Summary
Ileal lesions in Crohn’s disease (CD) patients are colonized by pathogenic adherent-invasive Escherichia coli (AIEC) able to invade and to replicate within intestinal epithelial cells. Recent genome-wide association studies have highlighted the autophagy pathway as being associated with CD risk. In the present study we investigated whether defects in autophagy enhance replication of commensal and pathogenic Escherichia coli and CD associated AIEC. We show that functional autophagy limits intracellular AIEC replication and that a subpopulation of the intracellular bacteria is located within LC3 positive autophagosomes. In IRGM and ATG16L1 deficient cells intracellular AIEC LF82 bacteria have enhanced replication. Surprisingly autophagy deficiency did not interfere with the ability of intracellular bacteria to survive and/or replicate for any other E. coli strains tested, including non pathogenic, environmental, commensal, or pathogenic strains involved in gastroenteritis. Together these findings demonstrate a central role for autophagy restraining Adherent-Invasive E. coli strains associated with ileal CD. AIEC infection in patients with polymorphisms in autophagy genes may have a significant impact on the outcome of intestinal inflammation.
Keywords: Crohn’s disease, autophagy, Adherent-Invasive E. coli, ATG16L1, IRGM
Introduction
Crohn’s disease (CD) and ulcerative colitis (UC) are two major forms of idiopathic inflammatory bowel disease (IBD), with a combined prevalence of about 150–200 cases per 100,000 in Western countries. They are multifactorial diseases, occurring in individuals with genetic predisposition in whom an environmental or microbial trigger causes an abnormal immune response (Strober et al., 2007; Xavier and Podolsky, 2007). Several lines of evidence suggest that bacteria play a role in the onset and perpetuation of IBD (Sartor, 2008). Intestinal bacteria are essential for the development of intestinal inflammation. In patients with CD, exposure of the terminal ileum post-surgically to luminal contents is associated with increased inflammation, and diversion of the faecal stream is associated with improvement (Rutgeerts et al., 1991).
Studies in CD patients have reported high concentrations of bacteria forming a biofilm on the surface of the gut mucosa (Swidsinski et al., 2002) and low numbers of anti-inflammatory bacteria (Sokol et al., 2008), and showed that high numbers of E. coli colonize the epithelial intestinal layer (Conte et al., 2006; Kotlowski et al., 2006; Martin et al., 2004; Darfeuille-Michaud et al., 1998). As a commensal, E. coli coexists with its mammalian host in good harmony and rarely causes disease except in immunocompromised hosts or when the normal gastrointestinal barriers are breached. However, some E. coli strains have acquired specific virulence factors that increase their ability to adapt to new niches and allow them to cause a broad spectrum of diseases. Among the E. coli strains that can cause intestinal gastroenteritis in humans, there are six well-characterized pathotypes: enteropathogenic E. coli (EPEC), enterohaemorrhagic E. coli (EHEC), enterotoxinogenic E. coli (ETEC), enteroaggregative E. coli (EAEC), enteroinvasive E. coli (EIEC) and diffusely adherent E. coli (DAEC) (for review, (Kaper et al., 2004)). Genotypic characterization of CD-associated E. coli showed that they do not possess the virulence factors of any of the above pathogenic E. coli strains. However, they are fully virulent, able to adhere to and to invade/replicate within intestinal epithelial cells, and also survive and replicate within macrophages thereby producing large amounts of TNF-α (Glasser et al., 2001; Boudeau et al., 1999). These pathogenic E. coli are called AIEC for Adherent-Invasive Escherichia coli. Their association with CD has been reported in independent studies performed in Europe and the United States (Martinez-Medina et al., 2009; Baumgart et al., 2007; Eaves-Pyles et al., 2007; Sasaki et al., 2007; Darfeuille-Michaud et al., 2004; Martin et al., 2004) and particularly with ileal CD owing to abnormal expression of the specific host receptor CEACAM6 (Barnich et al., 2007; Darfeuille-Michaud et al., 2004).
The theory of dysregulated host responses to intracellular micro-organisms is emerging as a contributing factor in CD pathogenesis. Many IBD susceptibility loci have been identified. The most well-replicated IBD genetic association is the NOD2 (nucleotide-binding oligomerization domain 2) gene occurring in CD (Hugot et al., 2001; Ogura et al., 2001). Recent genome-wide association (GWA) studies, in addition to identifying new susceptibility genes, have firmly established that some of them contribute to IBD, and in particular to CD (Hampe et al., 2007; Massey and Parkes, 2007; Rioux et al., 2007; Wellcome, 2007; Duerr et al., 2006). A highly significant and replicated association between CD and variants in two separate autophagy genes (ATG16L1 and IRGM) has been shown. Thr300Ala substitution within the ATG16L1 (Autophagy-related like 1) gene is mostly associated with ileal CD (Hampe et al., 2007; Rioux et al., 2007). A second autophagic gene has been identified in CD susceptibility: the IRGM (Immunity-related GTPase family M) (Parkes et al., 2007; Wellcome, 2007). Autophagy is a process by which eukaryotic cells maintain homeostasis by sequestering cytoplasm and degrading damaged organelles via the lysosomal pathway (Mizushima, 2007). In addition autophagy protects the cell by eliminating or limiting the growth of intracellular bacteria (Birmingham et al., 2006; Gutierrez et al., 2004). Thus a dysfunction in autophagy leads to persistent infection, as seen in Salmonella Typhimurium, Streptococcus pyogenes, and Mycobacterium tuberculosis (Kuballa et al., 2008; Birmingham et al., 2006; Singh et al., 2006; Nakagawa et al., 2004).
As defects in autophagy could predispose patients to CD by promoting prolonged survival and/or replication of intracellular microorganisms within host cells, the aim of the present study was to analyze whether the cellular autophagy mechanism can control bacterial survival and/or replication of various E. coli strains, including CD-associated strains, commensal, non pathogenic or pathogenic strains involved in gastroenteritis. We provide here the first evidence that intracellular replication of CD-associated AIEC bacteria is correlated with a loss of autophagy function mediated by either of the two CD-associated autophagy genes; ATG16L1 and IRGM.
Results
Intracellular behaviour of commensal, environmental, enteropathogenic and CD-associated E. coli strains within wild-type and autophagy deficient mouse embryonic fibroblasts
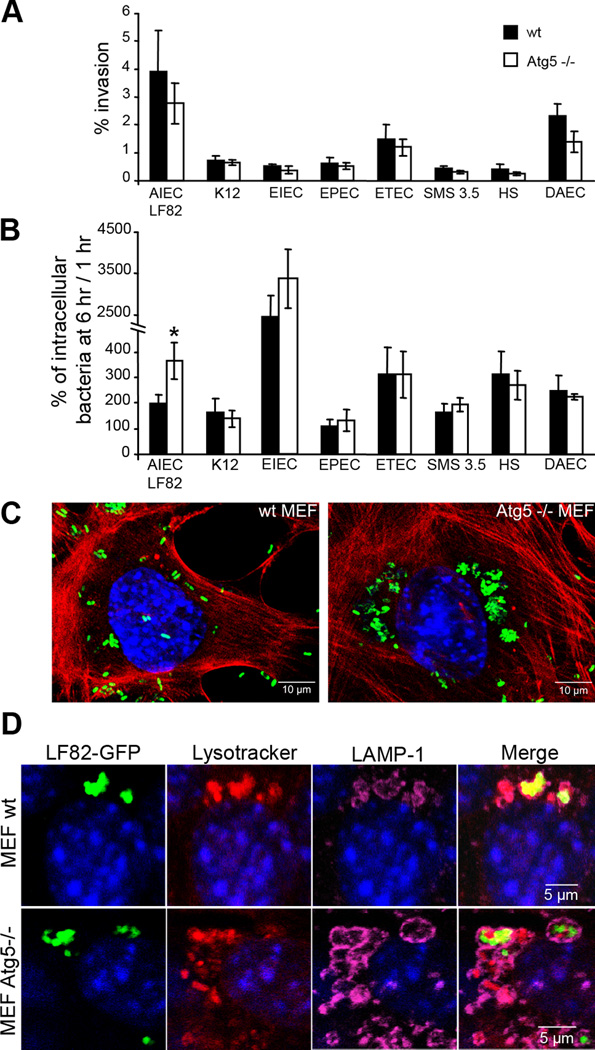
To test the role of autophagy in its ability to limit intracellular E. coli replication we performed time course invasion assays in wild-type (wt) mouse embryonic fibroblasts (MEFs) versus atg5−/− MEFs harboring a knockout of the atg5 locus (atg5−/−). The ability of all the E. coli strains tested to invade wt MEFs or atg5 −/− MEFs was compared (Fig 1A). As expected, the CD-associated AIEC strain LF82 was the most invasive strain with a percentage of intracellular bacteria at 1 h post-infection in wt MEFs and Atg5−/− MEFs corresponding respectively to 3.93 ± 1.47% in 2.77 ± 0.73% of the inoculum. ETEC strain H10407 and the DAEC strain C1845, known to be invasive, also invaded MEFs but with lesser efficiency than AIEC strain LF82. Surprisingly the EIEC strain E12860/0 did not efficiently invade MEFs but showed a high replication once internalized (Fig 1B). The non pathogenic E. coli K-12 strain MG1655, which is unable to replicate intracellularly, was similar in its behaviour to wild-type or atg5−/− MEFs (Fig 1B and Fig S1). Similarly, the numbers of intracellular bacteria for the environmental E. coli strain SMS 3.5 and the commensal E. coli strain HS were identical within wild-type and atg5−/− MEFs even though bacteria were able to replicate. Among the pathogenic E. coli strains responsible for gastroenteritis, only the AIEC strain LF82 showed a significant (P < 0.01) increase in the numbers of intracellular bacteria at 6 h post-infection in atg5−/− MEFs compared to wt MEFs. These findings demonstrate that the survival and/or replication of most of the E. coli strains, including non pathogenic K-12, commensal, environmental or pathogenic E. coli (ETEC, EPEC, EIEC and DAEC) is independent of the autophagy pathway. Confocal analysis showed the presence of large clusters of intracellular AIEC LF82 bacteria in atg5−/− MEFs, whereas only a few, mostly individual, bacteria were seen in wt MEFs (Fig 1C). Most of the LF82-containing phagosomes in WT or atg5−/− MEFs stained positive with both LAMP-1 antibody and Lysotracker, indicating that the bacteria clusters were located in mature acidic phagolysosomes (Fig 1D and Fig S2). Moreover to assess if a part of AIEC LF82 bacteria was in contact with cytosol, we permeabilized MEFs with digitonin prior to staining bacteria using an antibody to lipopolysaccharide O83. Both in MEFs wt and Atg5−/−, a subpopulation of AIEC LF82 bacteria were positive for O83 staining, indicating that some bacteria are cytosolic or in damaged vacuoles (Fig S3), a prerequisite for intracellular control by autophagy.
