screened a lentiviral shRNA library developed by The RNAi Consortium (Moffat et al., 2006) that targets most genes in the mouse genome (J. Gerdts, J. Milbrandt, unpublished observations). From this screen, we found that multiple independent shRNAs directed against the Toll and interleukin receptor adaptor molecule Sarm1 (SARM) suppress axotomy-induced axon degeneration. SARM is a member of a family of five intracellular Toll and interleukin receptor (TLR) adaptor molecules that includes MyD88, Ticam1/TRIF, Ticam2/TRAM, and Tollip/MAL (O'Neill and Bowie, 2007). We tested three independent shRNA vectors targeting SARM that suppress axon degeneration at 12 and 24 h postaxotomy with knockdown efficiency that correlates with axon protection (Fig. 1A). Myd88 knockdown does not suppress axon degeneration (Fig. 1A), nor does shRNA targeting other TLR adaptor molecules (data not shown). To confirm the specificity of the SARM shRNA effects on axon degeneration, we coinfected DRG neurons with lentiviruses expressing SARM shRNA and a human SARM cDNA that is not targeted by the murine shRNA. Human SARM reconstitutes the ability of these injured axons to degenerate (Fig. 1B), while Myd88 expression does not, confirming that SARM shRNAs protect axons via SARM knockdown. Notably, overexpression of SARM does not cause axon degeneration in the absence of injury, as indicated by a lack of axon degeneration in all treatment groups near the time of injury. These results indicate that SARM plays a critical role in the axon degeneration cascade and that, even when overexpressed, SARM requires an injury signal to promote axon degeneration.
To determine whether SARM mediates axon degeneration in vivo, we obtained SARM mutant mice (SARM−/−) that lack SARM expression in all tissues (Szretter et al., 2009). These SARM-deficient mice are developmentally normal with no overt abnormalities, although they do exhibit immune deficiencies when challenged with viral infection (Szretter et al., 2009). First, we tested whether cultured DRG neurons from these mutant animals manifested defects in axonal degeneration. DRG neurons from SARM−/− mice exhibit a dramatic delay in axotomy-induced axon degeneration that is restored by expression of SARM via lentivirus (Fig. 1C). To assess whether SARM is required for injury-induced axon degeneration in vivo, we severed the sciatic nerves of wild-type and SARM-deficient mice and examined the distal nerve segments by toluidine blue staining at 3, 7, and 14 d after transection. While cross sections of wild-type nerves reveal partial loss of intact axon profiles 3 d postaxotomy and near complete loss by 7 d, nerves from SARM mutant animals are fully preserved at 3 and 7 d, and exhibit only minor losses at 14 d (Fig. 1D). This robust axonal survival suggests that SARM is necessary for axonal degeneration in both sensory and motor fibers.
Axon degeneration is triggered by diverse stimuli, including mechanical trauma, chemotherapeutic agents, trophic factor withdrawal, and disease. It is widely believed that multiple signaling pathways are involved in axonal destruction and that different types of axonal damage activate specific pathways (Wang et al., 2012). To determine whether SARM is required for degeneration of axons damaged by insults other than axotomy, we first treated wild-type and SARM−/− DRG neurons with the microtubule destabilizing agent vincristine, a chemotherapeutic agent that causes neuropathic side effects in humans (Wang et al., 2004). Vincristine treatment of wild-type DRG neurons leads to axon fragmentation, whereas this effect is abrogated in SARM−/− axons (Fig. 1E).
SARM promotes axon degeneration in parallel with a soma-derived caspase-dependent pathway following trophic withdrawal
To further investigate the involvement of SARM in disparate axon degeneration paradigms, we subjected wild-type and SARM−/− DRG neurons to NGF deprivation, an insult known to be mechanistically distinct from axotomy (Nikolaev et al., 2009; Vohra et al., 2010; Simon et al., 2012). DRG neurons were cultured for 6 DIV, and then deprived of NGF. Within 36 h, wild-type neurons exhibit axon fragmentation and ethidium homodimer-positive staining of the soma indicative of apoptotic death (Fig. 2A,B). SARM−/− neurons deprived of NGF show no change in soma death but marked suppression of axon fragmentation at 36 h. This inhibition of axon fragmentation by SARM deficiency is reversed by coexpression of human SARM (Fig. 2A). Despite this protection, we noted that loss of SARM affords axon protection after NGF withdrawal that is of shorter duration than that observed after axon severing. For instance, axons from SARM-deficient neurons remain intact 72 h after axotomy, whereas SARM−/− axons are mostly fragmented after 72 h NGF deprivation, suggesting the existence of SARM-independent signals that mediate axon degeneration after trophic withdrawal. Because this late axon degeneration is preceded by apoptotic cell death of the soma, we considered the possibility that a soma-derived death signal might promote axon degeneration in a SARM-independent manner. To uncouple these somal and axonal signals, we severed SARM−/− axons immediately before depriving the neurons of NGF. After 72 h of NGF deprivation, nonsevered SARM−/− axons and axons proximal to the site of severing exhibit substantial fragmentation, whereas axons severed before NGF deprivation remain intact (Fig. 2C,D). Furthermore, consistent with a recent report (Chen et al., 2012), transcriptional inhibition with actinomycin D also blocks axon degeneration induced by trophic withdrawal in wild-type and SARM−/− neurons (Fig. 2C). These results confirm the existence of a soma-derived axon degeneration signal that requires de novo transcription and is SARM-independent. To determine whether this soma-derived, transcription-dependent signal is caspase-dependent, we performed immunostaining of wild-type and SARM−/− axons following 72 h NGF deprivation and found that degenerating SARM−/− axons exhibit cleaved caspase-3 immunoreactivity that is blocked by axon severing or transcriptional inhibition. Furthermore, caspase inhibition with inhibitor Z-VAD-FMK (Z-Val-Ala-Asp-fluoromethyl ketone; 10 μg/ml) fully blocks axon degeneration in SARM−/− axons deprived of NGF, while Z-VAD-FMK only partially suppresses degeneration of wild-type axons (Fig. 2C). Together, these results demonstrate a role for SARM in NGF-deprivation-induced axon degeneration as well as the existence of a second soma-derived and SARM-independent caspase-mediated pathway that drives a late phase of axonal degeneration after NGF deprivation.
