Abstract
Reprogramming of somatic cells into inducible pluripotent stem cells (iPSCs) provides an alternative to using embryonic stem cells (ESCs). Mesenchymal stem cells derived from human hair follicles (hHF-MSCs) are easily accessible, reproducible by direct plucking of human hairs. Whether these hHF-MSCs can be reprogrammed has not been previously reported. Here we report the generation of iPSCs from hHF-MSCs obtained by plucking several hairs. hHF-MSCs were isolated from hair follicle tissues and their mesenchymal nature confirmed by detecting cell surface antigens and multilineage differentiation potential towards adipocytes and osteoblasts. They were then reprogrammed into iPSCs by lentiviral transduction with Oct4, Sox2, c-Myc and Klf4. hHF-MSC-derived iPSCs appeared indistinguishable from human embryonic stem cells (hESCs) in colony morphology, expression of alkaline phosphotase, and expression of specific hESCs surface markers, SSEA-3, SSEA-4, Tra-1-60, Tra-1-81, Nanog, Oct4, E-Cadherin and endogenous pluripotent genes. When injected into immunocompromised mice, hHF-MSC-derived iPSCs formed teratomas containing representatives of all three germ layers. This is the first study to report reprogramming of hHF-MSCs into iPSCs.
Electronic supplementary material
The online version of this article (doi:10.1007/s12015-012-9420-5) contains supplementary material, which is available to authorized users.
Keywords: Induced pluripotent stem cells, Mesenchymal stem cells, Human hair follicle, Reprogrammed
Introduction
A major goal of stem cell research is the use of cells obtained from patient-derived pluripotent cells for therapeutic purposes. Embryonic stem cells (ESCs) are the most primitive stem cells. They are derived from pre-implantation embryos, and can differentiate into all three embryonic germ layers [1]. The characterization of ESCs as totipotent has made them a powerful tool for regenerative medicine. However, hESC-based therapeutic research is technically challenging and further complicated by ethical and legal concerns, significantly limiting its potential. Therefore, it has been the goal of the scientific community to explore new and innovative approaches to obtain hESC-like cells that do not destroy human embryos[2, 3].
Recently, iPSCs have been generated from both mouse and human fibroblasts following ectopic expression of the two transcription factors Oct4 and Sox2, combined with either c-Myc and Klf4 or Lin28 and Nanog [2–5]. These cells share many characteristics with ESCs, including the potential for pluripotent differentiation and capability for intensive proliferation in vitro. This result has not only facilitated disease research but also lays an important foundation for producing autologous cell therapies. Following this breakthrough, a variety of cells from mice, humans and other species have been used in reprogramming experiments, including fibroblasts[2–7], pancreatic β cells [8], hepatocytes [9], hematopoietic cells [10, 11], neural stem cells [12, 13], and mesenchymal stem cells (MSCs) [14–16]. MSCs derived from different tissues demonstrated various reactions to the Yamanaka factors (Oct4, Sox2, c-Myc, Klf4) but were not easier to reprogram because they are relatively primitive compared with other differentiated cells. Simply using Yamanaka factors failed to induce MSCs from human bone marrow to produce cells with ESC-like properties. The addition of human telomerase to catalyze subunit hTERT and SV40 large T-Antigen was also required to reprogram these MSCs [14]. Reprogramming mouse and human adipose tissue-derived MSCs into iPSCs has a higher efficiency compared with that for mouse and human fibroblasts [15]. MSCs or progenitor cells derived from teeth or dental pulp can also be reprogrammed into iPSCs [16]. Nevertheless, similar problems were encountered when attempting to reprogram these cells, for example: limited sources, difficulty in obtaining them, and the reality that the sampling procedure itself may cause injury to the individual donor.The ideal target somatic cells should be safely and easily accessible, methods for their retrieval reproducible, and the harvesting procedure relatively noninvasive.
Hair follicles are renewable organs, they undergo repeated cycles of growth (anagen), regression (catagen), and rest (telogen) throughout the life of mammals [17].
It has been reported that there is an essential role for the follicular dermal papilla and its related dermal sheath cells in hair follicle development and regeneration [18]. At present, the majority of skin biology research is focused on epidermal stem cells. However, the characteristics of MSCs also can be observed in hair follicle dermal cells [19, 20]. Our study first confirmed the existence of hair follicle derived MSCs obtained by direct plucking of human hairs. This method eliminates many problems associated with other procedures, and the resulting MSCs were found to be multipotent with potential to be directed toward differentiation into adipocytes and osteoblasts.
In this study, we sought to determine whether hHF-MSCs represent a cell type that may be reprogrammed to generate iPSCs. If so, this would provide a new source of reprogrammable target cells for research into personalized regenerative medicine.
Materials and Methods
Ethics Statement
After the approval of the study protocol by the Ethics committee of Jilin Province (China) and written informed consent, hHF-MSCs were obtained from the scalp of one of the authors by plucking the hairs.