Figure 1. Autophagy restricts the intracellular replication of AIEC LF82 strain but not that of commensal or pathogenic E coli strains.
Experiments were performed with wild-type mouse embryonic fibroblasts (black rectangle) and with autophagy deficient atg5−/− MEFs (white rectangle). Infections were performed with the non pathogenic E. coli K-12 strain MG1655, the environmental E. coli strain SMS 3.5, the commensal E. coli strain HS, enteropathogenic E. coli (EPEC) strain E2348/69 (D), the diffusely adhering E. coli (DAEC) strain C1845, the enterotoxigenic E. coli (ETEC) strain H10407, the enteroinvasive E. coli (EIEC) strain E12860/0, and the CD-associated adherent-invasive E. coli (AIEC) strain LF82. The numbers of intracellular bacteria were determined at various times post-infection ranging from 1 h to 6 h. Results are expressed as percent invasion corresponding to mean percentage of the inoculum retrieved as intracellular bacteria after 1 h of gentamycin treatment following 1 h of infection (A) or as percentage of intracellular bacteria after 6 h post-infection relative to that obtained at 1 h after gentamycin exposure, taken as 100% (B). Confocal microscopic examinations of AIEC LF82-GFP infected wt MEFs and autophagy deficient atg5−/− MEFs after 6 h post-infection (C), with bacteria expressing GFP in green, actin in red and nuclei in blue. Representative confocal micrographs of LAMP-1 (purple) and lysotracker (red) labelling of AIEC-LF82 infected wt MEFs and Atg5−/− MEFs at 6 h post infection (D).
AIEC LF82 bacteria induced autophagy in MEFs and were located within autophagosomes
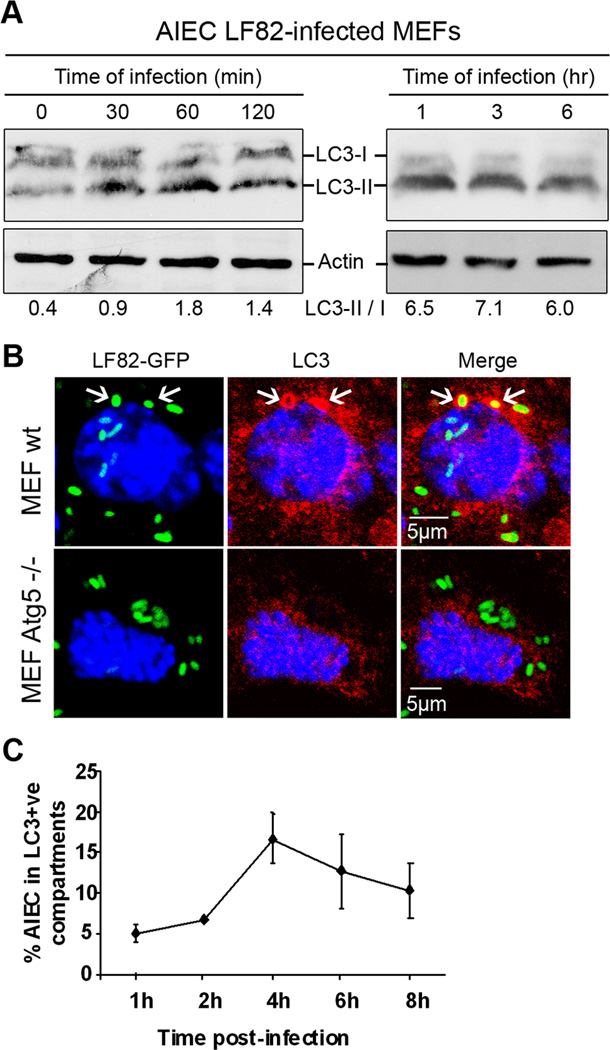
We next investigated whether autophagy is induced during infection of wt MEFs by AIEC strain LF82. We monitored autophagosome formation, as evidenced by the recruitment of LC3 protein (microtubule-associated protein light chain 3) (in its active form, LC3 II) to the autophagosome. We analyzed the shift of free cytosolic LC3-I towards the autophagosomal LC3-II by immunoblotting using an antibody raised against LC3 isoform B. In comparison to non-infected corresponding MEFs, the amount of LC3 conjugated form (LC3-II), which migrated slightly faster than the LC3-I, rapidly increased as of 30 min post infection, with a ratio LC3-II/LC3-I of 0.9 for LF82-infected MEFs compared to 0.4 for uninfected MEFs (Fig 2A). At 1 h of infection, this ratio was 1.8. Analysis of protein extracts from 1 h to 6 h post-infection revealed the presence of LC3-II, indicating an activation of the autophagic process during AIEC LF82 infection. Confocal analysis of immunostaining for LC3 protein indicated LC3 co-localization with AIEC LF82-containing vacuoles, indicating that bacteria were sequestrated in autophagosomal structures (Fig 2B). The percentages of colocalization of LC3 with intracellular bacteria increased from 5% at 1 h post-infection to 16.8% at 4 h post-infection (Fig 2C). Of note, the LC3 colocalized bacteria were mostly observed as single bacteria in a vacuole. Together these findings indicate that the replication of a subpopulation of intracellular AIEC LF82 bacteria could be hindered by the autophagy pathway in non-phagocytic cells.
Figure 2. AIEC LF82 bacteria induced autophagy in MEF and were located within autophagosomes.
A. Immunoblot analysis using antibodies against LC3 and actin to analyze the LC3 II/LC3 I ratio at early times of infection or post-infection with AIEC LF82. B. Confocal microscopic examinations of AIEC LF82-GFP infected wt MEFs and atg5−/− MEFs at 6 h post-infection to analyze LC3 colocalisation using monoclonal antibody to LC3 (red). Nuclei are in blue. C. Percentage of LC3 positive vacuole containing LF82-GFP bacteria determined by confocal microscopy at various times post-infection. Data shown represent means +/− SEM of three independent experiments, counting 50 cells per experiment.
AIEC LF82 bacteria interact with autophagy in epithelial cell lines
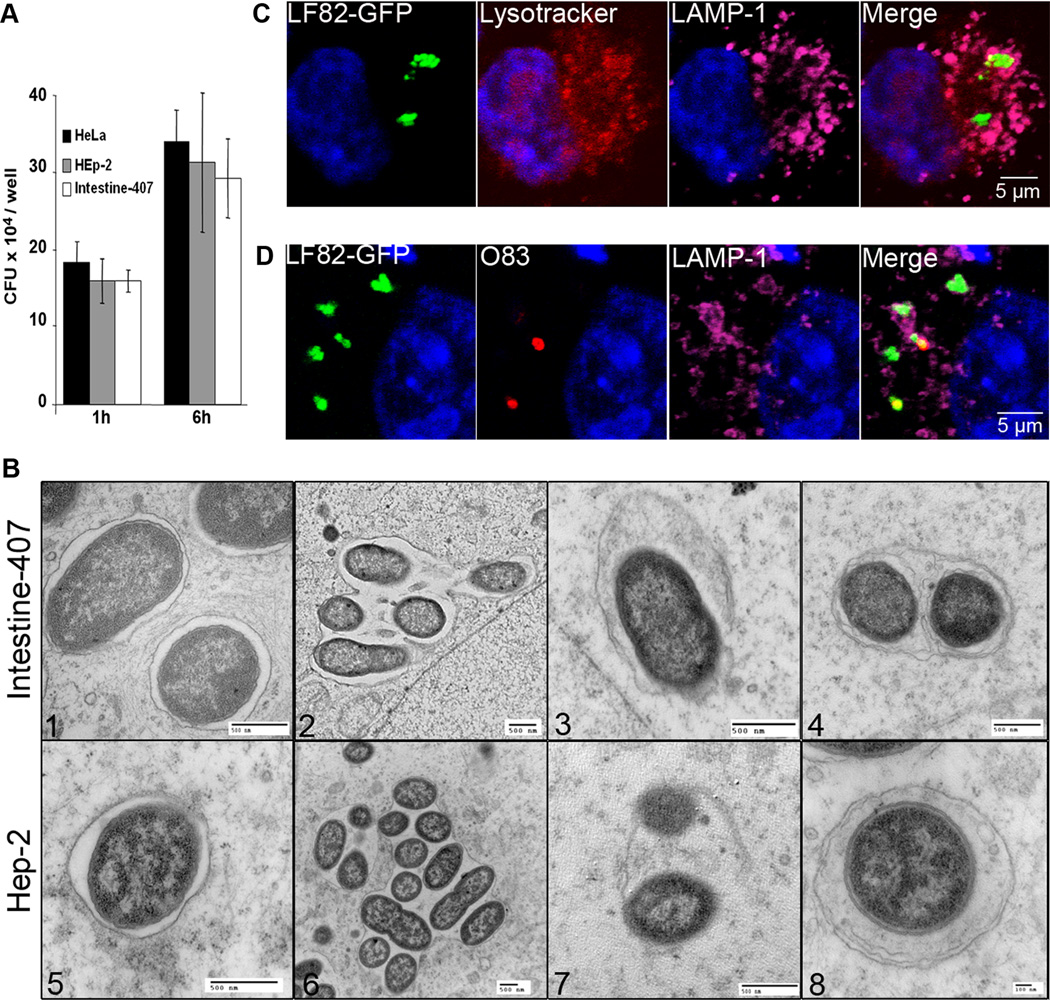
We next determined the intracellular behaviour of the reference AIEC strain LF82 in epithelial cell lines HeLa, Hep-2, and Intestine-407. Strain LF82 is able to strongly invade the three cell lines (Fig 3A) and, as previously described for Hep-2 cells (Boudeau et al., 2001), AIEC strain LF82 is also able to replicate within HeLa and Intestine-407 cells. Analysis of LF82 infected-epithelial cells by transmission electron microscopy showed various intracellular compartments for intracellular LF82 bacteria at 6 h infection. A subpopulation of bacteria was observed as single bacteria in single membrane vacuoles (Fig 3B 1 and 5). LF82 bacteria were also observed as several bacteria in a single membrane vacuole (Fig 3B 2 and 6). As observed in MEFs, most of vacuoles were both LAMP-1 and lysotracker positive (46.3±6.8%) (Fig 3C and Fig S4). In addition, some bacteria were observed by TEM in damaged vacuoles (Fig 3B 3 and 7) and confocal analysis after permeabilization with digitonin indicated the presence of bacteria positive for O83 labelling in contact with cytosol (Fig 3D). Interestingly, other individual bacteria were enclosed in a multilamellar membrane vacuole that also contained sequestered cytoplasm, the main morphological evidence of autophagosomes (Fig 3B 4 and 8). These results suggest that autophagy could be involved in handling a subpopulation of intracellular AIEC LF82 bacteria.
