Abstract
All microorganisms are exposed to periodic stresses that inhibit growth. Many bacteria and fungi weather these periods by entering a hardy, non-replicating state, often termed quiescence or dormancy. When this occurs during an infection, the resulting slowly-growing pathogen is able to tolerate both immune insults and prolonged antibiotic exposure. While the stresses encountered in a free-living environment may differ from those imposed by host immunity, these growth-limiting conditions impose common pressures and many of the corresponding microbial responses appear to be universal. In this review, we discuss the common features of these growth-limited states, which suggest new approaches for treating chronic infections such as tuberculosis.
A defining feature of Mycobacterium tuberculosis, the causative agent of tuberculosis, is its slow growth. The maximal doubling time of this bacterium is approximately twenty hours, and is significantly slower when exposed to stresses such as those encountered in the host. Indeed, the bacterial population found in chronically-infected animals replicates only once every 100 hours or more (Gill et al., 2009; Muñoz-Elías et al., 2005), and subpopulations of bacteria are thought to cease growth entirely for significant periods. The importance of this relatively quiescent behavior is difficult to overstate, as it likely underlies the chronicity of the infection as well as the requirement for extended antibiotic therapy.
Dormancy, latency, and persistence are conceptually related terms used to describe the propensity of M. tuberculosis to arrest its growth in response to host-imposed stress. Because this behavior is very different from well-studied model organisms or agents of acute infection, it is sometimes considered an unusual selective adaptation specific to the pathogenic mycobacteria. While this trait is likely adaptive, it is by no means unusual. In fact, slow to negligible replication is the norm in the microbial world where organisms often inhabit environments that are incompatible with rapid growth. In this review, we will consider mycobacterial dormancy in this broader ecological context.
Three strategies to weather the storm
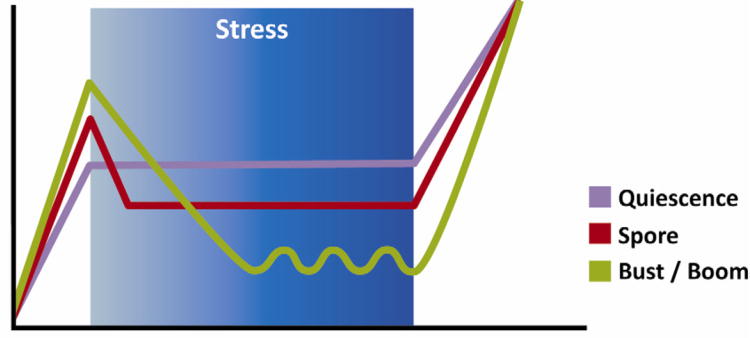
All microbes are subjected to changing environments, and the basic requirements for growth (carbon, nitrogen, phosphorus, water, etcetera) are not always available. The evolutionary success of virtually all microbial species requires the ability to weather these periods, and the spectrum of survival strategies used by different microbial species has been studied for decades (Steinhaus and Birkeland, 1939). In general, these strategies can be described as variations of three general themes (Figure 1).
Figure 1. Strategies to overcome growth-limiting stress.
All microorganisms encounter periods during which growth is impossible. Three fundamental themes describe the strategies used to weather these periods. “Bust and boom” describes a strategy that relies on the dynamic persistence of a small subpopulation, “sporulation” is defined by the production of metabolically-inactive spores, and “quiescence” describes a metabolically-active non-replicating cell that is resistant to many environmental insults. These strategies differ in several important respects including the population density of persistent organisms, the sensitivity of these cells to toxins and antibiotics, and the differential dependence on rapid growth to repopulate the niche.
Bust and boom
The physiology of organisms that evolved in consistently nutrient-rich environments, such as the bacteria Escherichia coli, are tuned to maximize growth rate (Neidhardt, 1999). Under nutrient-replete conditions in which bacterial metabolism is often studied, these organisms maximize their growth at the expense of economy by using relatively inefficient fermentative pathways to generate energy (Wolfe, 2005). Upon nutrient exhaustion the majority of these bacterial populations die, leaving a few viable organisms that subsist on the corpses of their siblings. Slow growth and cell death is balanced during this period (Finkel, 2006). When environmental conditions become more favorable, the few survivors resume growth. The ability to replicate rapidly is likely to be an essential component of this strategy, as these organisms must outcompete neighboring microbes to consume the newly introduced nutrients.
Cellular quiescence
A distinct strategy for surviving periods of growth limiting stress appears to be favored by both M. tuberculosis (Betts et al., 2002; Mitchison and Coates, 2004; Wayne, 1976) and many environmental bacteria (Lewis and Gattie, 1991). When these organisms are exposed to growth-limiting stress, the bulk of the bacterial population slows or arrests its growth and can persist in a viable non-replicating state for months or even years (Corper and Cohn, 1933). These “quiescent” cells can be differentiated from truly dormant spore-like forms because they display nominal metabolic capacity, maintain their membrane potential, and do not undergo obvious morphological differentiation (Gengenbacher et al., 2010; Rao et al., 2008). This strategy allows the viable bacterial population size to be maintained throughout the period of stress (Jones and Lennon, 2010), relieving the emphasis for rapid growth seen in the bust-and-boom model.
True dormancy
Sporulation is the purest form of microbial dormancy. When exposed to growth-restricting stress, some bacteria undergo an asymmetric cell division to produce a hardy metabolically-inactive daughter cell called a spore (Stragier and Losick, 1996). Upon exposure to favorable environmental conditions, a fraction of spores germinate and initiate rapid growth to reestablish the population. This strategy could be viewed as a combination of the first two. The spore, while fundamentally distinct, shares many structural and biochemical features with quiescent cells, which promote long-term survival. Upon germination, however, rapid growth may be advantageous to repopulate the niche. Indeed, the ten minute replication time of some spore-forming species of the clostridia bacteria are among the fastest known (Kreidl et al., 2002).
