Abstract
Homologous recombination between strains of the same alphaherpesvirus species occurs frequently both in vitro and in vivo. This process has been described between strains of herpes simplex virus type 1, herpes simplex virus type 2, pseudorabies virus, feline herpesvirus 1, varicella-zoster virus, and bovine herpesvirus 1 (BoHV-1). In vivo, the rise of recombinant viruses can be modulated by different factors, such as the dose of the inoculated viruses, the distance between inoculation sites, the time interval between inoculation of the first and the second virus, and the genes in which the mutations are located. The effect of the time interval between infections with two distinguishable BoHV-1 on recombination was studied in three ways: (i) recombination at the level of progeny viruses, (ii) interference induced by the first virus infection on β-galactosidase gene expression of a superinfecting virus, and (iii) recombination at the level of concatemeric DNA. A time interval of 2 to 8 h between two successive infections allows the establishment of a barrier, which reduces or prevents any successful superinfection needed to generate recombinant viruses. The dramatic effect of the time interval on the rise of recombinant viruses is particularly important for the risk assessment of recombination between glycoprotein E-negative marker vaccine and field strains that could threaten BoHV-1 control and eradication programs.
Bovine herpesvirus 1 (BoHV-1), a member of the Alphaherpesvirinae subfamily, causes two major disease syndromes in cattle: infectious bovine rhinotracheitis (IBR) and infectious pustular vulvovaginitis (42, 58, 61).
Homologous recombination between strains of the same alphaherpesvirus species frequently occurs, both in vitro and in vivo. This process has been described between strains of herpes simplex virus type 1 (HSV-1) and HSV-2, varicella-zoster virus, pseudorabies virus (PrV), feline herpesvirus 1, and BoHV-1 (14, 16, 20, 21, 25, 40, 49, 51, 52). The rise of recombinant viruses can be influenced by different factors, particularly those affecting the distribution of different viruses to common target cells, thereby limiting or increasing the likelihood of cellular coinfections. In vivo, some of these factors include (i) the dose of the inoculated viruses, (ii) the distance between inoculation sites, (iii) the time interval between inoculation of the first and the second virus, and (iv) the genes in which the mutations are located (19).
Although IBR, classified in list B of the Office International des Epizooties, was eradicated in several European countries, it still causes economic losses for the European and the U.S. beef industries: approximately $500 million yearly in the United States (according to the National Agricultural Statistics Service in 1996). In European nations where BoHV-1 has not been eradicated, BoHV-1 control and eradication programs are associated with the use of glycoprotein E (gE)-negative marker vaccines by analogy with the successful pseudorabies vaccination strategy (12, 56, 57). These marker vaccines, either inactivated or live attenuated, together with a serological detection of gE directed antibodies, allow differentiation between vaccinated and infected cattle (60).
The extensive use of gE-negative live attenuated vaccines for both PrV and BoHV-1 eradication programs led investigators to assess the risk of recombination between marker vaccines and field strains (49, 51) and to study factors involved in recombination, such as the interval between infections (19). A previous study of PrV showed that a time interval of 2 h allows recombination, but this effect was not investigated for longer time intervals (19). To occur, recombination needs the successful replication of the two viruses in the same cell (46). Recently, a study of PrV showed a very small time window for productive double infections (i.e., with a maximum time interval of 4 h) (2). This finding is of particular interest, especially because recombination between homologous viruses is usually studied in coinfection experiments. Nevertheless, a true cell coinfection must be a rare event in natural conditions. In such cases, the second infection is often delayed and the first virus has already started its replication cycle. Therefore, consecutive infections, leading to superinfection, can be considered as a more frequent event in both cell culture and infected animals. Although alphaherpesvirus recombination frequently occurs in coinfected cells, it can be assumed that the outcome is different when the second infection is delayed. Consequently, in the present study, we choose to further determine the effect of a temporal separation of two in vitro infections (including one with a BoHV-1 mutant with gE deleted) on the rise of BoHV-1 recombinants. The advantage of the in vitro system for studying recombination is that it is a well-defined entity that only contains viruses and cells, thereby avoiding the effects of other factors and particularly the immunological response of the host.
Our results clearly demonstrate that a time interval of 2 to 8 h between two consecutive infections of cells allows the establishment of a barrier that reduces or prevents any successful superinfection needed to generate recombinant viruses.
MATERIALS AND METHODS
Viruses and cell culture.
The four viruses used in the present study are designated BoHV-1 Lam gC−, Lam gE−, ST, and STBG. Lam gC− and Lam gE− mutants are derived from the BoHV-1 subtype 1 strain Lam (36). Lam gC− possesses a deletion in the gene encoding glycoprotein C (gC) (24), whereas the gene encoding gE is deleted in the Lam gE− mutant (59). The BoHV-1 subtype 2 strain ST was previously described (29). In the STBG mutant, derived from the ST strain, the BoHV-1 gE open reading frame (ORF) was replaced by the β-galactosidase (β-Gal) ORF (GenBank accession number U02451) under the control of the human cytomegalovirus immediate-early (CMV IE) promoter (28). Madin-Darby bovine kidney (MDBK; ATCC CCL-22) cells were grown in Earle minimum essential medium MEM (Gibco-BRL) supplemented with 2% penicillin (5,000 U/ml) and streptomycin (5,000 μg/ml) (PS medium; Gibco-BRL), and 5% heat-inactivated fetal bovine serum (BioWhittaker). Viral stocks were produced in MDBK cells as previously described (50).
Experimental design.
Monolayers of MDBK cells prepared in 24-well plates were coinfected with Lam gC− and Lam gE− mutants at a multiplicity of infection (MOI) of 10 PFU/cell for each mutant. In parallel, MDBK cells (four other monolayers) were infected with Lam gC− mutant (MOI of 10) and superinfected with Lam gE− mutant (MOI of 10) 2, 4, 6, and 8 h after infection with Lam gC− mutant. MDBK cells were infected with Lam gE− mutant (MOI of 10) and superinfected with Lam gC− mutant (MOI of 10) 2, 4, 6, and 8 h after Lam gE− infection. After the first infection (coinfection, the four Lam gC− mutant infections and the four Lam gE− mutant infections), virus attachment was allowed for 2 h at 4°C. Cells were then further incubated at 37°C and superinfections were performed. Two hours after the temperature shift and 2 h after each superinfection, cells were washed twice with minimal essential medium (MEM) and further incubated at 37°C in 1 ml of MEM supplemented with 2% PS medium and 2% heat-inactivated horse serum (Serolab/International Medical). At 24 h after each superinfection (when the monolayers showed extensive cytopathic effect), the culture medium was removed, clarified twice by centrifugation (1,000 × g), divided into aliquots, and stored at −80°C.