Figure 3. AIEC LF82 bacteria interact with autophagy in epithelial cell lines.
A. Intracellular replication of AIEC LF82 bacteria at 1 h and 6 h post-infection within HeLa, Hep-2 and Intestine-407 epithelial cells. Results are expressed as mean numbers +/− SEM of colony forming units (CFU) per well. Each point is the mean of at least three separate experiments. B. Ultrastructural analysis of Intestine-407 and Hep-2 epithelial cells infected with AIEC strain LF82 for 6 h by transmission electron microscopy showing various intracellular compartments for intracellular LF82 bacteria: single bacteria in monolayer membrane vacuoles (1 and 5), several bacteria in a single membrane vacuole (2 and 6), bacteria in damaged vacuole (3 and 7) and bacteria in a multilamellar membrane vacuole also containing sequestered cytoplasm (4 and 8). C. Confocal microscopic examinations of AIEC LF82 GFP-infected Hela cells at 6 h post-infection labelled with lysotracker (red) and for LAMP-1 (purple) to visualize the vacuoles. D. Confocal analysis after permeabilization of plasma membrane with digitonin and labelling of free cytosolic AIEC LF82 with O83 antibodies (red) and of vacuoles with LAMP-1 antibodies (purple).
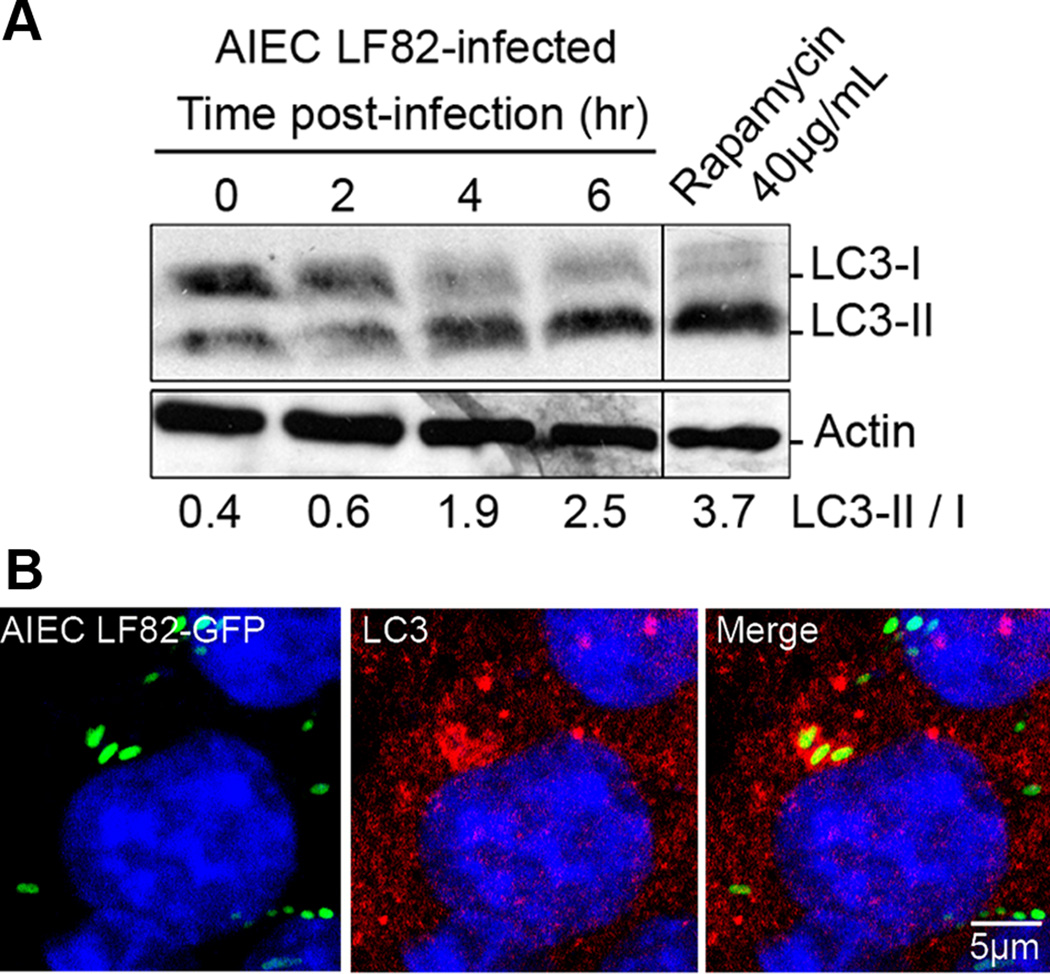
To provide further evidence of autophagic handling of internalized AIEC bacteria, we assessed endogenous LC3-I and LC3-II levels by immunoblotting. We first checked that LC3-II accumulated in cells treated with rapamycin, a drug known to induce autophagy (Fig 4A). LC3-II levels also greatly increased in AIEC LF82- infected cells, with ratios of LC3-II/LC3-I forms ranging from 0.6 to 2.5 at 2 h and 6 h post-infection, respectively (Fig 4A). To investigate whether the active LC3-II generated during infection was located on the vacuole surrounding intracellular LF82 bacteria, we performed immunofluorescence assays using a polyclonal antibody raised against LC3 protein. We observed endogenous LC3 positive labelling of a subpopulation of AIEC LF82-containing vacuoles (Fig 4B), indicating that autophagy plays a part in sensing AIEC LF82 bacteria. Thus the autophagy machinery of epithelial cells is able to target a subpopulation of intracellular AIEC bacteria.
Figure 4. AIEC LF82 infection induces autophagy in epithelial cell lines.
A. Immunoblot analysis using antibodies against LC3 B or actin to analyze the LC3 II/LC3 I ratio at various times post-infection with AIEC LF82. Induction of autophagy was performed with rapamycin treatment (40µg/ml for 2 h) as a positive control for LC3 II shift. B. Confocal microscopic examinations of AIEC LF82-GFP (green) infected HeLa at 6 h post-infection to analyze LC3 colocalisation using monoclonal antibody to LC3 (red). Nuclei are in blue.