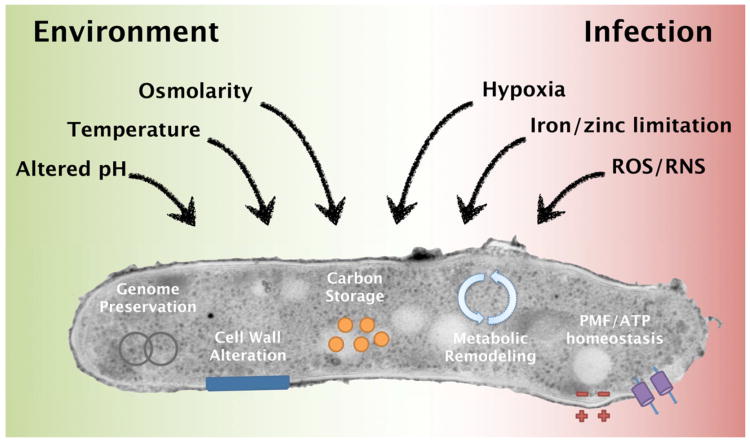
Historically, the strategies at either end of this spectrum have been most heavily studied. This is due to the experimental tractability of rapidly-growing organisms, not because these strategies are more common or important. Indeed, it has been estimated that 60% of the microbial biomass on earth exists in a quiescent state (Cole, 1999; Lewis and Gattie, 1991). Despite its ubiquity, we still know relatively little about the regulatory mechanisms and physiological changes that define microbial quiescence. While these cellular adaptations are not exactly the same for all organisms or under all conditions, common themes can be defined (Figure 2). In this review, we will consider the general adaptations that are required for quiescence in diverse microorganisms and discuss how these insights might be used to develop more effective therapies for chronic infections such as tuberculosis.
Figure 2. Common themes in microbial quiescence.
Growth-arrest can be induced by many stimuli and can have a variety of consequences on the cell. Common growth limiting stresses encountered by environmental microbes and pathogens are shown. With a few notable exceptions, most of the growth-limiting stresses encountered in these environments are similar. Some responses to these insults are linked to a particular stress. For example, specific DNA repair pathways are necessary to resist oxidative and nitrosative stress, and the remodeling of carbon metabolism will be different in hypoxic versus normoxic conditions. In contrast, other responses appear to be secondary to growth arrest per se. For example, a wide variety of growth-inhibiting stresses trigger carbon storage and cell wall remodeling, and the maintenance of energy homeostasis is universally required for viability. PMF: proton motive force, ROS: reactive oxygen species, RNI: reactive nitrogen intermediate.
Common features of quiescent cells
Carbon storage
An almost universal property of quiescent cells is the accumulation of carbon stores, although the chemical structure of the storage form can differ. During low growth states, the yeast Saccharomyces cerevisiae accumulates glycogen, trehalose, and triacylglycerols as the main forms of metabolizable carbon (Gray et al., 2004). The bacterial pathogen Vibrio cholerae accumulates glycogen in preparation for survival in nutrient-poor environments (Bourassa and Camilli, 2009). Additionally, many bacteria store fatty acids in the form of triglycerides (Daniel et al., 2004; Kalscheuer et al., 2007) and wax esters (Sirakova et al., 2012). Both triglycerides and wax esters also accumulate in plant seeds (Radunz and Schmid, 2000), indicating that this mode of storage is advantageous for organisms that represent vastly separated domains of life. In addition, linear plastic polymers like polyhydroxyalkanoates and poly-β-hydroxybutyric acid can serve as a carbon repository in a variety of bacteria living in the soil and the rhizosphere (Kadouri et al., 2005).
What is the purpose of carbon storage? The most intuitive answer is that these cells are simply “storing nuts for winter”, and these nutritional stores can be rapidly mobilized to fuel growth when environmental conditions improve. This role has been most clearly demonstrated in the S. cerevisiae cell, where the trehalose stores that accumulate in stationary cultures are immediately consumed upon addition of fresh media to fuel rapid regrowth (Shi et al., 2010). Glycogen may serve a similar role in V. cholerae, a bacterium whose life cycle relies on periodic switches from the nutrient-replete mammalian gut to nutrient-poor aquatic environments (Bourassa and Camilli, 2009).
Carbon storage has also been found to play an important role in remodeling cellular carbon fluxes and facilitating entry into the quiescent state. Diverse stresses, such as low oxygen, low pH, or low iron, all induce a storage response in M. tuberculosis through the activation of a common sensor-kinase system, DosRST (Bacon et al., 2007; Baek et al., 2011; Daniel et al., 2011). The DosS sensor likely responds to alterations in cellular redox state in these contexts (Honaker et al., 2010), and triggers the synthesis of triglycerides that are stored in large cytosolic inclusions (Garton et al., 2002). The impact of this response appears to extend beyond the generation of nutrient stores. That is, disruption of the triglyceride biosynthesis pathway in M. tuberculosis reverses the growth arrest that is normally caused by these stresses, but has little effect on the subsequent recovery of growth when the stress is relieved (Baek et al., 2011). This inverse relationship between growth and triglyceride production appears to result from the redirection of acetyl-CoA from the TCA cycle, where it is used to generate energy during aerobic respiration, into lipid synthesis where acetyl CoA serves as a building block for fatty acids. The growth-limiting effect of carbon storage is unlikely to be restricted to mycobacteria. For example, S. cerevisiae mutants that are unable to produce glycogen or trehalose consume more CO2 than the wild type strain during slow growth (Silljé et al., 1999), indicating higher TCA flux in the absence of carbon storage. The almost universal propensity of microorganisms to accumulate acetyl CoA-derived carbon stores under growth-limiting stresses suggests that this may represent a common strategy for reducing growth and metabolic rate.