Cell Culture
Several hairs with a full hair follicle were plucked, the root tissue cut with sterile ophthalmic scissors, and then placed into phosphate-buffered saline (PBS) containing penicillin and streptomycin. Hair follicle root tissues were rinsed three times in PBS containing penicillin and streptomycin, then placed into 96-well plates, cultured with hHF-MSCs medium: Dulbecco’s modified Eagle’s medium (DMEM)/Ham’s F-12 medium (DMEM/F12; Invitrogen, Carlsbad, CA, USA), 10 % fetal bovine serum (FBS; Hyclone, Victoria, Australia), 2 ng/ml basic fibroblast growth factor (bFGF; Invitrogen). Cells were passaged once the plate surface was covered with confluent cells. hHF-MSCs were frozen and stored in liquid nitrogen at passages 0–2. Cells were thawed and expanded for experimentation at passages 2–3. CF-1 mouse embryonic fibroblasts (MEFs) used in this report were derived from an embryonic day 12.5 embryo pool of CF-I mice. MEFs were cultured in DMEM (Invitrogen) supplemented with 10 % FBS. hESCs (X01) [21], obtained from the Xiaolei Group (Shanghai Institute of Biochemistry and Cell Biology, Shanghai Institutes for Biological Science, Chinese Academy of Sciences), and hHF-MSC-derived iPSCs were maintained in irradiated CF-1 MEFs in hESCs culture medium (80 % DMEM/F12 supplemented with 20 % KnockOut serum replacement, 1 % non-essential amino acids, 1 mM L-glutamine, 4 ng/ml human bFGF, 0.1 mM β-mercaptoethanol (all from Invitrogen). The hHF-MSC-derived iPSCs and hESCs were split with 1 mg/ml collagenase type IV (Invitrogen) for about 30 min at 37 °C, at a ratio of 1:6 every 6–7 days. Conditioned medium was collected according to Xu et al. [22].
Immunophenotyping of hHF-MSCs
Immunophenotyping was carried out as described previously [23]. Antibodies used were anti-CD29, anti-CD45, anti-CD105, anti-HLA-DR (Biolegend, USA), anti-CD90, anti-CD31, anti-CD73 (BD, USA) and anti-CD15 (Abcam, MA).
Differentiation Experiments
Adipogenic differentiation and detection was performed as previously described [23]. Osteogenic differentiation was induced by culturing cells at 80–90 % confluency in low glucose DMEM containing 10 % FBS, 10−8 μM dexamethasone, 10 mM β-glycerophosphate (Sigma-Aldrich, USA), and 0.2 mM L-ascorbic acid (Sigma-Aldrich, USA) for 3–4 weeks. Medium was replaced every 3 days. Mineralized bone nodules were detected using Alizarin Red S and Alkaline Phosphatase staining. Positive Alizarin Red S staining indicated calcium deposition, while Alkaline Phosphatase staining was black for bone nodules.
Lentivirus Production and Establishment of hHF-MSC-Derived iPSCs
We followed the protocol for lentiviral transduction as previously described [24]. Briefly, 3.6 μg of lentivirus vector, pLV-EF1α-CDNA-IRES-EGFP, was designed to carry one of the four transcription factor sequences encoding Oct4, Sox2, c-Myc or Klf4. 2.7 μg of lentiviral vector pCMVΔ8.91 and 1.8 μg of vesicular stomatitis virus G protein (VSV-G) were co-transfected into Human embryonic kidney (HEK) 293T cells (obtained from the Xiaolei Group, Shanghai Institute of Biochemistry and Cell Biology, Shanghai Institutes for Biological Science, Chinese Academy of Sciences) in T25 flask with Fugene HD transfection reagent (Roche, USA), respectively. Viral supernatants at 48 or 72 h post-transfection were collected for determination of viral titer. hHF-MSCs were transduced with a cocktail of lentiviruses carrying reprogramming factors at Day 0. At 24 h post-transduction, cells were harvested by trypsinization and plated onto MEFs (5 × 104 cells per well) in a six-well plate. The next day, medium was replaced with hESCs culture medium. On Day 25, iPS colonies were picked and plated onto new culture dishes.
Immunostaining
Immunostaining was carried out as previously described [25]. The primary antibodies used were: monoclonal anti-SSEA3 (1:200 dilution; Developmental Studies Hybridoma Bank); monoclonal anti-SSEA4 (1:400; Developmental Studies Hybridoma Bank); anti-Tra-1-60 (1:150; Chemicon, USA); anti-Tra-1-81 (1:150; Chemicon, USA); anti-Nanog (1:150; R&D Systems, USA); anti-Oct4 (1:100; Santa Cruz Biotechnology, Santa Cruz, CA, USA); and anti-E-Cadherin (1:100; Becton Dickinson, USA).
RNA Isolation and Quantitative Real-Time Polymerase Chain Reaction (qPCR)
Total RNA was extracted with TRIZOL reagent (Invitrogen) and used as a template for reverse transcription. The qPCR assays were performed in a Bio-Rad iQ5 real-time PCR detection system (Bio-Rad, Hercules, CA, USA) using a SYBR Green PCR Master mix (TOBOYO). The PCR primers are listed in Supplemental Table S1. Each assay was carried out in triplicate. Standard curves were acquired for both the gene of interest and the internal control, glyceraldehyde-3-phsosphate dehydrogenase (GAPDH). The CT data for the gene of interest and GAPDH were obtained from the qPCR assays then transformed into copy numbers using the standard curve. The expression value of each gene was normalized to the amount of GAPDH cDNA to calculate the relative amount of RNA present in each sample. The mRNA copy number of the gene of interest was defined as the number of copies per 106 copies of GAPDH.
