The generation of induced pluripotent stem cells (iPSCs) presents a challenge to normal developmental processes. The low efficiency and heterogeneity of most methods have hindered understanding of the precise molecular mechanisms promoting, and roadblocks preventing, efficient reprogramming. While several intermediate populations have been described1-7, it has proved difficult to characterize the rare, asynchronous transition from these intermediate stages to iPSCs. The rapid expansion of a minor population of reprogrammed cells can also obscure investigation of relevant processes. Understanding of the biological mechanisms essential for successful iPSC generation requires both accurate capture of cells undergoing the reprogramming process and identification of the associated global gene expression changes. Here we demonstrate that reprogramming follows an orderly sequence of stage transitions marked by changes in cell surface markers CD44 and ICAM1, and a Nanog-GFP reporter. RNA-sequencing (RNA-seq) analysis of these populations demonstrates two waves of pluripotency gene up-regulation, and unexpectedly, transient up-regulation of multiple epidermis-related genes, demonstrating that reprogramming is not simply the reversal of normal developmental processes. This novel high-resolution analysis enables the construction of a detailed reprogramming route map, and this improved understanding of the reprogramming process will lead to novel reprogramming strategies.
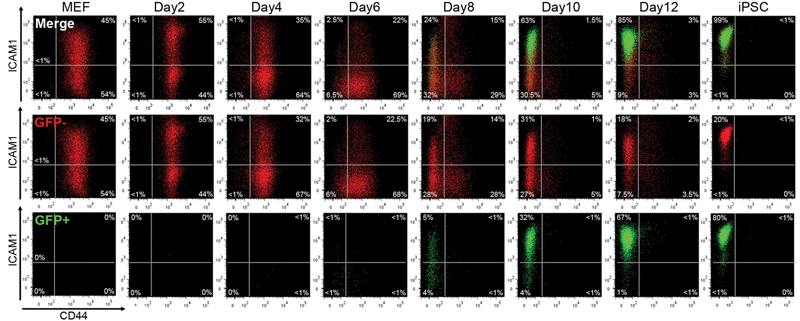
Several reports have suggested that reprogramming progresses in a somewhat ordered manner3,5,6,8-10. In order to identify markers whose expression changed concurrent with pluripotency gene expression we performed time course microarray analysis using a piggyBac (PB) transposon-based secondary (2°) reprogramming system3,11 (Supplementary Figure 2a). Of a number of candidate cell surface markers, Cd44 and Icam1 demonstrated the most dynamic expression changes throughout 2° MEF reprogramming (Supplementary Figure 2b). For further investigation we generated an efficient 2° reprogramming system in which doxycycline (dox) mediated induction of the reprogramming factors could be monitored via an mOrange reporter placed after the 2A-peptide-linked reprogramming cassette c-Myc-Klf4-Oct4-Sox2 (MKOS)12, and endogenous Nanog promoter activation could be followed by expression of EGFP13 (Supplementary Figures 3,4). Reprogramming cultures were supplemented with Vitamin C (VitC) and an Alk inhibitor (Alki) which enhance reprogramming efficiency10,14,15. In this 2° reprogramming system, Nanog-GFP+ cells appeared as early as day 6, and >60% of mOrange+ transgene expressing cells were found to be Nanog-GFP+ by day 12 (Supplementary Figure 5a). Most mOrange+ transgene expressing cells lost Thy1 and gained E-cadherin expression by day4 (Supplementary Figures 5b,c). SSEA-1 expression barely changed after day 8, with a gradual gain of Nanog-GFP+ cells in both SSEA-1+ and SSEA-1− populations (Supplementary Figure 5d). Consistent with heterogeneous expression of SSEA1 in iPSCs/ESCs, it was not possible to accurately delineate the reprogramming process using SSEA-1 (Supplementary Figure 6). In contrast, the appearance of CD44− and ICAM1+ cells at later time points closely correlated with Nanog-GFP expression (Supplementary Figures 5e,f). Double staining for CD44 and ICAM1 revealed that a distinct series of population changes occur during reprogramming (Figure 1). Initially, MEFs displayed high CD44 and broad ICAM1 expression with the majority becoming ICAM1− by day 6, along with the appearance of a minor CD44− ICAM1− population. By day 8, CD44− populations appeared enriched, and at day 12 almost all cells displayed an iPSC/ESC-like CD44− ICAM1+ profile, over 60% of which expressed Nanog-GFP. Consistent with an observation that Nanog expression is not necessarily a sign of completed reprogramming16, Nanog-GFP+ cells were observed even before cells obtained this iPSC/ESC-like phenotype (CD44− ICAM1+). Both ICAM1+ and ICAM1− sorted MEF demonstrated similar FACS profile changes during reprogramming (Supplementary Figure 7). Immunofluorescence for CD44 and ICAM1 revealed that reprogramming is not synchronized even within individual colonies (Supplementary Figure 8). Secondary reprogramming of non-polycistronic line 6c11,14 and primary reprogramming using MKOS and OSKM20 PB transposons resulted in similar ICAM1/CD44 profile changes indicating their sutability for use in other systems and contexts (Supplimentary Figure 9). These findings demonstrated the asynchronous but stepwise manner of reprogramming and highlighted the potential usefulness of CD44 and ICAM1 to isolate reprogramming intermediate subpopulations.