While the above experiments and the work of others (Osterloh et al., 2012) demonstrate a critical role for SARM in axon degeneration, the molecular function of SARM protein is not well understood. Human SARM1 is a 724 aa protein with two clearly defined conserved domains identified by the Pfam sequence analysis tool (Finn et al., 2010): a pair of tandem SAM domains, which commonly mediate intramolecular and intermolecular interactions (Kim and Bowie, 2003; Qiao and Bowie, 2005); and, a TIR domain, which in other TIR-containing proteins can homodimerize and interact with TIR domains from other molecules (O'Neill and Bowie, 2007). The N terminus of SARM contains HEAT/Armadillo repeat domains (Mink et al., 2001), which commonly mediate protein binding, as well as an N-terminal mitochondrial targeting sequence (residues 1–27; Panneerselvam et al., 2012); however, neuronal studies concerning SARM localization to mitochondria are not in agreement (Kim et al., 2007; Chen et al., 2011; Osterloh et al., 2012).
SARM peripherally associates with neuronal mitochondria
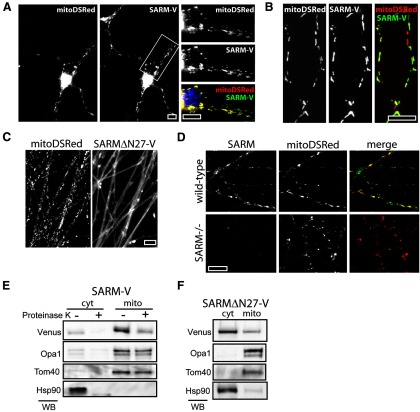
To assess SARM localization in neurons, we tagged human SARM with Venus fluorescent protein at the carboxy terminus (SARM-V) and expressed this SARM fusion protein in cortical and sensory neuron cultures. Importantly, SARM-V reconstitutes axon degeneration signaling in SARM−/− neurons (see Fig. 4C,D), verifying that the fluorescent tag does not interfere with protein function. Fluorescence microscopy demonstrates that SARM-V is strongly colocalizes with the mitochondria-targeted DsRed marker (Misko et al., 2012) in both cortical neurons and axons of DRG neurons (Fig. 3A,B). In contrast, SARM lacking the mitochondria-targeting residues 2–27 (SARMΔN27) reported by Panneerselvam et al. (2012) is diffusely localized in axons (Fig. 3C), confirming the importance of this region for mitochondrial targeting of SARM in neurons. To assess the localization of endogenous murine SARM, we generated a SARM-specific antibody for immunofluorescence and confirmed its specificity using SARM−/− neurons (Fig. 3D). In agreement with our localization studies with tagged human SARM, immunostaining of endogenous murine SARM reveals colocalization with mitoDSRed in axonal mitochondria (Fig. 3D). In addition, we performed subcellular fractionation studies that showed that SARM-V is enriched in the mitochondrial fraction of cortical neurons, although we consistently observe a pool of SARM-V in the cytosolic fraction as well (Fig. 3E). In contrast, SARMΔN27 resides predominantly in the cytosolic fraction (Fig. 3F). To determine whether SARM-V resides within the mitochondria or is associated with mitochondria, we treated these mitochondrial fractions with proteinase K protease to digest proteins outside the mitochondrial outer membrane (Komiya and Mihara, 1996). Proteinase K treatment of mitochondrial fractions of cortical neurons expressing SARM-V leads to proteolysis of mitochondrial SARM-V (Fig. 3E), indicating that most SARM-V is peripherally associated with mitochondria, perhaps associated with the outer mitochondrial membrane.
Figure 3.
SARM associates with neuronal mitochondria. A, A SARM-Venus fusion protein (SARM-V) expressed in cortical neurons colocalizes with mitochondria-targeted DSRed protein (mitoDSRed) in both soma and neurites. B, DRG axons expressing mitoDSRed and SARM-V show mitochondrial colocalization. C, Immunostaining of endogenous SARM in DRG axons shows colocalization with mitoDSRed. A lack of immunoreactivity in SARM−/− axons (bottom left) verifies the specificity of the antibody used. D, SARMΔN27 mutant is localized diffusely in DRG axons and does not colocalize with mitoDSRed protein. E, SARM-V cofractionates with mitochondrial resident proteins Opa1 and Tom40. Treatment of mitochondrial and cytosolic fractions with proteinase K leads to proteolysis of SARM but not Opa1 or Tom40, indicating SARM-V resides outside the mitochondria. F, SARM lacking residues 2–27 (SARMΔN27-V) predominantly fractionates with cytosolic protein (ex.Hsp90). Scale bars, 10 μm.
Structure–function analysis of SARM-mediated axon degeneration
We next assessed which domains of SARM are critical for its role in promoting axon degeneration. Because lentiviral delivery of SARM-V effectively restores injury-induced axon degeneration in SARM−/− neurons and allows visualization of expression level and localization, we constructed variants of this clone containing deletions of SAM, TIR, and N-terminal domains as well as multiple eight-residue alanine replacements (8×Ala) within in the SAM and TIR domains (Fig. 4A). During our lentivirus production of these SARM mutants, we observed that lentiviral packaging of the SAM-TIR construct, which lacks N-terminal residues 1–408, could not be achieved due to its toxicity in the HEK293 packaging cells. This mutant was thus analyzed separately (see below).