In the second experiment, coinfection with ST strain (MOI of 10) and STBG mutant (MOI of 10) was performed, whereas infections with ST strain and superinfections after 2, 4, 6, and 8 h with STBG mutants were carried out in a complementary assay. Cells were collected 6 h after STBG mutant infection (expression peak of β-Gal in cells infected with STBG mutant) and were analyzed by flow cytometry to measure β-Gal (STBG mutant) and gE (ST strain) expression as described below.
In the third experiment, ST strain and STBG mutant coinfection and superinfections were performed by using the same scheme. After an incubation of 30 h, cells were collected, and extracellular virion DNA and total cell DNA were prepared as previously described (50). Concatemeric DNA was analyzed by pulsed-field gel electrophoresis (PFGE).
Isolation and characterization of progeny viruses coming from co- and superinfection supernatants.
Isolation and characterization of progeny viruses as parental or recombinant viruses were performed as previously described (49). Progeny viruses were directly isolated by plaque picking and further propagated individually. Isolates were then characterized by using a PCR and an immunofluorescence based approach as previously described (48, 51). For each experimental condition (co- and superinfection), 50 progeny viruses were characterized. To ensure reproducibility of these results, this experiment was repeated three times.
Determination of β-Gal and gE levels of expression in co- and superinfected cells.
After collection, MDBK cells were washed twice with phosphate-buffered saline (PBS) and incubated for 30 min with the appropriate dilution of primary monoclonal antibody (anti-gE BH35) (3). Cells were washed twice with PBS containing 5% fetal calf serum (FCS), and further incubated with R-phycoerythrin (R-PE)-conjugated goat immunoglobulins anti-mouse immunoglobulin G1 (R-PE-GAM-IgG1; Imtech) for 30 min. After an additional wash with PBS containing 5% FCS, the β-Gal activity of cells infected with the STBG mutant was revealed by the procedure described by Nolan et al. (41). Briefly, cells were loaded with the fluorogenic substrate fluorescein di-β-d-galactopyranoside (FDG; Sigma) by using a short hypotonic shock. The hydrolytic cleavage of FDG by β-Gal releases fluorescein that is locked inside the cells kept on ice. The cells were then analyzed by flow cytometry for green (fluorescein, relative β-Gal activity) and red (gE expression) fluorescences.
The kinetics of β-Gal expression in virus-infected MDBK cells were determined by using the procedure described above (41) at 1, 3, 5, 7, and 9 h after infection with STBG virus.
Flow cytometry analysis was performed by using a Becton Dickinson fluorescence-activated cell sorter (Facstar Plus) equipped with an argon laser (ILT air cooled with 100-mW excitation lines at 488 nm). Debris were excluded from the analysis by the conventional scatter gating method. The cells or the nuclei doublets were excluded from analysis by using pulse processor boards (Becton Dickinson). Ten thousand events per sample were collected in a list mode, stored, and analyzed by the Consort 32 system (Becton Dickinson).
Analysis of concatemeric DNA of co- and superinfected cells by PFGE and Southern blotting.
PFGE material consisted of a CHEF-DRII drive module and model 200/2.0 power supply (Bio-Rad). Agarose plugs were placed into the wells containing a 1% PFGE-certified agarose (Bio-Rad) gel in 0.5× TBE (45 mM Tris base, 45 mM boric acid, 1 mM EDTA [pH 8]). Electrophoresis was performed at 14°C for 23 h at 200 V with ramped pulse conditions of 10 to 85 s. Restriction enzyme digestions of DNA embedded in agarose were performed by dialyzing plugs three times for 1 h each time with the appropriate restriction enzyme buffer and by incubation with 50 U of the XbaI restriction enzyme for 18 h at 37°C. The Southern blot analysis was performed as previously described (50). BoHV-1 DNA sequence from gD (nucleotides 118893 to 119444 of the published BoHV-1 sequence [accession number AJ004801]) gene was amplified by a previously described PCR method (48) with BoHV-1.2 strain ST DNA as a template. After purification, gD PCR product (551 bp) was cloned into the pGEM-T Easy vector (Promega) to produce pgD plasmid.
RESULTS
Rise of recombinants after simultaneous infections with two distinguishable BoHV-1 mutants.
It has been reported that coinfection with two different strains of the same herpesvirus led to the production of recombinant viruses both in vitro and in vivo (14, 20, 21, 25, 49, 51). To quantify the recombination between the two parental BoHV-1 used in the present study, MDBK cells were coinfected with both Lam gC− (gC−/gE+) and Lam gE− (gC+/gE−) mutants. To ensure the reproducibility of the results, this experiment was performed three times. After an incubation of 30 h, progeny viruses (n = 50) were isolated from the supernatant and characterized as parental mutants (gC−/gE+ and gC+/gE−) or recombinant viruses (gC+/gE+ and gC−/gE−). This methodology was used to determine the relative proportions, expressed as a percentage of the total number of isolates, of the four possible progeny populations (parental and recombinant) (Table 1). In our experimental conditions, 30 to 34% of progeny viruses were characterized as recombinant viruses (gC+/gE+ and gC−/gE−), thus confirming previous experiments that demonstrated that alphaherpesviruses recombine with high efficiency in vitro in a situation of coinfection (16, 20) (Table 1). As a consequence, this high recombination rate allowed us to study the effect of the time interval between infections on the recombination of BoHV-1.
TABLE 1.
Relative proportions of the four progeny populations (parental and recombinant) after coinfection of MDBK cells with BoHV-1 Lam gC−/gE+ and Lam gC+/gE−
| Expt no. (n = 50) | % Parental virus
|
% Recombinant virus
|
||
|---|---|---|---|---|
| gC−/gE+ | gC+/gE− | gC+/gE+ | gC−/gE− | |
| 1 | 36 | 30 | 34 | 0 |
| 2 | 38 | 32 | 32 | 0 |
| 3 | 34 | 36 | 28 | 2 |
Dramatic effect of increasing time interval between infections on the rise of BoHV-1 recombinants in vitro.
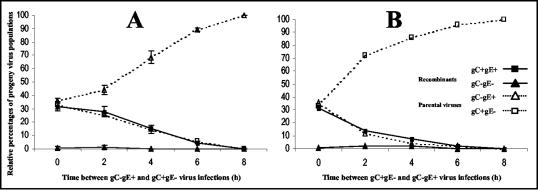
To investigate whether an increase of the time interval influences the rise of recombinant viruses, MDBK cells were coinfected with BoHV-1 Lam gC− (gC−/gE+) and Lam gE− (gC+/gE−) mutants or infected with Lam gC− mutant and superinfected with Lam gE− mutant 2, 4, 6, and 8 h later. Progeny viruses (n = 50) isolated from the supernatant 30 h after infection were characterized as parental (gC−/gE+ and gC+/gE−) or recombinant (gC+/gE+ and gC−/gE−) viruses. The results (Fig. 1A) indicated that recombinant viruses, mainly gC+/gE+, were still detected with a 2-h interval between the infections. The amount of recombinant viruses decreases when the time interval between infections increases, and we failed to detect them after an 8-h interval. MDBK cells were then infected with Lam gE− mutant (gC+/gE−) and superinfected with Lam gC− mutant (gC−/gE+). Results, presented in Fig. 1B, also demonstrated a decrease of recombinant viruses when the time interval between infections increases. In contrast to results presented in Fig. 1A, the decrease of recombinants was nevertheless more drastic when Lam gE− (gC+/gE−) was the first virus infecting the cells. Taken together, these results clearly demonstrate the dramatic effect of the time interval between infections on the rise of recombinant viruses and indicate the importance of this parameter in recombination between alphaherpesvirus strains.