Figure 1. FACS analysis during 2° reprogramming of MEF with CD44/ICAM1 double staining.
Loss of CD44 expression was rapidly followed by ICAM1 upregulation and Nanog-GFP expression. By day 12 the majority of cells displayed an ICAM+/CD44− ESC-like profile. Red; Nanog-GFP− cells, Green; Nanog-GFP+ cells.
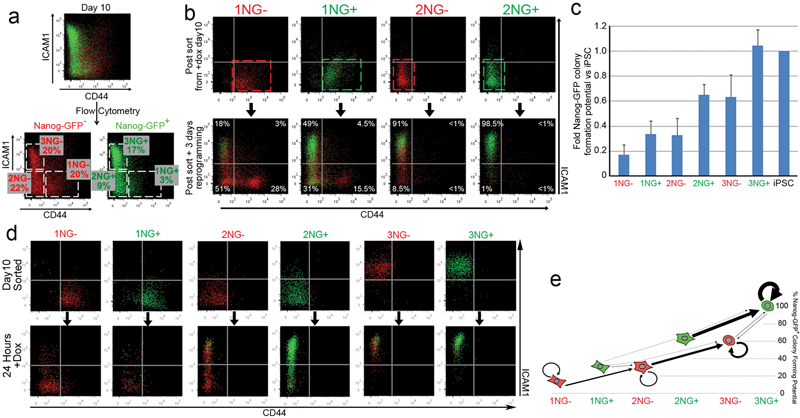
Next, we aimed to confirm that the observed CD44/ICAM1 profile changes reflected the transition of individual cells from one stage to the next, and not merely the loss of one major population and expansion of another minor population. CD44+ ICAM1− (Gate1), CD44− ICAM1− (Gate2) and CD44− ICAM1+ (Gate3) populations, either Nanog-GFP+ (NG+) or Nanog-GFP− (NG−) were isolated by cell-sorting at day 10 of reprogramming and re-plated in reprogramming conditions (Figure 2a). After 3 days both NG+ and NG− cells progressed in the order of gates 1→2→3 (Figure 2b). This progression correlated well with increased Nanog-GFP+ colony forming potential (cfp), with 3NG+ cells displaying similar clonogenicity to fully reprogrammed iPSCs (Figure 2c). Of cells with the same CD44/ICAM1 profile, Nanog-GFP expression correlated with a higher cfp (e.g. 1NG− vs 1NG+).
Figure 2. CD44/ICAM1 subpopulations represent distinct stages of reprogramming.
a. Nanog-GFP+ (NG+) and Nanog-GFP− (NG−) cells were subdivided into CD44+ ICAM1− (Gate1), CD44− ICAM1− (Gate 2) and CD44− ICAM1+ (Gate 3) populations at day 10 of reprogramming. b. FACS analysis of sorted subpopulations after 3 day culture in the presence of dox. c. Relative probability to generate Nanog-GFP+ iPSC colonies from each subpopulation compared to fully reprogrammed iPSCs. Error bars represent standard deviation, n=3 d. CD44/ICAM1/Nanog-GFP expression was re-analysed 24 hours after sorting. e. Major transitions (>500 cells) of each populationwithin 24 hours. Y axis indicates relative colony formation potential after an additional 10 days. Arrow size reflects relative cell numbers.