SAM domains of SARM are required for axon degeneration and SARM multimerization
To test the activity of these mutants, each was delivered via lentivirus to cultured SARM−/− DRG neurons and axon degeneration was assessed 36 h following axotomy. Because SARM−/− axons do not exhibit axon degeneration after injury, functional SARM mutants can be identified by their ability to restore a wild-type phenotype (i.e., axon degeneration in response to severing) in these cells (Fig. 4B). All SARM mutants that restore injury-induced axon degeneration to DI > 0.4 when expressed in SARM-deficient neurons were considered functional as they reconstitute the SARM-dependent axon destructive pathway, while those that fail to restore axon degeneration after axotomy (DI remained <0.2) were considered compromised in their prodegenerative function. The results of these functional studies for all SARM mutants tested are summarized in Tables 1 and 2. To ensure efficient transduction efficiency and adequate expression of each SARM mutant, we quantified Venus fluorescence in >100 DAPI-labeled DRG neurons at the experimental endpoint (Fig. 4C, box plot). Lentiviral transduction of all constructs shown in Figure 4C was highly efficient, and all the SARM mutants exhibited a median fluorescent signal greater than that of a positive control that functionally restored axon degeneration (Fig. 4C, dotted line). Intriguingly, SARM lacking the mitochondrial-targeting domain fully restores injury-induced degeneration in SARM-deficient neurons, indicating that mitochondrial localization of SARM is not essential for its role in axon degeneration (Fig. 4C,D). In contrast, multiple SARM mutants were found to be nonfunctional. First, we observed that disruption of the SAM domains leads to loss of function: SAM deletion (ΔSAM) and three of the four Ala replacement mutants in the SAM domains disrupt SARM function (Fig. 4C,D). SAM domains in other proteins mediate homo-oligomerization (Kim and Bowie, 2003; Qiao and Bowie, 2005); therefore, we investigated their potential role in mediating SARM multimerization. Coimmunoprecipitation experiments were performed to detect interaction between Flag-tagged full-length SARM and Venus-tagged SARM mutants lacking various domains. We detected strong SARM–SARM interactions indicating that SARM exists as a dimer or multimer (Fig. 5A). SARM mutants lacking SAM domains (ΔSAM, ΔSAMΔTIR, TIR) fail to interact with full-length SARM, whereas mutants lacking other domains (ΔTIR, SAM) retain the ability to coprecipitate full-length SARM (Fig. 5A), indicating that the SAM domains play an essential role in SARM multimerization. Together with our observations that SAM domains are required for axon degeneration and that overexpressed SAM is a potent dominant negative (see below), these data suggest that SARM multimerization is required for its ability to promote axon degeneration.
Table 1.
Summary of findings for SARM domain deletion mutants
| Mutant | Residues | SARM−/− rescue | Dominant negative | Toxicity | Localization | SARM binding |
|---|---|---|---|---|---|---|
| SARM-V | 1–724 | + | − | − | Mitochondria | + |
| ΔN27 | 28–704 | + | − | − | Cytosol | NE |
| ΔSAM | 1–409; 561–724 | − | − | − | Mitochondria | − |
| SAM | 409–560 | − | + | − | Cytosol | + |
| ΔTIR | 1–560 | − | + | − | Mitochondria | + |
| TIR | 561–724 | − | − | − | Cytosol | − |
| ΔSAMΔTIR | 1–408 | − | − | − | Mitochondria | NE |
| SAM/TIR | 409–724 | NEa | NEa | + | Cytosol | NEa |
| SAM/TIR [697-704A] | 409–724 | NE | NE | − | Cytosol | NE |
NE, Not evaluated.
aNot evaluated due to toxicity.
Table 2.
Summary of functional analyses of 8×Ala replacement mutations
| Replaced residues | SARM−/− rescue |
|---|---|
| 410-417A (SAM1) | − |
| 445-452A (SAM1) | − |
| 481-488A (SAM2) | − |
| 517-524A (SAM2) | + |
| 625-632A (TIR) | − |
| 661-668A (TIR) | − |
| 697-704A (TIR) | − |
| 715-721A (post-TIR) | + |
Figure 5.
SARM forms SAM-mediated complexes that require multiple TIR domains to promote axon degeneration. A, Coimmunoprecipitation of full-length Flag-tagged SARM (SARM-F) with Venus-tagged SARM and deletion mutants. Full-length SARM-V and mutants containing SAM domains (ΔTIR, SAM) interact strongly with full-length SARM (top, Flag blot), whereas little interaction is observed for mutants lacking SAM domains (ΔSAM, TIR). B, Axons of wild-type DRG neurons degenerate within 24 h postaxotomy. Degeneration is unaffected by expression of SARM-V, ΔSAM, ΔSAMΔTIR, or TIR, but is blocked by SARM mutants that lack a TIR domain but contain intact SAM domains (ΔTIR, SAM). C, Representative images of α-tubulin-stained axons from experiment shown in B. D, Time course measurement of axon degeneration following axotomy. While SARM-V expression does not affect the rate of axon degeneration, expression of SARM lacking a TIR domain (ΔTIR) completely blocks axon degeneration for ≥2 d. ***p < 0.001; error bars = SEM, scale bar, 50 μm.
SARM complexes require multiple TIR domains to mediate axon degeneration
The TIR domain is a signaling domain that is present in many components of the Toll-like receptor signaling pathways, where TIR dimerization often constitutes a critical step in signal transduction (O'Neill and Bowie, 2007). Deletion of the TIR domain as well as three mutations within the TIR domain each abolish SARM function (Fig. 4C,D). When the TIR domain alone is expressed in neurons, it fails to restore axon degeneration; thus, the TIR domain of SARM is necessary but not sufficient to mediate injury-induced axon degeneration. Our coexpression experiments demonstrate that, unlike the SAM domains, the TIR domain has no role in SARM–SARM binding; we found that TIR alone is not sufficient for SARM interaction and that SARM molecules lacking a TIR domain are still capable of interacting with full-length SARM (Fig. 5A) despite their lack of any degeneration-promoting function. Since TIR dimerization constitutes a critical signal transduction step in several other TIR-domain-containing proteins (O'Neill and Bowie, 2007) and because the TIR domain of SARM lies immediately adjacent the SARM–SARM interaction domain, we hypothesized that SARM complex assembly might mediate SARM signaling by allowing TIR–TIR association. In this model (Fig. 6I), the formation of complexes containing only a single TIR domain would be nonfunctional. We found that overexpression of full-length SARM-V does not affect the rate of axon degeneration after axotomy, whereas two SARM mutants containing SAM domains but no TIR domain (ΔTIR and SAM) strongly inhibit axon degeneration (Fig. 5B,C). These results indicate that SARM lacking a TIR domain acts as a dominant negative by forming inactive complexes with endogenous SARM, thus inhibiting injury-induced axon degeneration when expressed in wild-type neurons. Therefore, while the SARM TIR domain is dispensable for formation of SARM multimers, it is required to promote degeneration of injured axons. Furthermore, we infer that SARM protein complexes require multiple TIR domains to function in the axon degeneration pathway.
Figure 6.
