Summary
Limited clinical benefits derived from anti-VEGF therapy have driven the identification of new targets involved in tumor angiogenesis. Here, we report an integrative meta-analysis to define the transcriptional program underlying angiogenesis in human cancer. This approach identified ELTD1, an orphan G-protein-coupled receptor whose expression is induced by VEGF/bFGF and repressed by DLL4 signaling. Extensive analysis of multiple cancer types demonstrates significant upregulation of ELTD1 in tumor-associated endothelial cells, with a higher expression correlating with favorable prognosis. Importantly, ELTD1 silencing impairs endothelial sprouting and vessel formation in vitro and in vivo, drastically reducing tumor growth and greatly improving survival. Collectively, these results provide insight into the regulation of tumor angiogenesis and highlight ELTD1 as key player in blood vessel formation.
Highlights
-
•
A common core angiogenesis metasignature exists for different types of cancer
-
•
ELTD1 is upregulated in tumor endothelial cells and is a good prognostic marker
-
•
VEGF/DLL4 regulate ELTD1, which is important for tip cell function and angiogenesis
-
•
ELTD1 inhibition blocks tumor angiogenesis and substantially inhibits tumor growth
Significance
Angiogenesis represents a rational therapeutic target critical for solid tumor growth. Defining angiogenesis-related genes common to different cancer types enabled us to characterize the underlying core transcriptional program and its modulation in response to antiangiogenic therapies. A relatively unstudied G-protein-coupled receptor, ELTD1, was regulated by key angiogenic pathways and played an important role in blood vessel formation. Furthermore, ELTD1 silencing markedly impaired tumor growth in vivo despite the association between high expression in tumor-associated endothelial cells and good prognosis in multiple cancer types. These data advance our knowledge of the angiogenic process and identify ELTD1 as an important candidate for targeted therapies.
Introduction
Angiogenesis, defined as the formation of new blood vessels from pre-existing ones, is a key process in normal development and pathological conditions such as cancer. The proven role for angiogenesis in providing oxygen and nutrients necessary for tumor growth explains its clinical relevance and therapeutic potential (Hanahan and Weinberg, 2011; Li and Harris, 2009).
Despite great promise in preclinical models, approved antiangiogenic therapy has shown limited and transient efficacy due to initial or acquired resistance (Carmeliet and Jain, 2011a). Thus, the complex molecular programs regulating angiogenesis have been extensively studied to discover both new targets and predictive markers.
Several approaches have been exploited (van Beijnum and Griffioen, 2005), including in silico analysis of public databases (Herbert et al., 2008; Wallgard et al., 2008), expression profiling of cultured endothelial cells (ECs) (Engelse et al., 2008; Kutschera et al., 2011; Sana et al., 2005) or tumor-associated ECs from a limited number of primary ex vivo samples (Ghilardi et al., 2008; Seaman et al., 2007; St Croix et al., 2000; van Beijnum et al., 2006), and genetic approaches in animal models (del Toro et al., 2010; Sumanas et al., 2005; Weber et al., 2005).
Despite this, many components remain uncharacterized and could offer new avenues for cancer treatment. Our study goal was to identify regulators of tumor angiogenesis. Thus, we analyzed the expression profile of more than 1,000 primary human cancers to generate a vascular/angiogenesis core signature composed of genes whose expression jointly correlates with that of several well-recognized angiogenesis and/or EC “seed” genes in multiple cancers. To select uncharacterized genes potentially involved in angiogenesis, this analysis was complemented by in vivo expression profiling of antiangiogenic therapies and extensive characterization of the top-ranked candidate in different in vitro and in vivo model systems.
Results
Derivation of Cancer-Type-Specific Angiogenesis/EC Metasignatures in Different Primary Human Tumors
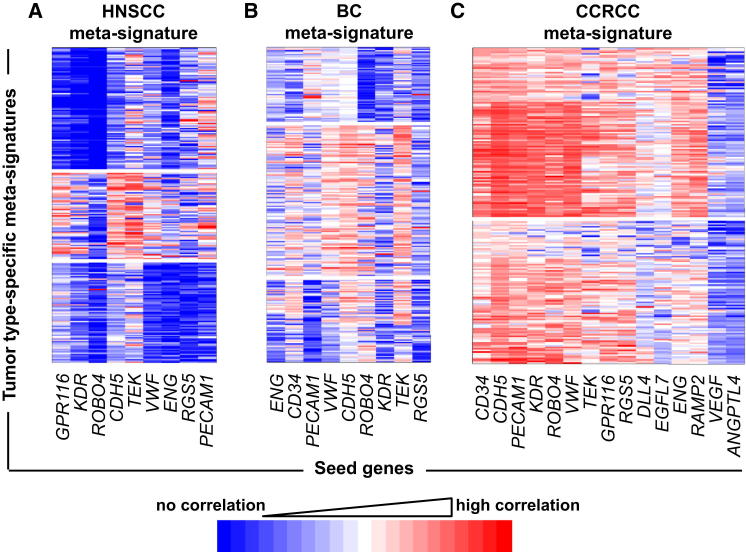
First, we developed an angiogenic/vascular metasignature of genes expressed in vivo in human tumor samples derived from different cancer types, namely 121 head and neck squamous cell carcinomas (HNSCCs), 959 breast cancers (BCs), and 170 clear cell renal cell carcinomas (CCRCCs). Coexpression networks were generated using, as initial seeds, multiple known angiogenesis/EC-related transcripts previously described as upregulated in cancer and/or involved in angiogenesis (see tables in Supplemental Experimental Procedures available online), thus representing “transcriptional markers” of angiogenesis/microvascular density (MVD). Seeds that passed filtering based on expression level, coexpression, and variability (see Experimental Procedures and Figures S1A–S1C) were used to derive cancer-type-specific angiogenesis metasignatures (Figure 1; Table S1), defining differences between tumor types. HNSCC and BC signatures were fairly heterogeneous and composed of three distinct gene clusters associated with different groups of seeds (Figures 1A and 1B; see Experimental Procedures). Different clusters might reflect the complexity/heterogeneity of the molecular program involved in tumor angiogenesis or different vasculature types. The CCRCC profile was more homogeneous, with only two subclusters (Figure 1C). This is consistent with the high angiogenic capacity of CCRCCs due to constitutive activation of the hypoxia-inducible factor pathway (Baldewijns et al., 2010).
Figure 1.
Derivation of Angiogenesis Metasignatures in Primary Human HNSCC, BC, and CCRCC
Angiogenic profiles of 121 HNSCC (A), 959 BC (B), and 170 CCRCC samples (C). The x axis represents seeds from highly overlapping clusters (see Figure S1), and the y axis shows genes whose expression clusters with the seeds; coloring indicates strength of membership (see color scale). Clustering on correlating expression (membership; y axis) revealed the existence of three distinct gene clusters associated with different patient groups in HNSCC (A) and BC (B) and two clusters with a more compact profile in CCRCC (C). See also Table S1.
Generation of a Common Angiogenesis Core Signature and Analysis of Its Modulation by Anti-VEGF and Anti-Notch Treatments In Vivo
A product rank score (Π) generated a common angiogenesis/EC metasignature of genes consistently highly ranked among HNSCC, BC, and CCRCC profiles. Of 471 genes whose expression positively correlated with that of the seeds (common overexpressed angiogenesis extended signature; top 20 in Table 1 and the full list in Table S2), the largest fraction includes genes encoding typical EC molecules (e.g., CDH5 and ESAM) or genes expressed by ECs, tumors, and other stromal cells (e.g., CXCL12 and PDGFD). Overall, we observed enrichment for receptor-coding genes and, to a lesser extent, extracellular matrix proteins and transcription factors. Underexpressed genes were heterogeneous between cancers (common underexpressed angiogenesis signature; Table S3) and were not further considered. A core signature of 43 overexpressed genes that were consistently top ranked in all cancers (Π > 0.9; Table S2 and top 20 in Table 1) and that did not exhibit subclusters was identified, potentially including genes highly expressed and similarly regulated in tumor angiogenesis. This was substantiated by analysis of an independent data set (Gene Expression Omnibus [GEO] accession number GSE9014) showing increased core signature expression in purified tumor-associated compared to normal breast stroma (Figures S2A and S2B), suggesting a role in tumor angiogenesis/vasculature.
Table 1.