To more accurately examine the progression of the reprogramming process, cells from each gate were sorted and their CD44/ICAM1/Nanog-GFP expression was re-analysed after 24 hours (Figure 2d). Based on total cell numbers in each gate after 24 hours (Supplementary Figure 10), we generated a reprogramming route map representing differences in the efficiency of these stage transitions and in Nanog-GFP+ cfp (Figure 2e). Similar results were obtained when each subpopulation was sorted at day8 (Supplementary Figure 11). This analysis revealed that reaching a Nanog-GFP+ state is a rate limiting step – as few cells overcame this barrier in the 24 hour assay – and those that do so reprogram more efficiently than their Nanog-GFP− counterparts, consistent with the role of Nanog as an accelerator of reprogramming and the gateway to pluripotency17,18.
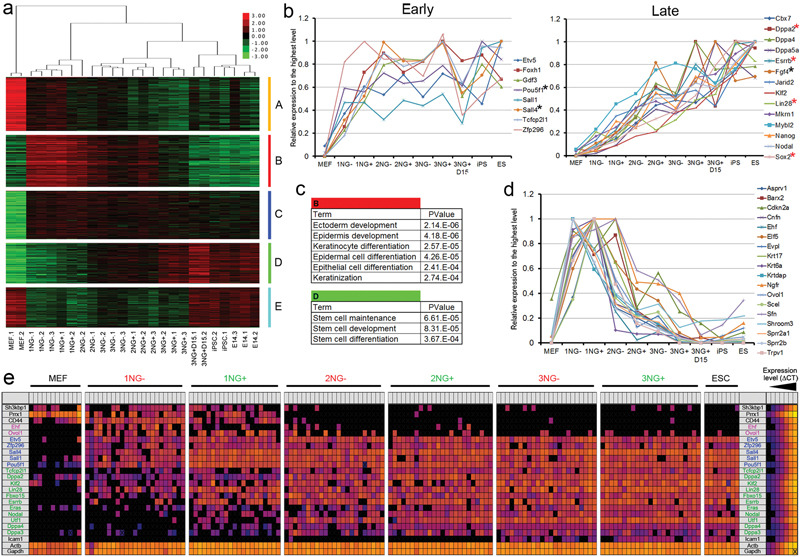
To elucidate global gene expression changes during these stage transitions, we carried out RNA-sequencing analysis using a highly multiplexed sample barcoding system19,20 (see Methods Online, Supplementary Table 1). Hierarchical clustering using the complete list of differentially expressed genes (DEGs) revealed four major branches; [MEFs], [1NG−/+ and 2NG−], [2NG−/+ and 3NG−/+], and [3NG+ sorted at day 15 (3NG+D15), iPSCs and ESCs] (Figure 3a). These clusters correlate well with similarities in cfp of the respective subpopulations (Figure 2c). There was a prominent gene expression difference between 3NG+ and 3NG+D15, with the latter being more similar to iPSCs and ESCs (Figure 3a, Supplementary Figures 12a,c), possibly reflecting the observed difference in cfp in the absence of dox (Supplementary Figure 13). The DEGs between these two populations may be involved in the establishment of an exogenous factor independent self-renewal state. Principal component analysis clearly distinguished 2NG+ from 3NG− cells, consistent with the higher probability of the former to reach the 3NG+ state within 24 hours (Supplementary Figures 11c and 12b). DEGs could be classified into five distinct expression pattern groups (A to E) (Figure 3a, Supplementary Tables 2,3). Group A contained readily down-regulated fibroblast-related genes. Group D comprised factors gradually up-regulated toward iPSCs, where ESC genes were highly enriched (P≤0.000367) (Figure 3c). However Group C, which contained genes up-regulated at early stages and maintained throughout reprogramming, also included some pluripotency related factors. To extend this finding, we examined the expression pattern of 22 pluripotency-related genes in our data set21,22. Interestingly, 8 pluripotency genes, including endogenous Oct4 (Pou5f1) were already up-regulated at the 1NG+/2NG− stages to the level found in 3NG+ cells (Figure 3b Early), while 14 pluripotency genes were more gradually up-regulated in the later stage reprogramming populations (Figure 3b Late, Supplementary Table 4). This early and late pluripotency gene up-regulation was confirmed at the single cell level5 (Figure 3e), highlighting the high resolution of the CD44/ICAM1 sorting system.
Figure 3. Global gene expression changes during the stage transition.
a. Hierarchical clustering of samples with DEGs and expression heat map. Groups A-E represent different expression patterns. b. Early and late up-regulation of pluripotency-related genes. Black and red asterisks indicate early and late pluripotency genes respectively as previously identified by single cell qPCR5. c. Epidermis gene and stem cell gene enrichment in gene list B and D respectively. d. Transient up-regulation of 18 epidermis/keratinocyte-related genes during reprogramming. e. Single-cell gene expression analysis. Each square represents one reaction chamber from one cell. Colour corresponds to ΔCT value as shown in legend at right.