Karyotype Analysis
Karyotyping was performed at the Xiangtan Center Hospital using standard protocols for high-resolution G-banding.
Teratoma Formation
The hHF-MSC-derived iPSCs were injected intramuscularly into non-obese diabetic/severe combined immune deficient (NOD/SCID) mice (approximately 5 × 106 cells per site). Two hHF-MSC-derived iPSC lines at a passage greater than 10 were used to produce teratomas. Three mice were injected for each cell line. Three tumors were generated when hHF-MSC-derived iPSCs 10-1 was used. Two tumors were generated when hHF-MSC-derived iPSCs 20-1 was used, (10-1 and 20-1 means the generated iPSCs as the multiplicity of infection of 10 or 20). After 8 weeks, tumors were processed for hematoxylin-eosin staining.
DNA Fingerprinting Analysis
To confirm the hHF-MSCs origins of the hHF-MSC-derived iPSCs, short tandem repeat (STR) analysis was performed by Beijing Microread Gene Technology Co., Ltd. (Beijing, China).
Results
Isolation and Characterization of hHF-MSCs
The hHF-MSCs migrated outwards from the human hair follicle root tissue and adhered to the surface of the culture plate (Fig. 1a, b). They were passaged every 3–4 days to a maximum of 12 passages without major morphological alteration. The primary and passaged cells all displayed typical fibroblast-like morphology (Fig. 1d, e, f). The hair follicle is a complex organ, and it is one of the few organs of the body that undergoes cyclic bouts of degeneration and regeneration throughout life [26]. It contains a variety of different types of cells. In the primary culture, in addition to hHF-MSCs, keratinocytes were also observed around the hair follicle (Fig. 1c), as described previously by Aasen et al. [27].
Fig. 1.
Isolation and characterization of human hair follicle mesenchymal stem cells (hHF-MSCs). The hHF-MSCs, resembling typical fibroblast-like cells, migrated out from the hair follicles (a Bars = 500 μm; b Bars = 100 μm). Keratinocytes also migrated out from the hair follicles (c Bars = 500 μm). hHF-MSCs from passage 2 (d), passage 8 (e) to passage 12 (F Bars = 200 μm). Flow cytometric analysis of cell surface markers on hHF-derived fibroblast-like cells. 2 × 105 cells were incubated with primary antibodies against CD29, CD73, CD105, CD90, HLA-DR, CD31, CK15 or CD45,respectively, followed by incubation with a secondary FITC-labeled antibody. Controls were incubated with secondary antibody only. Percentages indicate the fraction of cells that stained positive (g). Adipogenic differentiation of hHF-MSCs. Compared to non-induced control (h Bars = 100 μm), induction after 3 weeks, the number of intracellular lipid droplets was further increased (i Bars = 100 μm) and was detected by Oil-red O staining (j Bars = 100 μm). Osteogenic differentiation of hHF-MSCs. Compared to non-induced control (k Bars = 200 μm), calcium nodules were formed after induction for 4 weeks and was demonstrated by Alizarin red staining (l Bars = 200 μm) and Alkaline phosphatase staining (m Bars = 200 μm)
Flow cytometry (2 × 105 cells were used for each antibody) demonstrated that the majority of the hHF-MSCs expressed the MSC markers CD29, CD73, CD105, CD90 (99.8 %, 98.4 %, 84.4 % and 99.5 %, respectively). In contrast, hematopoietic (CD31, CD45) and keratinocyte (CK15) markers were not detected. Additionally, cell populations were negative for the major histocompatibility complex (MHC) class II (HLA-DR) antigen (Fig. 1g).
The hHF-MSCs had multilineage differentiation potential. After adipogenic induction for 4–5 days, the morphology of the cells changed from long spindle-shape into a round or polygonal shape. One week later, small bubble-shaped lipid droplets appeared in part of the cells (Supplemental Figure S1). After induction for 3 weeks, both the size and the number of lipid droplets were all increased, and most of the differentiated cells showed red lipid droplets throughout their cytoplasm (Fig. 1i, j).
When hHF-MSCs were cultured in osteogenic medium for 4 weeks, osteoblast-like cells were clearly demonstrated by alkaline phosphatase staining (Fig. 1m). In vitro mineralization could be shown by Alizarin red staining (Fig. 1l).
