Abstract
For this study, we used DNA-based immunizations to elicit gamma interferon-producing (Tc1) or interleukin 4 (IL-4)-producing (Tc2) CD8 T cells to the influenza virus nucleoprotein. We examined the response of these cells to an intranasal viral challenge. Both the Tc2- and Tc1-biased responses were present in mice with predominantly IL-4-producing (Th2) CD4 T cells. After viral challenge, Tc1 cells underwent more efficient expansion than did Tc2 cells, and only Tc1 cells were detected at the site of infection. In contrast, the CD4 response remained IL-4 biased. However, only a limited number of CD4 cells appeared in the postchallenge lung, and these were strongly enriched for the Th1 phenotype. Thus, the type of memory T-cell response induced by DNA vaccination does not determine the type of response that will predominate at the site of an infection.
Cellular immune responses are characterized by different patterns of lymphokine secretion that are determined by the interface between innate and acquired immune responses (for reviews, see references 10, 11, and 18). The nature of the microenvironment in which a T cell is activated by an antigen-presenting cell influences the differentiation of the T cell into different types of responses, which are defined and categorized based on the profile of the cytokines produced. The signature cytokine for type 1 cells is gamma interferon (IFN-γ), whereas the signature cytokine for type 2 cells is interleukin 4 (IL-4). Type 1 and type 2 responses were originally identified for clones of CD4 T-helper (Th) cells (9). Subsequent in vitro studies have revealed similar cytokine profiles for other lymphocyte populations, including CD8 cytotoxic T cells (Tc) (15), B cells (6), dendritic cells (17), natural killer cells (13), and macrophages (8).
Specialization of the immune response toward type 1 or type 2 influences how the immune system combats a pathogen. Type 1 responses are associated with high levels of IFN-γ-producing cytotoxic T cells, rearrangement of immunoglobulin (Ig) genes to complement-dependent antibody, and the activation of phagocytic cells such as macrophages (5, 18, 22). In contrast, type 2 responses are associated with rearrangements of Ig to complement-independent and IgE antibodies and with the mobilization of nonphagocytic defenses such as eosinophils that combat microbes by the release of toxic factors (5, 18, 22). Type 1 responses are the desired response for fighting intracellular viral or bacterial infections, whereas type 2 responses are favored for combating extracellular parasitic infections.
Recently, we demonstrated that DNA-based immunizations could be used to elicit responses that varied from type 1 responses with predominantly IFN-γ-producing CD4 and CD8 T cells and complement-dependent antibody, through mixed type 1 and type 2 responses, to type 2 responses in which most of the CD4 and CD8 T cells produced IL-4 and the associated antibody was complement independent (12). These studies used DNA vaccines expressing secreted and plasma membrane-associated forms of the influenza virus hemagglutinin (HA) glycoprotein. The most strongly biased type 1 responses were raised by intramuscular saline injections of DNA encoding the plasma membrane form of HA (tmHA), whereas the most extreme type 2 responses were raised by gene gun deliveries of the secreted form of HA (sHA). Herein, we initiated type 1 and type 2 CD8 T-cell responses against the influenza virus nucleoprotein and assessed the fates of these responses following a viral challenge.
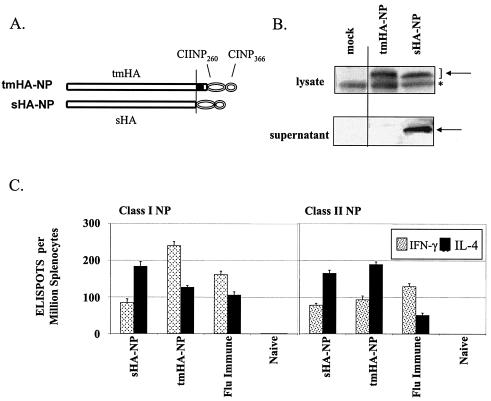
Immunizations for an immunodominant epitope in the influenza virus nucleoprotein (NP) were conducted using DNAs expressing the HA of influenza virus A/PR/8/34 (H1N1) fused at the C terminus to H-2b-restricted class II and class I NP epitopes (1, 19). These were termed tmHA-NP and sHA-NP, respectively (Fig. 1A). After transient transfections, Western blots detected the ∼75 kDa tmHA-NP in the cell lysates and the ∼70 kDa sHA-NP in the cell lysates and supernatants (Fig. 1B).