We also identified two additional gene expression patterns displaying transient up-regulation (Group B) or down-regulation (Group E) exclusively in the intermediate stages of reprogramming. This finding indicates that reprogramming from MEFs to iPSCs is not simply the loss of MEF genes and gain of ESC genes. Gene Ontology analysis revealed that genes related to ectoderm/epidermis development and keratinocyte differentiation were highly enriched in Group B (P≤0.000274) (Figures 3c,d, Supplementary Tables 3-5). While Sfn and Krt17 were barely detectable by immunofluorscence in MEFs and iPSCs, transient up-regulation was observed in the intermediate stages of reprogramming (Supplementary Figure 14). Single cell PCR confirmed co-expression of epidermis genes (Ehf, Ovol1) with early pluripotency genes in the 1NG−/+ stage (Figure 3e). Consistent with our data, analysis of three published microarray data sets incorporating partially reprogrammed iPSCs (piPSCs)1, a time course experiment3 and a subpopulation analysis with Thy1, SSEA-1 and Oct4-GFP6 confirmed transient epidermal gene expression during reprogramming (Supplementary Figures 15-17, Supplementary Tables 6-8). Partially reprogrammed cells from B cells also displayed similar epidermis gene expression4, while Oct4, Sox2 two factor-reprogramming of MEFs did not23. Therefore, this intermediate state could be a consequence of the use of Klf4 which is important for efficient reprogramming and demonstrates the reprogramming process is not simply reversion of normal differentiation (Summarized in Supplementary Figure 1). It would be intriguing to investigate if similar transient gene expression changes can be seen in reprogramming of ectoderm or endoderm lineages. Down-regulation of these epidermis genes coincided with up-regulation of “late” pluripotency genes. Future examination of this rapid switch in gene expression may provide a novel insight into the molecular mechanism of reprogramming.
The integrative data analysis described above demonstrated this CD44/ICAM1/Nanog-GFP marker system, uniquely, could provide high-resolution information during late pluripotency gene up-regulation enabling discrimination of ‘reprogramming’ from ‘expansion of reprogrammed cells’ (Figure 3b, Supplementary Figures 16b, 17f,). This system also refines investigation of the kinetics of reprogramming. It has recently been shown that VitC increases reprogramming efficiency by facilitating H3K9 demethylation7 and that reprogramming factors fail to bind trimethylated H3K9 rich regions in the initial stages of reprogramming24. We carried out reprogramming in the absence of VitC and observed not only a decrease in the iPSC colony number, but also a marked delay in the transition from one stage of reprogramming to the next (Supplementary Figure 18). Similar analyses can be performed using our marker system to investigate the mechanism of action of other factors that alter reprogramming efficiency. Isolation and analysis of subpopulations affected by these factors could reveal the downstream genes specifically involved in, and required for, successful reprogramming. Further studies using this high resolution analysis system have the potential to make a significant contribution towards revealing the molecular mechanisms of reprogramming.
METHODS
Vector construction
The piggyBac (PB) transposon PB-TAP containing the tetO2 promoter, an attR1R2 Gateway cloning cassette (Invitrogen) and rabbit β-globin poly A signal, was provided by A. Nagy. To minimize silencing of the reprogramming vector, a chicken β-globin insulator30, and a human lamin B2 (LMB2) replicator31 were inserted in PacI site between PB 3′TR and the tetO2, EcoRV site between rabbit β-globin poly A signal and PB 5′TR, respectively, to generate PB-TAP IRI. The BamHI fragment containing loxP flanked MKOS reprogramming cassette followed by ires-mOrange (2LMKOSimO) from pCAG2LMKOSimO12 were inserted into a Gateway entry vector pENTR 2B (Life Technologies), to generate attP2LMKOSimO pENTR. Finally the attP2LMKOSimO cassette was Gateway cloned into the PB-TAP IRI to yield reprogramming PB transposon PB-TAP IRI attP2LMKOSimO. Similarly, reprogramming PB transposon PB-TAP IRI 2LOSKMimO was generated after transferring the OSKM reprogramming cassette32 into attP2LMKOSimO pENTR replacing the MKOS cassette. Sequences of the plasmids are available upon request.