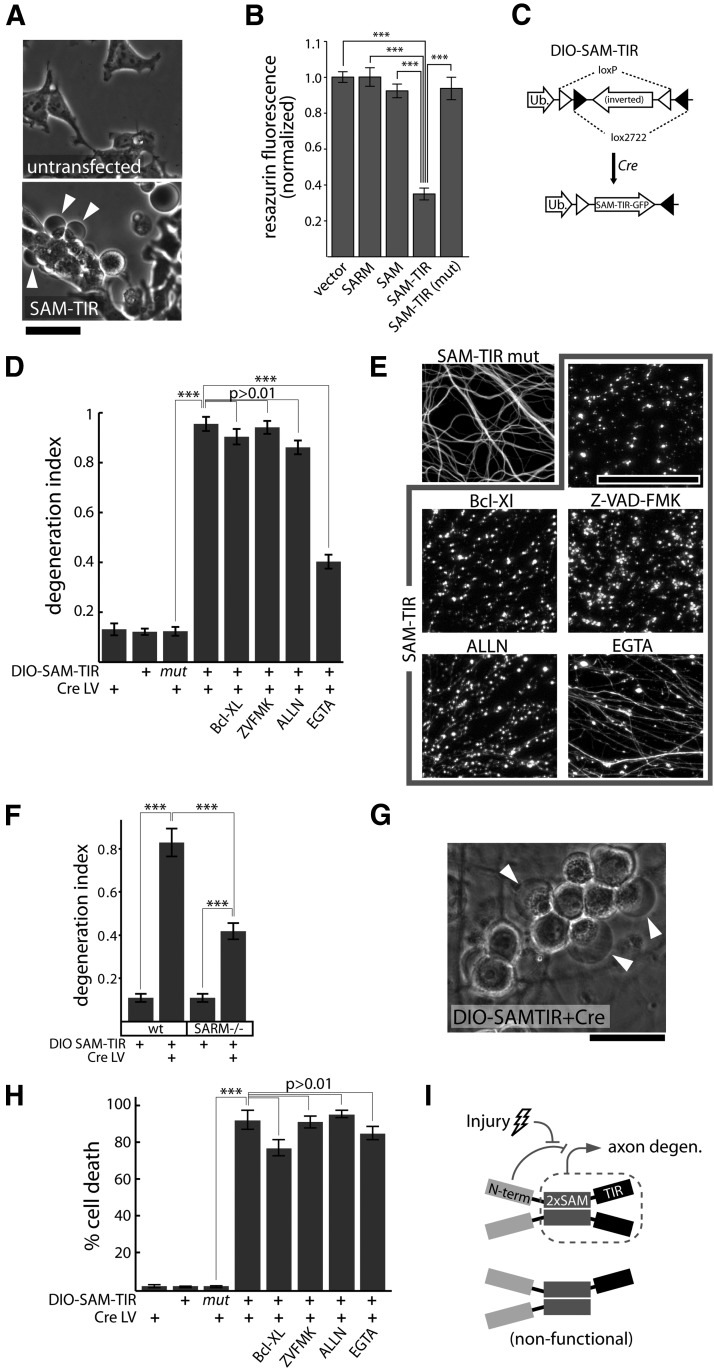
SAM-TIR expression induces axonal degeneration and nonapoptotic neuronal death. A, SAM-TIR expression induces pronounced blebbing (arrowheads) in HEK293 cells at 24 h post-transfection. B, SAM-TIR overexpression in HEK293 cells leads to a loss of cell viability measured by the metabolic resazurin assay at 36 h post-transfection. Cell viability is unaffected by expression of full-length SARM, SAM, or mutated SAM-TIR bearing an 8×Ala replacement mutation in the TIR domain that blocks SARM function in axon degeneration (SAM-TIR mut = [697-704A]). ***p < 0.001 unpaired t test with Bonferroni correction. C, Lentiviral packaging of SAM-TIR is made possible by a conditionally expressed DIO SAM-TIR: a lentivirus construct containing inverted SAM-TIR-GFP flanked by loxp and lox2722 sites is conditionally expressed in cells only upon Cre-dependent recombination. Ub, Human ubiquitin C promoter. D, SAM-TIR expression (DIO-SAM-TIR lentivirus plus Cre lentivirus) induces axon degeneration that is not blocked by the apoptosis-inhibiting protein Bcl-XL, pan-caspase inhibitor Z-VAD-FMK (ZVFMK; 10 μg/ml), or calpain inhibitor ALLN (25 μm). Chelation of extracellular Ca ions with EGTA (2.5 mm) provides partial suppression of axon degeneration. E, Representative α-tubulin-stained axons from experiment shown in D. F, SAM-TIR expression induces axon degeneration in both wild-type and SARM−/− axons measured 3 d postinfection; ***p < 0.001 unpaired t tests with Bonferroni correction. G, SAM-TIR expression induces large translucent neuronal swellings evident at 2 d postinfection. H, SAM-TIR expression induces neuronal death measured by ethidium homodimer staining at 3 d postinfection. Cell death is not blocked by the apoptosis-inhibiting protein Bcl-XL, pan-caspase inhibitor Z-VAD-FMK (ZVFMK; 10 μg/ml), calpain inhibitor ALLN (25 μm), or chelation of extracellular Ca ions with EGTA (2.5 mm). ***p < 0.001 unpaired t tests with Bonferroni correction. I, Proposed model, SARM forms SAM-mediated complexes that promote axon degeneration via intracomplex TIR interactions. Top, In uninjured cells, the N terminus of SARM suppresses the degeneration-promoting C terminus. Injury leads to release of this inhibition. Bottom, Incorporation of TIR-less molecules into SARM complexes renders them nonfunctional. Scale bars, 50 μm, error bars = SEM.