Top 20 Genes of the Common Angiogenesis Signature
| Rank | Symbol | Full Name | HNSCC Metascore | BC Metascore | CCRCC Metascore | Common Score |
|---|---|---|---|---|---|---|
| 1 | CDH5 | cadherin 5, type 2 (vascular endothelium) | 0.9996 | 0.9967 | 0.9978 | 0.9941 |
| 2 | ELTD1 | EGF, latrophilin and seven transmembrane domain containing 1 | 0.9993 | 0.9967 | 0.9965 | 0.9925 |
| 3 | CLEC14A | C-type lectin domain family 14, member A | 0.9989 | 0.9982 | 0.9924 | 0.9896 |
| 4 | LDB2 | LIM domain binding 2 | 0.9990 | 0.9967 | 0.9938 | 0.9895 |
| 5 | ECSCR | endothelial cell-specific chemotaxis regulator | 0.9995 | 0.9981 | 0.9913 | 0.9890 |
| 6 | MYCT1 | myc target 1 | 0.9969 | 0.9982 | 0.9937 | 0.9889 |
| 7 | RHOJ | ras homolog gene family, member J | 0.9989 | 0.9976 | 0.9910 | 0.9875 |
| 8 | VWF | von Willebrand factor | 0.9986 | 0.9910 | 0.9959 | 0.9855 |
| 9 | TIE1 | tyrosine kinase with immunoglobulin-like and EGF-like domains 1 | 0.9989 | 0.9982 | 0.9882 | 0.9853 |
| 10 | KDR | kinase insert domain receptor (a type III receptor tyrosine kinase) | 0.9972 | 0.9912 | 0.9967 | 0.9852 |
| 11 | ESAM | endothelial cell adhesion molecule | 0.9958 | 0.9971 | 0.9921 | 0.9850 |
| 12 | CD93 | CD93 molecule | 0.9984 | 0.9932 | 0.9929 | 0.9845 |
| 13 | PTPRB | protein tyrosine phosphatase, receptor type, B | 0.9966 | 0.9930 | 0.9936 | 0.9833 |
| 14 | GPR116 | G protein-coupled receptor 116 | 0.9989 | 0.9950 | 0.9872 | 0.9812 |
| 15 | SPARCL1 | SPARC-like 1 (hevin) | 0.9974 | 0.9956 | 0.9878 | 0.9809 |
| 16 | EMCN | endomucin | 0.9985 | 0.9967 | 0.9823 | 0.9776 |
| 17 | ROBO4 | roundabout homolog 4, magic roundabout (Drosophila) | 0.9895 | 0.9967 | 0.9876 | 0.9740 |
| 18 | ENG | endoglin | 0.9981 | 0.9910 | 0.9839 | 0.9732 |
| 19 | TEK | TEK tyrosine kinase, endothelial | 0.9992 | 0.9984 | 0.9754 | 0.9730 |
| 20 | S1PR1 | sphingosine-1-phosphate receptor 1 | 0.9862 | 0.9979 | 0.9885 | 0.9729 |
To effectively select genes involved in tumor angiogenesis, we studied modulation of the signature genes in response to antiangiogenic treatments in preclinical models. Gene expression analysis of treated xenograft tumors was performed using both human- and mouse-specific microarrays to discriminate human tumor versus murine stromal (including vascular/EC) reactions. Acute treatment with the anti-VEGF antibody bevacizumab caused a strong downregulation of the overexpressed signature in the stroma with a milder effect in tumor cells (Figures 2A and 2B; Table S4). The stromal effect may reflect a reduction in the number of cells expressing these genes (e.g., ECs), a direct effect of vascular endothelial growth factor (VEGF) blockade (for genes such as DLL4, ESM1, and CD93, where VEGF-induced regulation has been reported), or treatment-induced microenvironmental factors (e.g., hypoxia). Importantly, similar results were reported by Genentech when identifying bevacizumab-inhibited vascular/EC genes in tumor xenografts (Bais et al., 2011).
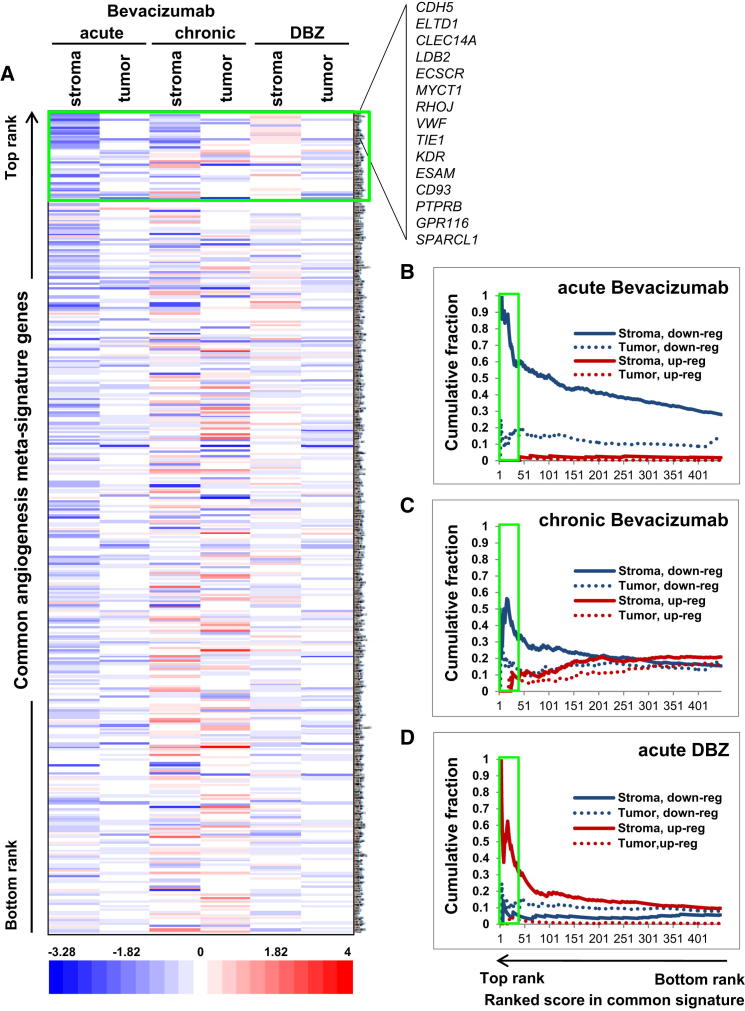
Figure 2.
Modulation of the Common Overexpressed Angiogenesis Signature by Anti-VEGF and Anti-Notch Treatment In Vivo
U87 xenografts comparing untreated tumors to bevacizumab- (acute and chronic) or DBZ-treated ones are shown. All overexpressed genes in the extended common signature are shown (the top 15 are listed). Green boxes indicate the core signature.
(A) Expression fold changes between treatments (x axis) and control are shown (see color scale). Genes (y axis) are ranked from highest to lowest common score.
(B–D) Cumulative plots show the fraction of downregulated and upregulated genes for each treatment, for both stroma and xenograft (tumor) expression. High values indicate a high fraction of downregulated or upregulated genes in the common signature after treatment. Genes are shown ranked from left to right. Plots are cumulative (that is, the fraction is calculated down to a given rank).
Interestingly, the stromal compartment exhibited a common pool of downregulated genes under both chronic and acute regimens (suggesting direct VEGF induction) but also several genes that were downregulated only by acute treatment, potentially indicating angiogenesis restoration upon chronic regimen. Under this last condition, stromal reaction also shows a small group of upregulated genes that may be involved in resistance (e.g., HTRA1, DCN, and MMP2 have been described as proangiogenic molecules) and tumor cells responded similarly (Figures 2A and 2C; Table S4).
Treatment with DBZ, a γ-secretase inhibitor, was also evaluated as its therapeutic effect is mediated primarily by inducing unproductive angiogenesis via Notch signaling inhibition in ECs (Li et al., 2011). As this treatment increases MVD, the general stromal signature upregulation may reflect both increased number of transcript-expressing cells (e.g., ECs) and inhibition of Notch signaling (Figures 2A and 2D; Table S4). DBZ treatment showed different effects on tumor signature, inducing a mild downregulation. Notably, both acute treatments induce hypoxia by reducing perfusion, but while the first inhibits angiogenesis, the second induces formation of nonfunctional vessels. This difference could explain why, despite similarly affecting the signature in tumor cells, they have opposite effects on the stromal compartment.
The Angiogenesis Core Signature Is Regulated by DLL4 In Vitro
As DBZ treatment regulated our common overexpressed signature in vivo and Dll4-Notch signaling is a key regulator of angiogenesis (Duarte et al., 2004; Gale et al., 2004; Krebs et al., 2004; Phng and Gerhardt, 2009), we evaluated the effects of this pathway on signature expression in primary human ECs using data from the FANTOM4 project (http://fantom.gsc.riken.jp/). Briefly, human umbilical vein endothelial cells (HUVECs) stimulated with recombinant human DLL4 (rhDLL4) were analyzed using cap analysis gene expression (CAGE). This confirmed that ECs express the signature genes and that ∼38% of these are regulated by DLL4 (18% upregulated and 19.9% downregulated; Table S4), increasing to 56% when considering the core signature (26% upregulated and 30% downregulated).
Notably, comparing in vivo DBZ treatment with in vitro DLL4 stimulation identified a gene subset whose expression was inversely regulated by the two approaches, as expected by their reciprocal regulation of the Notch pathway. Despite the in vivo signature derived from different stromal cells and the effects of microenvironmental factors, it is likely that these are Notch-regulated genes active in ECs in vivo. Regulation by DLL4 of six (of nine) interesting and/or uncharacterized EC genes identified by our signature and confirmed by CAGE analysis was validated by quantitative PCR (Figure S2C).
ELTD1 Selection and Expression by ECs and SMCs
We then focused on the top-ranked, relatively unstudied G-protein-coupled receptor (GPCR) EGF, latrophilin and seven transmembrane domain containing 1 (ELTD1). Excluding the seeds themselves, the gene encoding this receptor ranked first in our signature (Figure 2A; Table S2) and was significantly upregulated in purified BC stroma (Figure S2B). ELTD1 is an orphan receptor of the adhesion GPCR family; specifically, it belongs to the epidermal growth factor-seven-span transmembrane (EGF-TM7) receptor subfamily expressed in cardiomyocytes and vascular smooth muscle cells (VSMCs) of the developing rat heart (Bjarnadóttir et al., 2004; Kwakkenbos et al., 2004; Nechiporuk et al., 2001). Importantly, Eltd1 knockout (KO) mice develop normally but present increased cardiac hypertrophy in response to pressure overload (Xiao et al., 2012). ELTD1 upregulation in glioblastoma, both in tumor cells and ECs, has recently been reported (Dieterich et al., 2012; Towner et al., 2013). Here, Eltd1 expression was affected in stromal cells by both acute anti-VEGF and anti-Notch treatments in vivo (Table S4), with DBZ-induced upregulation being consistent with the reciprocal reduction observed in HUVEC stimulated by DLL4 in vitro (Table S4; Figure S2C), the latter confirming Eltd1 observed upregulation in retinal ECs from Dll4+/− mice (del Toro et al., 2010). Studies of the recombinant rat ortholog indicate that Eltd1 is expressed on the cell surface (Nechiporuk et al., 2001), and we confirmed this by fluorescence-activated cell sorting analysis of 293T cells overexpressing an N-terminally tagged human ELTD1 (Figure S5A). This is an important confirmation for potential therapeutic strategies.
Despite reports of vascular ELTD1 transcript (Herbert et al., 2008; Nechiporuk et al., 2001; Sumanas et al., 2005; Wallgard et al., 2008; Weber et al., 2005) and protein expression in human tissues (Sigma Human Protein Atlas) (Uhlen et al., 2010), expression patterns in both cancer and normal samples have only been reported for glioma and normal brain (Dieterich et al., 2012; Towner et al., 2013). Thus, we validated an antibody for ELTD1 detection by immunohistochemistry (IHC) and western blotting (Figures S3B and S3C).