Formation of hESCs-Like Colonies and Their Expansion
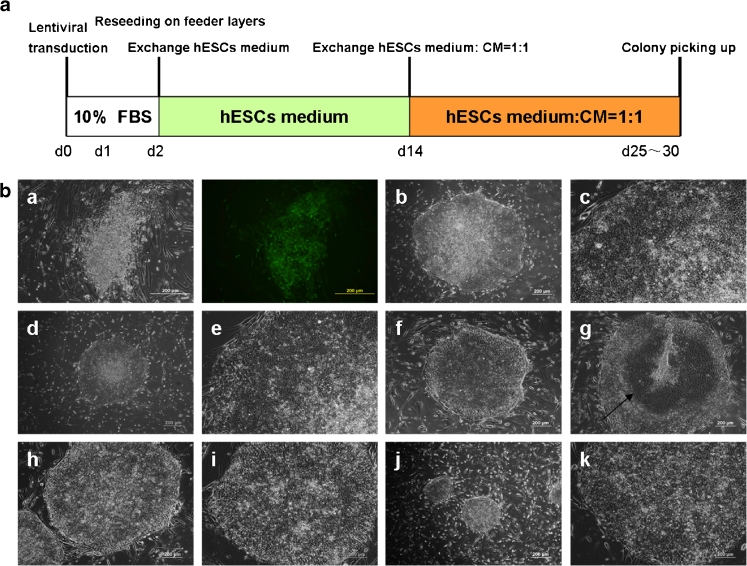
The schematic diagram of the reprogramming protocol is shown in Fig. 2a, and detailed methods are included in the “Materials and Methods” section. Viruses expressing a cocktail of reprogramming factors (OCT4, SOX2, C-MYC and KLF4) were used to transduce hHF-MSCs. 24 h after transduction, the cells were harvested by trypsinization and plated onto MEFs. Colonies with a hESCs-like morphology first became visible 7 days after transduction. When the colonies had been grown for 25–30 days, 131 colonies were picked mechanically according to the morphological characteristics of hESCs. Two hESCs-like colonies were finally selected on the basis of further characterization, and were named, hHF-MSC-derived iPSCs 10-1 and hHF-MSC-derived iPSCs 20-1 (Fig. 2b).
Fig. 2.
Generation of induced pluripotent stem cells from hHF-MSCs. (a) Schematic diagram of the reprogramming protocol used. (b) hHF-MSCs were transduced with four factors and seeded onto mouse embryonic fibroblasts until hESCs-like colonies emerged (representative colonies). (a) Typical non-hESCs-like colony. (b) hESCs colony, X01 at passage 24. (c) High magnification of the hESCs at passage 24. (d) Typical iPS colony, hHF-MSC-derived iPSCs 10-1 at passage 2. (e) High magnification of the hHF-MSC-derived iPSCs 10-1 at passage 2. (f) hHF-MSC-derived iPSCs 10-1 at passage 16. (g) Arrow indicates area of differentiation in the center of hHF-MSC-derived iPSCs 10-1 at passage 16. (h) hHF-MSC-derived iPSCs 20-1 at passage 3. (i) High magnification of hHF-MSC-derived iPSCs. at passage 3. (j) hHF-MSC-derived iPSCs 20-1 at passage 17. (k) High magnification of hHF-MSC-derived iPSCs 20-1 at passage 17. Bars = 200 μm
hHF-MSC-Derived iPSCs Exhibit Similar Morphology and Marker Expression to hESCs
hHF-MSC-derived iPSCs exhibited a similar morphology to that of hESCs, characterized by a high nucleus to cytoplasm ratio and prominent nucleoli. They have now been passaged continuously for more than 20 passages (>120 days). As observed in hESCs, the hHF-MSC-derived iPSCs expressed typical hESCs surface markers, including stage-specific embryonic antigen SSEA-3, SSEA-4, TRA-1-60, TRA-1-81 and E-Cadherin. They have also expressed alkaline phosphatase, Nanog (Fig. 3).
Fig. 3.
hESCs pluripotency marker expression by these iPSCs. a hESCs colonies, X01 at passage 25 were fixed and stained with antibodies against hESCs associated pluripotent proteins and examined under the fluorescence microscopy. b hHF-MSC-derived iPSCs 10-1 at passage 5. c hHF-MSC-derived iPSCs 20-1 at passage 6. Expressed proteins appeared red in the fluorescence microscope. Bars = 200 μm
We used quantitative PCR to analyze the expression of endogenous Oct4 and Sox2 and other pluripotency-associated genes, including Nanog, Cripto, FoxD3, Lin28, FGF4 and ESG1. Our results show that all of these genes were robustly induced. The expression of endogenous genes associated with pluripotency in hHF-MSC-derived iPSCs was effectively raised to a level similar to that observed in hESCs (Fig. 4a). These observations suggest that endogenous pluripotent genes can be fully induced by the reprogramming of hHF-MSCs. We used absolute quantitative PCR to calculate the mRNA copy number of Oct4 and Sox2 in these cells and found that the expression of exogenous genes Oct4 and Sox2 in hHF-MSC-derived iPSCs was downregulated compared to that in hESCs (Fig. 4b). Karyotype analysis showed that hHF-MSCs at passage 5, along with hHF-MSC-derived iPSCs 10-1 at passage 10 and hHF-MSC-derived iPSCs 20-1 at passage 11 had a normal male chromosome type (46XY) with no observable chromosome abnormalities (Fig. 5). DNA fingerprinting analysis confirmed the origin of the reprogrammed colonies. (Supplemental Table S2).