SAM-TIR fragment elicits axon degeneration and nonapoptotic neuronal death
As noted above, our efforts to produce lentiviral particles carrying the SAM-TIR fragment (SARM lacking amino acids 1–408) were hampered by apparent toxicity in the HEK viral packaging cells. SAM-TIR expression causes bleb formation (Fig. 6A) and decreased viability (Fig. 6B) in HEK293 cells. This toxicity is abolished by an eight-residue Ala replacement in the TIR domain that also abolishes SARM function in axon degeneration, indicating the toxicity is TIR-dependent and unlikely to be a consequence of protein misfolding. To study SAM-TIR expression in neurons, we created a conditional viral expression system using the double-floxed inverted ORF (DIO) strategy (Sohal et al., 2009; Fig. 6C). In this system, SAM-TIR is expressed only in cells that also express Cre recombinase, which is delivered via another lentivirus. Neither DIO-SAM-TIR nor Cre lentiviruses are toxic to neurons, but when combined these viruses promote robust axon degeneration (Fig. 6D,E). A TIR domain mutation that blocks SARM-mediated axon degeneration (697-704A) also blocks the ability of SAM-TIR to promote axon degeneration using this conditional expression strategy, indicating that SAM-TIR-induced axon degeneration is TIR-dependent. Axon degeneration elicited by SAM-TIR is not disrupted by apoptotic inhibitors, including Bcl-XL overexpression or caspase inhibitor Z-VAD-FMK (10 μg/ml). Calpain inhibitor ALLN (N-acetyl-Leu-Leu-Norleucinal; 25 μm) also does not inhibit SAM-TIR-induced axon degeneration; however, extracellular Ca2+ chelation with 2.5 mm EGTA provides significant, yet incomplete, axon preservation (Fig. 6D,E). SAM-TIR expression also causes axon fragmentation when expressed in SARM−/− neurons, indicating that SAM-TIR alone is sufficient to induce axon degeneration signaling in the absence of full-length SARM protein (Fig. 6F). These data demonstrate that while full-length SARM is required for injury-induced axon degeneration, an activated form of SARM is sufficient to elicit axon degeneration in the absence of injury.
While the above data and the work of Osterloh et al. (2012) define a clear role for SARM in axon degeneration, recent reports have also indicated a role for SARM in cell death. Two groups reported that SARM mediates oxygen/glucose-deprivation-induced neuronal death (Kim et al., 2007; Mukherjee et al., 2013). While SARM was shown to mediate T-cell death via an apoptotic pathway (Panneerselvam et al., 2013), the Caenohabditis elegans homolog of SARM, Tir-1, was found to be required for nonapoptotic programmed death of the linker cell during development (Blum et al., 2012). Consistent with a role for SARM in promotion of cell death, we also observed that SAM-TIR expression elicits neuronal death. DRG neurons expressing SAM-TIR exhibit large translucent swellings (Fig. 6G, arrowheads) before loss of membrane integrity that is evident by ethidium homodimer positivity (Fig. 6H). Similar to axon degeneration, SAM-TIR-induced neuronal death is independent of the apoptotic pathway; ethidium homodimer staining of SAM-TIR-transduced neurons reveals death of nearly 100% of treated neurons that is not blocked by Bcl-XL, Z-VAD-FMK, ALLN, or EGTA (Fig. 6H). Neuronal death is not induced by mutant SAM-TIR (Fig. 6H), or by full-length SARM, SAM, TIR, or any other SARM mutant analyzed (data not shown). These findings demonstrate that an activated form of SARM is sufficient to promote nonapoptotic neuronal death.
Discussion
While programmed cell death is a well appreciated mechanism for clearance of compromised and superfluous cells (Fuchs and Steller, 2011), there is now accumulating evidence for the existence of a signaling pathway regulating axon degeneration. This was first brought to light via studies of the Wlds mutant mice, in which axon degeneration is delayed after nerve transection, and in models of neurodegenerative disease (Coleman and Freeman, 2010). SARM deficiency also produces dramatic axon protection, demonstrating that SARM is essential for the mechanism by which injury leads to axon self-destruction. The identification of SARM in this study and in the report by Osterloh et al. (2012) strongly supports the existence of a signaling pathway that mediates the elimination of compromised axons and thus represents a therapeutic target for pathologic axon loss.
SARM is classified as one of five cytosolic “TIR adaptor” proteins that contain a TIR domain. The TIR adaptor proteins have been well studied as dynamic scaffold molecules promoting the downstream signaling of Toll-like receptors and have important roles in innate immunity (O'Neill and Bowie, 2007). SARM is unique among these proteins in that it has been reported to act as a negative regulator of innate immunity signaling (Carty et al., 2006; Akhouayri et al., 2011). Unlike the other TIR adaptors, SARM is preferentially expressed in the nervous system (Kim et al., 2007) and is linked to neural cell fate specification (Chuang and Bargmann, 2005), dendritic arborization (Chen et al., 2011), and microglial activation (Szretter et al., 2009). Thus, SARM has unique functions in the nervous system that have not been ascribed to other TIR-containing molecules. The discovery that SARM is required for injury-induced axon degeneration offers a fascinating link between innate immunity and neuronal damage sensing.
Structural requirements for SARM-mediated axon destruction
SARM has been reported to associate with mitochondria (Kim et al., 2007; Panneerselvam et al., 2012, 2013); accordingly, we found that GFP-tagged human SARM and endogenous murine SARM associate with neuronal mitochondria. Proteinase K treatment of neuronal mitochondrial fractions reveals that SARM mostly resides on the cytosolic surface of the mitochondria. While the genetic disruption of a previously reported SARM mitochondrial targeting sequence disrupts SARM mitochondrial localization in DRG axons, it does not affect the ability of SARM to promote axon degeneration. These findings suggest that the axonal degradation functions of SARM likely take place within the cytosol and that its association with mitochondria is not essential for this activity.
We have shown that the SAM and TIR domains of SARM are essential for its role in promoting elimination of injured axons. SARM mutants with disrupting mutations in either of these domains fail to restore injury-induced axon degeneration in SARM-deficient neurons. Interestingly, we found that the SAM domains, but not the TIR domain, are required to mediate SARM–SARM binding, indicating that self-association is required for SARM function. SARM lacking a TIR domain is capable of interacting with wild-type SARM but the resulting complexes are nonfunctional; indeed, this mutant acts as a potent dominant negative and prevents axonal degeneration in response to injury. These findings are consistent with a model in which SARM forms SAM-mediated multimers to create a complex that requires ≥2 TIR domains for active signaling (Fig. 6I). The requirement for multiple TIR domains to form a functional SARM complex suggests that SARM, like other TIR domain-containing proteins, may undergo TIR dimerization to facilitate interactions with other proteins. We note that if SAM-SAM and TIR interactions are independently required for SARM signaling, then pharmacologic agents that disrupt these protein interaction events could serve as potent inhibitors of axon degeneration and novel therapeutics for diseases involving axonopathy.