FIG. 1.
Effect of time interval between infections of two BoHV-1 strains on the detection of infectious recombinant viruses. (A) MDBK cells were either coinfected with Lam gC− mutant (gC−/gE+) and Lam gE− mutant (gC+/gE−) or infected with Lam gC− mutant and superinfected with Lam gE− mutant at 2, 4, 6, and 8 h after infection with Lam gC−. (B) MDBK cells were either coinfected with Lam gE− and Lam gC− mutants or infected with Lam gE− mutant and superinfected with Lam gC− mutant at 2, 4, 6, and 8 h after infection with Lam gE−. In each infection situation, progeny viruses isolated from the culture supernatant 30 h after infection with Lam gE− (A) or Lam gC− (B) were characterized as parental or recombinant viruses as described in Materials and Methods. Standard deviations of three independent experiments are indicated by vertical lines in panel A.
Superinfecting virus β-Gal gene expression is prevented in cells already infected with a first virus.
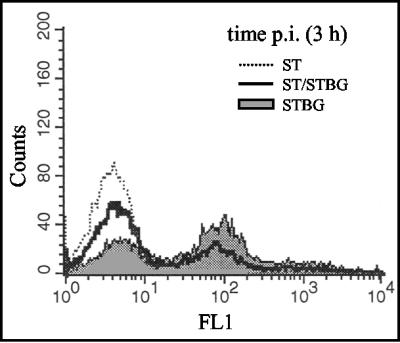
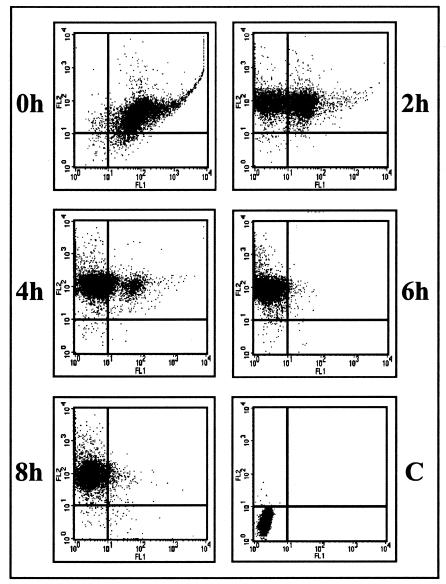
The experiment described above demonstrated the dramatic impact of time interval on the rise of recombinant viruses. Theoretically, prevention of recombination due to an increased time interval could be the consequence of a block at different levels of the viral cycle. Since the results presented above were based on the analysis of progeny viruses from extracellular virions, it did not bring us information to determine at what viral cycle stage the inhibition occurred. Consequently, we decided to measure the β-Gal gene expression, inserted in superinfecting virus genome, in cells that were already infected. The β-Gal activity was detected as early as 3 h infection and as late as 9 h after infection (Fig. 2), which indicates that the CMV IE promoter-regulated β-Gal gene of STBG virus is expressed at the beginning of the viral cycle as previously shown with a similar construction (8). However, these data did not allow us to conclude that CMV IE is regulated as a BoHV-1 IE gene in MDBK cells. To begin to address the issue of the stage at which inhibition occurred, cells coinfected with ST strain and STBG mutant or infected with ST strain and superinfected with STBG mutant were analyzed by flow cytometry (MOI of 10 for each virus). β-Gal (STBG mutant) and gE (ST strain) expressions in infected cells show that, when the two viruses were simultaneously applied, virtually all cells appeared to be double infected. In contrast, the results clearly demonstrate that infection in superinfected cells with the first virus (ST strain) prevents detection of β-Gal gene expression of the second virus (STBG mutant) (Fig. 3). In bovine cells infected with ST strain for 6 and 8 h, β-Gal gene expression of the superinfecting virus is totally undetected (Fig. 3). Similar results were observed when the same experiment was performed at an MOI of 1 (data not shown). Indeed, cells detected as ST virus infected were similarly resistant to STBG virus infection. However, since an MOI of 1 did not allow infection of all cells, numerous cells uninfected with ST virus could be infected with STBG virus applied 8 h after ST infection. Taken together, these results show the establishment of a fast and efficient barrier to superinfection, probably occurring prior to virus gene expressions, which indicates the importance of concomitant infections to allow productive infections. If the second infection takes place too late, interference effects directed to superinfecting virus are clearly detected.
FIG. 2.
β-Gal gene expression is detected at the beginning of the viral cycle. MDBK cells were infected with the ST strain of BoHV-1, STBG mutant (a modified ST strain in which the gE ORF was replaced by the β-Gal ORF under the control of the human CMV IE promoter), or both viruses at an MOI of 10 for each virus. Expression of β-Gal and gE was revealed at 3 h postinfection by Nolan's method (green fluorescence, FL1) (41). In the three situations, 104 cells were analyzed by flow cytometry.
FIG. 3.
Previous infection of MDBK cells with the ST strain of BoHV-1 prevented β-Gal gene expression of the superinfecting STBG mutant. MDBK cells were coinfected with ST and STBG BoHV-1 (a modified ST strain in which the BoHV-1 gE ORF was replaced by the β-Gal ORF under the control of the human CMV IE promoter) at an MOI of 10 for each virus. MDBK cells were infected with ST strain and superinfected with STBG at 0, 2, 4, 6, or 8 h after ST infection (MOI of 10 for each virus). The presence of β-Gal and gE was revealed by Nolan's method for β-Gal (green fluorescence, FL1) and R-PE fluorescence for glycoprotein E (FL2) (34). C, mock-infected cells. In each situation, 104 cells were analyzed by flow cytometry.
Detection of mixed concatemeric DNA resulting from recombination between two distinguishable types of BoHV-1.