Cell Wall Modification
Virtually all bacteria are surrounded by an elastic meshwork of peptidoglycan that maintains cellular integrity under changing environmental conditions. This structure is composed of glycan chains, consisting of N-acetylglucosamine (NAG) and N-acetylmuramic acid (NAM), cross-linked through short peptide moieties. Not surprisingly, the long-term survival of both spores and quiescent cells depends on specific alterations in the composition of this structure. For example, in stationary phase cultures, the gram-positive bacteria Staphylococcus aureus generates a cell wall that is structurally different from the peptidoglycan found during exponential phase growth, in that it contains fewer pentaglycine bridges, which cross-link the glycan chains, and is significantly thicker (Zhou and Cegelski, 2012). Similarly, the level and gradient of cross-linking are important for the formation of bacterial spores. In the spore peptidoglycan layer of the soil-dwelling bacteria Bacillus subtilis, the peptide side chains serving as cross-linkers are completely or partially removed from the NAM residues and replaced by muramic-δ-lactam, a specificity determinant for germination autolytic enzymes, at every second NAM position in the cortex glycan strands. As a consequence, overall levels of cross-linking are markedly decreased in the spore cortex as compared to the vegetative cell wall (Atrih et al., 1996). Thus, common features of the peptidoglycan in both quiescent cells and spores are reduced cross-links and increased peptidoglycan mass.
The regulation of these modifications is likely complex, but recent observations suggest that extracellular D-amino acids, such as D-methionine and D-leucine, could play an important role. D-amino acids accumulate to millimolar levels in the supernatants of stationary phase bacterial culture, where they regulate cell wall synthetic enzymes and are incorporated into the peptidoglycan polymer. The increased abundance of D-amino acids in cultures of non-growing cells and their ability to alter the osmotic sensitivity of V. cholerae (Lam et al., 2009) suggests a likely role in remodeling the cell wall for quiescence.
Like many other bacteria, M. tuberculosis may vary the cross-linking of its peptidoglycan in slow growth states (Lavollay et al., 2008). During exponential growth, M. tuberculosis peptidoglycan is cross-linked largely via linkages between the 3rd and 4th amino acids in the stem peptide, the chain of amino acids in peptidoglycan that crosslinks adjacent strands (i.e. 4→3 linkages). However, in stationary phase the cell wall primarily consists of 3→3 cross-links. While not all investigators have observe these changes (Kumar et al., 2012), altered cross-linking could significantly change the physical characteristics of the cell wall. In addition, 3→3 cross-links are made by transpeptidases that are insensitive to β-lactam antibiotics that inhibit cell wall synthesis, suggesting the reduction in 4→3 linkages may reduce antibiotic susceptibility. Indeed, when the L,D-transpeptidase (MT2594/Rv2518c) responsible for making the 3→3 linkages in M. tuberculosis was inactivated, the bacteria became more susceptible to the β-lactam antibiotic amoxicillin and persistence in animals was attenuated (Gupta et al., 2010).
The mycobacterial cell wall is much more complex than those surrounding the organisms discussed above, and the full complement of alterations that accompany quiescence have yet to be defined. Mycobacterial peptidoglycan is conjugated to an additional glycan layer and finally to a functional outer membrane composed of very long chain fatty acids called, mycolic acids. Surrounding this hydrophobic layer is a capsule that is largely comprised of the polysaccharide alpha-glucan. Thickening of the mycobacterial cell wall upon hypoxia-induced stasis was first demonstrated more than thirty years ago (Wayne, 1976). More recently, a computational model of the M. tuberculosis response to hypoxia was used to predict a large increase in production of cell wall components like mycolic acids and peptidoglycan (Fang et al., 2012). This prediction is consistent with electron microscopy studies that demonstrate thickening of the outer mycolic acid and/or capsule layers of the cell wall (Cunningham and Spreadbury, 1998). A major physiological outcome of these changes is decreased permeability of the cell wall, and the uptake of several classes of antibiotics into quiescent M. tuberculosis is significantly decreased relative to replicating cells (Sarathy et al., 2013).
In addition to its structural roles, cell wall metabolism also appears to play an important role in generating signals that regulate the germination of spores and the exit from quiescence. In B. subtilis, the PrkC Ser/Thr kinase responds to the presence of extracellular peptidoglycan fragments and induces spore germination (Shah et al., 2008). These fragments are released by growing cells, providing a mechanism by which the spore can sense the presence of a favorable growth environment using cues from neighboring bacteria. M. tuberculosis expresses a similar Ser/Thr kinase, PknB, which is also capable of binding extracellular peptidoglycan fragments (Mir et al., 2011) and regulates cell wall synthesis and growth (Gee et al., 2012). Activation of this kinase could explain the ability of spent culture medium to promote the regrowth of quiescent mycobacteria, as this activity depends on secreted lysozyme-like proteins (Mukamolova et al., 1998) that could act by liberating peptidoglycan-derived PknB ligands.