Fig. 4.
a Quantitative reverse transcription-polymerase chain reaction (PCR) analyses of endogenous Oct4, Sox2, Nanog and other pluripotency genes expression in hESCs and hHF-MSC-derived iPSCs relative to parental somatic cell populations. b Quantitative RT-PCR analyses of exogenous gene Oct4 and Sox2 expression
Fig. 5.
Karyotype analysis. hHF-MSCs at passage 5 (a), hHF-MSC-derived iPSCs 10-1 at passage 10 (b) and hHF-MSC-derived iPSCs 20-1 at passage 11(c) all showed a normal 46XY karyotype
Teratoma Formation
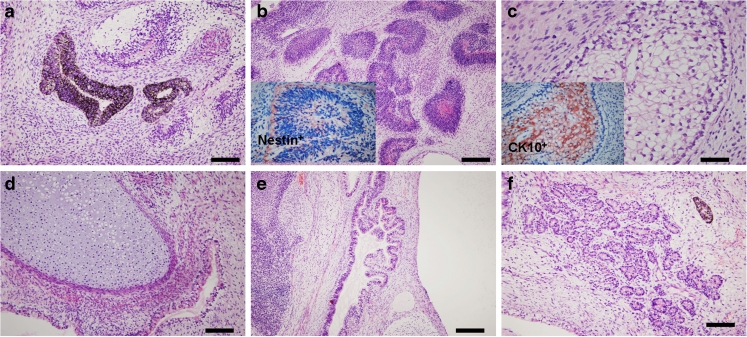
hHF-MSC-derived iPSCs were injected intramuscularly into NOD/SCID mice. At 4–5 weeks after injection we observed tumor formation. After 8 weeks, tumors had formed in three mice injected with passage 13 hHF-MSC-derived iPSCs 10-1. Tumors also formed in two mice injected with passage 14 hHF-MSC-derived iPSCs 20-1, one of which died. Histological examination revealed that the tumors contained various tissues of the three germ layers, including pigmented epithelium (ectoderm), neural tissues (ectoderm), cartilage (mesoderm), gut-like epithelial tissues (endoderm) and respiratory epithelium (endoderm) (Fig. 6). Tumors were not observed in mice injected with hHF-MSCs only.
Fig. 6.
Hematoxylin-eosin staining of teratomas derived from hHF-MSC-derived iPSCs. Teratomas is composed of various type of tissues: (a) Pigmented epithelium (ectoderm). b Rosettes of the neural epithelium (ectoderm). c squamous epithelium (ectoderm). d cartilage tissue (mesoderm). e Respiratory epithelium (endoderm). f Gland-like structures (endoderm). Bars = 500 μm
Discussion
It is possible to use a terminally differentiated somatic cell in order to produce a whole animal or individual via transfer of the cell nucleus or cloning [28, 29]. Cell nucleus transfer technology has shown that differentiated cells can return to their more immature state if provided with the appropriate conditions. Recently, somatic cells have been reprogrammed directly to a pluripotent state by forced expression of four or less transcription factors to generate personalized iPSCs [2–16]. This approach not only bypasses immune rejection but also circumvents barriers of technical, ethical and legal issues which apply to the use of embryonic materials.
At present, a wide variety of cells from many different species, including humans, have been reprogrammed into iPSCs [2–16]. However, the methods for obtaining target cells for reprogramming are invasive and painful, often accompanied by restrictions on time, place and quantity of available tissues. In our study we attempted to establish a simple, universally applicable (any age, sex and physical condition) and noninvasive sampling method. We also sought to determine whether hHF-MSCs represent a reprogrammable cell type for use in generating iPSCs. This is the first report to demonstrate that hHF-MSCs can be reprogrammed into iPSCs.
Multipotent MSCs were originally found in the bone marrow, and were considered to be the inherent stem cells at this location [30]. Later, these MSCs or MSC-like cells have been separated from, and identified in various tissues, including white adipose tissue, pancreas, skin, and the umbilical cord blood [31–34]. In recent years, studies have shown the presence of MSCs in the dermal sheath and dermal papillae of the hair follicle: In 2006, a comparative study investigated the properties of follicular dermal stem cells from whisker hairs of Wistar rats and bone marrow MSCs were isolated from femora of the same animals [35]. The results from that study showed that dermal stem cells from hair follicles have a similar morphology and population doubling time, and express the same cell-surface markers as the MSCs. Following exposure to appropriate induction stimuli, both cell populations have the capacity to differentiate into various mesenchymal lineages, such as osteoblasts, adipocytes, chondrocytes and myocytes.