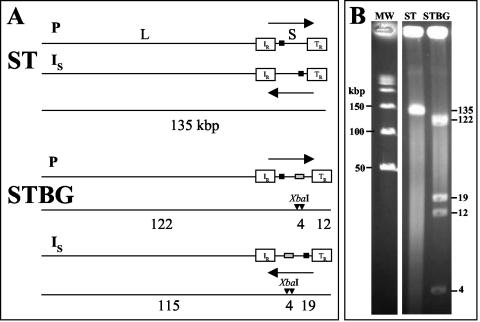
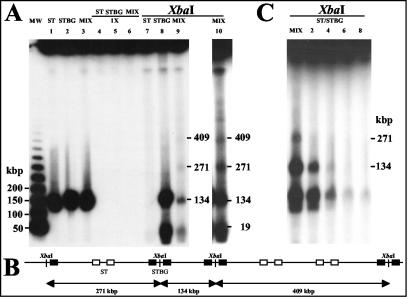
DNA recombination and replication of herpesviruses are two processes that are intimately linked (46). DNA replication takes place in the nucleus, where the genomes circularize (17, 35) and are replicated by a poorly understood mechanism, resulting in the formation of concatemeric molecules in which the termini of the viral genomes are fused in a head-to-tail arrangement (46). As previously demonstrated (53), recombination between two strains of the same herpesvirus can be studied at the concatemeric level. Briefly, the methodology reported by Slobedman et al. (53) requires the use of two distinguishable strains: the first possessing a unique restriction site within the genome and a second that does not possess this unique restriction site. In the case of separate inoculations, digestion of viral concatemeric DNA of the strain containing the unique restriction site then generates unit-length genomes, whereas no fragment is generated from concatemeric DNA of the strain devoid of the unique restriction site. After coinoculation between these two strains, digestion of concatemeric DNA will generate not only unit-length genomes (due to the presence of the strain containing the unique restriction site) but also higher-molecular-weight intermediates of replication (having, for example, the length of two or three genomes) due to recombination between genomes of the two strains within or between concatemeric DNA. Then, our first goal was to identify an endonuclease that does not cut into the BoHV-1 genome. Virion DNA from different BoHV-1 strains was then prepared and digested with different endonucleases (data not shown). We found that the genome of strain ST did not possess any XbaI restriction site (as depicted in Fig. 4A) since a unique fragment of the unit length genome size was observed after XbaI digestion of virion DNA (Fig. 4B). Virion DNA from STBG mutant (in which the entire gE ORF was replaced by the β-Gal ORF) was then prepared and digested with XbaI because two XbaI sites are present in the β-Gal ORF. As depicted in Fig. 4A, the location of the β-Gal sequence inserted into the BoHV-1 genome of strain ST predicted fragment sizes of 123 and 12 kbp (if the S segment of the genome is in the prototype [P] orientation) and of 115 and 19 kbp (if the S segment is in an inverted [IS] orientation) after XbaI digestion. The resulting fragments (Fig. 4B) were in precise agreement with the fragment sizes shown in Fig. 4A, allowing us to study recombination between ST strain and STBG mutant at the concatemeric level. Agarose plugs containing total DNA from cells infected with ST or STBG or coinfected with both viruses were then subjected to PFGE to remove unit-length linear genomes (Fig. 5, lanes 1 to 3). These samples were then either mock digested or digested with XbaI, separated by PFGE, and hybridized by using pgD as probe. A single PFGE separation was sufficient to fully remove unit length genomes because no 135-kbp DNA was detected in lanes containing mock-digested plugs (Fig. 5A, lanes 4 to 5). After XbaI digestion of ST concatemeric DNA, we failed to detect any fragment (Fig. 5A, lane 7), thus confirming the absence of XbaI sites within the genome of strain ST. In contrast, when STBG concatemeric DNA was XbaI digested, a fragment of ca. 135-kbp was detected (Fig. 5A, lane 8) due to the presence of the two XbaI restriction sites present within the β-Gal ORF. If recombination occurs in cells coinfected with ST strain and STBG mutant, XbaI fragments of different sizes must be generated, as depicted in Fig. 5B. Indeed, digestion of two adjacent STBG genomes within concatemeric DNA will generate a fragment of the unit length genome size (135 kbp), whereas digestion of two adjacent ST or STBG genomes within concatemeric DNA will generate fragments of higher-molecular-weight sizes (for example, 270 and 405 kbp). The observed fragments (Fig. 5A, lanes 9 and 10) were in precise agreement with the theoretical situation depicted in Fig. 4B, indicating that recombination generated mixed concatemeric DNA after coinfection with ST and STBG viruses.
FIG. 4.
β-Gal ORF brings two restriction sites into the genome of BoHV-1 ST strain. (A) XbaI restriction maps of the BoHV-1 ST strain and STBG mutant with the S segment of the genome in either prototype (P) or inverted S (IS) orientation. The horizontal arrows indicate orientations of S segments, and white boxes represent internal (IR) and terminal (TR) inverted repeats. Locations of predicted unique restriction sites based on the β-Gal ORF sequence are represented by black vertical arrows. Black boxes indicate the location of sequence contained within hybridization probe pgD. Gray boxes indicate the location of β-Gal ORF. Below each map are illustrated the expected fragments generated by XbaI digestions. The predicted fragment sizes are given in kilobase pairs. (B) Virion DNA from the two strains was digested with XbaI. DNA fragments were separated by PFGE, and the gel was stained with ethidium bromide. The sizes (in kilobase pairs) and the locations of molecular weight markers are indicated to the left of lane MW, and the estimated sizes of specific restriction fragments are indicated to the right of lane STBG.
FIG. 5.
Detection of recombination between ST and STBG BoHV-1 at the concatemeric level. (A) MDBK cells were either singly infected with ST or STBG or coinfected with both viruses. Plugs were prepared 30 h after infection and subjected to PFGE to produce concatemeric DNA free of unit length genomes (lanes 1 to 3). Plugs that migrated once were then either mock digested (lanes 4 to 6) or digested with XbaI (lanes 7 to 9). Lane 10 is an overexposure of lane 9. (B) Schematic representation of mixed concatemeric DNA and potential high-molecular-weight fragments generated after XbaI digestion. Inverted repeated sequences (IR and TR) derived from STBG and ST strains are represented by black and white boxes, respectively. Locations of predicted XbaI restriction sites are represented by black vertical arrows. Below the map are illustrated the expected fragments generated by XbaI digestion. (C) MDBK cells were either coinfected with ST and STBG viruses or infected with ST and superinfected with STBG mutant at 2 (lane 2), 4 (lane 3), 6 (lane 4), or 8 (lane 5) h after ST infection as indicated above each lane. Plugs were prepared 30 h after STBG infection and subjected to PFGE to produce concatemeric DNA free of unit length genomes. Plugs were then digested with XbaI. Samples were separated by PFGE, transferred to a nylon membrane, and hybridized with the probe pgD.
Progressive disappearance of STBG and mixed concatemeric DNA with the increase of time interval between ST and STBG BoHV-1 infections.