Autophagy can restrict the replication of intracellular AIEC LF82 bacteria
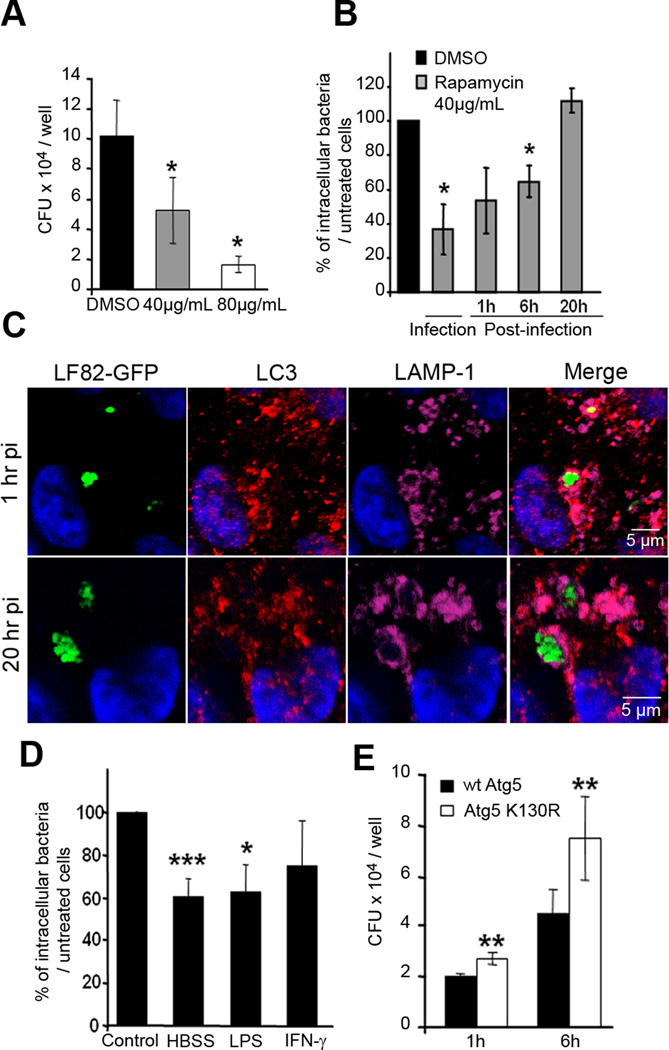
We next determined whether autophagy is required for controlling AIEC replication in epithelial cells, as previously demonstrated for some other pathogenic bacteria (Birmingham et al., 2006; Singh et al., 2006; Nakagawa et al., 2004). To address this point we modulated autophagy either by inducing or by blocking its mechanism. Treatment of HeLa cells with rapamycin to induce autophagy was performed concomitantly with infection by AIEC LF82 bacteria, and after 1 h post-infection the number of intracellular LF82 bacteria was determined. As shown in Fig.5A, a 3 h rapamycin treatment of HeLa cells significantly reduced the number of intracellular AIEC LF82 bacteria in a dose dependent-manner, with 48 ± 16% and 22 ± 5% of bacteria retrieved at 1 h post-infection in HeLa cells treated with 40 and 80µg/mL of rapamycin, respectively, compared to that retrieved in untreated cells, taken as 100%. Trypan blue exclusion assay showed that this effect was not due to cytotoxicity (data not shown). Interestingly, the time points at which the induction of autophagy was performed after infection with LF82 bacteria impacted on the numbers of intracellular bacteria. Autophagy induced by immediate rapamycin treatment achieved far greater control of intracellular bacterial load (Fig 5B), than treatment performed at 1 h or 6 h post-infection. In contrast, when the autophagy process was induced at 20 h post-infection, there was no effect on the ability of LF82 bacteria to survive and replicate within epithelial cells. Confocal analysis of AIEC LF82 colocalization with endogeneous LC3 showed that rapamycin treatment at an early time (1 h post-infection) highly increased the number of bacteria in LC3 positive compartments from 6±0.6% in untreated cells to 20.5±7.2% in rapamycin treated cells (Fig 5C and S5). In contrast, rapamycin treatment at late time (20h post-infection) did not modify colocalization of bacteria with endogeneous LC3 (Fig 5C and S5). In addition, induction of autophagy by amino acid starvation performed by incubating HeLa cells for 2 h in HBSS medium, gave similar results, with a decrease in the number of intracellular LF82 bacteria at 1 h post-infection compared to untreated cells (Fig. 5D). Similarly when autophagy was induced by stimulating HeLa cells with physiologic inducers, such as lipopolysaccharide (LPS) and interferon-γ (IFN-γ), similar results were observed since a decrease in the numbers of intracellular LF82 bacteria in cells with induced autophagy occured (Fig 5D). In addition to induce autophagy, HBSS, LPS and IFN-γ treatments can be imagined to affect the entry of bacteria. However, we validated that autophagy plays a key role in limiting AIEC bacterial replication by performing experiments using HeLa cells transfected with plasmid pEGFP-Atg5K130R encoding an Atg5K130R dominant negative. We observed a significant increase in the numbers of intracellular AIEC LF82 bacteria at 1 h post-infection, but with a more pronounced effect at 6 h post-infection, in Atg5K130R expressing transfected HeLa cells compared to those transfected with pEGFP-Atg5 encoding wild-type Atg5 (Fig 5E). Thus, by modulating autophagy, our experiments confirm that autophagy limits the number of intracellular AIEC LF82 bacteria in human epithelial cells.
Figure 5. Pharmacological- and physiological-induced autophagy can restrict the replication of intracellular AIEC LF82 bacteria.
A. Rapamycin dose-dependent decrease in the numbers of intracellular AIEC LF82 bacteria. Cells were treated concomitantly with bacterial infection. Results are expressed as mean numbers +/− SEM of colony forming units (CFU) per well at 1 h post-infection. Each point is the mean of at least three separate experiments. B. Comparison of early or late induced autophagy by rapamycin treatment on the ability of AIEC LF82 bacteria to survive and/or to replicate intracellularly. Rapamycin was added during the 3 h infection, or at 1, 6 or 20 h post-infection. Results are expressed as the number of intracellular bacteria in treated cells relative to that obtained in untreated cells, taken as 100%. C. Confocal microscopic examinations of rapamycin treatment of Hela cells infected with AIEC LF82-GFP at 1 h and 20 h post-infection labelled with LC3 (red) and for LAMP-1 (purple). D. Effects of induced autophagy in HeLa cells by starvation or by LPS (100 ng/mL) and IFN-γ(1000U/mL) stimulations on the numbers of intracellular AIEC LF82 bacteria. Results are expressed as the number of intracellular bacteria in treated cells relative to that obtained in untreated cells, taken as 100%. E. Expression of the Atg5K130R dominant negative in HeLa cells leads to increased numbers of intracellular AIEC LF82 bacteria. HeLa cells transfected with plasmid pEGFP-Atg5 encoding wild-type Atg5 (black bars) or with pEGFP-Atg5K130R encoding the Atg5K130R dominant negative (white bars) were infected for 3 h. Results are expressed as mean numbers +/− SEM of colony forming units (CFU) per well. Each point is the mean of at least three separate experiments.
Intracellular AIEC LF82 bacteria replicate within cells having impaired ATG16L1 expression
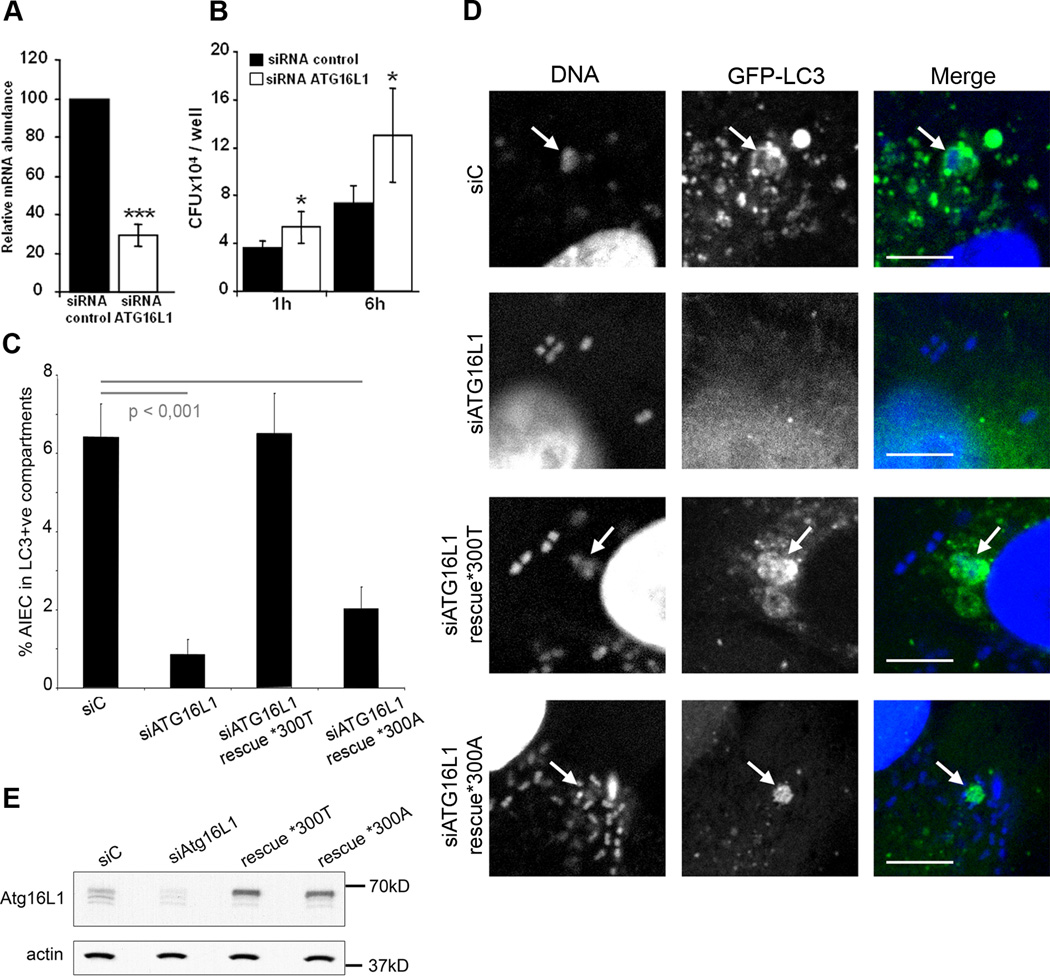
Since a coding polymorphism in the autophagy gene ATG16L1 was identified as a risk factor in CD, we assessed the effect of endogenous ATG16L1 knockdown in HeLa cells by specific siRNA. The efficiency of knockdown was evaluated by real time RT-PCR at 48 h post-transfection. Compared to control siRNA (siC), ATG16L1 siRNA (siATG16L1) yielded a 71% reduction in ATG16L1 transcripts (Fig 6A). Following a reduction in ATG16L1 expression, we observed significant (p<0.04) increases in the numbers of intracellular AIEC LF82 bacteria at 1 h and 6 h post-infection (Fig 6B). To confirm these results we performed a quantitative microscopic analysis to determine the percentage of LC3 positive vacuoles containing intracellular AIEC bacteria. In LC3-GFP expressing HeLa cells transfected with control siRNA a percentage of 6.4 (+/− 0.9) of internalised AIEC were observed within LC3 positive vacuoles after three hours of infection (Fig 6C and D). This was reduced to 0.9% (+/− 0.4%) in cells treated with siRNA targeting ATG16L1. Rescue of ATG16L1 expression using constructs containing synonymous base changes resulted in complete rescue in the case of the *300T variant with 6.5% (+/− 1%) of AIEC within LC3-positive compartments. However, the Crohn’s disease-associated allele *300A was unable to mediate full phenotypic rescue, despite equal levels of protein expression (Fig 6E), with only 2% (+/− 0.6%) of AIEC within LC3 positive vacuoles. Thus, as in the Salmonella Typhimurium model, the Crohn’s disease-associated ATG16L1 variant is unable to mediate fully effective antibacterial autophagy (Kuballa et al., 2008).