FIG. 1.
DNA vaccines, expression, and immune responses. (A) DNA expression vectors encoding transmembrane and secreted forms of HA coupled with the H-2Db-restricted class I NP366-374 (CINP366)- and class II NP260-283 (CIINP260)-designated tmHA-NP and sHA-NP, respectively (12). (B) Western blot of the expressed proteins. The Western blot used polyclonal mouse sera that recognized influenza A (H1N1). The specific bands are denoted by the arrows, and a nonspecific band is represented by an asterisk. (C) Ex vivo ELISPOT analyses of splenocytes harvested 4 weeks after gene gun vaccination of C57BL/6 mice with 2 μg of DNA at 0 and 4 weeks (for a review of the methods, see reference 12). Responding cells were stimulated with peptides representing amino acids 366 to 374 (Class I) and amino acids 260 to 283 (Class II) of NP. The immunizing DNA and the lymphokine tested for in the assays are indicated below the bars. Bars indicate the average response ± the standard deviation for groups of five mice.
Gene gun inoculations of sHA-NP were used to elicit a Tc2-biased response, and gene gun inoculations of tmHA-NP were used to elicit a Tc1-biased response against NP (Fig. 1C) (12). Gene gun vaccinations were delivered at 0 and 4 weeks, and splenocytes were harvested at 8 weeks for ex vivo enzyme-linked immunosorbent assays (ELISPOTs) for IL-4- and IFN-γ-producing NP-specific T cells (Fig. 1C). The ELISPOTs used epitope-specific NP class I and class II peptides for stimulation. Gene gun delivery of the sHA-NP DNA elicited a Tc2/Th2-biased response with about twice as many IL-4- as IFN-γ-producing cells for both CD8 and CD4 T cells. In contrast, gene gun delivery of the tmHA-NP DNA elicited a Tc1/Th2 response with twice as many IFN-γ- as IL-4-producing CD8 T cells and more IL-4- than IFN-γ-producing CD4 T cells. As expected, splenocytes from a mouse that had survived an influenza virus infection had a Tc1/Th1-biased immune response, with NP-specific CD8 and CD4 T cells having higher frequencies of IFN-γ- than IL-4-producing cells.
To evaluate the responses of the NP-specific Tc2 and Tc1 cells to a challenge infection, we challenged mice intranasally with an influenza virus. To prevent neutralizing antibody from blocking the challenge infection, we used hemagglutinin of the H1 subtype for the immunizations and used the A/HK/X-31 H3N2 reassortant virus (A/Aichi/2/68 × A/PR/8/34) for the challenge. Neutralizing antibodies raised to these subtypes do not cross-react. The challenge was administered 4 weeks after the second DNA inoculation at week 8 in the study.