To investigate the impact on recombination of an increase of the time interval between ST and STBG infections, agarose plugs containing total DNA from cells coinfected with both viruses or infected with ST and superinfected with STBG were subjected to PFGE to remove unit length linear genomes and then digested with XbaI, separated by PFGE, and hybridized by using pgD as probe. As described above, fragments indicative of recombination between these two viruses were observed (see the 271-kbp fragment in Fig. 5C, lane 1). In contrast, these fragments progressively disappeared as the time interval between ST infection and STBG superinfection increases (Fig. 5C, lanes 2 to 5). A faint band was still observed after a time interval of 4 h, whereas no band was detected when the time interval was of 6 h. The disappearance of mixed concatemeric DNA fragments correlates with an absence of replication of STBG genomes since no 135-kbp fragments were observed when the time interval between infections was 6 h. These results confirm at the concatemeric level that recombination is prevented when the time interval between infections increases.
DISCUSSION
Our results demonstrate that simultaneous infections or infections separated by short periods (maximum of 2 h) with two distinguishable BoHV-1 lead to the production of a large amount of recombinant viruses in vitro. With an increasing time interval between infections, productive superinfection was progressively prevented and, consequently, the rise of recombinant viruses declined. The impact of the time interval between infection and superinfection on recombination process was analyzed and confirmed at three steps of the replication cycle of the second virus. First, after replication, production of the progeny of the second virus and recombinant viruses became undetectable with the increase of the time interval between infections. Second, before replication, an increase of the time interval between infections prevented β-Gal gene expression by the second virus. Third, replication of the second virus and its consequences, i.e., the formation of concatemeric structures, were prevented in cells already infected with the first virus. Recombinant viruses were barely detectable after a 6-h interval and not detectable at all after an 8-h interval. These results show the fast establishment of the inhibition of superinfection between Lam gC− and Lam gE− mutants of BoHV-1 and its deep impact on the production of recombinant viruses. A 2-h time interval between infections of the same PrV strain allows recombination events and the rise of recombinant viruses (19). Our results show that increasing time interval progressively prevents the rise of recombinant viruses.
The results relative to experiments focusing on the stage when inhibition of superinfection occurs show that the latter takes place before or during replication and/or before or during viral gene expression of the second virus. Therefore, these results allow us to postulate that superinfection inhibition could occur at different stages of the viral cycle. These stages include attachment and entry, migration of the capsid to the cell nucleus, gene expression, and DNA replication. Additional investigations are required to demonstrate when and where the inhibition occurs. For example, it will be of particular interest to construct, as already described for PrV (54), a green fluorescent protein (GFP) capsid BoHV-1 as a superinfecting virus and to trace it. However, results must be interpreted cautiously since superinfection inhibition could be multifactorial and/or occur at different stages (37, 63).
Inhibition of superinfection is one of several mechanisms that can be invoked in the interference between related and unrelated viruses. Six types of interference, which all cover the possible mechanisms, are usually described: interferons (IFNs), incompatibility of heterologous viruses, superinfection inhibition, defective interfering particles, dominant-negative mutants, and RNA interference (37, 63). Interference in alphaherpesvirus superinfections has already been studied with HSV-1, HSV-2, PrV, and the equid herpesviruses 1 and 2 (15, 26, 27, 34, 43, 45, 64). In some cases, the first infecting virus acts as a helper and accelerates the replication of the second superinfecting virus (43, 45). In other cases, a previous infection with a homologous strain inhibits the replication of a superinfecting HSV-1 strain (15, 26, 27, 34, 43, 64). An interference effect of gD, which is essential for viral penetration, was described for both HSV-1 and BoHV-1 (4, 5, 22, 55). This interference results from the cellular expression of gD, which interferes with alphaherpesvirus entry, probably by blocking ligand-binding sites of the gD receptors used for entry. Moreover, cross-interference (e.g., gD of HSV-1 against PrV) can occur because different forms of alphaherpesvirus gD can compete for shared entry receptors (18). Nevertheless, all of these observations were carried out with gD-expressing cell lines and not in situations of natural superinfection (5-7, 10, 11, 22, 23). It is therefore not clear whether the interference observed in a situation of superinfection is mainly due to the expression of gD in lytically infected cells or not. On the other hand, kinetic expression of BoHV-1 gD, expressed as an early protein (32; V. Keuser, B. Detry, F. Schynts, P.-P. Pastoret, A. Vanderplasschen, and E. Thiry, unpublished data), matches well with the superinfection inhibition reported above. IFNs, especially beta IFN (IFN-β), formerly known as fibroblast IFN, could be implicated in the observed interference (38). However, early establishment of the inhibition of superinfection in infected cells (already detected 4 h postinfection) (33, 62) and the relative resistance of BoHV-1 to IFN-β are not consistent with this hypothesis (1, 9, 47). Additional investigations are required to clarify its possible implication. Thus, IFN-β, dominant-negative mutants (Lam gC− and Lam gE−), superinfection inhibition, e.g., gD-mediated interference, or the three mechanisms together cannot be excluded to explain the interference effect observed in our experiments. On the other hand, defective interfering particles cannot be the cause of the phenomenon since viral stocks were prepared by passing the virus at a low MOI. Moreover, receptor saturation cannot explain the interference since an MOI of 1, as well as a very high MOI (10), prevented superinfection. Whatever the mechanism involved in interference with the second virus, the observed phenomenon is important in the assessment of the risk of recombination between homologous alphaherpesviruses.
When prior infection and superinfection are carried out, respectively, with the BoHV-1 mutants Lam gE− (gC+/gE−) and Lam gC− (gC−/gE+) instead of Lam gC− as the first virus and Lam gE− as the superinfecting virus, superinfection inhibition is observed sooner. A possible explanation could be that the absence of gC in the superinfecting Lam gC− mutant partly impairs viral attachment to the target cells (30, 31).
Detection of Lam gC−gE− (gC−/gE−) recombinant was surprisingly low compared to that of double-positive recombinant (gC+/gE+). Previous studies with PrV indicated that the simultaneous deletion of gC and gE has an important impact on the ability of these mutants to replicate both in vitro and in vivo (39, 65). We hypothesize that, because Lam gC−gE− recombinants rise only in a cell coinoculated with both parental mutants (gC−/gE+ and gC+/gE−), they should be complemented phenotypically for both gC and gE, allowing them to interact with their environment as efficiently as does the wild-type virus. In particular, after egress from the cell, these recombinants possess a viral envelope containing gC, which plays a role in the primary attachment of the virus to target cell (30, 31, 51). Moreover, because gE is required for efficient cell-to-cell spread, the synthesis of gE in cells in which Lam gC−gE− recombinants were generated could contribute to the survival of these recombinants by an easier propagation to uninfected cells (44).