Figure 6. Intracellular AIEC LF82 bacteria replicate within cells impaired in ATG16L1 expression.
The siRNA experiments against endogeneous ATG16L1 transcripts were performed in HeLa cells transfected with ATG16L1 specific siRNA oligonucleotides. A. Endogenous ATG16L1 mRNA knockdown by specific siRNA in HeLa cells after 48 h of transfection was assessed by real-time quantitative RT-PCR normalized to GAPDH, compared with control duplexes. RT-PCR was performed in triplicate. Results represent independent experiments. B. ATG16L1 knockdown increases the ability of the AIEC LF82 bacteria to replicate intracellularly. HeLa cells transfected with control (black bar) or ATG16L1 (white bar) -directed siRNA oligos were infected for 3 h. Results are expressed as mean numbers +/− SEM of colony forming units (CFU) per well. Each point is the mean of at least three separate experiments. C. ATG16L1 knockdown and allele-specific rescue reveals compromised autophagy of AIEC mediated by the CD-associated *300A allele. HeLa-GFP-LC3 cells were transfected with control (siC) or ATG16L1-targeting siRNA duplexes, along with empty plasmid (siATG16L1), or allele-specific rescue constructs prior to infection with AIEC. After three hours of infection, cells were fixed and stained for microscopic analysis. Data shown represent means +/− SEM of three independent experiments, counting 50 cells per experiment. Significance was determined using two-tailed Student’s T-tests with Bonferroni correction for multiple comparisons. D. Confocal microscopic examination of colocalization of intracellular AIEC LF82 bacteria with GFP-LC3 in HeLa-GFP-LC3 cells transfected with control or ATG16L1-targeting siRNA duplexes, along with empty plasmid, or allele-specific rescue constructs prior to infection with AIEC. E. ATG16L1 knockdown and rescue results in similar levels of allele-specific expression. Expression of both rescue constructs was equal, as determined by Western blotting with ATG16L1-specific antibodies, using actin as a loading control.
IRGM, a CD-associated genetic marker, controls the fate of intracellular AIEC LF82 bacteria
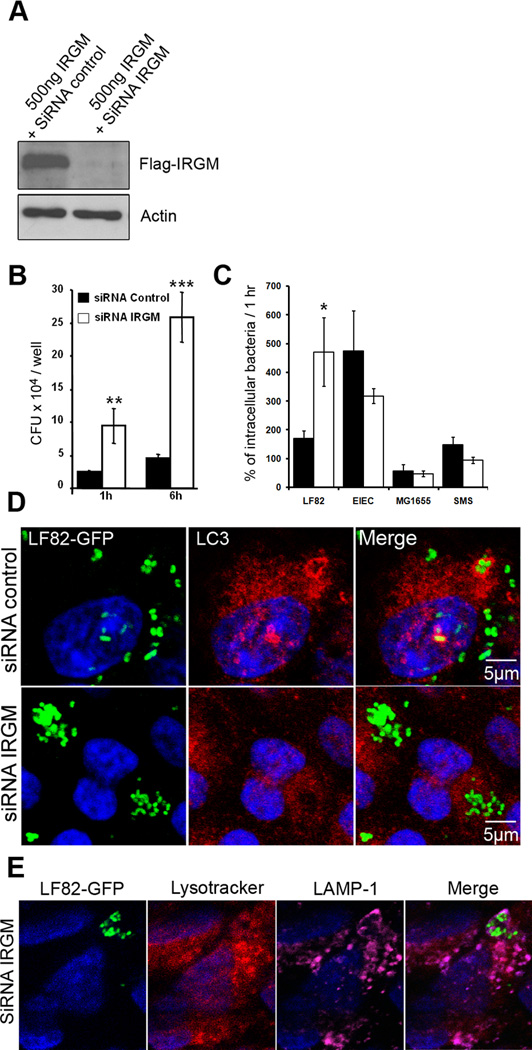
The human IRGM gene product stimulates the early stages of autophagy, and we therefore performed siRNA experiments against endogenous IRGM transcripts in HeLa cells, which mainly express the CD protective C allele (Huett et al., 2009). Using co-transfection of a Flag-tagged IRGM expression plasmid and siRNA oligonucleotide, we verified specific knockdown of IRGM in HeLa cells (Fig 7A). In HeLa cells treated with IRGM siRNA the numbers of intracellular LF82 bacteria were significantly higher than those in control siRNA treated cells (Fig 7B). At 6 h post-infection, a large difference in intracellular bacterial counts was measured, with 258,000 ± 38,056 CFU per well retrieved in IRGM siRNA treated cells and only 44,733 ± 7,130 CFU per well in control siRNA treated cells. The comparison of the behaviour of AIEC strain LF82 with that of the diarrheagenic enteroinvasive E. coli strain E12860/0, the non pathogenic E. coli strains K-12 MG1655 and the environmental strain SMS 3.5 indicated that IRGM-mediated autophagy only controlled the bacterial replication of the CD-associated LF82 bacteria (Fig 7C). To investigate whether IRGM siRNA treatment modified the intracellular niche of AIEC LF82 bacteria, we examined infected cells at 6 h post-infection using GFP-expressing LF82, and anti-LC3 antibodies. In control siRNA treated cells, most of the intracellular bacteria were seen as small clusters enclosed in LC3 positive vacuoles. When cells were treated with specific siRNA against IRGM transcripts, numerous AIEC LF82 bacteria were found as large clusters of bacteria in LAMP-1-positive phagosomes, negative for LC3 and without any accumulation of the lysotracker (Fig 7D and E). This suggests that IRGM, and hence autophagy, plays an important role in restricting AIEC LF82 replication in epithelial cells and that decreased levels of IRGM modified phagosome maturation, that lead to the formation of large intracellular microcolonies, which were not observed in atg5−/− MEFs or in ATG16L1 depleted HeLa cells.
Figure 7. IRGM, a CD-associated genetic marker, controls the number of intracellular AIEC LF82 bacteria within epithelial cells.
The siRNA experiments against endogeneous IRGM transcripts were performed in HeLa cells cotransfected with the Flag-tagged IRGM expression plasmid pCMV-3xFlag and with IRGM specific siRNA oligonucleotides. A. Immunoblot using anti-Flag antibody to analyze IRGM expression. B. IRGM knockdown increases the ability of the AIEC LF82 bacteria to replicate intracellularly. HeLa cells transfected with control (black bar) or IRGM (white bar) -directed siRNA oligos were infected for 3 h. Results are expressed as mean numbers +/− SEM of colony forming units (CFU) per well. Each point is the mean of at least three separate experiments. C. IRGM knockdown does not modify the survival/replication of non pathogenic E. coli. Results are expressed as the number of intracellular bacteria at 6 h post-infection relative to that obtained at 1 h post-infection, taken as 100%. D. Confocal microscopic examination reveals an increased number of intracellular AIEC LF82 bacteria in IRGM knockdown cells. Control siRNA-treated (siControl) and IRGM siRNA-treated (siRNA IRGM) cells were infected with AIEC LF82-GFP bacteria and imaged by confocal microscopy. Control cells show intracellular GFP-bacteria (green) as small clusters enclosed in LC3 positive (red) vacuoles. E. Confocal microscopic examinations of IRGM siRNA-treated Hela cells infected with AIEC LF82 GFP at 6 h post-infection labelled with lysotracker (red) and for LAMP-1 (purple) to visualize the vacuoles.
Discussion
IBD involves a dysregulation of the normal co-evolved homeostatic relationship between the gut microbiota and the host immune system. Microorganisms, including pathogens and members of the indigenous microbiota, may participate in multiple roles, including initiation and/or propagation of the inflammatory processes. Recent studies have demonstrated that among the enteric microflora adherent-invasive E. coli (AIEC) strains are found more commonly in IBD patients than in control patients (Martinez-Medina et al., 2009; Baumgart et al., 2007; Eaves-Pyles et al., 2007; Sasaki et al., 2007; Darfeuille-Michaud et al., 2004; Martin et al., 2004). Genome-wide association studies have identified new susceptibility genes ATG16L1 and IRGM, which are involved in bacterial clearance and antigen processing via autophagy. As autophagy is an innate defence mechanism acting as a cell-autonomous system for elimination of intracellular pathogens, these findings lead weight to the notion that intracellular bacteria including AIEC might play a role in CD pathogenesis. Thus, the aim of the present study was to determine whether AIEC bacteria can take advantage of defects in autophagy to replicate within host epithelial cells, thereby establishing a link between genetic CD predisposition and the abnormal presence of AIEC colonizing ileal mucosa.