IHC of tissue microarrays (TMAs) containing primary human tumor samples (65 head and neck, 157 renal, 120 colorectal, 200 ovarian, and a few other tumor types), normal tissues (148 renal and 120 colorectal; of these, 126 and 120 were matched to tumor samples, respectively), and reactive tonsil sections was performed to study ELTD1 protein expression.
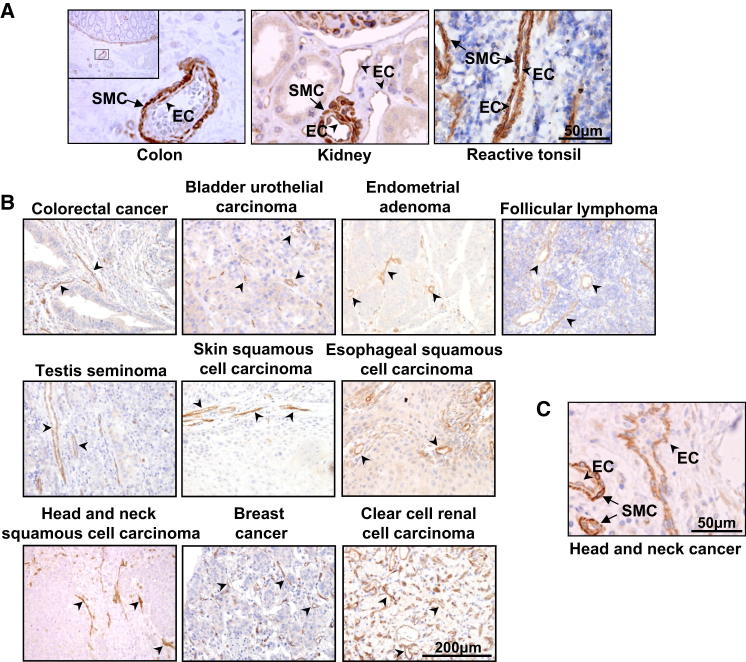
ELTD1 was detected in the majority of ECs in tumor and normal tissues (Figures 3A and 3B). Despite some variability, stronger positivity was observed in tumor-associated compared to normal ECs (Figures 3B, 4A, 4B, 4H, and 4I; Figures S4C and S4D; see below). Arteries and arterioles, occasionally seen in tumors, showed high ELTD1 staining in VSMCs (Figures 3A and 3C), generally stronger than in neighboring ECs. Vascular ELTD1 protein expression is consistent with existing data, although a comparative analysis between ECs and VSMCs/pericytes was not previously reported. Immunofluorescence analysis for ELTD1 and CD34/α-SMA (EC and SMC marker, respectively) confirmed expression by both cell types (Figure S3D). ELTD1 labeling of neoplastic cells was observed in a minority of cases and generally as weak cytoplasmic positivity. However, strong tumoral ELTD1 expression was observed in some tumor types (Figure S3E).
Figure 3.
ELTD1 Vascular Expression in Primary Human Tissues
(A) Pictures of normal tissues showing ELTD1 expression in ECs (arrowheads) and pericytes/SMCs (arrows).
(B) IHC analysis of different primary human tumors invariably shows vascular/EC expression (arrowheads).
(C) High-magnification picture showing ELTD1 expression by both ECs (arrowheads) and SMCs/pericytes (arrows) in a tumor sample.
See also Figure S3.
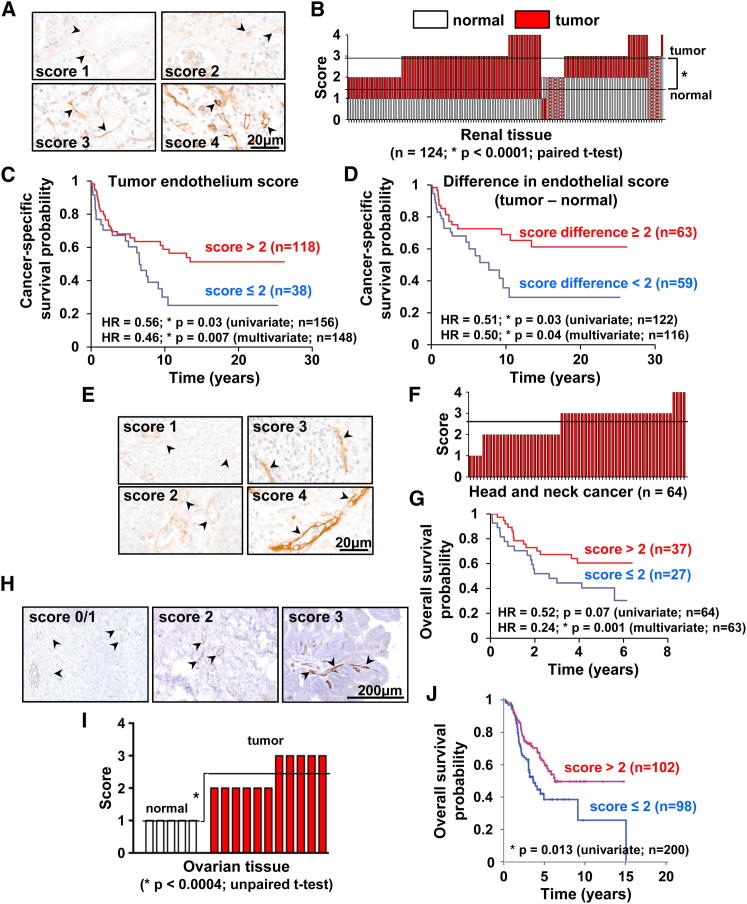
Figure 4.
Endothelial ELTD1 Expression in Cancer and Normal Matched Tissues and Its Prognostic Value
(A) Representative IHC pictures of ELTD1 scoring in renal tissue (scores 1 and 2 are from normal kidney while 3 and 4 are from renal cancer).
(B) ELTD1 expression in renal cancer and normal matched kidney. Each column represents a patient with red and white bars showing the score in tumor-associated and normal ECs, respectively. Horizontal lines represent score averages.
(C and D) Significant correlation between cancer-specific survival and both tumor-associated EC ELTD1 expression (C) and the differential EC score (tumor minus normal) (D).
(E) Representative IHC pictures of ELTD1 score categories in head and neck cancer.
(F) Summary of ELTD1 expression. Each column represents a patient and the horizontal line represents the average.
(G) Significant correlation between tumor-associated EC ELTD1 expression and overall survival. HR, hazard ratio.
(H) Representative IHC pictures of ELTD1 score categories in human ovarian tissue (scores 0 and 1 are from normal ovary while scores 2 and 3 are from ovarian cancer).
(I) Summary of EC ELTD1 expression in normal and neoplastic ovarian tissue shows upregulation in tumor samples.
(J) Significant correlation between tumor-associated EC ELTD1 levels and overall survival. Arrowheads indicate blood vessels.
See also Figure S4 and Tables S5–S7.
Clinical Relevance of EC ELTD1 in Renal, Head and Neck, Colorectal, and Ovarian Cancer
Extensive analysis in head and neck, renal, and colorectal cancers showed predominantly negative/weak positivity in tumor cells. Interestingly, on comparing tumor and normal matched tissues, we observed a clear increase in ELTD1 expression in tumor-associated ECs. Indeed, 89% of renal (110/124 samples) and 82% of colorectal cases (98/119 samples) showed increased EC ELTD1 staining (Figures 4A and 4B; Figures S4C and S4D). EC upregulation was also observed in ovarian cancer, albeit in a small series (Figures 4H and 4I). These findings are consistent with data previously reported in glioma (Dieterich et al., 2012).
Interestingly, higher EC ELTD1 levels significantly correlated with increased MVD in renal, head, neck, and colorectal cancer (Figure S4F), suggesting an involvement in tumor angiogenesis. Furthermore, expression profiling of HNSCC cases showed a significant inverse correlation between ELTD1 messenger RNA (mRNA) levels and a published hypoxia signature (Buffa et al., 2010) or the hypoxia-inducible gene CA9 (Figure S4G), possibly suggesting better perfusion of tumors with high ELTD1.
In renal cancer, higher ELTD1 scores in tumor-associated ECs correlated with smaller tumor size (Table S5; p = 0.03) and improved cancer-specific (Figure 4C) and overall survival (Figure S4A). Notably, analyzing the differential score between tumor-associated and normal ECs gave similar results, with bigger differences correlating with smaller tumor size (p = 0.02) and improved cancer-specific survival (Figure 4D). In HNSCC (Figures 4E and 4F), higher EC ELTD1 expression identified less aggressive tumors, correlating with lower UICC stage (p = 0.02), less perineural invasion (p = 0.03), borderline trend toward lower pT value (p = 0.08; Table S6), and improved overall survival (Figure 4G). Colorectal cancer showed borderline significant associations between the highest EC ELTD1 scores (score 4) and tumor differentiation status (p = 0.05; Table S7) and improved overall survival (Figure S4E). Finally, higher EC ELTD1 significantly correlated with improved overall survival in ovarian cancer (Figure 4J). These results demonstrate clinical relevance across four different tumor types and identify a potential role for EC ELTD1 in cancer biology.