Macromolecular synthesis and stability
It may seem intuitive that RNA and protein synthesis will proceed at negligible rates in the quiescent cell. However, the dynamics of macromolecular synthesis are more complicated than they appear and vary during the entry, maintenance, and exit from quiescence. During entry and exit, protein synthesis accelerates. Protein turnover increases five-fold in famished E. coli cells due to proteases that are produced in early stationary phase. This enhanced protein turnover during the transition to the growth-limited state facilitates de novo protein synthesis in the absence of an exogenous carbon source (Shaikh et al., 2010) and the required amino acids are provided by peptidase-dependent autophagy, in which amino acids are produced via protein hydrolysis and degradation (Reeve et al., 1984). Similarly, increased protein turnover may also be required for exiting the quiescent state. Regrowth of M. tuberculosis from hypoxia-induced stasis is accompanied by an increase of the ClgR regulatory protein, which induces expression of the ClpXP protease that uses ATP hydrolysis to unfold proteins for subsequent degradation (Sherrid et al., 2010). Here, increased protein turnover is a likely indicator of the wholesale metabolic remodeling necessary to shift between growth states.
Once quiescence is established, however, it is reasonable to assume that the synthesis of RNA and protein will slow considerably. Indeed, quiescence in S. cerevisiae is accompanied by a three- to five-fold decrease in overall transcription rate (Choder, 1991), and a twenty-fold decrease in protein synthesis (Fuge et al., 1994). The mechanisms underlying the reduction of macromolecular synthesis in slowly growing E. coli have been explored in great detail. While the rate of nascent RNA and polypeptide chain elongation remains relatively constant, the number of synthetic sites decreases (Pedersen, 1986). As approximately half of the mass of the rapidly-growing E. coli cell is comprised of protein synthesis machinery, the economy realized by this strategy is evident (Neidhardt, 1999). The same analysis has not been performed on non-replicating cells, and it remains likely that both initiation and elongation rate slow. This could be the result of specific regulatory systems, such as the stringent response, which controls ribosomal RNA transcription during stress (Stallings et al., 2009), and is critical for M. tuberculosis survival in both hypoxia- and starvation-induced stasis (Primm et al., 2000). In addition, low levels of nucleotide triphosphates and amino acids could nonspecifically limit elongation rate. This would be consistent with the proposed mechanism by which the drug pyrazinamide kills non-replicating M. tuberculosis by inhibiting trans-translation, a mechanism for recycling stalled ribosomes (Shi et al., 2011).
The apparent reduction in RNA synthesis upon mycobacterial entry into quiescence is coupled to a fifteen-fold increase in mRNA stability (Rustad et al., 2013), and these stable mRNAs are required to sustain pools of essential proteins (Rao and Li, 2009). This phenomenon is not unique to mycobacteria. In both S. aureus and S. cerevisiae, mRNA transcripts are globally stabilized in response to stationary phase and stress (Anderson et al., 2006; Jona et al., 2000). In addition, slow growth induces the preferential stabilization of a set of transcripts in E. coli (Goergellis et al., 1993), indicating that survival requires the continual synthesis of select proteins. In support of this model, studies have shown that stationary phase E. coli requires the continual expression of a subset of genes that are controlled by the σS subunit of the RNA polymerase, including many genes involved in cellular stress responses (Talukder et al., 1996). Additionally a subset of mRNAs are strongly induced in quiescent S. cerevisiae (Werner-Washburne et al., 1996). In sum, while the rate of macromolecular synthesis clearly decreases in quiescent cells, continual transcription and translation occurs. The ability of RNA polymerase and DNA gyrase inhibitors to kill non-replicating mycobacteria indicates that these activities may also be required for survival (Betts et al., 2002; Sala et al., 2010).
Energetics and metabolism during quiescence
Maintenance of membrane potential and ATP synthesis are not required for sustaining the viability of spores, even though a repertoire of ATPases and ATP-dependent regulatory proteins is utilized during the initiation of germination (Errington, 2003). In contrast, quiescent bacteria maintain their membrane potential (Pernthaler and Amann, 2004; Rao et al. 2008), and energy homeostasis appears to be critical for survival. In non-replicating M. tuberculosis cells starved for oxygen or nutrients, ATP levels are maintained at a steady level, which is only five-fold lower than replicating cells (Gengenbacher et al., 2010; Rao et al., 2008). This maintenance of ATP homeostasis is clearly important, as disruption of the proton motive force or chemical inhibition of the F0F1 ATP synthase involved in ATP synthesis induces cell death in nutrient-starved or hypoxic cultures (Rao et al., 2008; Sala et al., 2010). Diverse strategies can be used to maintain energy homeostasis. Both respiratory and fermentative pathways can support the long-term survival of bacteria in stationary phase (Duwat et al., 2001). Similarly, M. tuberculosis harbors a number of respiratory systems utilizing both oxygen and nitrate as terminal electron acceptors (Boshoff and Barry, 2005). When respiration is not possible, recent data suggest that fermentation can lead to succinate secretion, which plays an important role in maintaining membrane potential (Watanabe et al., 2011, Eoh and Rhee, 2013). This general strategy is used by a number of gram-positive and gram-negative organisms under respiration-limited conditions (Engel et al., 1994; Schnorpfeil et al., 2001).