To test the hypothesis that ineffective autophagy may enable persistent intracellular survival, we compared AIEC infection of autophagy-deficient (Atg5−/−) versus wild-type mouse embryonic fibroblasts. We observed that autophagy can restrict the replication of AIEC LF82 intracellular bacteria and therefore constitutes an efficient host innate defense mechanism. We next observed that, as in S. Typhimurium infection (Birmingham et al., 2006), a subpopulation of intracellular AIEC bacteria is targeted by autophagy. Interestingly, when we examined the behavior of related E. coli strains, including non pathogenic, environmental, commensal or pathogenic ETEC, EPEC, DAEC, EIEC bacteria with regard to autophagy, we found that functional autophagy limited AIEC bacterial replication and that there was no significant differences in survival/replication of all the other E. coli strains tested between wt MEFs and atg5−/− MEFs. The findings obtained in experiments using MEFs were confirmed in human epithelial cell lines. Confocal analysis of specific autophagosomal marker LC3 immunostaining showed that a subset of AIEC LF82 bacteria were in a LC3 positive vacuole, as reported for S. Typhimurium (Birmingham et al., 2006). Moreover, transmission electron microscopy of AIEC LF82-infected Hep-2 and Intestine-407 cells revealed that these bacteria were sequestered as single bacteria in multilamellar membrane vacuoles, corresponding to autophagosomes (Klionsky et al., 2008). TEM or confocal images suggested that the bacteria were not able to replicate in a LC3 positive compartment since we only observed clusters of bacteria in LC3 negative spacious phagosomes or in autophagy deficient epithelial cells. Furthermore dominant negative of ATG5 enhanced bacterial replication whereas induction of autophagy attenuated replication. A fraction of intracellular AIEC is observed free in the cytosol or in damaged vacuoles and could represent the preferential target of autophagy machinery. Indeed, in addition to free bacteria in the cytosol like group A Streptococci that are handled by the autophagy process (Nakagawa et al., 2004), endocytic phagosome-damaging by bacteria represents a strong signalling node to induce autophagic response as very recently reported for Shigella flexneri (Dupont et al., 2009). However, in contrast to Shigella flexneri (Ogawa et al., 2005), AIEC bacteria did not subvert early autophagy. While AIEC bacteria at late time post-infection are mostly in vacuoles presenting features of mature phagolysosomes they are not targeted by the autophagy process. This finding is consistent with previous reports for S. Typhimurium and Streptococcus pyogenes (Birmingham et al., 2006; Nakagawa et al., 2004).
So far, the bacterial pathogens demonstrated to be controlled by autophagy exhibit significant adaptations to intracellular modes of survival and pathogenesis. Indeed, Shigella, Salmonella and AIEC have all been characterised as intracellular pathogens, in contrast to EPEC, EHEC, DAEC and ETEC whose lifestyles are primarily extracellular, despite their ability to enter cells in limited numbers. Also recent data indicates that both the bacteria and their vacuolar remnants are targeted by autophagy upon vacuolar lysis and bacterial escape (Dupont et al., 2009; Birmingham et al., 2007; Birmingham et al., 2006). Since we found that pathogens without such direct intracellular adaptations (EPEC, ETEC, DAEC) or commensal bacteria, do not trigger autophagy this raises the question of whether it is the specific intracellular replication strategies of pathogens that renders them susceptible to autophagy. Bacteria which attempt vacuolar escape appear susceptible to autophagy when effecting exit of their entry vacuoles, and even those which attempt to maintain the vacuolar niche can fail and succumb to autophagy. Intracellular bacteria have conflicting needs, the vacuole is a secure environment, but both nutrition and space are extremely limited. They must either escape and replicate in the cytoplasm, or permeabilise/grow the vacuole to provide sufficient nutrition for replication. However, these routes to growth all expose them to autophagy, through signals yet to be defined. Thus vacuolar exit must be either rapid and complete, such as seen in Shigella or Listeria (Cossart and Lecuit, 1998; High et al., 1992), or significant effort must be expended to safely grow the vacuole to allow slow, but continued replication as shown for Salmonella and AIEC (Brumell et al., 2001; Glasser et al., 2001). Both approaches result in sufficient replication to yield a viable intracellular pathogenic strategy, but both require specific and elaborate mechanisms to be effective.
The host also is subject to strict requirements, signalling to trigger autophagy must be both specific and rapid, to avoid unnecessary degradation of organelles and cytoplasm, yet restrict pathogen escape and replication. To this end, any defects in autophagy or upstream signalling are likely to yield impaired control of infections, especially at sites where innate immune surveillance is restricted due to high bacterial load, as in the gut. In this report we investigated the impact of expression alteration of the two autophagy-related genes identified as susceptibility genes for CD, ATG16L1 and IRGM. A functional knockdown of ATG16L1 by siRNA abrogates autophagy of AIEC strain LF82. This loss was fully rescued by expression of ATG16L1, but failed to be rescued by expression of the CD-associated ATG16L1 variant *300A. This result is similar to that observed in the S. Typhimurium model, where *300A is also less able to mediate anti-bacterial autophagy (Kuballa et al., 2008). This suggests that autophagy may be a general feature of the epithelial cell autophagic response to infection with an intracellular bacterium and that the CD-associated *300A ATG16L1 allele results in a diminished and ineffective autophagic response to intracellular pathogens. IRGM protein, which is highly expressed in response to infection, seems to be critical for clearance of intracellular bacteria such as Salmonella spp. by autophagy (Taylor, 2007). Mice rendered deficient in the homologue of IRGM gene have impaired ability to eliminate the intracellular pathogens Toxoplasma gondii, Listeria monocytogenes and S. Typhimurium (Henry et al., 2007; Collazo et al., 2001). In addition, experiments in murine macrophages showed that IRGM1-mediated autophagy generated large autolysosomal organelles as a mechanism for the elimination of intracellular Mycobacterium tuberculosis, and experiments in human macrophages reported that knockdown of IRGM leads to markedly prolonged survival of Mycobacterium tuberculosis (Singh et al., 2006). In the present study, we observed that when intracellular IRGM expression was reduced by specific siRNA there was a great increase in the number of AIEC LF82 intracellular bacteria, indicating a key role of IRGM in controlling intracellular AIEC replication. The absence of LC3 co-localization with bacteria was accompanied by huge clusters of intracellular AIEC LF82 bacteria mostly located in non acidic compartments. Such huge intracellular microcolonies were not observed in LF82-infected Atg5−/− MEFs or in ATG16L1 depleted HeLa cells, indicating that the role of IRGM may not be restricted to autophagy since this GTPase protein has been described to be associated to the membrane of the phagosomes. In accordance, defective bacterial killing in LRG-47 −/− (a murine ortholog of IRGM) macrophages due to impaired maturation of bacteria-containing phagosomes was reported (MacMicking et al., 2003).
ATG16L1 polymorphism T300A predisposes to ileal disease and polymorphisms of the IRGM locus might predispose to fistulizing behaviour in CD (Latiano et al., 2009; Prescott et al., 2007; Rioux et al., 2007). In parallel, AIEC are mainly associated with severe ileal CD, and high levels of antibodies against E. coli OmpC are present in patients with severe CD, characterized by small bowel involvement, frequent disease progression, longer disease duration, and greater need for intestinal surgery (Mow et al., 2004; Landers et al., 2002). Thus, the CD risk-conferring large upstream deletion at the IRGM locus and the ATG16L1 mutation observed in a CD patient population associated with the presence of pathogenic AIEC bacteria colonizing the intestinal mucosa may lead to functional alterations in the ability of intestinal cells to initiate and sustain autophagy to control AIEC replication. Thus, AIEC infection in patients with polymorphisms in autophagy genes may have a significant impact on the outcome of intestinal inflammation. From the data presented here, we suggest that it would be of interest to investigate the presence of mucosa-associated AIEC bacteria and ATG16L1 or IRGM polymorphism in CD patients in order to define CD patients at high risk of developing structuring and fistulizing CD.
Experimental procedure
Bacterial strains and cell culture
E. coli strain LF82 was isolated from a chronic ileal lesion of a patient with CD. Non pathogenic E. coli K-12 strain MG1655, the environmental E. coli strain SMS 3.5, the commensal E. coli strain HS, enteropathogenic E. coli (EPEC) strain E2348/69 (D), the diffusely adhering E. coli (DAEC) strain C1845, the enterotoxigenic E. coli (ETEC) strain H10407, and the enteroinvasive E. coli (EIEC) strain E12860/0 were tested in parallel. All the strains were grown in Luria Bertani (LB) broth or plated on LB agar. Human epihelial cell lines HeLa, Hep-2 and Intestine-407 were obtained from ATCC and cultured as described elsewhere (Boudeau et al., 1999). Stable HeLa-GFP-LC3 cells generated by lentiviral transduction were used for quantitative microscopy (Kuballa et al., 2008). Mouse embryonic fibroblasts (MEF) deficient in atg5 and the corresponding wild type cells, kindly provided by Dr. Naboru Mizuschima, were maintained as previously described (Nakagawa et al., 2004).
Antibodies and reagents
Rabbit polyclonal anti-LC3B, anti-Flag, and anti-Actin for Western blot experiments were purchased from Sigma, and anti-ATG16L1 from Affinity Bioreagents. Rabbit polyclonal anti-LC3 for immunofluorescence analysis was purchased from MBL. Mouse monoclonal anti-LAMP-1 human and Rat monoclonal anti-LAMP-1 mouse were from DSBH Iowa. Rabbit antibody raised against E. Coli LPS O83 was generously provided by Lothar Beutin (Department of biological safety, Robert Koch Institut, Berlin, Germany). TRITC-labeled phalloidin for vizualization of actin cytoskeleton in MEF and Hoechst 33342 for labelling nuclei were purchased from Sigma, Lysotracker probe DND-99 was purchased from Invitrogen. Rapamycin (LC laboratories) and Hank’s balanced salt solution (Sigma), LPS (Calbiochem) and Interferon γ (R&D) were used as autophagy inducers.