Surprisingly, we found that expression of SAM-TIR (SARM lacking amino acids 1–408), but not other SARM mutant molecules, is strongly toxic when expressed in HEK293 cells and neurons. Neuronal SAM-TIR expression elicits axon degeneration and neuronal death that are unaffected by apoptosis inhibition. In both HEK cells and neurons, the observed toxicity is abolished by an eight-residue alanine replacement within the TIR domain that also disrupts SARM-mediated axon degeneration, suggesting that SAM-TIR elicits toxicity by a mechanism shared with that of SARM-mediated axon degeneration. Moreover, unlike any other SARM constructs we analyzed, the SAM-TIR fragment elicits degeneration of wild-type and SARM−/− axons independent of injury, providing the first evidence that SARM may be not only necessary but sufficient to drive axon degeneration.
Together, our findings indicate that SARM is likely to be regulated at the post-translational level. First, SARM is expressed in uninjured neurons at levels that permit axon degeneration in response to injury without de novo transcription (this is true because severed axons are incapable of nuclear transcription). Therefore SARM-mediated axon degeneration cannot involve de novo transcription of SARM. This leaves two possible general models for SARM-mediated axon degeneration: (1) SARM expression is required before injury to maintain axon degeneration competence via regulation of other (axonal) factors, or (2) SARM is post-translationally activated within the severed axon and elicits a local self-destruction pathway. In support of the latter model, we have identified a “constitutive” degeneration-promoting form of SARM that is sufficient to cause axon degeneration and neuronal death in the absence of injury (SAM-TIR). SARM thus possesses an intrinsic TIR-dependent degeneration-inducing capability; however, this activity is suppressed in full-length SARM, indicating full-length SARM exists in an inactive state. Finally, the ability of SARM to trigger a death pathway argues in favor of direct involvement of SARM as an axonal self-destruction switch. We speculate that SARM exists in axons in an autoinhibited state and becomes post-translationally activated upon injury via an unknown mechanism. This autoinhibition involves suppression by the N terminus of TIR domain interactions that comprise a death signal. Understanding the suppressive function of the N terminus and the degeneration-promoting signal mediated by TIR domain interactions will be an important goal of future research.
The role of SARM in trophic-withdrawal-induced axon degeneration
We found that SARM ablation delays axon degeneration but not apoptotic death of DRG neurons deprived of NGF. This delay in degeneration demonstrates that SARM is involved in an early phase of axonal degeneration following NGF withdrawal; however, our finding that these axons ultimately degenerate is largely in agreement with the recent report that SARM ablation does not afford protection against this insult (Osterloh et al., 2012). In the previous study, DRG explant cultures deprived of NGF underwent very rapid degeneration (∼12 h) that was unaffected by SARM ablation. In dissociated cultures, we find that NGF deprivation induces degeneration of wild-type neurons after 24–36 h that is blocked in SARM−/− neurons and rescued by reintroduction of SARM cDNA. We observe that NGF-deprived SARM−/− neurons do eventually degenerate within 72 h of NGF deprivation, consistent with the existence of a second SARM-independent signal. Degeneration of SARM−/− neurons is blocked by severing the axons or by transcriptional inhibition, indicating that the SARM-independent degeneration signal derives from the nucleus and is anterogradely transported to the axonal compartment. Pharmacologic and immunostaining experiments reveal that this SARM-independent signal is caspase-mediated, consistent with multiple reports that caspases mediate axon degeneration in NGF deprivation (Nikolaev et al., 2009; Schoenmann et al., 2010; Vohra et al., 2010; Simon et al., 2012). Our findings support a model in which NGF deprivation elicits separate SARM-dependent and Caspase-dependent axon-destruction pathways, consistent with a proposal by Schoenmann et al. (2010) that parallel pathways promote NGF-withdrawal-induced axon degeneration. Because blockade of the apoptotic pathway affords complete protection against NGF withdrawal-induced degeneration in wild-type neurons, it is possible a SARM-dependent pathway becomes activated secondary to these signals.
SARM-mediated neuronal death
Recently, multiple groups have reported initial evidence that SARM is involved in cell death. Loss of Tir-1, the SARM homolog in C. elegans, conferred protection against anoxic death (Hayakawa et al., 2011) and programmed nonapoptotic death of the linker cell during development (Blum et al., 2012). In mice, SARM ablation suppressed neuronal death in brain slices (Kim et al., 2007) deprived of oxygen and glucose. SARM was also found to contribute to apoptotic neuronal death elicited by bunyavirus infection (Mukherjee et al., 2013) and apoptotic death of T cells following their expansion (Panneerselvam et al., 2013). In our studies, we found that expression of an activated SARM mutant lacking amino acids 1–408, but not full-length SARM, induces neuronal death, indicating SARM activation is sufficient to trigger a death program. SAM-TIR-induced neuronal death is not inhibited by Bcl-XL overexpression or caspase inhibition and is thus nonapoptotic. The involvement of SARM in nonapoptotic neuronal death in the contexts of development, injury, and disease is thus a promising new area of study.
In summary, SARM is an integral component of the pathway that elicits the dismantling of damaged axons. Exploiting this pathway for treatment of neurological disorders is an important goal and will require additional studies to identify other molecules that work with SARM to activate and execute the destructive pathway leading to axonal elimination and neuronal death.
Footnotes
This work was supported by National Institutes of Health (NIH) Grants AG013730 (J.M.), NS078007, and NS65053. J.G. is supported by NIH National Research Service Award Grant NS074517. D.W.S. is supported by Training Grant 2T32CA9547-26. We thank Tim Fahrner for assistance with molecular cloning and we thank Amy Strickland for assistance with tissue sectioning and microscopy. SARM−/− mice were a kind gift from Marco Colonna. Silencing RNA lentiviral vectors were obtained from the Washington University RNAi Core, which is sponsored by the Washington University Children's Discovery Institute.