ELTD1 Is Glycosylated in the Extracellular Domain and Expressed as Monomer and Multimers
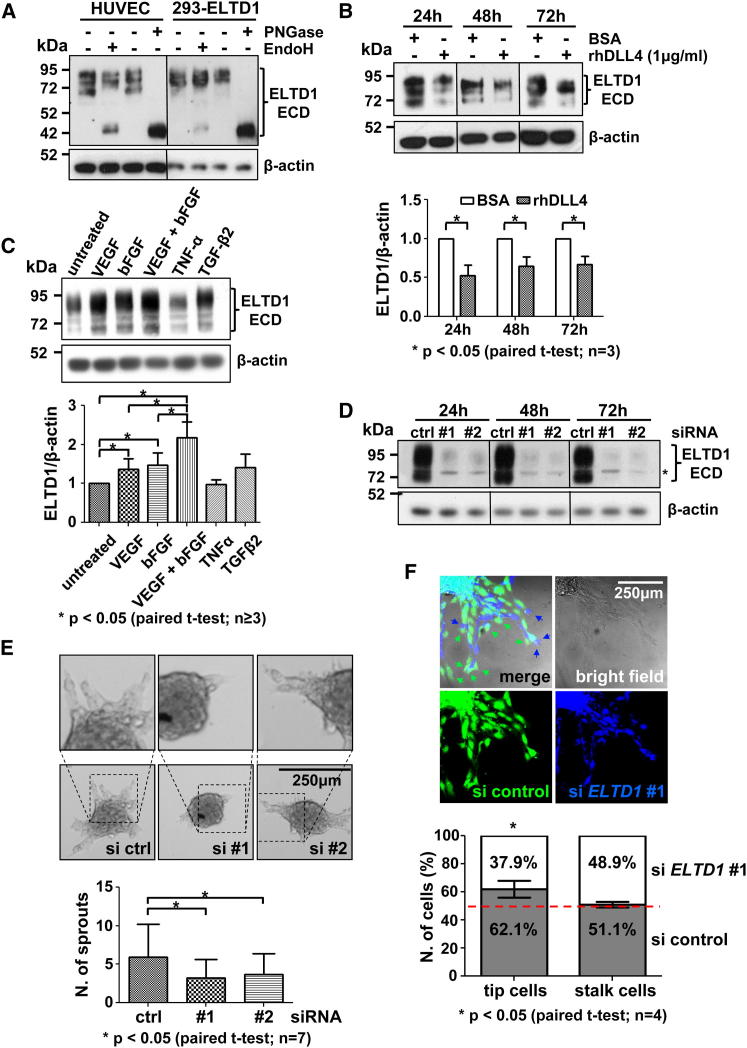
Analysis of HUVEC and ELTD1-transfected 293 cell lysates showed multiple bands, with molecular weights (MWs) ranging between 95 and 70 kDa (the expected MW was 78 kDa; Figure 5A). To assess if this reflected posttranslational modifications (e.g., N-glycosylation) or alternative splicing, lysates were subjected to glycosidase digestion prior to analysis. ELTD1 MW was sensitive to endoglycosidase H (EndoH), shifting the lowest band to ∼44 kDa, while Peptide:N-glycosidase F (PNGase) treatment shifted all forms to a similar 44 kDa band (Figure 5A). Thus, different forms derive from glycosylation of a single protein. The antibody used binds the ELTD1 extracellular domain (ECD) and the MW observed after deglycosylation matches that predicted for the ECD alone (Figure S5C). This is consistent with reports that rat eltd1 is expressed as a noncovalent heterodimer composed of the ECD and transmembrane domains (Nechiporuk et al., 2001) (Figure S5B). Accordingly, a human ELTD1-ECD-FC fusion protein was expressed as a single form with comparable MW to the highest glycosylated molecule in HUVEC plus the tag (Figure S5D). PNGase treatment confirmed glycosylation of the recombinant ECD-FC (Figure S5E), indicating that only the ECD is detected under denaturing conditions. GPCRs are known to form homomers and heteromers (Casadó et al., 2009), and analysis of cell lysates under nondenaturing conditions identified four ELTD1 main forms, consistent with the MWs of a glycosylated homomonomer, homodimer (predominant), homotrimer, and homotetramer (Figure S5F), although interaction with other molecules cannot be excluded.
Figure 5.
ELTD1 Expression, Regulation, and Function in Primary Human Endothelial Cells In Vitro
(A) HUVEC- and ELTD1-overexpressing 293 cell lysates were treated with deglycosylating enzymes before western blotting (WB) analysis. Untreated samples show multiple bands that are lost after treatment with contemporary appearance of a lower MW band corresponding to the deglycosylated ECD.
(B) WB analysis of DLL4-stimulated HUVECs shows ELTD1 downregulation at different time points (bars represent average ±SD of band densitometric analysis).
(C) Similar analysis on HUVECs treated with different cytokines for 24 hr (bars represent average ±SD of band densitometric analysis).
(D) WB validation of ELTD1 silencing with two different siRNAs. ∗Nonspecific band appearing with long exposure.
(E) HUVEC sprouting is reduced by ELTD1 silencing (bars represent mean ±SD). Representative spheroids are shown.
(F) In vitro cell fate assay. Sprouting analysis from spheroids composed of a 1:1 ratio between prelabeled control and siELTD1 cells. ELTD1 silencing impairs the ability to take the tip cell position (arrows) without affecting stalk cell formation (graph shows the number of tip and stalk cells as the average percentage ±SD).
See also Figure S5.
ELTD1 Is Regulated by Both DLL4 Signaling and Proangiogenic Cytokines
ELTD1 transcript expression is regulated by DLL4-induced signaling in vitro (Table S4; Figure S2C), and analysis of DLL4-stimulated HUVECs confirmed that protein levels are also significantly reduced (Figure 5B). Although the DLL4-NOTCH pathway plays a relevant role in angiogenesis, this cannot explain ELTD1 upregulation observed in tumor-associated ECs. Factors such as VEGF and basic fibroblast growth factor (bFGF) are additional key players in tumor angiogenesis (Carmeliet and Jain, 2011a), and both increased ELTD1 mRNA (not shown) and protein levels in HUVECs, having an additive effect in combination (Figure 5C), possibly contributing to increased ELTD1 expression in tumor-associated ECs.
ELTD1 Silencing Reduces HUVEC Adhesion and Inhibits Sprouting and Tip Cell Phenotype In Vitro
To study ELTD1’s role in EC biology, in vitro gene silencing was functionally evaluated in HUVECs using two different small interfering RNAs (siRNAs) (Figure 5D). ELTD1 function is unknown and initial experiments indicated silencing had no effect on EC viability in normal culture conditions (Figure S5H). Other members of this receptor family are involved in cell adhesion (Kwakkenbos et al., 2004) but are low or undetectable in ECs (Figure S5G). As the ELTD1 ligand is unknown, Matrigel was used to evaluate the role of ELTD1 in adhesion. Gene knockdown slightly, but significantly, reduced EC adherence (Figure S5I), although this is insufficient to demonstrate direct involvement in cell adhesion. General migratory ability, evaluated by the wound-healing assay, showed no change (data not shown). Using an in vitro three-dimensional spheroid-based angiogenesis assay, ELTD1-silenced HUVECs showed reduced angiogenic ability, generating fewer sprouts (50%–60%) compared to control (Figure 5E). This phenotype represents a functional explanation for both VEGF/bFGF-induced ELTD1 upregulation and DLL4-induced repression, as VEGF and bFGF are known to induce EC sprouting while Dll4 is a negative regulator of this process through the restriction of tip cell phenotype/specification (Hellström et al., 2007; Phng and Gerhardt, 2009). Hypothesizing a role in tip cell biology, a cell fate assay was performed using spheroids composed of a 1:1 mixture of control and ELTD1-silenced HUVECs, both prelabeled with different fluorescent dyes. Consistent with our previous experiments showing reduced sprouting, ELTD1-silenced HUVEC showed a reduced ability to take the tip position compared to control cells (37.9% ± 5.8% versus 62.1% ± 5.8%, respectively; Figure 5F). This demonstrates that ELTD1 plays an intrinsic role during the sprouting process. Importantly, although stalk cell number was expected to increase, gene knockdown did not affect this cell subpopulation, possibly because of back migration to the spheroid (Figure 5F). This finding confirms ELTD1’s role only in tip cell function/specification.
ELTD1 Is Involved Human Umbilical Vein Smooth Muscle Cell Viability and Adhesion
As VSMC express ELTD1, we evaluated silencing consequences (Figure S5J) in this cell type in vitro. Opposite to observations in ECs, ELTD1 knockdown caused a significant reduction in cell viability 72 hr after transfection (Figure S5K). Similarly to EC, adhesion to Matrigel was significantly reduced while no effect on general migratory ability was observed (Figure S5L and data not shown).