Our research group tried to determine whether MSCs can be obtained from human follicles through plucking methods. Research on hair follicle stem cells indicates that most hair follicle samples have been derived from animal backskins, or the human scalp by surgical means. If we could obtain MSCs through simple plucking, it would have great significance for stem cell research. Recently, it has been shown that skin keratinocytes can also be obtained by the same approach and they have already been reprogrammed into iPSCs [27]. Since the specific location where hHF-MSCs reside remains unclear [36], we postulate that there may be a niche in the dermal sheath and/or hair papilla. In the process of reprogramming hHF-MSCs, we observed iPS cell colony formation about 26 days after transduction, resulting in a reprogramming efficiency of ∼0.001 %. hHF-MSCs possess lower reprogramming efficiencies than do cells from human hair follicle dermal papilla which have a reprogramming efficiency of ∼0.02 %, as shown by Christiano’s research group [37]. We suggest that the possible reason for this observation is that the endogenous expression of Sox2 and Nanog in dermal papilla cells is higher than in hHF-MSCs, which express low levels of endogenous Sox2 and Nanog (Supplemental Figure S2, Fig. 4a). Possibly these represent different cell populations of hHF-MSCs. Rendl et al., using only the transcription factors Oct4 or Oct4/Klf4 successfully reprogrammed mouse dermal papilla cells into iPSCs [38, 39]. This is in contrast to hHF-MSCs, which are incapable of being reprogrammed using less than four factors or use Oct4 alone. This is the same observation as made when reprogramming human hair follicle dermal papilla cells by Christiano et al. [37]. There are other noninvasive methods to obtain cells. Pei et al. [40] have successfully derived iPSCs from exfoliated renal tubular cells present in urine samples. This maybe a preferred souce for generating iPSCs.
Although we have a relatively better way to obtain cells, the lentivirus we used for gene delivery will integrate into the genome anyway, and may cause insertional mutagenesis risks that could be resolved via usage of a different approach, such as synthetic mRNA [41] or any of the other integration-free approaches that are currently being performed in the field.
We conclude that the hHF-MSCs provides an accessible source for making iPSCs, and that hHF-MSC-derived iPSCs are morphological similar to hESCs in that they exhibit a high nuclear/cytoplasmic ratio and express specific surface markers of hESCs. They also expressed pluripotency-associated transcription factor genes at similar levels as hESCs. They were able to form teratomas in vivo containing tissues of all three germ layers.
Conclusion
In summary, MSCs were isolated from human hair follicle tissues by noninvasive means. The differentiation potential of hHF-MSCs was increased by reprogramming with Yamanaka factors. The method used to access hHF-MSCs was very convenient and enabled quick establishment of a large-scale hHF-MSCs library, which was then used to create an hHF-MSC-derived iPSCs bank. This work lays the foundation for further research into how these hHF-MSC-derived iPSCs can be applied to personalized regenerative medicine.
Electronic supplementary material
Below is the link to the electronic supplementary material.
(DOC 42 kb)
(DOC 31 kb)
Adipogenic differentiation of hHF-MSCs. Compared to non-induced control (A), induction after 1 week, small bubble-shaped lipid droplets appeared in part of the cells (B). Induction after 2 weeks, an increase in the number of lipid droplets was observed (C) and was detected by Oil-red O staining (D). Bars=100 μm (JPEG 41 kb)
Quantitative reverse transcription-polymerase chain reaction (PCR) analyses of gene (Oct4, Sox2, c-Myc and Klf4) expression in hHF-MSCs. Compared to expression of four genes in hESCs (X01), the expression of Klf4 in hHF-MSCs was at high levels,, and the expression of Oct4, Sox2 and c-Myc was absent. RNA was isolated from hHF-MSCs at passage 5; hESCs (X01) at passage 25 (JPEG 22 kb)
Acknowledgments
We thank Xiao Lei and Wu Zhao at the Shanghai Academy of Sciences for their technical assistance and Dr William Orr, Department of Pathology, University of Manitoba, Canada, for revising the language of this manuscript. We acknowledge the assistance given by Li Hui, Director of Reproductive Center at Xiangtan Central Hospital, for the karyotype analysis. This study was supported in part by the Major State Basic Research Development Program of China (973 Program) (2011CB606201). This work was also partially supported by National Natural Science Foundation of China (30930026/C100101) and Science and Technology Development Plan of the Office of Science and Technology Project in Jilin Province (200905180).
Conflicts of interest
The authors declare no potential conflict of interest.