Invasion assay
Bacterial invasion of human epithelial cells and MEFs was performed using gentamycin protection assay (Boudeau et al., 1999). Epithelial cell monolayers were infected for 3 h and MEFs for 1 h at a multiplicity of infection (MOI) of 10 bacteria per cell. The number of intracellular bacteria was determined by counting the number of colony forming units (CFU). AIEC within autophagosomes in stable HeLa-GFP-LC3 cells was counted as previously described (Kuballa et al., 2008). Random fields were counted with data from 50 cells collected for each condition.
Differential permeabilization with digitonin
This assay was performed as previously described by Checroun et al. 2006 PNAS). Briefly, infected Hela cells or MEFs were washed 2 x with KHM buffer (110 mM potassium acetate, 20 mM Hepes and 2 mM MgCl2), incubated with KHM containing 25µg/mL digitonin (Sigma) for 1 min at room temperature and then washed with KHM buffer. Cells were then incubated with O83 antibody diluted in KHM buffer for 12 minutes at 37°C, washed with PBS and fixed with 4% PFA.
Autophagy induction
Autophagy was induced by using rapamycin treatment, with dose ranging from 20µg to 80µg/mL, during 3 h. Autophagy was also induced by incubation of cells monolayer during 2 h in HBSS minimum medium before infection. For LPS (100 ng/mL) and IFN-γ (1000U/mL) stimulations, cells were treated for 48 h before infection.
Plasmids and transfection
The pEGFP-Atg5 or pEGFP-Atg5K130R expression vectors were a kind gift from T. Yoshimori (Osaka University, Japan). Plasmid pCMV-3xFlag-IRGM and ATG16L1 rescue constructs have been previously described (Kuballa et al., 2008; McCarroll et al., 2008). HeLa cells were transfected with plasmids and Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol.
SiRNA directed against ATG16L1 or IRGM
siRNA experiments were performed using stealth RNAi (Invitrogen). The sequences used were as previously described (McCarroll et al., 2008; Rioux et al., 2007). Transfections were performed using Lipofectamine 2000 (Invitrogen). To evaluate knock-down efficiency, total mRNA from RNAi-treated HeLa cells was isolated using TRIzol reagent (Invitrogen) and analysed by real-time quantitative RT-PCR assays. Analysis of protein extract from RNAi-treated HeLa cells by Western blot confirmed knock-down.
Immunoblot analysis
Whole-cell protein extracts were prepared by adding lysis buffer (2% Triton X-100, 50mM Tris-Hcl, 150mM NaCl, 1mM EDTA). Proteins were separated on SDS/15% PAGE gels, transferred to polyvinylidene difluoride membrane, blocked 2 h in tris-buffered saline solution containing 2% BSA, probed overnight with primary antibodies, and 2 h with secondary HRP-coupled antibodies. Anti-actin was used to normalize protein quantity. After membrane revelation using the ECL detection kit (Amersham), quantification was done with Phoretix 1D software.
Electron Microscopy
Cross sections of Hep2 or Intestine-407 were prepared as previously described (Boudeau et al., 1999). Ultrathin sections stained with uranyl acetate and lead citrate were then processed for electron microscopy with a Hitachi H-7650 transmission electron microscope at 80 kV.
Fluorescence microscopy
After bacterial infection HeLa cells or MEFs were fixed with 3% paraformaldehyde and immunostained overnight at 4°C, with the indicated specific primary antibodies. A 1 h incubation with secondary antibodies was done. The slides were examined with a Zeiss LSM 510 Meta confocal microscope. For quantitative microscopy stable HeLa-GFP-LC3 cells were transfected with 20pmol of siRNA duplex and 500ng of appropriate plasmid using Lipofectamine 2000. Following a media change after 6 hours, cells were allowed to grow for 48 h and subsequently infected or lysed for Western blotting.
Statistical analysis
Experiments were independently carried out at least three times and one representative data set out of the three independent experiments was presented where appropriate. The results were evaluated for statistical significance by student t-test. Error bars were marked as the standard deviation (S.D.) of the mean. p values less than 0.05 were regarded as significant.
Supplementary Material
Acknowledgments
This study was supported by the Ministère de la Recherche et de la Technologie (JE2526), INRA (USC-2018) and by grants from the Association F. Aupetit (AFA), and European Commission through FP7 IBDase project. We thank Pr. Noboru Mizushima (Department of Physiology and Cell Biology, Tokyo Medical and Dental University, Tokyo 113–8519, Japan) for providing mouse embryonic fibroblasts (MEFs) deficient in Atg5, Pr Tamotsu Yoshimori (Department of Cellular Regulation, Research Institute for Microbial Diseases, Osaka University, Suita, Japan) who kindly provided GFP-LC3, GFP-Atg5 and mAtg5 plamids. We also thank Christelle Blavignac and Claire Sczepaniak (CICS, Université d’Auvergne, France) for technical assistance with electron microscopy and the CICS platform for confocal microscopy.
Nonstandard abbreviations used
- AIEC
Adherent-Invasive Escherichia coli
- CD
Crohn’s Disease
- CEACAM
carcinoembryonic antigen-related cell adhesion molecule
- IRGM
Immunity-related GTPase family M
- ATG16L1
Autophagy-related like 1
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Author contributions
Conceived and designed experiments: PL, ALG, RX, ADM. Performed the experiments: PL, AH. Analyzed the data: PL, ALG, AH, RX, ADM. Wrote the paper: PL, ALG, RX, ADM.
References
- Barnich N, Carvalho FA, Glasser AL, Darcha C, Jantscheff P, Allez M, et al. CEACAM6 acts as a receptor for adherent-invasive E. coli, supporting ileal mucosa colonization in Crohn disease. J Clin Invest. 2007;117:1566–1574. doi: 10.1172/JCI30504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgart M, Dogan B, Rishniw M, Weitzman G, Bosworth B, Yantiss R, et al. Culture independent analysis of ileal mucosa reveals a selective increase in invasive Escherichia coli of novel phylogeny relative to depletion of Clostridiales in Crohn’s disease involving the ileum. Isme J. 2007;1:403–418. doi: 10.1038/ismej.2007.52. [DOI] [PubMed] [Google Scholar]
- Birmingham CL, Smith AC, Bakowski MA, Yoshimori T, Brumell JH. Autophagy controls Salmonella infection in response to damage to the Salmonella-containing vacuole. J Biol Chem. 2006;281:11374–11383. doi: 10.1074/jbc.M509157200. [DOI] [PubMed] [Google Scholar]
- Birmingham CL, Canadien V, Gouin E, Troy EB, Yoshimori T, Cossart P, et al. Listeria monocytogenes evades killing by autophagy during colonization of host cells. Autophagy. 2007;3:442–451. doi: 10.4161/auto.4450. [DOI] [PubMed] [Google Scholar]
- Boudeau J, Barnich N, Darfeuille-Michaud A. Type 1 pili-mediated adherence of Escherichia coli strain LF82 isolated from Crohn’s disease is involved in bacterial invasion of intestinal epithelial cells. Mol Microbiol. 2001;39:1272–1284. doi: 10.1111/j.1365-2958.2001.02315.x. [DOI] [PubMed] [Google Scholar]
- Boudeau J, Glasser AL, Masseret E, Joly B, Darfeuille-Michaud A. Invasive ability of an Escherichia coli strain isolated from the ileal mucosa of a patient with Crohn’s disease. Infect Immun. 1999;67:4499–4509. doi: 10.1128/iai.67.9.4499-4509.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brumell JH, Tang P, Mills SD, Finlay BB. Characterization of Salmonella-induced filaments (Sifs) reveals a delayed interaction between Salmonella-containing vacuoles and late endocytic compartments. Traffic. 2001;2:643–653. doi: 10.1034/j.1600-0854.2001.20907.x. [DOI] [PubMed] [Google Scholar]
- Collazo CM, Yap GS, Sempowski GD, Lusby KC, Tessarollo L, Woude GF, et al. Inactivation of LRG-47 and IRG-47 reveals a family of interferon gamma-inducible genes with essential, pathogen-specific roles in resistance to infection. J Exp Med. 2001;194:181–188. doi: 10.1084/jem.194.2.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conte MP, Schippa S, Zamboni I, Penta M, Chiarini F, Seganti L, et al. Gut-associated bacterial microbiota in paediatric patients with inflammatory bowel disease. Gut. 2006;55:1760–1767. doi: 10.1136/gut.2005.078824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cossart P, Lecuit M. Interactions of Listeria monocytogenes with mammalian cells during entry and actin-based movement: bacterial factors, cellular ligands and signaling. Embo J. 1998;17:3797–3806. doi: 10.1093/emboj/17.14.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darfeuille-Michaud A, Neut C, Barnich N, Lederman E, Di Martino P, Desreumaux P, et al. Presence of adherent Escherichia coli strains in ileal mucosa of patients with Crohn’s disease. Gastroenterology. 1998;115:1405–1413. doi: 10.1016/s0016-5085(98)70019-8. [DOI] [PubMed] [Google Scholar]
- Darfeuille-Michaud A, Boudeau J, Bulois P, Neut C, Glasser AL, Barnich N, et al. High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn’s disease. Gastroenterology. 2004;127:412–421. doi: 10.1053/j.gastro.2004.04.061. [DOI] [PubMed] [Google Scholar]