Generation of a primary iPSC line D6s4B5
12.5 d.p.c. embryos were obtained from Rosa-rtTA/rtAT, Nanog-GFP/+, Col1a1+/+ mice which were derived by a crossing TNG mice13 and B6;129-Gt(ROSA)26Sortm1(rtTA*M2)Jae Col1a1tm2(tetO-Pou5f1)Jae/J (The Jackson Laboratory). The embryos were decapitated, eviscerated, dissociated with 0.25% trypsin, 0.1% EDTA and plated in MEF medium (GMEM, 10% FBS, penicillin-streptomycin, 1× Non-Essential Amino Acids (Invitrogen), 1 mM Sodium Pyruvate, 0.05 mM 2-Mercaptoethanol ). The PB-TAP IRI attP2LMKOSimO (500 ng) and pCyL43 PB transposase expression vector33 (2 μg) were introduced into the MEFs via Nucleofection (Amaxa) as before11, and cells were cultured in the ES cell medium (MEF medium supplemented with 1000 U ml−1 Leukemia inhibiting factor (LIF)) in the presence of 1.0 μ ml−1 doxycycline (dox) (Sigma) for an initial 8 days and thereafter 0.5 μg ml−1 dox. Pluripotency of a clonal iPSC line D6 was confirmed by teratoma formation and a subclone D6s4B5 was used for secondary reprogramming. For comparing CD44 and ICAM1 profiles of primary reprogramming with PB-TAP IRI attP2LMKOSimO and PB-TAP IRI 2LOSKMimO vectors, MEF were nucleofected as above and cultured in the presence of 1.0 μ ml−1 dox, Vitamin C (10 μg ml−1) (Sigma) and Alk inhibitor (500 nM) (A83-01, TOCRIS Bioscience).
Secondary reprogramming
Each chimeric embryo were harvested at 12.5 d.p.c., dissociated and cultured in MEF medium. One twentieth of the dissociated cells were exposed to dox (300 ng ml−1) for 2 days and the proportion of transgenic (Tg) MEFs was measured by FACS analysis of mOrange expression. For FACS time course and colony counting experiments secondary Tg MEFs were diluted to 5% and 30% by addition of wild type 129 MEFs and plated in gelatinized 6 well-plate at 1×105 cells per well (5,000 and 30,000 Tg MEFs per well, respectively). For sorting experiments MEFs were plated at 2×105 cells per gelatinized 100 mm plate (1×104 Tg MEFs per plate). Cells were cultured in Reprogramming medium, which is ES cell medium supplemented with dox (300 ng ml−1), Vitamin C (10 μg ml−1) and Alki (500 nM). Medium was changed every 2 days.
Flow Cytometry and cell sorting
Surface marker analysis was performed with the following eBioscience antibodies (catalogue number; dilution): CD54/ICAM-1-biotin (13-0541; 1/100), CD44-biotin (17-0441; 1/100), CD44-APC (17-0441; 1/300), Streptavidin-PE-Cy7 (25-4317-82; 1/1500), SSEA-1-647 (51-8813; 1/50), CD324/E-Cadherin-biotin (13-3249; 1/100), Thy1-APC (17-0902, 1/300), CD2-biotin (13-0029; 1/100). For sorting experiments dead cells were excluded using DAPI nucleic acid stain (Invitrogen) (0.5ng/mL). Cells were incubated in 0.25% Trypsin, 1mM EDTA (Life Technologies) for 1-2 minutes at 37°C, harvested in GMEM containing 10% FCS and counted. Staining was carried out in FACS buffer (2% FCS in PBS) at ~1×106 cells ml−1 for 15-30 min at 4°C, and which was followed by washing with FACS buffer, sorting and/or analysis with FACSAriaII and LSRFortessa (both BD Biosciences), respectively. Excitation laser lines and filters used for each fluorophore are summarized in Supplementary Table 9. Data were analysed using FlowJo software (Tree Star). Intact cells were identified based on forward and side light scatter, and subsequently, analysed for fluorescence intensity. Additional gating was carried out as outlined in Supplementary Figure 2. For colony formation assays, sorted cells were plated on γ-irradiated MEFs in 12-well plates at 3.5×103 cells per well. Nanog-GFP+ colonies were quantified 10 days post sort. For 24 hour or time course analysis, sorted cells were plated in gelatinized 48-well plate at 1×104 cells per well. In both cases, cells were cultured in Reprogramming medium after sort.