References
- Akhouayri I, Turc C, Royet J, Charroux B. Toll-8/Tollo negatively regulates antimicrobial response in the Drosophila respiratory epithelium. PLoS Pathogens. 2011;7:e1002319. doi: 10.1371/journal.ppat.1002319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki T, Sasaki Y, Milbrandt J. Increased nuclear NAD biosynthesis and SIRT1 activation prevent axonal degeneration. Science. 2004;305:1010–1013. doi: 10.1126/science.1098014. [DOI] [PubMed] [Google Scholar]
- Babetto E, Beirowski B, Russler EV, Milbrandt J, Diantonio A. The Phr1 ubiquitin ligase promotes injury-induced axon self-destruction. Cell Rep. 2013;3:1422–1429. doi: 10.1016/j.celrep.2013.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrientos SA, Martinez NW, Yoo S, Jara JS, Zamorano S, Hetz C, Twiss JL, Alvarez J, Court FA. Axonal degeneration is mediated by the mitochondrial permeability transition pore. J Neurosci. 2011;31:966–978. doi: 10.1523/JNEUROSCI.4065-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya MR, Gerdts J, Naylor SA, Royse EX, Ebstein SY, Sasaki Y, Milbrandt J, DiAntonio A. A model of toxic neuropathy in Drosophila reveals a role for MORN4 in promoting axonal degeneration. J Neurosci. 2012;32:5054–5061. doi: 10.1523/JNEUROSCI.4951-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum ES, Abraham MC, Yoshimura S, Lu Y, Shaham S. Control of nonapoptotic developmental cell death in Caenorhabditis elegans by a polyglutamine-repeat protein. Science. 2012;335:970–973. doi: 10.1126/science.1215156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carty M, Goodbody R, Schröder M, Stack J, Moynagh PN, Bowie AG. The human adaptor SARM negatively regulates adaptor protein TRIF-dependent Toll-like receptor signaling. Nat Immunol. 2006;7:1074–1081. doi: 10.1038/ni1382. [DOI] [PubMed] [Google Scholar]
- Chen CY, Lin CW, Chang CY, Jiang ST, Hsueh YP. Sarm1, a negative regulator of innate immunity, interacts with syndecan-2 and regulates neuronal morphology. J Cell Biol. 2011;193:769–784. doi: 10.1083/jcb.201008050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Maloney JA, Kallop DY, Atwal JK, Tam SJ, Baer K, Kissel H, Kaminker JS, Lewcock JW, Weimer RM, Watts RJ. Spatially coordinated kinase signaling regulates local axon degeneration. J Neurosci. 2012;32:13439–13453. doi: 10.1523/JNEUROSCI.2039-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang CF, Bargmann CI. A Toll-interleukin 1 repeat protein at the synapse specifies asymmetric odorant receptor expression via ASK1 MAPKKK signaling. Genes Dev. 2005;19:270–281. doi: 10.1101/gad.1276505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman MP, Freeman MR. Wallerian degeneration, wld(s), and nmnat. Annu Rev Neurosci. 2010;33:245–267. doi: 10.1146/annurev-neuro-060909-153248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman MP, Conforti L, Buckmaster EA, Tarlton A, Ewing RM, Brown MC, Lyon MF, Perry VH. An 85-kb tandem triplication in the slow Wallerian degeneration (Wlds) mouse. Proc Natl Acad Sci U S A. 1998;95:9985–9990. doi: 10.1073/pnas.95.17.9985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta B, Milbrandt J. Resveratrol stimulates AMP kinase activity in neurons. Proc Natl Acad Sci U S A. 2007;104:7217–7222. doi: 10.1073/pnas.0610068104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, Holm L, Sonnhammer EL, Eddy SR, Bateman A. The Pfam protein families database. Nucleic Acids Res. 2010;38:D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs Y, Steller H. Programmed cell death in animal development and disease. Cell. 2011;147:742–758. doi: 10.1016/j.cell.2011.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdts J, Sasaki Y, Vohra B, Marasa J, Milbrandt J. Image-based screening identifies novel roles for IkB kinase and glycogen synthase kinase 3 in axonal degeneration. J Biol Chem. 2011;286:28011–28018. doi: 10.1074/jbc.M111.250472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayakawa T, Kato K, Hayakawa R, Hisamoto N, Matsumoto K, Takeda K, Ichijo H. Regulation of anoxic death in Caenorhabditis elegans by mammalian apoptosis signal-regulating kinase (ASK) family proteins. Genetics. 2011;187:785–792. doi: 10.1534/genetics.110.124883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter DA, Moradzadeh A, Whitlock EL, Brenner MJ, Myckatyn TM, Wei CH, Tung TH, Mackinnon SE. Binary imaging analysis for comprehensive quantitative histomorphometry of peripheral nerve. J Neurosci Methods. 2007;166:116–124. doi: 10.1016/j.jneumeth.2007.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kammann M, Laufs J, Schell J, Gronenborn B. Rapid insertional mutagenesis of DNA by polymerase chain reaction (PCR) Nucleic Acids Res. 1989;17:5404. doi: 10.1093/nar/17.13.5404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim CA, Bowie JU. SAM domains: uniform structure, diversity of function. Trends Biochem Sci. 2003;28:625–628. doi: 10.1016/j.tibs.2003.11.001. [DOI] [PubMed] [Google Scholar]
- Kim Y, Zhou P, Qian L, Chuang JZ, Lee J, Li C, Iadecola C, Nathan C, Ding A. MyD88-5 links mitochondria, microtubules, and JNK3 in neurons and regulates neuronal survival. J Exp Med. 2007;204:2063–2074. doi: 10.1084/jem.20070868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komiya T, Mihara K. Protein import into mammalian mitochondria. J Biol Chem. 1996;271:22105–22110. doi: 10.1074/jbc.271.36.22105. [DOI] [PubMed] [Google Scholar]
- Luo L, O'Leary DD. Axon retraction and degeneration in development and disease. Annu Rev Neurosci. 2005;28:127–156. doi: 10.1146/annurev.neuro.28.061604.135632. [DOI] [PubMed] [Google Scholar]