Abbreviations
- iPSCs
Induced pluripotent stem cells
- ESCs
Embryonic stem cells
- hESCs
Human embryonic stem cells
- MSCs
Mesenchymal stem cells
- hHF-MSCs
Human hair follicle mesenchymal stem cells
- MEFs
Mouse embryonic fibroblasts
References
- 1.Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 2.Takahashi K, Tanabe K, Ohnuki M, et al. Induction of pluriptent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 3.Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 4.Wenig M, Meissner A, Foreman R, et al. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:318–324. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- 5.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 6.Liu H, Zhu F, Yong J, Zhang P, Hou P, Li H, et al. Generation of induced pluripotent stem cells from adult rhesus monkey fibroblasts. Cell Stem Cell. 2008;3:587–590. doi: 10.1016/j.stem.2008.10.014. [DOI] [PubMed] [Google Scholar]
- 7.Esteban MA, Xu J, Yang J, Peng M, Qin D, Li W, et al. Generation of induced pluripotent stem cell lines from Tibetan miniature pig. Journal of Biological Chemistry. 2009;284:17634–17640. doi: 10.1074/jbc.M109.008938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stadtfeld M, Brennand K, Hochedlinger K. Reprogramming of pancreatic beta cells into induced pluripotent stem cells. Current Biology. 2008;18:890–894. doi: 10.1016/j.cub.2008.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu H, Ye Z, Kim Y, Sharkis S, Jang YY. Generation of endoderm-derived human induced pluripotent stem cells from primary hepatocytes. Hepatology. 2010;51:1810–1819. doi: 10.1002/hep.23626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hanna J, Markoulaki S, Schorderet P, Carey BW, Beard C, et al. Direct reprogramming of terminally differentiated mature B lymphocytes to pluripotency. Cell. 2008;133:250–264. doi: 10.1016/j.cell.2008.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eminli S, Foudi A, Stadtfeld M, Maherali N, Ahfeldt T, et al. Differentiation stage determines potential of hematopoietic cells for reprogramming into induced pluripotent stem cells. Nature Genetics. 2009;41:968–976. doi: 10.1038/ng.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim JB, Zaehres H, Wu G, Gentile L, Ko K, et al. Pluripotent stem cells induced from adult neural stem cells by reprogramming with two factors. Nature. 2008;454:646–650. doi: 10.1038/nature07061. [DOI] [PubMed] [Google Scholar]
- 13.Kim JB, Greber B, AraÚzo-Bravo MJ, Meyer J, Park KI, et al. Direct reprogramming of human neural stem cells by OCT4. Nature. 2009;461:649–653. doi: 10.1038/nature08436. [DOI] [PubMed] [Google Scholar]
- 14.Park IH, Zhao R, West JA, Yabuuchi A, Huo H, et al. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- 15.Sugii S, Kida Y, Kawamura T, Suzuki J, Vassena R, et al. Human and mouse adipose-derived cells support feeder independent induction of pluripotent stem cells. Proceedings of the National Academy of Sciences. 2010;107:3558–3563. doi: 10.1073/pnas.0910172106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yan X, Qin H, Qu C, Tuan RS, Shi S, et al. iPS cells reprogrammed from human mesenchymal-like stem/progenior cells of dental tissue origin. Stem Cells and Development. 2010;19:469–480. doi: 10.1089/scd.2009.0314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoffman RM. The hair follicle as a gene therapy target. Nature Biotechnology. 2000;18:20–1. doi: 10.1038/71866. [DOI] [PubMed] [Google Scholar]
- 18.Ohyama M, Zheng Y, Paus R, Stemm KS. The mesenchymal component of hair follicle neogenesis: background, methods and molecular characterization. Experimental Dermatology. 2009;19:89–99. doi: 10.1111/j.1600-0625.2009.00935.x. [DOI] [PubMed] [Google Scholar]
- 19.Richardson GD, Arnott EC, Whitehouse CJ, Lawrence CM, Hole N, et al. Cultured cells from the adult human hair follicle demis can be directed toward adipogenic and osteogenic differentiation. The Journal of Investigative Dermatology. 2005;124:1090–1091. doi: 10.1111/j.0022-202X.2005.23734.x. [DOI] [PubMed] [Google Scholar]
- 20.Reynolds AJ, Lawrence C, Cserhalmi-Friedman PB, Christiano AM, Jahoda CA. Trans-gender induction of hair follicles. Nature. 1999;402:33–34. doi: 10.1038/46938. [DOI] [PubMed] [Google Scholar]
- 21.Wu Z, Li H, Rao L, He L, Bao L, et al. Derivation and characterization of human embryonic stem cell lines from the Chinese population. Journal of Genetics and Genomics. 2011;38:13–20. doi: 10.1016/j.jcg.2010.12.006. [DOI] [PubMed] [Google Scholar]
- 22.Xu C, Inokuma MS, Denham J, Golds K, Kundu P, et al. Feeder-free growth of undifferentiated human embryonic stem cells. Nature Biotechnology. 2001;19:971–974. doi: 10.1038/nbt1001-971. [DOI] [PubMed] [Google Scholar]
- 23.Yang XF, He X, He J, Zhang LH, Xj S, et al. High efficient isolation and systematic identification of human adipose-derived mesenchymal stem cells. Journal of Biomedical Science. 2011;18:59. doi: 10.1186/1423-0127-18-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liao J, Wu Z, Wang Y, Cheng L, Cui C, et al. Enhanced efficiency of generating induced pluripotent stem (iPS) cells from human somatic cells by a combination of six transcription factors. Cell Research. 2008;18:600–603. doi: 10.1038/cr.2008.51. [DOI] [PubMed] [Google Scholar]