While the overall rate of carbon utilization decreases in quiescence, a few metabolic pathways display enhanced flux (Sauer et al., 1999, Zhang et al., 2009). The specific pathways used to maintain the quiescent cell are diverse and their relative importance depends on the peculiarities of both the organism and the environment. However, common themes have emerged, such as a central role for the glyoxylate shunt in adapting to the growth-limited state. This pathway consists of isocitrate lyase and malate synthase, and is responsible for the conversion of isocitrate and acetyl-CoA into malate and succinate. This bypasses a segment of the TCA cycle that is normally siphoned to produce biosynthetic precursors. While initially considered simply as an anapleurotic system that is used to replenish TCA intermediates during growth on non-glycolytic carbon sources such as lipids (Muñoz-Elías and McKinney, 2005), recent studies indicate that these reactions are an essential component of metabolic cycles that sustain diverse bacterial species in slowly-growing states. The glyoxylate shunt is an essential component of the phosphoenolpyruvate-glyoxylate cycle in E. coli (Fischer and Sauer, 2003), and the “GAS” pathway in mycobacteria (Beste et al. 2011). These pathways are important for the utilization of glucose, in contrast to the canonical role or the glyoxylate shunt in lipid catabolism. In both cases, these cycles are used to uncouple glucose oxidation from the production of reducing equivalents, and may function to maintain the redox state of the cytosol. In the mycobacterial case, inhibition of isocitrate lyase causes cell death in both hypoxia- and starvation-induced quiescence, supporting the importance of these pathways in non-growing states (Gengenbacher et al., 2010, Eoh and Rhee, 2013).
Preservation of genome integrity
Maintaining genome fidelity when little or no metabolic capacity is available for canonical DNA repair mechanisms is a challenge faced by both quiescent cells and dormant spores. One strategy common to both types of cells is altering chromosomal structure to a more chemically-stable form. The chromosome of stationary phase E. coli assumes an extremely compact structure. A nucleoid-associated protein called Dps, which is expressed only in stationary phase, mediates biocrystallization of the nucleoid and protects DNA from damage (Martinez and Kolter, 1997). This compaction of DNA can be very dynamic as bacteria enter and exit different growth states. In the photosynthetic cyanobacterium, Synechococcus elongates, a circadian clock-controlled mechanism induces periodic chromosome compaction during the night (Smith and Williams, 2006), and the resulting alterations in DNA supercoiling control global gene expression patterns (Vijayan et al., 2009). M. tuberculosis might use a similar mechanism to protect its chromosome. A mycobacterial histone-like protein, Lsr2, mediates chromosome compaction and protection from reactive oxygen and nitrogen species (Summers et al., 2012). The mechanisms underlying this protective ability are unclear, but could be related to the reported association between Lsr2 and FAD-binding flavoprotein that mediates general oxidative stress resistance (Du et al., 2012). The compact packaging of bacterial DNA can be also facilitated by cationic metabolites, such as polyamines, which have been implicated in the protection of DNA from chemical damage (Baeza et al., 1991). M. tuberculosis synthesizes a repertoire of polyamines that facilitate transcription and DNA replication (Marton and Pegg, 1995), although the roles of these compounds during cellular quiescence remain unknown.
Truly dormant spores are not able to actively maintain their chromosome, but depend on the induction of DNA repair systems upon exit from the dormant state. While the nominal metabolic capacity of the quiescent cell likely allows a subset of DNA repair mechanisms to operate continuously, the relative activity of different systems in growing and non-growing states remains uncertain and distinct organisms favor different strategies. Some microorganisms arrest growth with a single chromosome (Valcourt et al., 2012), while others, such as M. tuberculosis, exit the cell cycle with two chromosomal copies (Wayne, 1977). Thus, high-fidelity recombinational repair mechanisms, which often dominate in growing cells, are only available to a subset of quiescent organisms. Despite the apparent presence of a recombinational template in non-replicating M. tuberculosis, this organism still appears to utilize more error-prone repair systems. For example, error-prone translesion polymerases, which replicate past DNA damage lesions, are important for the survival of slowly-growing M. tuberculosis in chronically infected animals (Boshoff et al., 2003). Similarly, mycobacteria rely on nonhomologous end joining (NHEJ), in which double strand breaks are imprecisely rejoined, to repair double stranded breaks in quiescent states (Shuman and Glickman, 2007). The use of the low fidelity NEHJ system under these conditions could be due to its dependence on ribonucleotides as opposed to deoxyribonucleotide triphosphates, which may be limiting in non-replicating cells (Gong et al., 2005).
The particular DNA repair pathways used by quiescent organisms have significant implications for genome evolution. While the mutation rate of rapidly-growing organisms is largely determined by the error rate of the replisome (Kunkel, 2004), the fidelity of DNA repair systems might dominate in organisms that spend a significant portion of their existence in slowly-growing states. Consistent with this model, it was recently estimated that the M. tuberculosis genome accumulates mutations at a similar rate in active and latent infection states (Ford et al., 2011), and mutations accumulate in a time-dependent and not replication-dependent manner (Ford et al., 2013). As drug resistance in M. tuberculosis is the product of spontaneous mutation, the specific DNA repair pathways that are operational in the quiescent state could determine the rate at which resistance emerges.