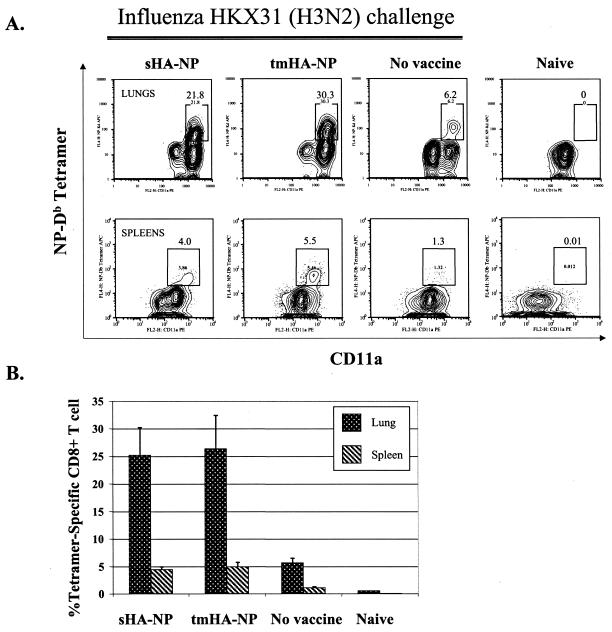
Following the intranasal challenge, the NP-specific CD8 T cells underwent rapid expansion (Fig. 2). At 12 days postchallenge, a major histocompatibility complex tetramer that recognizes T-cell receptors specific for the H-2Db-restricted NP366-374 epitope was used to quantify the frequencies of specific CD8 cells in the lungs and spleens of the challenged mice. NP-specific CD8 cells underwent expansion in both the sHA-NP- and tmHA-NP-immunized mice. As expected, the expansion of NP-specific CD8 T cells was more marked in the lung than in the spleen. About 25% of the total lung CD8 T cells were NP specific in both the sHA-NP- and tmHA-NP-immunized mice. These levels of NP-specific cells were higher than those in the lungs of unimmunized control mice, which were about 6% of the total CD8 T cells. In the spleens, the tmHA-NP and sHA-NP mice had 5.0 to 6.0% tetramer-specific CD8 T cells, in comparison with just under 1% in the unimmunized control mice. The tetramer-positive cells were activated CD4−, CD19−, CD8+, CD11ahi, NP-specific T cells.
FIG. 2.
Postchallenge H2-Db-NP tetramer-specific cells (A) Data are for tetramer-specific T cells in the lungs and spleens of individual mice vaccinated with the indicated DNAs and then challenged with 0.1 hemagglutinating unit of influenza A/HK/x31 (H3N2) at 12 days postchallenge. Lungs were harvested and processed for white blood cells in the same manner as were spleens (12). Cells were gated on CD8+, CD4−, and CD19− cells and were cell surface stained for activated (CD11ahi) NP-specific (Db-NP tetramer+) cells. (B) Percentage of tetramer-positive CD8 T cells for each group. Values represent the averages and standard deviations of the results for five mice per group and are representative of the results for two independent experiments. Groups are indicated below the bars.
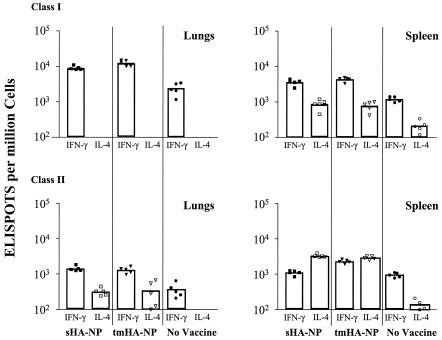
Examination of the cytokines produced by the postchallenge CD8 T cells revealed primarily IFN-γ- and not IL-4-producing cells in the lung (Fig. 3) (Table 1). At both 12 days and earlier times postchallenge, the high frequencies of NP-specific CD8 T cells in the lung appeared to be exclusively IFN-γ secreting. Before challenge, no NP-specific CD8 cells were detected in the lung, with the entire population of IFN-γ-specific T cells appearing to have been recruited to the lung in response to the challenge infection. In the spleen, IFN-γ-producing NP-specific T cells also predominated. Prior to challenge, the spleens of the sHA-NP-vaccinated mice had contained higher frequencies of IL-4-producing than of IFN-γ-producing cells. However, the frequencies of IFN-γ-producing cells underwent much greater expansion (20- to 30-fold) than the IL-4-producing T cells (4- to 7-fold) in response to the challenge.
FIG. 3.
ELISPOT analyses of responding T cells. Lung and spleen cells from influenza-challenged mice were stimulated with class I or class II NP peptide, and ELISPOTS for both IFN-γ and IL-4 were performed, as previously described (12). Symbols indicate data for individual mice, and the bars show data for the average of five mice per group. Immunogens are indicated at the bottom of the figure, and tested lymphokines are shown below the data bars.
TABLE 1.