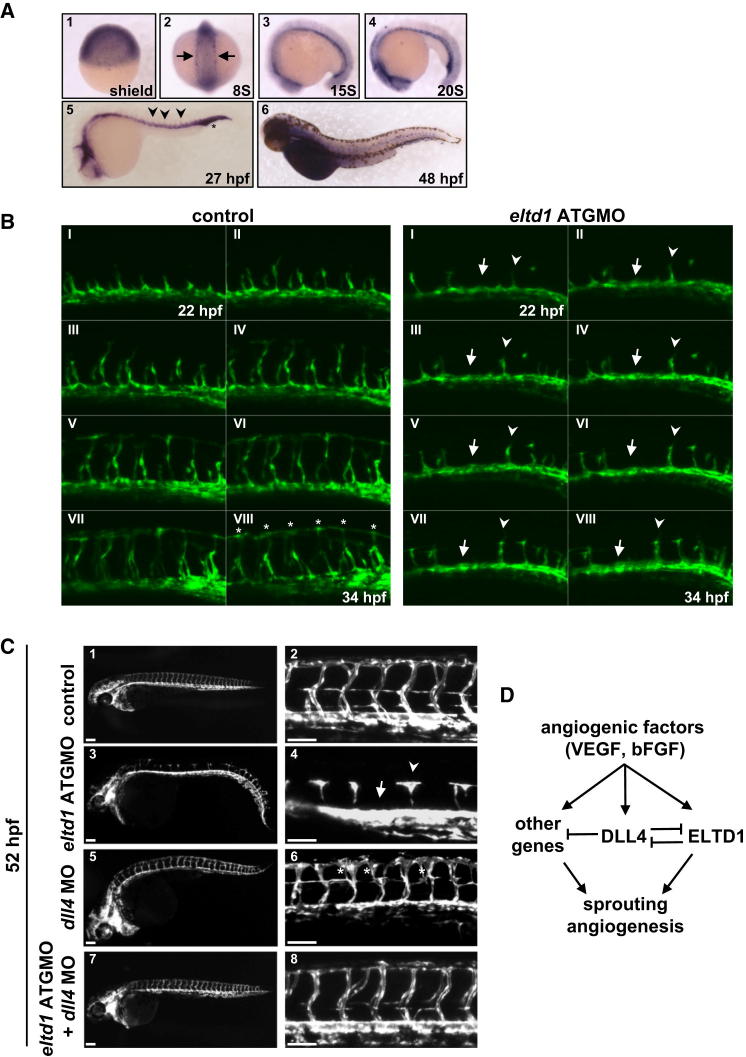
Eltd1 Is Expressed in the Zebrafish Embryo Vasculature and Has a Role Antagonizing dll4 in Blood Vessel Development
To investigate ELTD1’s role in vivo, we exploited the developing zebrafish embryo model, being the only member of its receptor family conserved in this species (Kwakkenbos et al., 2004). Eltd1 expression was assessed by in situ hybridization. During gastrulation, expression was ubiquitously detected throughout the embryo (Figure 6A1) while by eight somites it was observed in the posterior lateral plate mesoderm in bilateral stripes (Figure 6A2, arrows). This group of cells has been described as the posterior hemangioblast population, containing cells that give rise to both ECs and blood cells (Gering et al., 1998). As these cells migrate to the midline (Figure 6A3) and form the dorsal aorta (DA) and the posterior cardinal vein (PCV) (Figure 6A4), eltd1 expression was still detectable. By 27 hr postfertilization (hpf), the distinction between the DA and PCV is complete and intersegmental vessels (ISV), formed by EC sprouting from the DA, also express eltd1 (Figure 6A5, arrowheads). By 48 hpf, weaker eltd1 expression is still detected in ECs (Figure 6A6). The lack of eltd1 in the cloche zebrafish mutant (Figure S6A), which has severe defects in blood and endothelium formation (Xiong et al., 2008), confirms its EC association.
Figure 6.
Eltd1 Expression and Function during Vascular Development in Zebrafish
(A) Eltd1 in situ hybridization at different developmental stages shows expression in ECs and blood precursor cells (arrow) and developing vessels: DA, PCV, ISV (arrowheads), and vascular plexus (asterisk).
(B) Time-lapse confocal microscopy of Tg(kdrl:GFP) uninjected and eltd1 morphants every 90 min. In eltd1 morphants, the formation of ISV sprouts is blocked (arrows) or severely reduced (arrowheads), never reaching the dorsal position observed in WT siblings (asterisks).
(C) Lateral views of uninjected control (C1 and C2) and different morphants (C3–C8) 52hpf Tg(kdrl:GFP) embryos. Trunk vessels imaging revealed a failure to form ISV in eltd1 morphants (C3 and C4, arrow and arrowhead), which is rescued in eltd1+dll4 double morphants (C7 and C8). The dll4 morphant arterial hyperbranching phenotype (C5 and C6, asterisks) is also rescued in the eltd1+dll4 double morphants (C7 and C8). Scale bars: 100 μm.
(D) Working hypothesis.
To study the role of eltd1 during development, we designed two antisense morpholinos (MOs) targeting the start codon (ATGMO) and a splicing junction (spliceMO) of the eltd1 pre-mRNA, respectively (Figure S6B), and injected them into single-cell embryos. RT-PCR analysis confirmed eltd1 knockdown by the spliceMO (Figure S6C, compare lanes 4 and 5). By using Tg(kdrl:GFP) transgenic embryos, in which the vasculature expresses GFP, we consistently observed severe vascular defects during the formation of the ISV at 30 hpf when eltd1 ATGMO (Figure S6D) or eltd1 spliceMO was injected. Although both MOs gave the same phenotype, here we show data for the eltd1 ATGMO where approximately 90% of embryos showed the phenotype. Confocal time lapse microscopy imaging of uninjected and eltd1 ATGMO-injected Tg(kdrl:GFP) embryos allowed us to follow the trunk development in detail. In eltd1 morphants, defects start as early as 22 hpf, the time when the primary sprouts from the DA giving rise to ISV start to develop (Isogai et al., 2003) (Figure 6B; Movies S1 and S2). In eltd1 morphants, the formation of ISV sprouts is blocked (Figure 6B, arrows) or severely reduced (Figure 6B, arrowheads), never reaching the dorsal position observed in wild-type (WT) siblings (Figure 6B, asterisks). Similar defects were observed by assessing expression of another vascular-related gene, fli1, by in situ hybridization (Figure S6E, 1 and 4).
As ELTD1 is repressed by DLL4 in vitro (Figure 5B; Figure S2C; Table S4) and loss of eltd1 function in zebrafish leads to impaired sprouting, a phenotype opposite to that associated with dll4 inhibition (Hellström et al., 2007; Siekmann and Lawson, 2007), we explored this potential connection by disrupting dll4 function in embryos and analyzing eltd1 expression as well as by assessing dll4 expression in eltd1 morphants (Figure S6E). As these genes negatively regulated each other (Figures S6E2, S6E5, S6E3, and S6E6), we attempted to rescue the ISV phenotype observed in eltd1 morphants by dll4 silencing. Tg(kdrl:GFP) embryos were injected with eltd1 MO, dll4 MO, or both and analyzed at 52 hpf (Figure 6C). Eltd1 morphants showed severely impaired ISV formation, from preventing ISV formation (arrow) to severe growth retardation (Figures 6C3 and 6C4, arrowhead), as observed at earlier stages (Figure 6B; Figures S6D and S6E; Movies S1 and S2), while dll4 MO recapitulated the previously reported arterial hyperbranching phenotype (Figures 6C5 and 6C6, asterisks) (Leslie et al., 2007). However, coinjection of eltd1 and dll4 MOs, at concentrations causing ISV defects and arterial hyperbranching, respectively, rescued ISV development in 52% of embryos (Figures 6C7 and 6C8; Figure S6F). Moreover, the excessive arterial angiogenesis caused by dll4 depletion was also rescued in 55% of embryos (Figures 6C7 and 6C8; Figure S6F). A concordant finding was observed at the histological level, since tip cell/sprouting structures were increased in dll4 MO embryos but were almost normalized in double morphants (Figure S6G).
Effects of In Vivo Eltd1 Gene Silencing during Tumor Growth
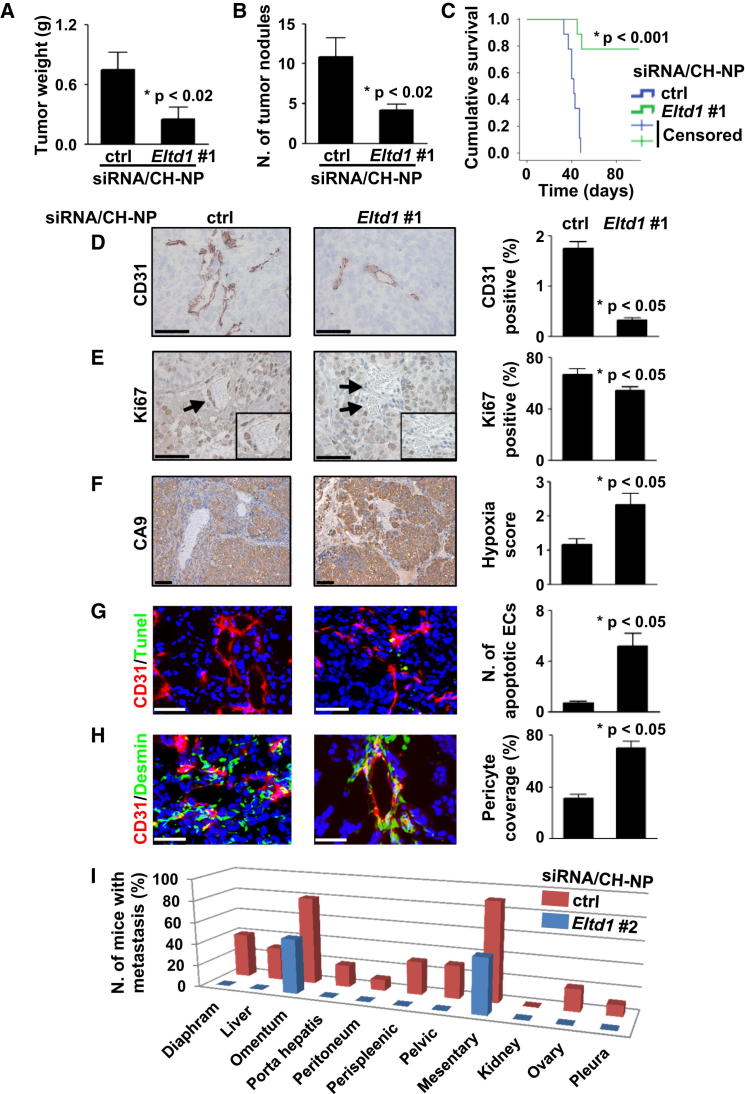
Based on our findings, we hypothesized that ELTD1 played a role in tumor angiogenesis. Using two mouse Eltd1-specific siRNAs (Figures S7A and S7B), we silenced murine Eltd1 in an orthotopic model of ovarian carcinoma by intravenous injection of Eltd1 siRNA incorporated into chitosan nanoparticles (CH-NP). This approach was highly efficient for in vivo EC gene silencing (Lu et al., 2010) and did not trigger an interferon response (Figure S7C). One week after intraperitoneal injection of SKOV3ip1 cells, mice were randomly allocated to treatments (control siRNA/CH-NP or Eltd1 #1 siRNA/CH-NP). Treatment with Eltd1 #1 siRNA/CH-NP resulted in a significant decrease in tumor weight (67% reduction; Figure 7A) and nodule number (Figure 7B); results were confirmed by a second Eltd1 siRNA (siRNA #2; Figures S7E and S7F). Effects on tumor growth were associated with dramatic increase in survival (Figure 7C) and reduced metastatic dissemination (Figure 7I). Importantly, in the survival experiment, seven out of nine Eltd1 siRNA/CH-NP-treated mice were sacrificed while still healthy to finish the experiment on day 101 after tumor cell injection (44 day mean survival for the control group). These mice showed very small nodules (n = 5) or undetectable tumors (n = 2), confirming that Eltd1 silencing maintained the tumor growth inhibition observed in previous short-term experiments.