Under the conditions described here, there is only a very small window (0 to 6 h) in which superinfection efficient for recombination occurs. These results are in line with a very recent study on PrV in which the authors show a very small time window for productive double infections (i.e., with a maximum time interval of 4 h) (2). If the time interval between infections is longer than 6 h, incoming viruses cannot provide genomes for recombination or progeny production. Successful recombination is therefore closely dependent on simultaneous infections. This finding is of particular interest when the risk of recombination between BoHV-1 gE− marker vaccines and field strains is assessed. Indeed, during IBR epidemic events intranasal gE deletion vaccination is frequently carried out and consequently co- and superinfections with wild and gE− vaccine strains of BoHV-1 can occur with the possibility to generate virulent viruses from which gE has been deleted. These recombinant viruses could endanger control and eradication programs. Recently, the isolation in the field of a BoHV-1 strain with gE deleted, a strain homologous to the Difivac marker vaccine, strengthened the requirement of this risk assessment (13). The results obtained in that study and the low likelihood of cellular coinfections in natural conditions allow us to conclude that recombination and its potential consequences are rare events. However, a single recombinant that retains virulence and acquires the gE− genotype is enough to severely impair control programs based on vaccination.
In conclusion, the present study is the first to extensively investigate the impact of the time parameter on the recombination between alphaherpesviruses. The results emphasize the crucial importance of the small time window between infections and highlights its consequences in the context of the extensive use of marker vaccines with gE deleted as a tool in BoHV-1 eradication programs.
Acknowledgments
We thank J. Letchworth (University of Wisconsin-Madison) for providing monoclonal antibodies and J. T. van Oirschot and F. A. M. Rijsewijk (Institute for Animal Science and Health, Lelystad, The Netherlands) for providing viruses. We thank J. C. Audonnet (Merial, Lyon, France) for providing the STBG mutant of BoHV-1. We thank D. Sifakakis for excellent technical assistance. We thank Régine Denaegel, Luc Bauret, and Michel Lambot for careful reading of the manuscript.
This study was financially supported by the Ministère des Classes Moyennes et de l'Agriculture, Administration Recherche, et Développement and by the Fonds National Belge de la Recherche Scientifique (FNRS; FRFC 2.4508.02 and grant 1.5.105.03). F. Meurens and B. Muylkens are Research Fellows of the FNRS. A. Vanderplasschen is a Senior Research Associate of the FNRS. The confocal microscope was purchased with funds from the following grants: FRFC 2.4532.98 from the FNRS, FNRS LOTTO 9.4592.97 from the Belgian National Lottery, and ARC 98/03-220 from the French Community of Belgium.
REFERENCES
- 1.Babiuk, L. A., H. B. Ohmann, G. Gifford, C. W. Czarniecki, V. T. Scialli, and E. B. Hamilton. 1985. Effect of bovine alpha 1 interferon on bovine herpesvirus type 1-induced respiratory disease. J. Gen. Virol. 66:2383-2394. [DOI] [PubMed] [Google Scholar]
- 2.Banfield, B. W., J. D. Kaufman, J. A. Randall, and G. E. Pickard. 2003. Development of pseudorabies virus strains expressing red fluorescent proteins: new tools for multisynaptic labeling applications. J. Virol. 77:10106-10112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baranowski, E., J. Dubuisson, P.-P. Pastoret, and E. Thiry. 1993. Identification of 108K, 93K, and 42K glycoproteins of bovine herpesvirus-1 by monoclonal antibodies. Arch. Virol. 133:97-111. [DOI] [PubMed] [Google Scholar]
- 4.Campadelli-Fiume, G., F. Cocchi, L. Menotti, and M. Lopez. 2000. The novel receptors that mediate the entry of herpes simplex viruses and animal alphaherpesviruses into cells. Rev. Med. Virol. 10:305-319. [DOI] [PubMed] [Google Scholar]
- 5.Chase, C. C., K. Carter-Allen, C. Lohff, and G. J. Letchworth. 1990. Bovine cells expressing bovine herpesvirus 1 (BHV-1) glycoprotein IV resist infection by BHV-1, herpes simplex virus, and pseudorabies virus. J. Virol. 64:4866-4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chase, C. C., C. Lohff, and G. J. Letchworth. 1993. Resistance and susceptibility of bovine cells expressing herpesviral glycoprotein D homologs to herpesviral infections. Virology 194:365-369. [DOI] [PubMed] [Google Scholar]
- 7.Chase, C. C., and G. J. Letchworth. 1994. Bovine herpesvirus 1 gIV-expressing cells resist virus penetration. J. Gen. Virol. 75:177-181. [DOI] [PubMed] [Google Scholar]
- 8.Chowdhury, S. I., C. S. D. Ross, B. J. Lee, V. Hall, H. J. Chu. 1999. Construction and characterization of a glycoprotein E gene-deleted bovine herpesvirus type 1 recombinant. Am. J. Vet. Res. 60:227-232. [PubMed] [Google Scholar]
- 9.Czarniecki, C. W., E. B. Hamilton, C. W. Fennie, and R. L. Wolf. 1986. In vitro biological activities of Escherichia coli-derived bovine interferon-alpha, -beta, and -gamma. J. Interferon Res. 6:29-37. [DOI] [PubMed] [Google Scholar]
- 10.Dasika, G. K., and G. J. Letchworth. 1999. Cellular expression of bovine herpesvirus 1 gD inhibits cell-to-cell spread of two closely related viruses without blocking their primary infection. Virology 254:24-36. [DOI] [PubMed] [Google Scholar]
- 11.Dasika, G. K., and G. J. Letchworth. 2000. Homologous and heterologous interference requires bovine herpesvirus-1 glycoprotein D at the cell surface during virus entry. J. Gen. Virol. 81:1041-1049. [DOI] [PubMed] [Google Scholar]
- 12.De Wit, W. J., J. J. Hage, J. Brinkhof, and F. Westenbrink. 1998. A comparative study of serological tests for use in the bovine herpesvirus 1 eradication program in The Netherlands. Vet. Microbiol. 61:153-163. [DOI] [PubMed] [Google Scholar]
- 13.Dispas, M., F. Schynts, M. Lemaire, C. Letellier, E. Vanopdenbosch, E. Thiry, and P. Kerkhofs. 2003. Isolation of a glycoprotein E deleted bovine herpesvirus-1 strain in the field. Vet. Rec. 153:209-212. [DOI] [PubMed] [Google Scholar]
- 14.Dohner, D. E., S. G. Adams, and L. D. Gelb. 1988. Recombination in tissue culture between varicella-zoster virus strains. J. Med. Virol. 24:329-341. [DOI] [PubMed] [Google Scholar]
- 15.Dutta, S. K., A. C. Myrup, and S. R. Thaker. 1986. In vitro interference between equine herpesvirus types 1 and 2. Am. J. Vet. Res. 47:747-750. [PubMed] [Google Scholar]