NP-specific ELISPOT data for vaccinated mice pre- and postchallengea
| T-cell type, vaccine, and cytokine | Data for spleen
|
Data for lungs
|
|||||
|---|---|---|---|---|---|---|---|
| Pre | Post 1 | Post 2 | Avg increase (n-fold) | Pre | Post 1 | Post 2 | |
| CD4 | |||||||
| sHA-NP | |||||||
| IL-4 | 166 ± 12 | 3,744 ± 158 | 3,307 ± 370 | 21 | <b | NDc | 316 ± 78 |
| IFN-γ | 76 ± 7 | 1,238 ± 121 | 1,096 ± 155 | 15 | < | ND | 1,213 ± 236 |
| tmHA-NP | |||||||
| IL-4 | 181 ± 12 | 3,540 ± 157 | 2,902 ± 317 | 18 | < | ND | 341 ± 250 |
| IFN-γ | 97 ± 4 | 1,200 ± 101 | 2,300 ± 259 | 18 | < | ND | 1,296 ± 259 |
| CD8 | |||||||
| sHA-NP | |||||||
| IL-4 | 179 ± 11 | 641 ± 54 | 882 ± 276 | 4 | < | < | < |
| IFN-γ | 85 ± 2 | 2,100 ± 141 | 3,624 ± 701 | 34 | < | 6,321 ± 210 | 9,016 ± 1,125 |
| tmHA-NP | |||||||
| IL-4 | 116 ± 5 | 861 ± 23 | 781 ± 237 | 7 | < | < | < |
| IFN-γ | 237 ± 11 | 4,294 ± 153 | 4,355 ± 552 | 18 | < | 15,124 ± 226 | 12,487 ± 2,243 |
Values represent the average numbers of NP-specific T cells producing the indicated cytokine (per million white blood cells) as detected by ELISPOT for three mice per group. Pre is prechallenge data, and Post 1 and Post 2 are postchallenge data for two subsets of a group of mice that were immunized at the same time but challenged at different times. Postchallenge, the total number of cells recovered from the lungs increased approximately fourfold, whereas no increase in the total number of recovered cells occurred for spleens.
<, below detection threshold.
ND, not done.
In contrast, the lymphokine profiles of the postchallenge CD4 T cells revealed that this response was predominantly Th1 biased in the lungs but remained predominantly an IL-4-biased Th2 response in the spleen (Fig. 3). The lungs contained about four times more IFN-γ-producing than IL-4-producing CD4 T cells. In the spleens, the IL-4-producing CD4 T cells, which had predominated prechallenge, underwent expansions comparable to those of the IFN-γ-producing CD4 T cells, and the Th2 bias of the CD4 response was maintained. The tmHA-NP spleens had an 18-fold expansion in both IFN-γ- and IL-4-producing class II-restricted responses. The sHA-NP spleens had 18-fold and 21-fold expansions in the IFN-γ- and IL-4-producing class II-restricted responses, respectively. A control influenza virus challenge in mice that had not been DNA vaccinated exhibited the expected type 1 bias for both CD8 and CD4 cells in lungs and spleen.
All of the mice survived the sublethal challenge, with the most severe weight loss occurring in the unvaccinated controls. A trend toward increased weight loss in the group primed to have an IL-4-producing CD8 T-cell response compared to the weight loss in the group primed to have an IFN-γ-producing CD8 response was not significant (data not shown).
Our results suggest that Th2 and Tc2 cells differ in their expansion following an influenza virus challenge. In our study, NP-specific Tc1 cells underwent more effective expansion than did NP-specific Tc2 cells in the spleen (Table 1). This finding was in contrast to that for the NP-specific Th1 and Th2 cells that underwent similar expansions in the spleen (Table 1). Our study does not address whether the postchallenge change from an IL-4-biased CD8 T-cell response to an IFN-γ-biased CD8 response was due to a conversion of Tc2 to Tc1 cells or the preferential expansion of Tc1 cells.
Our data also suggest a difference in trafficking and/or retention of type 1 and type 2 cells to a pleural influenza infection. For both CD4 and CD8 T cells, type 1 cells were preferentially enriched in the infected lung. This hypothesis is supported by Wohlleben et al., who demonstrated previously that infection with influenza virus not only induced Th1 responses but also suppressed the recruitment of Th2 cells in the lung airways of mice (23). Evidence that retention plays a role in the differential presence of type 1 and type 2 cells in the lung has been provided by Cerwenka et al., who demonstrated that although influenza-specific Tc2 and Tc1 cells trafficked to flu-infected lung epithelium, only Tc1 cells were able to remain within the infected tissues (3). The different retention kinetics were explained by differences in β chemokine receptor (CCR) expression. CCR3 and CCR4 are expressed on Th2 cells, whereas CCR1 and CCR5 are expressed on Th1 effector populations (1, 16).