Figure 7.
Effects of Eltd1 Silencing on Tumor Growth and Angiogenesis in an Orthotopic Ovarian Cancer Model
(A and B) Eltd1 siRNA/CH-NP treatment reduces tumor weight (A) and nodule number (B) in a SKOV3ip1 orthotopic mouse model.
(C) Reduced tumor growth was associated with a strong improvement in mice survival.
(D–H) Analysis of tumor tissue sections shows that Eltd1 silencing reduces MVD (D; CD31 IHC) and tumor tissue proliferation (E; Ki67 IHC) but also increases hypoxic areas (F; CA9 IHC), EC apoptosis (G; CD31/TUNEL immunofluorescence [IF]), and vascular pericyte coverage (H; CD31/desmin IF). Representative pictures are shown. Arrows indicate blood vessels within insets. Scale bars: 50 μm.
(I) Drastic reduction in metastatic spread upon Eltd1 silencing was also observed. All graph bars represent average ±SEM (unpaired t test).
See also Figure S7.
To identify mechanisms by which stromal Eltd1 silencing could reduce tumor growth, we examined different histologic markers. Eltd1 knockdown significantly reduced MVD (Figure 7D) compared to control treatment, consistent with increased hypoxia (Figure 7F), significant reduction in cell proliferation (Figure 7E), and increased EC apoptosis (Figure 7G). Pericyte coverage was increased (Figure 7H). No effect on macrophage infiltration was observed (Figure S7D).
To exclude model-specific effects, Eltd1 silencing (siRNA #1) was also performed in a subcutaneous colorectal cancer model. Eltd1 knockdown (Figure S7K) in HCT116 xenografts caused significant growth reduction associated with histologic changes similar to those observed in SKOV3ip1 tumors (Figures S7J and S7M).
Anti-Eltd1 treatment was well tolerated (Figure S7G) and did not cause observable heart changes, which were described in Eltd1 KO mice under cardiac pressure overload (Xiao et al., 2012) (Figures S7G, S7I, and S7L).
Discussion
Our approach is based on global expression profile analysis of a large number of unfractionated primary human tumors to infer cancer-type-specific angiogenesis/vascular signatures composed of genes whose expression correlated with that of multiple seeds (vascular/angiogenesis genes used as transcriptional markers of MVD/angiogenesis). Interestingly, multiple subsignatures were observed, suggesting the existence of different angiogenic/vascular programs in distinct patient subsets, further demonstrating interindividual variability, possibly explained by different “stage or type of angiogenesis” (e.g., vessel maturation) or triggering condition (e.g., different proangiogenic factors) that might have clinical relevance. While HNSCC and BC presented three distinct clusters, CCRCC presented a more compact profile composed of only two subclusters. Notably, two main types of blood vessels with opposite prognostic value have been reported in CCRCC (Yao et al., 2007).
A 43-gene compact common signature was extracted from the cancer-type-specific ones. This core signature included known angiogenesis molecules (e.g., KDR, ROBO4, RHOJ, GPR124, CLEC14A, TIE1, ENG, and TEK/TIE2), proteins reported to be upregulated in tumor angiogenesis (e.g., CLEC14A, CD93, ROBO4, ENG, TEK/TIE2, RGS5, and ACVRL1/ALK1), and genes not yet implicated in vascular biology or generally not well characterized (e.g., ELTD1, GPR116, and MYCT1). This signature is enriched for typical EC molecules (e.g., CDH5, VWF, and ESAM) but also contains transcripts likely to be derived from tumor and other stromal cells (e.g., PDGFD).
We described upregulation of this signature in the stromal component of primary BC and hypothesize that this might represent a core program in tumor angiogenesis/vasculature whose expression is unrelated to the cancer type or proangiogenic stimulus, therefore explaining its compactness. Confirmation would offer future targets for therapy that might be less sensitive to interindividual variability. Indirect validation of the involvement of these genes in tumor angiogenesis was also indicated by their altered expression in antiangiogenesis therapeutic tumor models. Both anti-VEGF and anti-Notch therapies induced changes in signature expression mainly in the murine stroma rather than the human xenograft.
This core signature was also evaluated in ECs stimulated by DLL4, a key regulator of angiogenesis (Phng and Gerhardt, 2009). Nearly two-thirds of the signature is modulated by DLL4-NOTCH signaling, further suggesting its relevance in EC biology. As simple expression does not prove functional activity, we focused on the top-ranked, relatively unstudied molecule ELTD1, a GPCR with transcript expression in ECs (Herbert et al., 2008; Wallgard et al., 2008) that is lost in zebrafish embryos lacking blood vessel development (Sumanas et al., 2005; Weber et al., 2005) and upregulated in glioblastoma ECs and tumor cells (Dieterich et al., 2012; Towner et al., 2013). Recent data from Eltd1 KO mice also proved that this gene is dispensable for normal development but has a role in heart adaptation to pathologic conditions such as pressure overload (Xiao et al., 2012).
Here, we report ELTD1 protein expression in both ECs and VSMCs, as the other members of this receptor family are primarily expressed by immune cells (Kwakkenbos et al., 2004). ELTD1 is expressed as a highly glycosylated molecule, as suggested by Liu et al. (2005), in monomeric and predominantly dimeric forms. EC ELTD1 expression is reduced by Dll4-Notch signaling in vitro and in vivo but induced by VEGF and bFGF. Functionally, we prove that ELTD1 plays a key role in angiogenesis both in vitro and in vivo, and our data suggest the mechanism may be via regulation of the sprouting process. The dramatic vascular impairment in zebrafish embryos lacking eltd1 while Eltd1 KO mice do not present any developmental defect (Xiao et al., 2012) might reflect compensatory mechanisms acquired during evolution, consistent with receptor family expansion in higher vertebrates (Kwakkenbos et al., 2004). However, a potential caveat of the KO strategy of Kwakkenbos et al. is the retention of the entire Eltd1 extracellular domain. Thus, we cannot exclude that functionally active soluble Eltd1 or aberrant membrane receptor forms may be expressed. Nevertheless, taken together with the rest of our data, the simplest interpretation of our in vivo siRNA data is that Eltd1 is important for promoting tumor growth and metastasis by playing a role in pathologic angiogenesis. Further insights into the specific cell types and mechanisms contributing to impaired tumor growth following Eltd1 silencing might be gained with conditional Eltd1 KO mice. Importantly, systemically given Eltd1 siRNAs were nontoxic in mice as assessed by body weight, heart to body weight ratio, and heart histology, indicating a differential role of Eltd1 in normal versus tumor vasculature and safety of its inhibition in animals without pre-existing cardiac pathologies.
ELTD1 modulates vascular sprouting by regulating tip cell specification/activity, possibly in part by negatively affecting DLL4 expression (Figure 6D). Functional interplay between dll4 and eltd1 pathways is also proven by their reciprocal phenotypic rescue in double-MO zebrafish embryos. Importantly, eltd1 plays a dll4-independent role during vascular development since dll4 MO-mediated functional rescue in eltd1-deficient embryos only partially restores normal phenotype.
Analysis of human samples demonstrated ELTD1 was upregulated in tumor-associated ECs in renal, colorectal, and ovarian cancers compared to their normal corresponding control tissues. Despite being extensively used in research and clinical settings, IHC is semiquantitative and further studies are required to accurately quantify ELTD1 levels in the normal and tumor endothelium. However, limited mRNA analyses (breast and renal cancer data not shown; Dieterich et al., 2012) indicate a 2- to 4-fold increase in ELTD1 transcripts in tumor versus normal ECs. This fits with the VEGF/bFGF induction of ELTD1, as these cytokines are often upregulated in cancer, playing a key role in tumor angiogenesis. Importantly, the positive correlation between ELTD1 levels and MVD observed in patient samples represents an indirect validation of the proangiogenic function described in vitro and in vivo. Clinically, higher EC ELTD1 levels correlated with less aggressive cancer features and better prognosis in all tumor types analyzed. Although many angiogenic molecules correlate with poor survival, clearly some must also be associated with the vasculature of those with better outcome. In most studies of tumor vessels, no note is made of protein expression level in adjacent normal vessels from the same patient, although it likely there will be substantial variability from person to person. In our study, the differences between tumor and normal expression translate into a greater difference in outcome, suggesting the importance of assessing the host background in future studies. The concept of “vascular normalization” in response to anti-VEGF therapy has been well described (Carmeliet and Jain, 2011b). However, if tumors were better perfused prior to therapy (because of higher MVD, more differentiated vessels, and/or reduced leakiness), then there would be less hypoxia and intratumoral pressure and potentially less aggressive cancer. Higher ELTD1 levels, promoting EC sprouting, may improve tumor vascularization and perfusion, thus aiding drug delivery. This hypothesis needs further validation but is consistent with the findings that high EC ELTD1 levels correlate with higher MVD and longer survival.
Therapeutic potential is suggested by ELTD1 upregulation in tumor ECs and the dramatic impairment in tumor growth upon its silencing. This latter is probably caused by MVD reduction, consistent with the positive correlation observed between ELTD1 levels and MVD in clinical samples and functionally explained by ELTD1’s proangiogenic role identified in vitro and in vivo during zebrafish development.