- Duerr RH, Taylor KD, Brant SR, Rioux JD, Silverberg MS, Daly MJ, et al. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science. 2006;314:1461–1463. doi: 10.1126/science.1135245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont N, Lacas-Gervais S, Bertout J, Paz I, Freche B, Van Nhieu GT, et al. Shigella phagocytic vacuolar membrane remnants participate in the cellular response to pathogen invasion and are regulated by autophagy. Cell Host Microbe. 2009;6:137–149. doi: 10.1016/j.chom.2009.07.005. [DOI] [PubMed] [Google Scholar]
- Eaves-Pyles T, Allen CA, Taormina J, Swidsinski A, Tutt CB, Eric Jezek G, et al. Escherichia coli isolated from a Crohn’s disease patient adheres, invades, and induces inflammatory responses in polarized intestinal epithelial cells. Int J Med Microbiol. 2007 doi: 10.1016/j.ijmm.2007.05.011. [DOI] [PubMed] [Google Scholar]
- Glasser AL, Boudeau J, Barnich N, Perruchot MH, Colombel JF, Darfeuille-Michaud A. Adherent invasive Escherichia coli strains from patients with Crohn’s disease survive and replicate within macrophages without inducing host cell death. Infect Immun. 2001;69:5529–5537. doi: 10.1128/IAI.69.9.5529-5537.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez MG, Master SS, Singh SB, Taylor GA, Colombo MI, Deretic V. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell. 2004;119:753–766. doi: 10.1016/j.cell.2004.11.038. [DOI] [PubMed] [Google Scholar]
- Hampe J, Franke A, Rosenstiel P, Till A, Teuber M, Huse K, et al. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat Genet. 2007;39:207–211. doi: 10.1038/ng1954. [DOI] [PubMed] [Google Scholar]
- Henry SC, Daniell X, Indaram M, Whitesides JF, Sempowski GD, Howell D, et al. Impaired macrophage function underscores susceptibility to Salmonella in mice lacking Irgm1 (LRG-47) J Immunol. 2007;179:6963–6972. doi: 10.4049/jimmunol.179.10.6963. [DOI] [PubMed] [Google Scholar]
- High N, Mounier J, Prevost MC, Sansonetti PJ. IpaB of Shigella flexneri causes entry into epithelial cells and escape from the phagocytic vacuole. Embo J. 1992;11:1991–1999. doi: 10.1002/j.1460-2075.1992.tb05253.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huett A, McCarroll SA, Daly MJ, Xavier RJ. On the level: IRGM gene function is all about expression. Autophagy. 2009;5:96–99. doi: 10.4161/auto.5.1.7263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugot JP, Chamaillard M, Zouali H, Lesage S, Cezard JP, Belaiche J, et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature. 2001;411:599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- Kaper JB, Nataro JP, Mobley HL. Pathogenic Escherichia coli . Nat Rev Microbiol. 2004;2:123–140. doi: 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- Klionsky DJ, Abeliovich H, Agostinis P, Agrawal DK, Aliev G, Askew DS, et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4:151–175. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotlowski R, Bernstein CN, Sepehri S, Krause DO. High prevalence of Escherichia coli belonging to the B2 +D phylogenetic group in inflammatory bowel disease. Gut. 2006 doi: 10.1136/gut.2006.099796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuballa P, Huett A, Rioux JD, Daly MJ, Xavier RJ. Impaired autophagy of an intracellular pathogen induced by a Crohn’s disease associated ATG16L1 variant. PLoS ONE. 2008;3:e3391. doi: 10.1371/journal.pone.0003391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landers CJ, Cohavy O, Misra R, Yang H, Lin YC, Braun J, Targan SR. Selected loss of tolerance evidenced by Crohn’s disease-associated immune responses to auto- and microbial antigens. Gastroenterology. 2002;123:689–699. doi: 10.1053/gast.2002.35379. [DOI] [PubMed] [Google Scholar]
- Latiano A, Palmieri O, Cucchiara S, Castro M, D’Inca R, Guariso G, et al. Polymorphism of the IRGM gene might predispose to fistulizing behavior in Crohn’s disease. Am J Gastroenterol. 2009;104:110–116. doi: 10.1038/ajg.2008.3. [DOI] [PubMed] [Google Scholar]
- MacMicking JD, Taylor GA, McKinney JD. Immune control of tuberculosis by IFN-gamma-inducible LRG-47. Science. 2003;302:654–659. doi: 10.1126/science.1088063. [DOI] [PubMed] [Google Scholar]
- Martin HM, Campbell BJ, Hart CA, Mpofu C, Nayar M, Singh R, et al. Enhanced Escherichia coli adherence and invasion in Crohn’s disease and colon cancer. Gastroenterology. 2004;127:80–93. doi: 10.1053/j.gastro.2004.03.054. [DOI] [PubMed] [Google Scholar]
- Martinez-Medina M, Aldeguer X, Lopez-Siles M, Gonzalez-Huix F, Lopez-Oliu C, Dahbi G, et al. Molecular diversity of Escherichia coli in the human gut: New ecological evidence supporting the role of adherent-invasive E. coli (AIEC) in Crohn’s disease. Inflamm Bowel Dis. 2009 doi: 10.1002/ibd.20860. [DOI] [PubMed] [Google Scholar]
- Massey DC, Parkes M. Genome-wide association scanning highlights two autophagy genes, ATG16L1 and IRGM, as being significantly associated with Crohn’s disease. Autophagy. 2007;3:649–651. doi: 10.4161/auto.5075. [DOI] [PubMed] [Google Scholar]
- McCarroll SA, Huett A, Kuballa P, Chilewski SD, Landry A, Goyette P, et al. Deletion polymorphism upstream of IRGM associated with altered IRGM expression and Crohn’s disease. Nat Genet. 2008;40:1107–1112. doi: 10.1038/ng.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N. Autophagy: process and function. Genes Dev. 2007;21:2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- Mow WS, Vasiliauskas EA, Lin YC, Fleshner PR, Papadakis KA, Taylor KD, et al. Association of antibody responses to microbial antigens and complications of small bowel Crohn’s disease. Gastroenterology. 2004;126:414–424. doi: 10.1053/j.gastro.2003.11.015. [DOI] [PubMed] [Google Scholar]
- Nakagawa I, Amano A, Mizushima N, Yamamoto A, Yamaguchi H, Kamimoto T, et al. Autophagy defends cells against invading group A Streptococcus . Science. 2004;306:1037–1040. doi: 10.1126/science.1103966. [DOI] [PubMed] [Google Scholar]
- Ogawa M, Yoshimori T, Suzuki T, Sagara H, Mizushima N, Sasakawa C. Escape of intracellular Shigella from autophagy. Science. 2005;307:727–731. doi: 10.1126/science.1106036. [DOI] [PubMed] [Google Scholar]
- Ogura Y, Bonen DK, Inohara N, Nicolae DL, Chen FF, Ramos R, et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn’s disease. Nature. 2001;411:603–606. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- Parkes M, Barrett JC, Prescott NJ, Tremelling M, Anderson CA, Fisher SA, et al. Sequence variants in the autophagy gene IRGM and multiple other replicating loci contribute to Crohn’s disease susceptibility. Nat Genet. 2007;39:830–832. doi: 10.1038/ng2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott NJ, Fisher SA, Franke A, Hampe J, Onnie CM, Soars D, et al. A nonsynonymous SNP in ATG16L1 predisposes to ileal Crohn’s disease and is independent of CARD15 and IBD5. Gastroenterology. 2007;132:1665–1671. doi: 10.1053/j.gastro.2007.03.034. [DOI] [PubMed] [Google Scholar]
- Rioux JD, Xavier RJ, Taylor KD, Silverberg MS, Goyette P, Huett A, et al. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat Genet. 2007;39:596–604. doi: 10.1038/ng2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutgeerts P, Goboes K, Peeters M, Hiele M, Penninckx F, Aerts R, et al. Effect of faecal stream diversion on recurrence of Crohn’s disease in the neoterminal ileum. Lancet. 1991;338:771–774. doi: 10.1016/0140-6736(91)90663-a. [DOI] [PubMed] [Google Scholar]
- Sartor RB. Microbial influences in inflammatory bowel diseases. Gastroenterology. 2008;134:577–594. doi: 10.1053/j.gastro.2007.11.059. [DOI] [PubMed] [Google Scholar]
- Sasaki M, Sitaraman SV, Babbin BA, Gerner-Smidt P, Ribot EM, Garrett N, et al. Invasive Escherichia coli are a feature of Crohn’s disease. Lab Invest. 2007;87:1042–1054. doi: 10.1038/labinvest.3700661. [DOI] [PubMed] [Google Scholar]
- Singh SB, Davis AS, Taylor GA, Deretic V. Human IRGM induces autophagy to eliminate intracellular mycobacteria. Science. 2006;313:1438–1441. doi: 10.1126/science.1129577. [DOI] [PubMed] [Google Scholar]
- Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermudez-Humaran LG, Gratadoux JJ, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci U S A. 2008;105:16731–16736. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strober W, Fuss I, Mannon P. The fundamental basis of inflammatory bowel disease. J Clin Invest. 2007;117:514–521. doi: 10.1172/JCI30587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swidsinski A, Ladhoff A, Pernthaler A, Swidsinski S, Loening-Baucke V, Ortner M, et al. Mucosal flora in inflammatory bowel disease. Gastroenterology. 2002;122:44–54. doi: 10.1053/gast.2002.30294. [DOI] [PubMed] [Google Scholar]
- Taylor GA. IRG proteins: key mediators of interferon-regulated host resistance to intracellular pathogens. Cell Microbiol. 2007;9:1099–1107. doi: 10.1111/j.1462-5822.2007.00916.x. [DOI] [PubMed] [Google Scholar]
- Wellcome. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xavier RJ, Podolsky DK. Unravelling the pathogenesis of inflammatory bowel disease. Nature. 2007;448:427–434. doi: 10.1038/nature06005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.