Previous studies have demonstrated that different antigen-presenting cell populations induce different types of responses, especially at peripheral sites. Dendritic cells in the respiratory tract are specialized for mobilizing a default Th2 immunity (19). In contrast, alveolar macrophages are associated with the development of Th1 cells and the attenuation of Th2 responses (2, 4, 7, 21). This effect is believed to occur through the production of IL-12 by macrophages (20). Alveolar macrophages are highly susceptible to infection by influenza virus and express viral antigens upon infection (14). Consequently, these cells may be mediating the type 1 T-cell responses in the lung in our experiments.
In conclusion, we have tested the responses of type 1 and type 2 NP-specific CD8 and CD4 T cells to a sublethal influenza virus infection and found that the expansion of the memory response at the site of infection was composed almost exclusively of type 1 cells. Thus, for an influenza virus infection, type 1 CD8 T cells predominate at the pleural site of inflammation irrespective of whether Tc1- or Tc2-biased responses were elicited by DNA immunization.
Acknowledgments
We are indebted to J. Katz for discussion and critical comments on the manuscript and to H. Drake-Perrow for excellent administrative assistance.
This research was supported by PHS grant number R01 AI34946 to H. Robinson and by grant number P51 RR00165, base grant support to the Yerkes National Primate Research Center.
REFERENCES
- 1.Bonecchi, R., G. Bianchi, P. P. Bordignon, D. D'Ambrosio, R. Lang, A. Borsatti, S. Sozzani, P. Allavena, P. A. Gray, A. Mantovani, and F. Sinigaglia. 1998. Differential expression of chemokine receptors and chemotactic responsiveness of type 1 T helper cells (Th1s) and Th2s. J. Exp. Med. 187:129-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brewer, J. M., J. Richmond, and J. Alexander. 1994. The demonstration of an essential role for macrophages in the in vivo generation of IgG2a antibodies. Clin. Exp. Immunol. 97:164-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cerwenka, A., T. M. Morgan, A. G. Harmsen, and R. W. Dutton. 1999. Migration kinetics and final destination of type 1 and type 2 CD8 effector cells predict protection against pulmonary virus infection. J. Exp. Med. 189:423-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chattergoon, M. A., T. M. Robinson, J. D. Boyer, and D. B. Weiner. 1998. Specific immune induction following DNA-based immunization through in vivo transfection and activation of macrophages/antigen-presenting cells. J. Immunol. 160:5707-5718. [PubMed] [Google Scholar]
- 5.Fearon, D. T., and R. M. Locksley. 1996. The instructive role of innate immunity in the acquired immune response. Science 272:50-53. [DOI] [PubMed] [Google Scholar]
- 6.Harris, D. P., L. Haynes, P. C. Sayles, D. K. Duso, S. M. Eaton, N. M. Lepak, L. L. Johnson, S. L. Swain, and F. E. Lund. 2000. Reciprocal regulation of polarized cytokine production by effector B and T cells. Nat. Immunol. 1:475-482. [DOI] [PubMed] [Google Scholar]
- 7.Hsieh, C. S., S. E. Macatonia, C. S. Tripp, S. F. Wolf, A. O'Garra, and K. M. Murphy. 1993. Development of TH1 CD4+ T cells through IL-12 produced by Listeria-induced macrophages. Science 260:547-549. [DOI] [PubMed] [Google Scholar]
- 8.Mills, C. D., K. Kincaid, J. M. Alt, M. J. Heilman, and A. M. Hill. 2000. M-1/M-2 macrophages and the Th1/Th2 paradigm. J. Immunol. 164:6166-6173.10843666 [Google Scholar]