Immunofluorescence and confocal microscopy imaging
Images of cells stained with CD54/ICAM-1-biotin (1/100 dilution), CD44-APC (1/300 dilution) antibodies and Streptavidin-PE-Cy7 (1/1500 dilution) described above were captured with a confocal microscope (Leica TSC SP2) and Leica confocal software. Cells stained with anti-Krt17 (LifeSpan BioSciences), anti-Sfn (Sigma) antibodies and anti-Rabbit IgG CF633 secondary antibody (Sigma) were imaged with a fluorescence microscopy (Olympus).
Multiplexed RNA Sequencing and data analysis
RNA was isolated with TRI reagent (Sigma) following the manufacturer’s instructions. RNA quality and concentration was determined using the Agilent 2100 Bioanalyzer (Agilent Technologies). Using 10ng RNA, reverse transcription with barcoded primers, cDNA amplification and sequencing with Illumina HiSeq 2000 was performed as previously described19,20. Quality control of the obtained reads and alignment to the mouse reference genome (NCBI37/mm9) was performed using the GeneProf web-based analysis suite with default parameters25. Gene expression read counts were exported and analysed in R to identify differentially expressed genes (DEGs), using the edgeR and DESeq Bioconductor libraries26-28. For both methods, low expression transcripts (less than 13 reads in all samples) were filtered out and P-values were adjusted using a threshold for false discovery rate (FDR) ≤ 0.05. Genes listed as DEGs by both methods in any two subpopulation comparison indicated in Supplementary Figure 6a (total 3,171) were used for further analysis. Hierarchical clustering and K-means clustering (K=5) was performed using Cluster 3.0 and Java Treeview was used for visualisation34,35. This multiplexed RNA-sequencing technology reads only the 5′ end of transcript, thus detecting only endogenous Oct4 and Sox2. Nanog expression was detectable in Nanog-GFP− populations due to the reporter system. Principal Components Analysis (PCA) was performed in R and plotted with the scatterplot3d library36. Gene ontology (GO) enrichment was calculated using the DAVID functional annotation bioinformatics tool29. GO term enrichment analysis was carried out with a modified Fisher Exact p-value. The three additional published studies1,3,6 (GEO accession number GSE21757, GSE14012, GSE42379) were analysed in a similar way. For the time course data the analysis was performed as following: data were RMA37 normalised using the Expression Console from Affymetrix and, since no replicates were provided, fold changes (FC) between each two samples were calculated in Excel. Genes with more than 1.5 FC were classified as DEGs. For the Plath and Polo dataset, data were RMA normalised using the ‘affy’ package38 in R and DEGs were identified using the ‘limma’ package38 in R with FC>=1.5 and FDR<=0.05 or FC>=1.5 where no replicates were available. Subsequently, K-means clustering of the identified DEGs was performed for all studies. Selected gene expression data shown as relative expression against the highest signal among the samples using an averaged signal value (reads per million) of duplicates/triplicates.
Single-cell gene expression analysis
Single cell qPCR was performed as described previously5 with slight modifications. Briefly, 22 sets of TaqMan Gene Expression assays (Applied Biosystems, Supplementary Table 9) were pooled at a final concentration of 180 nM per primer set and 50 μM per probe. Individual cells were sorted directly into 10 μl RT-PreAmp Master Mix (5 μl of CellsDirect Reaction Mix (Invitrogen), 2.5 μl of pooled assays, 0.2 μl of SuperScript III (Invitrogen), 1.3 μl of water) using FACSAria II. Cell lysis and sequence-specific reverse transcription were performed at 50°C for 15 min. The reverse transcriptase was inactivated by heating to 95°C for 2 min. Subsequently, in the same tube, cDNA went through sequence-specific amplification by denaturing at 95C for 15 s, and annealing and amplification at 60°C for 4 min for 22 cycles. Preamplified products were diluted 5-fold with water and analysed in 48.48 Dynamic Arrays on a BioMark System (Fluidigm) following the Fluidigm protocol. Ct values were calculated and visualized using BioMark Real-time PCR Analysis software (Fluidigm). Each assay was performed in replicate.
Supplementary Material
Acknowledgements
We thank A. Nagy and K. Woltjen for providing 6c iPSC line, I. Chambers for providing TNG MEFs, S. Monard and O. Rodrigues for assistance with flow cytometry, T. Kunath, T. Burdon, S. Lowell, N. Festuccia for discussions and comments on the manuscript. We also thank L.Robertson for technical assistance, Biomed unit staff for mouse husbandry. This work was supported by an ERC starting grant and the Anne Rowling Regenerative Neurology Clinic. J.O’M. and T.R. are funded by an MRC PhD Studentship and a Darwin Trust of Edinburgh Scholarship, respectively.