Strategies to eradicate quiescent bacteria
Arguably, the most important factor limiting tuberculosis control efforts is the exceptionally-long treatment course that is required to prevent relapse. While the standard regimen of antitubercular drugs rapidly kills replicating bacteria in vitro, the same drugs must be administered for at least six months to effectively treat an active TB infection. The reduced activity of antibiotics in the in vivo environment is not specific to mycobacteria and is often attributed to slowly-replicating or quiescent populations of the pathogen (Eagle, 1952; McDermott, 1958). The antibiotic sensitivity of quiescent bacterial populations has been investigated extensively in the context of environmentally-induced stasis in M. tuberculosis (Mitchison and Coates, 2004; Rao et al., 2008; Xie et al., 2005), drug tolerant populations found in biofilms (Brown et al.,1988) and non-replicating “persister” subpopulations that exist even in rapidly-growing cultures (Balaban et al., 2004). In all situations, quiescent bacterial populations are less sensitive to existing antibiotics that target functions necessary for cell growth. In principle, the identification of cellular functions that are important for the regulation or maintenance of quiescence should suggest new strategies for eliminating non-replicating bacterial populations and could be used to accelerate the treatment of chronic infections. Three general approaches have been pursued to this end:
Inhibit pathways that are essential in the quiescent state
In the case of M. tuberculosis there is an anecdotal correlation between drugs that retain activity against non-replicating cells in vitro and those that have the strongest sterilizing activity in vivo (Mitchison and Coates, 2004), although this correlation is not absolute. As a result, new drug candidates are generally tested for their ability to kill quiescent bacteria. Among the several new regimens under development, a combination that includes drugs retaining activity against non-replicating cells, bedaquiline, PA-824, and moxifloxacin, showed the highest early bactericidal activity in a recent human trial (Diacon et al., 2012). While none of these drugs were developed specifically to kill in a growth rate-independent manner, all three might have been predicted to have this ability based on their mechanism of action. Bedaquiline inhibits the F0F1 ATP synthase, a function required for energy maintenance (Andries et al., 2005), PA-824 is a nitroimidazole that kills via nonspecific nitrosative (nitrogen radical-mediated) damage (Singh et al., 2008), and moxifloxacin produces DNA breaks at sites of ongoing transcription (Drlica et al., 2008). These observations suggest that specifically targeting other functions necessary for survival, such as those maintaining carbon flux, energy generation, or redox maintenance, might represent a productive strategy to produce more effective therapies (Table 1).
Table 1. Strategies to eradicate quiescent bacterial populations.
| Pathways that may be essential during quiescence | Known drugs/inhibitors | References |
|---|---|---|
| RNA synthesis | Rifampin | Betts et al., 2002; Mitchison and Coates, 2004 |
| Proton motive force/ATP generation | Bedaquiline, possibly pyrazinamide | Gengenbacher et al., 2010; Rao et al., 2008; Zhang et al., 2003 |
| DNA gyrase/DNA integrity | Fluorquinolones | Hussain et al., 2009; Sala et al., 2010 |
| Trans-translation | Pyrazinamide | Mitchison and Coates, 2004; Shi et al., 2011 |
| Cell wall remodeling | Carbapenem | Hugonnet et al., 2009 |
| Glyoxylate shunt | None | Beste et al. 2011; Munoz-Elias and McKinney, 2005 |
| Reductive TCA branch | None | Eoh and Rhee, 2013; Watanabe et al., 2011 |
| Strategies to resensitize quiescent cells | Examples | References |
| Inhibit stringent response | Increases antibiotic sensitivity in P. aeruginosa biofilm | Nguyen et al., 2011 |
| Modulate toxin/antitoxin systems | Determines the proportion of quiescent cells in a population | Lewis, 2007 |
| Enhance antibiotic uptake | Metabolite supplementation increases aminoglycoside uptake | Allison et al., 2011 |
| Impose nonspecific nitrosative damage | Nitromidazoles (e.g. Metronatazole, PA-824, Delamanid) | Singh et al., 2008 |
| Enhance TCA activity | Sensitizes to multiple antibiotics | Baek et al., 2011; Kohanski et al., 2007 |
| Strategies to remove quiescence-inducing cues | References | |
| Biofilm dispersion | Disrupts starvation-induced quiescence | Potera, 2010 |
| TNF modulation | Enhances anti-TB therapy | Bourigault et al., 2013 |
| Immunosuppression | Accelerates antibacterial therapy in animal models | Assfalg et al., 2010; Lenaerts et al., 2003 |
Sensitize the quiescent cell to existing antibiotics
As an alternative to targeting what is likely a restricted set of pathways that are necessary for maintaining viability in the absence of replication, it may also be possible to alter the physiology of the quiescent cell to render it more antibiotic susceptible. Two recent studies have focused on increasing antibiotic activity by altering flux through central carbon metabolism. In M. tuberculosis, antibiotic efficacy is reduced as a result of carbon storage responses that lower TCA flux. Genetic mutations or nutritional supplements that inhibit triglyceride production, and thus drive activity of the TCA cycle, or directly enhance TCA flux were found to enhance antibiotic activity in quiescent cells in vitro and in animal models (Baek et al., 2011). A similar approach for enhancing TCA-dependent antibiotic activity by metabolite supplementation was subsequently found to enhance aminoglycoside activity in E. coli and S. aureus (Allison et al., 2011). Another promising approach is inhibiting the stringent response. This system coordinates cellular physiology during slow growth states in many bacteria and genetic inhibition of the stringent response sensitizes Pseudomonas aeruginosa biofilms to antibiotics (Nguyen et al., 2011). While metabolite supplementation might not be practical for many infections of the deep tissues, and small molecule modulators of these pathways are not currently available, these studies provide proof for the concept that quiescent cells can be rendered antibiotic susceptible.
Alter the growth-limiting stress
Instead of directly modulating bacterial metabolism, it may be possible to enhance drug efficacy by removing the specific environmental pressures that induce quiescence. For example, non-replicating, antibiotic tolerant cells are found in relatively high numbers in bacterial biofilms (Lewis, 2007), and the disruption of the biofilm architecture increases antibiotic activity at least in part by reversing this differentiation (Musk and Hergenrother, 2006). Similarly, it may be possible to modulate the host immune pressures that induce the antibiotic tolerant state. Overt immunosuppression can enhance antibiotic activity in a number of models (Assfalg et al., 2010; Lenaerts et al., 2003). Even more subtle chemical modulation of tumor necrosis factor (TNF) signaling, which plays central roles in the inflammatory response, has been shown to accelerate anti-tuberculosis therapy in animals (Bourigault et al., 2013).