- 16.Fujita, K., K. Maeda, N. Yokoyama, T. Miyazawa, C. Kai, and T. Mikami. 1998. In vitro recombination of feline herpesvirus type 1. Arch. Virol. 143:25-34. [DOI] [PubMed] [Google Scholar]
- 17.Garber, D. A., S. M. Beverley, and D. M. Coen. 1993. Demonstration of circularization of herpes simplex virus DNA following infection using pulsed field gel electrophoresis. Virology 197:459-462. [DOI] [PubMed] [Google Scholar]
- 18.Geraghty, R. J., C. R. Jogger, and P. G. Spear. 2000. Cellular expression of alphaherpesvirus gD interferes with entry of homologous and heterologous alphaherpesviruses by blocking access to a shared gD receptor. Virology 268:147-158. [DOI] [PubMed] [Google Scholar]
- 19.Glazenburg, K. L., R. J. M. Moorman, T. G. Kimman, A. L. J. Gielkens, and B. P. H. Peeters. 1994. In vivo recombination of pseudorabies virus strains in mice. Virus Res. 34:115-126. [DOI] [PubMed] [Google Scholar]
- 20.Henderson, L. M., J. B. Katz, G. A. Erickson, and J. E. Mayfield. 1990. In vivo and in vitro genetic recombination between conventional and gene-deleted vaccine strains of pseudorabies virus. Am. J. Vet. Res. 51:1656-1662. [PubMed] [Google Scholar]
- 21.Javier, R. T., F. Sedarati, and J. G. Stevens. 1986. Two avirulent herpes simplex viruses generate lethal recombinants in vivo. Science 234:746-748. [DOI] [PubMed] [Google Scholar]
- 22.Johnson, R. M., and P. G. Spear. 1989. Herpes simplex virus glycoprotein D mediates interference with herpes simplex virus infection. J. Virol. 63:819-827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson, D. C., R. L. Burke, and T. Gregory. 1990. Soluble forms of herpes simplex virus glycoprotein D bind to a limited number of cell surface receptors and inhibit virus entry into cells. J. Virol. 64:2569-2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaashoek, M. J., F. A. Rijsewijk, R. C. Ruuls, G. M. Keil, E. Thiry, P.-P. Pastoret, and J. T. Van Oirschot. 1998. Virulence, immunogenicity and reactivation of bovine herpesvirus 1 mutants with a deletion in the gC, gG, gI, gE, or in both the gI and gE gene. Vaccine 16:802-809. [DOI] [PubMed] [Google Scholar]
- 25.Katz, J. B., L. M. Henderson, and G. A. Erickson. 1990. Recombination in vivo of pseudorabies vaccine strains to produce new virus strains. Vaccine 8:286-288. [DOI] [PubMed] [Google Scholar]
- 26.Keywan, K., and E. Katz. 1999. Co-infection of acyclovir-resistant and acyclovir-sensitive herpes simplex type 2 virus strains in BS-C-1 cells. Intervirology 42:247-251. [DOI] [PubMed] [Google Scholar]
- 27.Kim, J. S., L. W. Enquist, and J. P. Card. 1999. Circuit-specific coinfection of neurons in the rat central nervous system with two pseudorabies virus recombinants. J. Virol. 73:9521-9531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leung-Tack, P. 1993. L'herpèsvirus bovin de type 1: caractérisation de la région US du génome viral (souche ST) et étude de mutants délétés. Ph.D. thesis. University Louis Pasteur, Strasbourg, France.
- 29.Leung-Tack, P., J. C. Audonnet, and M. Rivière. 1994. The complete DNA sequence and the genetic organization of the short unique region (US) of the bovine herpesvirus type 1 (ST strain). Virology 199:409-421. [DOI] [PubMed] [Google Scholar]
- 30.Liang, X. P., L. A. Babiuk, L. A., S. van Drunen Little-van Den Hurk, D. R. Fitzpatrick, and T. J. Zamb. 1991. Bovine herpesvirus 1 attachment to permissive cells is mediated by its major glycoproteins gI, gIII, and gIV. J. Virol. 65:1124-1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liang, X., L. A. Babiuk, and T. J. Zamb. 1992. An in vivo study of a glycoprotein gIII-negative bovine herpesvirus 1 (BHV-1) mutant expressing β-galactosidase: evaluation of the role of gIII in virus infectivity and its use as a vector for mucosal immunization. Virology 189:629-639. [DOI] [PubMed] [Google Scholar]
- 32.Ludwig, G. V., and G. J. Letchworth III. 1987. Temporal control of bovine herpesvirus 1 glycoprotein synthesis. J. Virol. 61:3292-3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marcus, P. I. 1999. Interferons, p. 854-862. In A. Granoff and R. G. Webster (ed.), Encyclopedia of virology. Academic Press, Inc., San Diego, Calif.
- 34.Matis, J., M. Kudelova, and J. Rajcani. 1999. Interference of the low-pH inactivated herpes simplex virus type 1 (HSV-1) strain HSZP with the early shutoff function of superinfecting HSV-1 strain KOS. Virus Res. 60:81-86. [DOI] [PubMed] [Google Scholar]
- 35.McVoy, M. A., and S. P. Adler. 1994. Human cytomegalovirus DNA replicates after early circularization by concatemer formation, and inversion occurs within the concatemer. J. Virol. 68:1040-1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Metzler, A. E., H. Matile, U. Gassmann, M. Engels, and R. Wyler. 1985. European isolates of bovine herpesvirus 1: a comparison of restriction endonuclease sites, polypeptides, and reactivity with monoclonal antibodies. Arch. Virol. 85:57-69. [DOI] [PubMed] [Google Scholar]
- 37.Meurens, F., B. Muylkens, F. Schynts, I. Bourgot, A. Billiau, and E. Thiry. 2003. L'interférence virale chez les Alphaherpesvirinae. Virologie 7:319-328. [Google Scholar]
- 38.Mossman, K. L. 2002. Activation and inhibition of virus and interferon: the herpesvirus story. Viral Immunol. 15:3-15. [DOI] [PubMed] [Google Scholar]
- 39.Nauwynck, H. J., G. G. Labarque, and M. B. Pensaert. 1999. Efficacy of an intranasal immunization with gEgC and gEgI double-deletion mutants of Aujeszky's disease virus in maternally immune pigs and the effects of a successive intramolecular booster with commercial vaccines. J. Vet. Med. B 46:713-722. [DOI] [PubMed] [Google Scholar]
- 40.Nishiyama, Y., H. Kimura, and T. Daikoku. 1991. Complementary lethal invasion of the central nervous system by nonneuroinvasive herpes simplex virus types 1 and 2. J. Virol. 65:4520-4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nolan, G. P., S. Fiering, J. F. Nicolas, and L. A. Herzenberg. 1988. Fluorescence activated cell analysis and sorting of viable mammalian cells based on β-d-galactosidase activity after transduction of Escherichia coli lacZ. Proc. Natl. Acad. Sci. USA 85:2603-2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pastoret, P.-P., E. Thiry, B. Brochier, G. Derboven, and H. Vindevogel. 1984. The role of latency in the epizootiology of infectious bovine rhinotracheitis, p. 221-227. In G. Wittmann, R. M. Gaskell, and H. J. Rziha (ed.), Latent herpesvirus infections in veterinary medicine. Kluwer Academic Publishers, Boston, Mass.