Footnotes
Supplementary Information is linked to the on line version of the paper at www.nature.com/nature.
Author Contribution J.O’M. designed and performed flow cytometry analysis and sorting experiments, prepared RNA for sequencing, carried out immunofluorescence imaging, and collected, analysed, interpreted data, and wrote the manuscript. S.S. analysed RNA-sequencing and published microarray data sets. K.I. carried out single cell PCR analysis. E.C. performed primary reprogramming and FACS analysis. T.R. carried out immunofluorescence and confocal imaging. S.R.T. performed microarray analysis to identify cell surface marker candidates. A.J. and S.L. performed multiplexed RNA-sequencing and collected data. K.K. conceived the study, identified the surface markers, generated D6s4B5 iPSC line, analysed RNA-sequencing data, supervised experiment design and data interpretation, and wrote the manuscript.
Author Information RNA-sequencing data are deposited in the ArrayExpress under accession number E-MTAB-1654. Reprints and permissions information is available online at www.nature.com/reprints. The authors declare no competing financial interests.
REFERENCES
- 1.Sridharan R, et al. Role of the murine reprogramming factors in the induction of pluripotency. Cell. 2009;136:364–377. doi: 10.1016/j.cell.2009.01.001. doi:10.1016/j.cell.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Golipour A, et al. A late transition in somatic cell reprogramming requires regulators distinct from the pluripotency network. Cell stem cell. 2012;11:769–782. doi: 10.1016/j.stem.2012.11.008. doi:10.1016/j.stem.2012.11.008. [DOI] [PubMed] [Google Scholar]
- 3.Samavarchi-Tehrani P, et al. Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell stem cell. 2010;7:64–77. doi: 10.1016/j.stem.2010.04.015. doi:10.1016/j.stem.2010.04.015. [DOI] [PubMed] [Google Scholar]
- 4.Mikkelsen TS, et al. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008;454:49–55. doi: 10.1038/nature07056. doi:10.1038/nature07056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buganim Y, et al. Single-cell expression analyses during cellular reprogramming reveal an early stochastic and a late hierarchic phase. Cell. 2012;150:1209–1222. doi: 10.1016/j.cell.2012.08.023. doi:10.1016/j.cell.2012.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Polo JM, et al. A molecular roadmap of reprogramming somatic cells into iPS cells. Cell. 2012;151:1617–1632. doi: 10.1016/j.cell.2012.11.039. doi:10.1016/j.cell.2012.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen J, et al. H3K9 methylation is a barrier during somatic cell reprogramming into iPSCs. Nature genetics. 2013;45:34–42. doi: 10.1038/ng.2491. doi:10.1038/ng.2491. [DOI] [PubMed] [Google Scholar]
- 8.Stadtfeld M, Maherali N, Breault DT, Hochedlinger K. Defining molecular cornerstones during fibroblast to iPS cell reprogramming in mouse. Cell stem cell. 2008;2:230–240. doi: 10.1016/j.stem.2008.02.001. doi:10.1016/j.stem.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brambrink T, et al. Sequential expression of pluripotency markers during direct reprogramming of mouse somatic cells. Cell stem cell. 2008;2:151–159. doi: 10.1016/j.stem.2008.01.004. doi:10.1016/j.stem.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li R, et al. A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell stem cell. 2010;7:51–63. doi: 10.1016/j.stem.2010.04.014. doi:10.1016/j.stem.2010.04.014. [DOI] [PubMed] [Google Scholar]
- 11.Woltjen K, et al. piggyBac transposition reprograms fibroblasts to induced pluripotent stem cells. Nature. 2009;458:766–770. doi: 10.1038/nature07863. doi:10.1038/nature07863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaji K, et al. Virus-free induction of pluripotency and subsequent excision of reprogramming factors. Nature. 2009;458:771–775. doi: 10.1038/nature07864. doi:10.1038/nature07864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chambers I, et al. Nanog safeguards pluripotency and mediates germline development. Nature. 2007;450:1230–1234. doi: 10.1038/nature06403. doi:10.1038/nature06403. [DOI] [PubMed] [Google Scholar]
- 14.Esteban MA, et al. Vitamin C enhances the generation of mouse and human induced pluripotent stem cells. Cell stem cell. 2010;6:71–79. doi: 10.1016/j.stem.2009.12.001. doi:10.1016/j.stem.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 15.Maherali N, Hochedlinger K. Tgfbeta signal inhibition cooperates in the induction of iPSCs and replaces Sox2 and cMyc. Current biology : CB. 2009;19:1718–1723. doi: 10.1016/j.cub.2009.08.025. doi:10.1016/j.cub.2009.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chan EM, et al. Live cell imaging distinguishes bona fide human iPS cells from partially reprogrammed cells. Nature biotechnology. 2009;27:1033–1037. doi: 10.1038/nbt.1580. doi:10.1038/nbt.1580. [DOI] [PubMed] [Google Scholar]