Thus, both the host and pathogen can be manipulated to increase antibiotic efficacy. A more detailed understanding of the specific pressures that limit growth during infection and the pathogen's adaptations to these stresses could lead to the rational development of new synergistic therapies that accelerate antibacterial treatment.
Acknowledgments
This work was supported the NIH (grant #AI064282 to CMS) and the Howard Hughes Medical Institute (CMS). We thank Jarukit Edward Long for thoughtful review of this manuscript, Annaliese R. Brauman for assistance in graphic design, and K. Atmakuri for the electron micrograph shown in figure 2.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Allison KR, Brynildsen MP, Collins JJ. Metabolite-enabled eradication of bacterial persisters by aminoglycosides. Nature. 2011;473:216–220. doi: 10.1038/nature10069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson KL, Roberts C, Disz T, Vonstein V, Hwang K, Overbeek R, Olson PD, Projan SJ, Dunman PM. Characterization of the Staphylococcus aureus heat shock, cold shock, stringent, and SOS responses and their effects on log-phase mRNA turnover. J Bacteriol. 2006;188:6739–6756. doi: 10.1128/JB.00609-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andries K, Verhasselt P, Guillemont J, Gohlmann HW, Neefs JM, Winkler H, Van Gestel J, Timmerman P, Zhu M, Lee E, et al. A diarylquinoline drug active on the ATP synthase of Mycobacterium tuberculosis. Science. 2005;307:223–227. doi: 10.1126/science.1106753. [DOI] [PubMed] [Google Scholar]
- Assfalg V, Huser N, Reim D, Kaiser-Moore S, Rossmann-Bloeck T, Weighardt H, Novotny AR, Stangl MJ, Holzmann B, Emmanuel KL. Combined immunosuppressive and antibiotic therapy improves bacterial clearance and survival of polymicrobial septic peritonitis. Shock. 2010;33:155–162. doi: 10.1097/SHK.0b013e3181ab9014. [DOI] [PubMed] [Google Scholar]
- Atrih A, Zollner P, Allmaier G, Foster SJ. Structural analysis of Bacillus subtilis 168 endospore peptidoglycan and its role during differentiation. J Bacteriol. 1996;178:6173–6183. doi: 10.1128/jb.178.21.6173-6183.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacon J, Dover LG, Hatch KA, Zhang Y, Gomes JM, Kendall S, Wernisch L, Stoker NG, Butcher PD, Besra GS, et al. Lipid composition and transcriptional response of Mycobacterium tuberculosis grown under iron-limitation in continuous culture: identification of a novel wax ester. Microbiology. 2007;153:1435–1444. doi: 10.1099/mic.0.2006/004317-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek SH, Li AH, Sassetti CM. Metabolic regulation of mycobacterial growth and antibiotic sensitivity. PLoS Biol. 2011;9:e1001065. doi: 10.1371/journal.pbio.1001065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baeza I, Ibanez M, Wong C, Chavez P, Gariglio P, Oro J. Possible prebiotic significance of polyamines in the condensation, protection, encapsulation, and biological properties of DNA. Orig Life Evol Biosph. 1991;21:225–242. doi: 10.1007/BF01809858. [DOI] [PubMed] [Google Scholar]
- Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler S. Bacterial persistence as a phenotypic switch. Science. 2004;305:1622–1625. doi: 10.1126/science.1099390. [DOI] [PubMed] [Google Scholar]
- Beste DJ, Bonde B, Hawkins N, Ward JL, Beale MH, Noack S, Noh K, Kruger NJ, Ratcliffe RG, McFadden J. (1)(3)C metabolic flux analysis identifies an unusual route for pyruvate dissimilation in mycobacteria which requires isocitrate lyase and carbon dioxide fixation. PLoS Pathog. 2011;7:e1002091. doi: 10.1371/journal.ppat.1002091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betts JC, Lukey PT, Robb LC, McAdam RA, Duncan K. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol Microbiol. 2002;43:717–731. doi: 10.1046/j.1365-2958.2002.02779.x. [DOI] [PubMed] [Google Scholar]
- Boshoff HI, Barry CE., 3rd Tuberculosis - metabolism and respiration in the absence of growth. Nat Rev Microbiol. 2005;3:70–80. doi: 10.1038/nrmicro1065. [DOI] [PubMed] [Google Scholar]
- Boshoff HI, Reed MB, Barry CE, 3rd, Mizrahi V. DnaE2 polymerase contributes to in vivo survival and the emergence of drug resistance in Mycobacterium tuberculosis. Cell. 2003;113:183–193. doi: 10.1016/s0092-8674(03)00270-8. [DOI] [PubMed] [Google Scholar]
- Bourassa L, Camilli A. Glycogen contributes to the environmental persistence and transmission of Vibrio cholerae. Mol Microbiol. 2009;72:124–138. doi: 10.1111/j.1365-2958.2009.06629.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourigault M, Vacher R, Rose S, Olleros ML, Janssens J, Quesniaux VF, Garcia I. Tumor necrosis factor neutralization combined with chemotherapy enhances Mycobacterium tuberculosis clearance and reduces lung pathology. Am J Clin Exp Immunol. 2013;2:124–134. [PMC free article] [PubMed] [Google Scholar]
- Brown MR, Allison DG, Gilbert P. Resistance of bacterial biofilms to antibiotics: a growth-rate related effect? J Antimicrob Chemother. 1988;22:777–780. doi: 10.1093/jac/22.6.777. [DOI] [PubMed] [Google Scholar]