- Miller BR, Press C, Daniels RW, Sasaki Y, Milbrandt J, DiAntonio A. A dual leucine kinase-dependent axon self-destruction program promotes Wallerian degeneration. Nat Neurosci. 2009;12:387–389. doi: 10.1038/nn.2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mink M, Fogelgren B, Olszewski K, Maroy P, Csiszar K. A novel human gene (SARM) at chromosome 17q11 encodes a protein with a SAM motif and structural similarity to Armadillo/beta-catenin that is conserved in mouse, Drosophila, and Caenorhabditis elegans. Genomics. 2001;74:234–244. doi: 10.1006/geno.2001.6548. [DOI] [PubMed] [Google Scholar]
- Misko AL, Sasaki Y, Tuck E, Milbrandt J, Baloh RH. Mitofusin2 mutations disrupt axonal mitochondrial positioning and promote axon degeneration. J Neurosci. 2012;32:4145–4155. doi: 10.1523/JNEUROSCI.6338-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffat J, Grueneberg DA, Yang X, Kim SY, Kloepfer AM, Hinkle G, Piqani B, Eisenhaure TM, Luo B, Grenier JK, Carpenter AE, Foo SY, Stewart SA, Stockwell BR, Hacohen N, Hahn WC, Lander ES, Sabatini DM, Root DE. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 2006;124:1283–1298. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- Mukherjee P, Woods TA, Moore RA, Peterson KE. Activation of the innate signaling molecule MAVS by bunyavirus infection upregulates the adaptor protein SARM1, leading to neuronal death. Immunity. 2013;38:705–716. doi: 10.1016/j.immuni.2013.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolaev A, McLaughlin T, O'Leary DD, Tessier-Lavigne M. APP binds DR6 to trigger axon pruning and neuron death via distinct caspases. Nature. 2009;457:981–989. doi: 10.1038/nature07767. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- O'Neill LA, Bowie AG. The family of five: TIR-domain-containing adaptors in Toll-like receptor signalling. Nat Rev Immunol. 2007;7:353–364. doi: 10.1038/nri2079. [DOI] [PubMed] [Google Scholar]
- Osterloh JM, Yang J, Rooney TM, Fox AN, Adalbert R, Powell EH, Sheehan AE, Avery MA, Hackett R, Logan MA, MacDonald JM, Ziegenfuss JS, Milde S, Hou YJ, Nathan C, Ding A, Brown RH, Jr, Conforti L, Coleman M, Tessier-Lavigne M, et al. dSarm/Sarm1 is required for activation of an injury-induced axon death pathway. Science. 2012;337:481–484. doi: 10.1126/science.1223899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panneerselvam P, Singh LP, Ho B, Chen J, Ding JL. Targeting of pro-apoptotic TLR adaptor SARM to mitochondria: definition of the critical region and residues in the signal sequence. Biochem J. 2012;442:263–271. doi: 10.1042/BJ20111653. [DOI] [PubMed] [Google Scholar]
- Panneerselvam P, Singh LP, Selvarajan V, Chng WJ, Ng SB, Tan NS, Ho B, Chen J, Ding JL. T-cell death following immune activation is mediated by mitochondria-localized SARM. Cell Death Differ. 2013;20:478–489. doi: 10.1038/cdd.2012.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao F, Bowie JU. The many faces of SAM. Sci STKE STKE. 2005;2005:re7. doi: 10.1126/stke.2862005re7. [DOI] [PubMed] [Google Scholar]
- Sasaki Y, Vohra BP, Lund FE, Milbrandt J. Nicotinamide mononucleotide adenylyl transferase-mediated axonal protection requires enzymatic activity but not increased levels of neuronal nicotinamide adenine dinucleotide. J Neurosci. 2009a;29:5525–5535. doi: 10.1523/JNEUROSCI.5469-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki Y, Vohra BP, Baloh RH, Milbrandt J. Transgenic mice expressing the Nmnat1 protein manifest robust delay in axonal degeneration in vivo. J Neurosci. 2009b;29:6526–6534. doi: 10.1523/JNEUROSCI.1429-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena S, Caroni P. Mechanisms of axon degeneration: from development to disease. Prog Neurobiol. 2007;83:174–191. doi: 10.1016/j.pneurobio.2007.07.007. [DOI] [PubMed] [Google Scholar]
- Schoenmann Z, Assa-Kunik E, Tiomny S, Minis A, Haklai-Topper L, Arama E, Yaron A. Axonal degeneration is regulated by the apoptotic machinery or a NAD+-sensitive pathway in insects and mammals. J Neurosci. 2010;30:6375–6386. doi: 10.1523/JNEUROSCI.0922-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon DJ, Weimer RM, McLaughlin T, Kallop D, Stanger K, Yang J, O'Leary DD, Hannoush RN, Tessier-Lavigne M. A caspase cascade regulating developmental axon degeneration. J Neurosci. 2012;32:17540–17553. doi: 10.1523/JNEUROSCI.3012-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohal VS, Zhang F, Yizhar O, Deisseroth K. Parvalbumin neurons and gamma rhythms enhance cortical circuit performance. Nature. 2009;459:698–702. doi: 10.1038/nature07991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szretter KJ, Samuel MA, Gilfillan S, Fuchs A, Colonna M, Diamond MS. The immune adaptor molecule SARM modulates tumor necrosis factor alpha production and microglia activation in the brainstem and restricts West Nile Virus pathogenesis. J Virol. 2009;83:9329–9338. doi: 10.1128/JVI.00836-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vohra BP, Sasaki Y, Miller BR, Chang J, DiAntonio A, Milbrandt J. Amyloid precursor protein cleavage-dependent and -independent axonal degeneration programs share a common nicotinamide mononucleotide adenylyltransferase 1-sensitive pathway. J Neurosci. 2010;30:13729–13738. doi: 10.1523/JNEUROSCI.2939-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakatsuki S, Saitoh F, Araki T. ZNRF1 promotes Wallerian degeneration by degrading AKT to induce GSK3B-dependent CRMP2 phosphorylation. Nat Cell Biol. 2011;13:1415–1423. doi: 10.1038/ncb2373. [DOI] [PubMed] [Google Scholar]
- Wang JT, Medress ZA, Barres BA. Axon degeneration: molecular mechanisms of a self-destruction pathway. J Cell Biol. 2012;196:7–18. doi: 10.1083/jcb.201108111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang MS, Davis AA, Culver DG, Wang Q, Powers JC, Glass JD. Calpain inhibition protects against Taxol-induced sensory neuropathy. Brain. 2004;127:671–679. doi: 10.1093/brain/awh078. [DOI] [PubMed] [Google Scholar]