- 9.Mosmann, T. R., H. Cherwinski, M. W. Bond, M. A. Giedlin, and R. L. Coffman. 1986. Two types of murine helper T cell clone. I. Definition according to profiles of lymphokine activities and secreted proteins. J. Immunol. 136:2348-2357. [PubMed] [Google Scholar]
- 10.Mosmann, T. R., and S. Sad. 1996. The expanding universe of T-cell subsets: Th1, Th2 and more. Immunol. Today 17:138-146. [DOI] [PubMed] [Google Scholar]
- 11.Mosmann, T. R., S. Sad, L. Krishnan, T. G. Wegmann, L. J. Guilbert, and M. Belosevic. 1995. Differentiation of subsets of CD4+ and CD8+ T cells. Ciba Found. Symp. 195:42-50. [DOI] [PubMed] [Google Scholar]
- 12.Oran, A. E., and H. L. Robinson. 2003. DNA vaccines, combining form of antigen and method of delivery to raise a spectrum of IFN-γ and IL-4 CD4+ and CD8+ T cells. J. Immunol. 171:1999-2005. [DOI] [PubMed] [Google Scholar]
- 13.Peritt, D., S. Robertson, G. Gri, L. Showe, M. Aste-Amezaga, and G. Trinchieri. 1998. Differentiation of human NK cells into NK1 and NK2 subsets. J. Immunol. 161:5821-5824. [PubMed] [Google Scholar]
- 14.Rodgers, B., and C. A. Mims. 1981. Interaction of influenza virus with mouse macrophages. Infect. Immun. 31:751-757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sad, S., R. Marcotte, and T. R. Mosmann. 1995. Cytokine-induced differentiation of precursor mouse CD8+ T cells into cytotoxic CD8+ T cells secreting Th1 or Th2 cytokines. Immunity 2:271-279. [DOI] [PubMed] [Google Scholar]
- 16.Sallusto, F., C. R. Mackay, and A. Lanzavecchia. 1997. Selective expression of the eotaxin receptor CCR3 by human T helper 2 cells. Science 277:2005-2007. [DOI] [PubMed] [Google Scholar]
- 17.Sato, M., K. Iwakabe, S. Kimura, and T. Nishimura. 1999. Functional skewing of bone marrow-derived dendritic cells by Th1- or Th2-inducing cytokines. Immunol. Lett. 67:63-68. [DOI] [PubMed] [Google Scholar]
- 18.Seder, R. A., and W. E. Paul. 1994. Acquisition of lymphokine-producing phenotype by CD4+ T cells. Annu. Rev. Immunol. 12:635-673. [DOI] [PubMed] [Google Scholar]
- 19.Stumbles, P. A., J. A. Thomas, C. L. Pimm, P. T. Lee, T. J. Venaille, S. Proksch, and P. G. Holt. 1998. Resting respiratory tract dendritic cells preferentially stimulate T helper cell type 2 (Th2) responses and require obligatory cytokine signals for induction of Th1 immunity. J. Exp. Med. 188:2019-2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takeuchi, M., P. Alard, and J. W. Streilein. 1998. TGF-β promotes immune deviation by altering accessory signals of antigen-presenting cells. J. Immunol. 160:1589-1597. [PubMed] [Google Scholar]
- 21.Tang, C., J. M. Rolland, C. Ward, F. Thien, X. Li, S. Gollant, and E. H. Walters. 1998. Differential regulation of allergen-specific TH2- but not TH1-type responses by alveolar macrophages in atopic asthma. J. Allergy Clin. Immunol. 102:368-375. [DOI] [PubMed] [Google Scholar]
- 22.Ward, S. G., K. Bacon, and J. Westwick. 1998. Chemokines and T lymphocytes: more than an attraction. Immunity 9:1-11. [DOI] [PubMed] [Google Scholar]
- 23.Wohlleben, G., J. Muller, U. Tatsch, C. Hambrecht, U. Herz, H. Renz, E. Schmitt, H. Moll, and K. J. Erb. 2003. Influenza A virus infection inhibits the efficient recruitment of Th2 cells into the airways and the development of airway eosinophilia. J. Immunol. 170:4601-4611. [DOI] [PubMed] [Google Scholar]