- 43.Purifoy, D. J., and K. L. Powell. 1977. Interference between strains of type 1 and type 2 herpes simplex virus. Virology 77:84-94. [DOI] [PubMed] [Google Scholar]
- 44.Rebordosa, X., J. Pinol, J. A. Perez-Pons, J. Lloberas, J. Naval, X. Serra-Hartmann, E. Espuna, and E. Querol. 1996. Glycoprotein E of bovine herpesvirus type 1 is involved in virus transmission by direct cell-to-cell spread. Virus Res. 45:59-68. [DOI] [PubMed] [Google Scholar]
- 45.Roizman, B. 1963. The programming of herpes virus multiplication in doubly-infected and in puromycin-treated cells. Proc. Natl. Acad. Sci. USA 49:165-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roizman, B., and D. M. Knipe. 2001. Herpes simplex viruses and their replication, p. 2399-2461. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott/The Williams & Wilkins Co., Philadelphia, Pa.
- 47.Savan, M., A. B. Angulo, and J. B. Derbyshire. 1979. Interferon, antibody responses and protection induced by an intranasal infectious bovine rhinotracheitis vaccine. Can. Vet. J. 20:207-210. [PMC free article] [PubMed] [Google Scholar]
- 48.Schynts, F., E. Baranowski, M. Lemaire, and E. Thiry. 1999. A specific PCR to differentiate between gE negative vaccine and wild-type bovine herpesvirus type 1 strains. Vet. Microbiol. 66:187-195. [DOI] [PubMed] [Google Scholar]
- 49.Schynts, F., A. Vanderplasschen, E. Hanon, F. A. Rijsewijk, J. T. Van Oirschot, and E. Thiry. 2001. Use of PCR and immunofluorescence to detect bovine herpesvirus 1 recombinants. J. Virol. Methods 92:99-104. [DOI] [PubMed] [Google Scholar]
- 50.Schynts, F., M. A. McVoy, F. Meurens, B. Detry, A. L. Epstein, and E. Thiry. 2003. The structures of bovine herpesvirus 1 virion and concatemeric DNA: implications for cleavage and packaging of herpesvirus genomes. Virology 314:326-335. [DOI] [PubMed] [Google Scholar]
- 51.Schynts, F., F. Meurens, B. Detry, A. Vanderplasschen, and E. Thiry. 2003. Rise and survival of bovine herpesvirus 1 recombinants after primary infection and reactivation from latency. J. Virol. 77:12535-12542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sedarati, F., R. T. Javier, and J. G. Stevens. 1988. Pathogenesis of a lethal mixed infection in mice with two nonneuroinvasive herpes simplex virus strains. J. Virol. 62:3037-3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Slobedman, B., X. Zhang, and A. Simmons. 1999. Herpes simplex virus genome isomerization: origins of adjacent long segments in concatemeric viral DNA. J. Virol. 73:810-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smith, G. A., S. T. Gross, and L. W. Enquist. 2001. Herpesviruses use bidirectionnal fast-axonal transport to spread in sensory neurons. Proc. Natl. Acad. Sci. USA 98:3466-3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Spear, P. G., R. J. Eisenberg, and G. H. Cohen. 2000. Three classes of cell surface receptors for alphaherpesvirus entry. Virology 275:1-8. [DOI] [PubMed] [Google Scholar]
- 56.Stegeman, A. 1995. Pseudorabies virus eradication by area-wide vaccination is feasible. Ph.D. thesis. University of Utrecht, Utrecht, The Netherlands. [DOI] [PubMed]
- 57.Strube, W., S. Auer, W. Block, E. Heinen, D. Kretzdorn, C. Rodenbach, and N. Schmeer. 1996. A gE deleted infectious bovine rhinotracheitis marker vaccine for use in improved bovine herpesvirus 1 control programs. Vet. Microbiol. 53:181-189. [DOI] [PubMed] [Google Scholar]
- 58.Tikoo, S. K., M. Campos, and L. A. Babiuk. 1995. Bovine herpesvirus 1 (BHV-1): biology, pathogenesis, and control. Adv. Virus Res. 45:191-223. [DOI] [PubMed] [Google Scholar]
- 59.Van Engelenburg, E. F., M. J. Kaashoek, F. A. Rijsewijk, D. B. van den Burg, A. Moerman, A. L. Gielkens, and J. T. Van Oirschot. 1994. A glycoprotein E deletion mutant of bovine herpesvirus 1 is avirulent in calves. J. Gen. Virol. 75:2311-2318. [DOI] [PubMed] [Google Scholar]
- 60.Van Oirschot, J. T., M. J. Kaashoek, M. A. Maris-Veldhuis, K. Weerdmeester, and F. A. Rijsewijk. 1997. An enzyme-linked immunosorbent assay to detect antibodies against glycoprotein gE of bovine herpesvirus 1 allows differentiation between infected and vaccinated cattle. J. Virol. Methods 67:23-34. [DOI] [PubMed] [Google Scholar]
- 61.Wyler, R., M. Engels, and M. Schwyzer. 1989. Infectious bovine rhinotracheitis/vulvovaginitis (BHV-1), p. 1-72. In G. Wittmann (ed.), Herpesvirus diseases of cattle, horses and pig. Kluwer Academic Publishers, Boston, Mass.
- 62.Xiao, C. Z., Z. L. Chen, and B. Z. Wu. 1989. Kinetics of antiviral state in cells activated by interferons in vitro. Arch. Immunol. Ther. Exp. 37:508-513. [PubMed] [Google Scholar]
- 63.Youngner, J. S., and P. Whitaker-Dowling. 1999. Interference, p. 850-862. In A. Granoff and R. G. Webster (ed.), Encyclopedia of virology. Academic Press, Inc., San Diego, Calif.
- 64.Zelena, D., J. Roubal, and V. Vonka. 1976. Inhibitory effect of herpes simplex virus type 1 on type 2 virus replication. J. Gen. Virol. 33:249-257. [DOI] [PubMed] [Google Scholar]
- 65.Zsak, L., F. Zuckermann, N. Sugg, and T. Ben-Porat. 1992. Glycoprotein gI of pseudorabies virus promotes cell fusion and virus spread via direct cell-to-cell transmission. J. Virol. 66:2316-2325. [DOI] [PMC free article] [PubMed] [Google Scholar]