Analyzing ELTD1 expression in prospective randomized studies of antiangiogenic therapies will determine if there is any modulation during vascular normalization and if it is an intrinsic mechanism of resistance to anti-VEGF therapy, possibly offering combined antiangiogenic treatments. The latter may help explain the lack of efficacy of antiangiogenic therapies in the adjuvant situation if smaller earlier lesions have a different type of vasculature. The observation that higher ELTD1 levels correlate with better outcome does not obviate it as a therapeutic target as observed for other good prognostic markers (ALK in T cell lymphomas, estrogen receptor in relapsed BC, and MVD in renal cancer). Of note, even within the high-expressing group of patients, a relevant fraction die of their disease. Furthermore, a good prognosis may relate to a more mature vasculature resistant to metastasis. Clinically, ELTD1 represents a relevant cell-surface candidate for targeted antiangiogenic approaches whose safety is suggested by target upregulation in tumor ECs, normal development of KO mice (Xiao et al., 2012), and lack of visible toxic effects in our in vivo experiments. However, safety in humans would need to be verified in clinical trials, possibly focusing on patients with high EC ELTD1 levels and no cardiac impairment. Furthermore, if subsequent studies demonstrate ELTD1 involvement in vessel maturation, then its inhibition might also be considered for combination therapies to sensitize vasculature to anti-VEGF or anti-Notch therapies. Our work shows clear therapeutic effects of Eltd1 targeting and a role in vascular sprouting.
Experimental Procedures
Gene Expression Profiling and Derivation of Cancer Type-Specific and Core Angiogenesis Signatures
A coexpression network was constructed using seed clustering and metasignatures derived as described previously (Buffa et al., 2010). Prototype genes (seeds) were selected on the basis of published evidence of involvement in angiogenesis, and/or expression in vessels (table in Supplemental Experimental Procedures). Patients treated in Oxford were analyzed and other data sets were retrieved from the National Center for Biotechnology Information’s GEO (http://www.ncbi.nlm.nih.gov/geo/) (table in Supplemental Experimental Procedures) for a total of 121 HNSCCs, 959 BCs, and 170 CCRCCs.
Patients and Tissue Samples
Formalin-fixed, paraffin-embedded tumor tissues from 157 patients with renal cancer (plus 148 normal kidney samples; 126 matched to tumor cases), 120 with colorectal cancer (plus 120 matched normal tissues), 65 with head and neck cancer, and 21 mixed tumor types were used to assemble TMAs (Bubendorf et al., 2001). Demographic and pathological details of renal, head and neck, and colorectal cancer cases are reported in Supplemental Information. Ovarian cancer TMA and patient data (n = 200) were previously reported (Lu et al., 2010). Whole tissue sections from 11 cases of ovarian serous adenocarcinoma, five normal ovaries, and reactive tonsil were also analyzed. Samples were obtained in accordance with the National Research Ethics Service South Central Oxford B Research Ethics Committee (project reference number C02.216). Deidentified samples were accessed through the Oxford Centre for Histopathology Research according to UK regulatory requirements.
Orthotopic In Vivo Model of Ovarian Cancer
Female athymic nude mice (NCr-nu) purchased from the National Cancer Institute Frederick Cancer Research and Development Center (Frederick, MD) were maintained as previously described (Lu et al., 2010). All mouse studies were approved by the MD Anderson Cancer Center Institutional Animal Care and Use Committee. The mice used for in vivo experiments were 8 to 12 weeks old. SKOV3ip1 (1 × 106 cells per mouse) or HCT116 (1 × 106 cells per mouse) cells were injected intraperitoneally or subcutaneously, respectively. Mice were monitored daily for adverse effects and were sacrificed when any of the mice seemed moribund.
To assess tumor growth, treatment began 1 week after cell injection. Each siRNA (Sigma-Aldrich) was incorporated into chitosan nanoparticles (Han et al., 2010; Lu et al., 2010) and given twice weekly (150 μg/kg body weight) through intravenous injection. After screening (Figures S7A and S7B), mouse eltd1-specific siRNA 85 (#1) and 53 (#2) were selected (#1: 5′-GTTGCATTCCTCTGCTATA-3′; #2: 5′-GTTGAAAGGAGTACACATA-3′). For tumor growth studies (n = 10 per group), treatment continued until any experimental group became moribund (control, typically 4 or 5 weeks), while for the survival experiment (n = 9 per group) treatment continued for 101 days, with individual mice being sacrificed as they became moribund. Mouse, heart, and tumor weight, number of nodules, and distribution/number of metastasis were recorded at the time of sacrifice. Mouse weight was also recorded weekly during treatment. Tissue specimens were fixed with formalin or optimum cutting temperature (Miles) or were snap frozen. To exclude treatment-induced interferon response, healthy animals (n = 3 per group) were treated with a single dose of saline, naked CH-NP, or Eltd1 #1 siRNA CH-NP. Blood sampled 12 hr after injection was tested with ELISA for immunostimulatory cytokines (R&D Systems).
Statistical Analysis
Survival was measured from the day of surgery. The log-rank test was used in univariate survival analyses. Prognostic factors were evaluated in Cox proportional hazards regression including relevant clinical covariates (see Tables S5–S7); backward stepwise likelihood was used for selection. Other paired or unpaired parametric or nonparametric tests were used as necessary; methods, statistics, and significance are cited in the text and figures. Stata 11.0, GraphPad Prism 5.00, and R were used.
Acknowledgments
This study was supported by a Cancer Research UK Program grant A10702 (AHB/ALH) and the p-medicine EU FP7 network (to F.M.B.). Further support included grants from the National Institutes of Health (P50 CA083639, P50 CA098258, and U54 CA151668), DOD (W81XWH-10-1-0158), and the Gilder and RGK Foundations. Primary human material was sourced from the NIHR Biomedical Research Centre, Oxford Radcliffe Biobank, Oxford, UK. The study was supported by the National Institute for Health Research (NIHR) Oxford Biomedical Research Centre Programme. The views expressed are those of the authors and not necessarily those of the NHS, the NIHR, or the Department of Health. The authors thank Jenny Greig, Joy Winter, and Helen Turley for technical assistance and Drs. Helen Sheldon and Duncan Gascoyne for helpful discussions.
Published: July 18, 2013
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-No Derivative Works License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
Supplemental Information includes Supplemental Experimental Procedures, seven figures, seven tables, and two movies and can be found with this article online at http://dx.doi.org/10.1016/j.ccr.2013.06.004.
Contributor Information
Adrian L. Harris, Email: adrian.harris@oncology.ox.ac.uk.
Francesca M. Buffa, Email: francesca.buffa@oncology.ox.ac.uk.
Accession Numbers
Microarray data produced in this study have been deposited to the GEO with the accession numbers GSE37956, GSE3922, GSE39223, and GSE39413.
Supplemental Information
Genes are ranked based on the correlation of their expression with that of the seeds across 5 breast cancer.
Genes in common between the head and neck squamous cell carcinoma (HNSCC), breast cancer.
Fold change (FC) and significance (Q value) are reported. See Experimental Procedures for details.
References
- Bais, C., Singh, M., Kaminker, J., and Brauer, M. May 2011. Biological markers for monitoring patient response to vegf antagonists. U.S. Patent 20,110,117,083.
- Baldewijns M.M., van Vlodrop I.J., Vermeulen P.B., Soetekouw P.M., van Engeland M., de Bruïne A.P. VHL and HIF signalling in renal cell carcinogenesis. J. Pathol. 2010;221:125–138. doi: 10.1002/path.2689. [DOI] [PubMed] [Google Scholar]
- Bjarnadóttir T.K., Fredriksson R., Höglund P.J., Gloriam D.E., Lagerström M.C., Schiöth H.B. The human and mouse repertoire of the adhesion family of G-protein-coupled receptors. Genomics. 2004;84:23–33. doi: 10.1016/j.ygeno.2003.12.004. [DOI] [PubMed] [Google Scholar]
- Bubendorf L., Nocito A., Moch H., Sauter G. Tissue microarray (TMA) technology: miniaturized pathology archives for high-throughput in situ studies. J. Pathol. 2001;195:72–79. doi: 10.1002/path.893. [DOI] [PubMed] [Google Scholar]
- Buffa F.M., Harris A.L., West C.M., Miller C.J. Large meta-analysis of multiple cancers reveals a common, compact and highly prognostic hypoxia metagene. Br. J. Cancer. 2010;102:428–435. doi: 10.1038/sj.bjc.6605450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmeliet P., Jain R.K. Molecular mechanisms and clinical applications of angiogenesis. Nature. 2011;473:298–307. doi: 10.1038/nature10144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmeliet P., Jain R.K. Principles and mechanisms of vessel normalization for cancer and other angiogenic diseases. Nat. Rev. Drug Discov. 2011;10:417–427. doi: 10.1038/nrd3455. [DOI] [PubMed] [Google Scholar]
- Casadó V., Cortés A., Mallol J., Pérez-Capote K., Ferré S., Lluis C., Franco R., Canela E.I. GPCR homomers and heteromers: a better choice as targets for drug development than GPCR monomers? Pharmacol. Ther. 2009;124:248–257. doi: 10.1016/j.pharmthera.2009.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Toro R., Prahst C., Mathivet T., Siegfried G., Kaminker J.S., Larrivee B., Breant C., Duarte A., Takakura N., Fukamizu A. Identification and functional analysis of endothelial tip cell-enriched genes. Blood. 2010;116:4025–4033. doi: 10.1182/blood-2010-02-270819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieterich L.C., Mellberg S., Langenkamp E., Zhang L., Zieba A., Salomäki H., Teichert M., Huang H., Edqvist P.H., Kraus T. Transcriptional profiling of human glioblastoma vessels indicates a key role of VEGF-A and TGFβ2 in vascular abnormalization. J. Pathol. 2012;228:378–390. doi: 10.1002/path.4072. [DOI] [PubMed] [Google Scholar]