- 17.Hanna J, et al. Direct cell reprogramming is a stochastic process amenable to acceleration. Nature. 2009;462:595–601. doi: 10.1038/nature08592. doi:10.1038/nature08592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Silva J, et al. Nanog is the gateway to the pluripotent ground state. Cell. 2009;138:722–737. doi: 10.1016/j.cell.2009.07.039. doi:10.1016/j.cell.2009.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Islam S, et al. Characterization of the single-cell transcriptional landscape by highly multiplex RNA-seq. Genome research. 2011;21:1160–1167. doi: 10.1101/gr.110882.110. doi:10.1101/gr.110882.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Islam S, et al. Highly multiplexed and strand-specific single-cell RNA 5′ end sequencing. Nature protocols. 2012;7:813–828. doi: 10.1038/nprot.2012.022. doi:10.1038/nprot.2012.022. [DOI] [PubMed] [Google Scholar]
- 21.Kim J, Chu J, Shen X, Wang J, Orkin SH. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132:1049–1061. doi: 10.1016/j.cell.2008.02.039. doi:10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu H, Lemischka IR, Ma’ayan A. SVM classifier to predict genes important for self-renewal and pluripotency of mouse embryonic stem cells. BMC systems biology. 2010;4:173. doi: 10.1186/1752-0509-4-173. doi:10.1186/1752-0509-4-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nemajerova A, Kim SY, Petrenko O, Moll UM. Two-factor reprogramming of somatic cells to pluripotent stem cells reveals partial functional redundancy of Sox2 and Klf4. Cell death and differentiation. 2012;19:1268–1276. doi: 10.1038/cdd.2012.45. doi:10.1038/cdd.2012.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Soufi A, Donahue G, Zaret KS. Facilitators and impediments of the pluripotency reprogramming factors’ initial engagement with the genome. Cell. 2012;151:994–1004. doi: 10.1016/j.cell.2012.09.045. doi:10.1016/j.cell.2012.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
REFERENCES
- 25.Halbritter F, Vaidya HJ, Tomlinson SR. GeneProf: analysis of high-throughput sequencing experiments. Nature methods. 2012;9:7–8. doi: 10.1038/nmeth.1809. doi:10.1038/nmeth.1809. [DOI] [PubMed] [Google Scholar]
- 26.Anders S, Huber W. Differential expression analysis for sequence count data. Genome biology. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. doi:10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. doi:10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gentleman RC, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome biology. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. doi:10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols. 2009;4:44–57. doi: 10.1038/nprot.2008.211. doi:10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 30.Gaszner M, Felsenfeld G. Insulators: exploiting transcriptional and epigenetic mechanisms. Nature reviews. Genetics. 2006;7:703–713. doi: 10.1038/nrg1925. doi:10.1038/nrg1925. [DOI] [PubMed] [Google Scholar]
- 31.Fu H, et al. Preventing gene silencing with human replicators. Nature biotechnology. 2006;24:572–576. doi: 10.1038/nbt1202. doi:10.1038/nbt1202. [DOI] [PubMed] [Google Scholar]
- 32.Carey BW, et al. Reprogramming of murine and human somatic cells using a single polycistronic vector. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:157–162. doi: 10.1073/pnas.0811426106. doi:10.1073/pnas.0811426106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang W, et al. Chromosomal transposition of PiggyBac in mouse embryonic stem cells. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:9290–9295. doi: 10.1073/pnas.0801017105. doi:10.1073/pnas.0801017105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saldanha AJ. Java Treeview--extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. doi:10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- 35.de Hoon MJ, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics. 2004;20:1453–1454. doi: 10.1093/bioinformatics/bth078. doi:10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- 36.Ligges U, Maechler M. scatterplot3d - An R Package for Visualizing Multivariate Data. J Stat Softw. 2003;8:1–20. [Google Scholar]
- 37.Irizarry RA, et al. Summaries of Affymetrix GeneChip probe level data. Nucleic acids research. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gautier L, Cope L, Bolstad BM, Irizarry RA. affy--analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. doi:10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.