- 25.Xiao L, Yuan X, Sharkis SJ. Activin A maintains self-renewal and regulates fibroblast growth factor, Wnt, and bone morphogenic protein pathways in human embryonic stem cells. Stem Cells. 2006;24:1476–1486. doi: 10.1634/stemcells.2005-0299. [DOI] [PubMed] [Google Scholar]
- 26.Fuchs E. Scratching the surface of skin development. Nature. 2007;445:834–842. doi: 10.1038/nature05659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aasen T, Izpisúa Belmonte JC. Isolation and cultivation of human keratinocytes from skin or plucked hair for the generation of induced pluripotent stem cells. Nature Protocols. 2010;5:371–382. doi: 10.1038/nprot.2009.241. [DOI] [PubMed] [Google Scholar]
- 28.Solter D. Mammalian cloning: advances and limitations. Nature Reviews Genetics. 2000;1:199–207. doi: 10.1038/35042066. [DOI] [PubMed] [Google Scholar]
- 29.Rideout WM, Eggan K, Jaenisch R. Nuclear cloning and epigenetic reprogramming of the genome. Science. 2001;293:1093–1098. doi: 10.1126/science.1063206. [DOI] [PubMed] [Google Scholar]
- 30.Uccelli A, Moretta L, Pistoia V. Mesenchymal stem cells in health and disease. Nature Reviews Immunology. 2008;8:726–736. doi: 10.1038/nri2395. [DOI] [PubMed] [Google Scholar]
- 31.Zuk PA, Zhu M, Ashjian P, De Ugarte DA, Huang JI, et al. Human adipose tissue is a source of multipotent stem cells. Molecular Biology of the Cell. 2002;13:4279–4295. doi: 10.1091/mbc.E02-02-0105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baertschiger RM, Bosco D, Morel P, Serre-Beinier V, Berney T, et al. Mesenchymal stem cells derived from human exocrine pancreas express transcription factors implicated in beta-cell development. Pancreas. 2008;37:75–84. doi: 10.1097/MPA.0b013e31815fcb1e. [DOI] [PubMed] [Google Scholar]
- 33.Metcalfe AD, Ferguson MW. Skin stem and progenitor cells: using regeneration as a tissue-engineering strategy. Cellular and Molecular Life Sciences. 2008;65:24–32. doi: 10.1007/s00018-007-7427-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Markov V, Kusumi K, Tadesse MG, William DA, Hall DM, et al. Identification of cord blood-derived mesenchymal stem/stromal cell populations with distinct growth kinetics, differentiation potentials, and gene expression profiles. Stem Cells and Development. 2007;16:53–73. doi: 10.1089/scd.2006.0660. [DOI] [PubMed] [Google Scholar]
- 35.Hoogduijn MJ, Gorjup E, Genever PG. Comparative characterization of hair follicle dermal stem cells and bone marrow mesenchymal stem cells. Stem Cell and Development. 2006;15:49–60. doi: 10.1089/scd.2006.15.49. [DOI] [PubMed] [Google Scholar]
- 36.Toma JG, Akhavan M, Fernandes KJ, Barnabé-Heider F, Sadikot A, et al. Isolation of multipotent adult stem cells from the dermis of mammalian skin. Nature Cell Biology. 2001;3:778–784. doi: 10.1038/ncb0901-778. [DOI] [PubMed] [Google Scholar]
- 37.Higgins CA, Itoh M, Inoue K, Richardson GD, Jahoda CA, et al. Reprogramming of human hair follicle dermal papilla cells into induced pluripotent stem cells. The Journal of Investigative Dermatology. 2012;132:1725–1727. doi: 10.1038/jid.2012.12. [DOI] [PubMed] [Google Scholar]
- 38.Tsai SY, Bouwman BA, Ang YS, Kim SJ, Lee DF, et al. Single transcription factor reprogramming of hair follicle dermal papilla cells to induced pluripotent stem cells. Stem Cells. 2011;29:964–971. doi: 10.1002/stem.649. [DOI] [PubMed] [Google Scholar]
- 39.Tsai SY, Clavel C, Kim S, Ang YS, Grisanti L, et al. Oct4 and klf4 reprogram dermal papilla cells into induced pluripotent stem cells. Stem Cells. 2010;28:221–228. doi: 10.1002/stem.281. [DOI] [PubMed] [Google Scholar]
- 40.Zhou T, Benda C, Duzinger S, Huang Y, Li X, et al. Generation of induced pluripotent stem cells from urine. Journal of the American Society of Nephrology. 2011;22:1221–1228. doi: 10.1681/ASN.2011010106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Warren L, Manos PD, Ahfeldt T, Loh YH, Lau F, et al. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell. 2010;7:618–630. doi: 10.1016/j.stem.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC 42 kb)
(DOC 31 kb)
Adipogenic differentiation of hHF-MSCs. Compared to non-induced control (A), induction after 1 week, small bubble-shaped lipid droplets appeared in part of the cells (B). Induction after 2 weeks, an increase in the number of lipid droplets was observed (C) and was detected by Oil-red O staining (D). Bars=100 μm (JPEG 41 kb)
Quantitative reverse transcription-polymerase chain reaction (PCR) analyses of gene (Oct4, Sox2, c-Myc and Klf4) expression in hHF-MSCs. Compared to expression of four genes in hESCs (X01), the expression of Klf4 in hHF-MSCs was at high levels,, and the expression of Oct4, Sox2 and c-Myc was absent. RNA was isolated from hHF-MSCs at passage 5; hESCs (X01) at passage 25 (JPEG 22 kb)