- Duarte A., Hirashima M., Benedito R., Trindade A., Diniz P., Bekman E., Costa L., Henrique D., Rossant J. Dosage-sensitive requirement for mouse Dll4 in artery development. Genes Dev. 2004;18:2474–2478. doi: 10.1101/gad.1239004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelse M.A., Laurens N., Verloop R.E., Koolwijk P., van Hinsbergh V.W. Differential gene expression analysis of tubule forming and non-tubule forming endothelial cells: CDC42GAP as a counter-regulator in tubule formation. Angiogenesis. 2008;11:153–167. doi: 10.1007/s10456-007-9086-9. [DOI] [PubMed] [Google Scholar]
- Gale N.W., Dominguez M.G., Noguera I., Pan L., Hughes V., Valenzuela D.M., Murphy A.J., Adams N.C., Lin H.C., Holash J. Haploinsufficiency of delta-like 4 ligand results in embryonic lethality due to major defects in arterial and vascular development. Proc. Natl. Acad. Sci. USA. 2004;101:15949–15954. doi: 10.1073/pnas.0407290101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gering M., Rodaway A.R., Göttgens B., Patient R.K., Green A.R. The SCL gene specifies haemangioblast development from early mesoderm. EMBO J. 1998;17:4029–4045. doi: 10.1093/emboj/17.14.4029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghilardi C., Chiorino G., Dossi R., Nagy Z., Giavazzi R., Bani M. Identification of novel vascular markers through gene expression profiling of tumor-derived endothelium. BMC Genomics. 2008;9:201. doi: 10.1186/1471-2164-9-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han H.D., Mangala L.S., Lee J.W., Shahzad M.M., Kim H.S., Shen D., Nam E.J., Mora E.M., Stone R.L., Lu C. Targeted gene silencing using RGD-labeled chitosan nanoparticles. Clin. Cancer Res. 2010;16:3910–3922. doi: 10.1158/1078-0432.CCR-10-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Hellström M., Phng L.K., Hofmann J.J., Wallgard E., Coultas L., Lindblom P., Alva J., Nilsson A.K., Karlsson L., Gaiano N. Dll4 signalling through Notch1 regulates formation of tip cells during angiogenesis. Nature. 2007;445:776–780. doi: 10.1038/nature05571. [DOI] [PubMed] [Google Scholar]
- Herbert J.M., Stekel D., Sanderson S., Heath V.L., Bicknell R. A novel method of differential gene expression analysis using multiple cDNA libraries applied to the identification of tumour endothelial genes. BMC Genomics. 2008;9:153. doi: 10.1186/1471-2164-9-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isogai S., Lawson N.D., Torrealday S., Horiguchi M., Weinstein B.M. Angiogenic network formation in the developing vertebrate trunk. Development. 2003;130:5281–5290. doi: 10.1242/dev.00733. [DOI] [PubMed] [Google Scholar]
- Krebs L.T., Shutter J.R., Tanigaki K., Honjo T., Stark K.L., Gridley T. Haploinsufficient lethality and formation of arteriovenous malformations in Notch pathway mutants. Genes Dev. 2004;18:2469–2473. doi: 10.1101/gad.1239204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutschera S., Weber H., Weick A., De Smet F., Genove G., Takemoto M., Prahst C., Riedel M., Mikelis C., Baulande S. Differential endothelial transcriptomics identifies semaphorin 3G as a vascular class 3 semaphorin. Arterioscler. Thromb. Vasc. Biol. 2011;31:151–159. doi: 10.1161/ATVBAHA.110.215871. [DOI] [PubMed] [Google Scholar]
- Kwakkenbos M.J., Kop E.N., Stacey M., Matmati M., Gordon S., Lin H.H., Hamann J. The EGF-TM7 family: a postgenomic view. Immunogenetics. 2004;55:655–666. doi: 10.1007/s00251-003-0625-2. [DOI] [PubMed] [Google Scholar]
- Leslie J.D., Ariza-McNaughton L., Bermange A.L., McAdow R., Johnson S.L., Lewis J. Endothelial signalling by the Notch ligand Delta-like 4 restricts angiogenesis. Development. 2007;134:839–844. doi: 10.1242/dev.003244. [DOI] [PubMed] [Google Scholar]
- Li J.L., Harris A.L. Crosstalk of VEGF and Notch pathways in tumour angiogenesis: therapeutic implications. Front. Biosci. 2009;14:3094–3110. doi: 10.2741/3438. [DOI] [PubMed] [Google Scholar]
- Li J.L., Sainson R.C., Oon C.E., Turley H., Leek R., Sheldon H., Bridges E., Shi W., Snell C., Bowden E.T. DLL4-Notch signaling mediates tumor resistance to anti-VEGF therapy in vivo. Cancer Res. 2011;71:6073–6083. doi: 10.1158/0008-5472.CAN-11-1704. [DOI] [PubMed] [Google Scholar]
- Liu T., Qian W.J., Gritsenko M.A., Camp D.G., 2nd, Monroe M.E., Moore R.J., Smith R.D. Human plasma N-glycoproteome analysis by immunoaffinity subtraction, hydrazide chemistry, and mass spectrometry. J. Proteome Res. 2005;4:2070–2080. doi: 10.1021/pr0502065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C., Han H.D., Mangala L.S., Ali-Fehmi R., Newton C.S., Ozbun L., Armaiz-Pena G.N., Hu W., Stone R.L., Munkarah A. Regulation of tumor angiogenesis by EZH2. Cancer Cell. 2010;18:185–197. doi: 10.1016/j.ccr.2010.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nechiporuk T., Urness L.D., Keating M.T. ETL, a novel seven-transmembrane receptor that is developmentally regulated in the heart. ETL is a member of the secretin family and belongs to the epidermal growth factor-seven-transmembrane subfamily. J. Biol. Chem. 2001;276:4150–4157. doi: 10.1074/jbc.M004814200. [DOI] [PubMed] [Google Scholar]
- Phng L.K., Gerhardt H. Angiogenesis: a team effort coordinated by notch. Dev. Cell. 2009;16:196–208. doi: 10.1016/j.devcel.2009.01.015. [DOI] [PubMed] [Google Scholar]
- Sana T.R., Janatpour M.J., Sathe M., McEvoy L.M., McClanahan T.K. Microarray analysis of primary endothelial cells challenged with different inflammatory and immune cytokines. Cytokine. 2005;29:256–269. doi: 10.1016/j.cyto.2004.11.003. [DOI] [PubMed] [Google Scholar]
- Seaman S., Stevens J., Yang M.Y., Logsdon D., Graff-Cherry C., St Croix B. Genes that distinguish physiological and pathological angiogenesis. Cancer Cell. 2007;11:539–554. doi: 10.1016/j.ccr.2007.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siekmann A.F., Lawson N.D. Notch signalling limits angiogenic cell behaviour in developing zebrafish arteries. Nature. 2007;445:781–784. doi: 10.1038/nature05577. [DOI] [PubMed] [Google Scholar]
- St Croix B., Rago C., Velculescu V., Traverso G., Romans K.E., Montgomery E., Lal A., Riggins G.J., Lengauer C., Vogelstein B., Kinzler K.W. Genes expressed in human tumor endothelium. Science. 2000;289:1197–1202. doi: 10.1126/science.289.5482.1197. [DOI] [PubMed] [Google Scholar]
- Sumanas S., Jorniak T., Lin S. Identification of novel vascular endothelial-specific genes by the microarray analysis of the zebrafish cloche mutants. Blood. 2005;106:534–541. doi: 10.1182/blood-2004-12-4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towner R.A., Jensen R.L., Colman H., Vaillant B., Smith N., Casteel R., Saunders D., Gillespie D.L., Silasi-Mansat R., Lupu F. ELTD1, a potential new biomarker for gliomas. Neurosurgery. 2013;72:77–90. doi: 10.1227/NEU.0b013e318276b29d. discussion 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlen M., Oksvold P., Fagerberg L., Lundberg E., Jonasson K., Forsberg M., Zwahlen M., Kampf C., Wester K., Hober S. Towards a knowledge-based Human Protein Atlas. Nat. Biotechnol. 2010;28:1248–1250. doi: 10.1038/nbt1210-1248. [DOI] [PubMed] [Google Scholar]
- van Beijnum J.R., Griffioen A.W. In silico analysis of angiogenesis associated gene expression identifies angiogenic stage related profiles. Biochim. Biophys. Acta. 2005;1755:121–134. doi: 10.1016/j.bbcan.2005.06.001. [DOI] [PubMed] [Google Scholar]
- van Beijnum J.R., Dings R.P., van der Linden E., Zwaans B.M., Ramaekers F.C., Mayo K.H., Griffioen A.W. Gene expression of tumor angiogenesis dissected: specific targeting of colon cancer angiogenic vasculature. Blood. 2006;108:2339–2348. doi: 10.1182/blood-2006-02-004291. [DOI] [PubMed] [Google Scholar]
- Wallgard E., Larsson E., He L., Hellström M., Armulik A., Nisancioglu M.H., Genove G., Lindahl P., Betsholtz C. Identification of a core set of 58 gene transcripts with broad and specific expression in the microvasculature. Arterioscler. Thromb. Vasc. Biol. 2008;28:1469–1476. doi: 10.1161/ATVBAHA.108.165738. [DOI] [PubMed] [Google Scholar]
- Weber G.J., Choe S.E., Dooley K.A., Paffett-Lugassy N.N., Zhou Y., Zon L.I. Mutant-specific gene programs in the zebrafish. Blood. 2005;106:521–530. doi: 10.1182/blood-2004-11-4541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J., Jiang H., Zhang R., Fan G., Zhang Y., Jiang D., Li H. Augmented cardiac hypertrophy in response to pressure overload in mice lacking ELTD1. PLoS ONE. 2012;7:e35779. doi: 10.1371/journal.pone.0035779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong J.W., Yu Q., Zhang J., Mably J.D. An acyltransferase controls the generation of hematopoietic and endothelial lineages in zebrafish. Circ. Res. 2008;102:1057–1064. doi: 10.1161/CIRCRESAHA.107.163907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao X., Qian C.N., Zhang Z.F., Tan M.H., Kort E.J., Yang X.J., Resau J.H., Teh B.T. Two distinct types of blood vessels in clear cell renal cell carcinoma have contrasting prognostic implications. Clin. Cancer Res. 2007;13:161–169. doi: 10.1158/1078-0432.CCR-06-0774. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genes are ranked based on the correlation of their expression with that of the seeds across 5 breast cancer.
Genes in common between the head and neck squamous cell carcinoma (HNSCC), breast cancer.
Fold change (FC) and significance (Q value) are reported. See Experimental Procedures for details.