- Choder M. A general topoisomerase I-dependent transcriptional repression in the stationary phase in yeast. Genes Dev. 1991;5:2315–2326. doi: 10.1101/gad.5.12a.2315. [DOI] [PubMed] [Google Scholar]
- Cole JJ. Aquatic Microbiology for Ecosystem Scientists: New and Recycled Paradigms in Ecological Microbiology. Ecosystems. 1999;2:215–225. [Google Scholar]
- Corper HJ, Cohn ML. The viability and virulence of old cultures of tuercule bacilli. Am Rev Tuberc. 1933;28:856–874. [Google Scholar]
- Cunningham AF, Spreadbury CL. Mycobacterial stationary phase induced by low oxygen tension: cell wall thickening and localization of the 16-kilodalton alpha-crystallin homolog. J Bacteriol. 1998;180:801–808. doi: 10.1128/jb.180.4.801-808.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel J, Deb C, Dubey VS, Sirakova TD, Abomoelak B, Morbidoni HR, Kolattukudy PE. Induction of a novel class of diacylglycerol acyltransferases and triacylglycerol accumulation in Mycobacterium tuberculosis as it goes into a dormancy-like state in culture. J Bacteriol. 2004;186:5017–5030. doi: 10.1128/JB.186.15.5017-5030.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel J, Maamar H, Deb C, Sirakova TD, Kolattukudy PE. Mycobacterium tuberculosis uses host triacylglycerol to accumulate lipid droplets and acquires a dormancy-like phenotype in lipid-loaded macrophages. PLoS Pathog. 2011;7:e1002093. doi: 10.1371/journal.ppat.1002093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diacon AH, Dawson R, von Groote-Bidlingmaier F, Symons G, Venter A, Donald PR, van Niekerk C, Everitt D, Winter H, Becker P, et al. 14-day bactericidal activity of PA-824, bedaquiline, pyrazinamide, and moxifloxacin combinations: a randomised trial. Lancet. 2012;380:986–993. doi: 10.1016/S0140-6736(12)61080-0. [DOI] [PubMed] [Google Scholar]
- Drlica K, Malik M, Kerns RJ, Zhao X. Quinolone-mediated bacterial death. Antimicrob Agents Chemother. 2008;52:385–392. doi: 10.1128/AAC.01617-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y, Zhang H, He Y, Huang F, He ZG. Mycobacterium smegmatis Lsr2 physically and functionally interacts with a new flavoprotein involved in bacterial resistance to oxidative stress. J Biochem. 2012;152:479–486. doi: 10.1093/jb/mvs095. [DOI] [PubMed] [Google Scholar]
- Duwat P, Sourice S, Cesselin B, Lamberet G, Vido K, Gaudu P, Le Loir Y, Violet F, Loubiere P, Gruss A. Respiration capacity of the fermenting bacterium Lactococcus lactis and its positive effects on growth and survival. J Bacteriol. 2001;183:4509–4516. doi: 10.1128/JB.183.15.4509-4516.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eagle H. Experimental approach to the problem of treatment failure with penicillin. I. Group A streptococcal infection in mice. Am J Med. 1952;13:389–399. doi: 10.1016/0002-9343(52)90293-3. [DOI] [PubMed] [Google Scholar]
- Engel P, Kramer R, Unden G. Transport of C4-dicarboxylates by anaerobically grown Escherichia coli. Energetics and mechanism of exchange, uptake and efflux. Eur J Biochem. 1994;222:605–614. doi: 10.1111/j.1432-1033.1994.tb18903.x. [DOI] [PubMed] [Google Scholar]
- Eoh H, Rhee K. Multifunctional essentiality of succinate metabolism in adaptation to hypoxia in Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 2013 doi: 10.1073/pnas.1219375110. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errington J. Regulation of endospore formation in Bacillus subtilis. Nat Rev Microbiol. 2003;1:117–126. doi: 10.1038/nrmicro750. [DOI] [PubMed] [Google Scholar]
- Fang X, Wallqvist A, Reifman J. Modeling phenotypic metabolic adaptations of Mycobacterium tuberculosis H37Rv under hypoxia. PLoS Comput Biol. 2012;8:e1002688. doi: 10.1371/journal.pcbi.1002688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel SE. Long-term survival during stationary phase: evolution and the GASP phenotype. Nat Rev Microbiol. 2006;4:113–120. doi: 10.1038/nrmicro1340. [DOI] [PubMed] [Google Scholar]
- Fischer E, Sauer U. A novel metabolic cycle catalyzes glucose oxidation and anaplerosis in hungry Escherichia coli. J Biol Chem. 2003;278:46446–46451. doi: 10.1074/jbc.M307968200. [DOI] [PubMed] [Google Scholar]
- Ford CB, Lin PL, Chase MR, Shah RR, Iartchouk O, Galagan J, Mohaideen N, Ioerger TR, Sacchettini JC, Lipsitch M, et al. Use of whole genome sequencing to estimate the mutation rate of Mycobacterium tuberculosis during latent infection. Nat Genet. 2011;43:482–486. doi: 10.1038/ng.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford CB, Shah RR, Maeda MK, Ganeux S, Murray MB, Cohen T, Johnston JC, Gardy J, Lipsitch M, Fortune SM. Mycobacterium tuberculosis mutation rate estimates from different lineages predict substantial differences in the emergence of drug resistant tuberculosis. Nat Genet. 2013 doi: 10.1038/ng.2656. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuge EK, Braun EL, Werner-Washburne M. Protein synthesis in long-term stationary-phase cultures of Saccharomyces cerevisiae. J Bacteriol. 1994;176:5802–5813. doi: 10.1128/jb.176.18.5802-5813.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garton NJ, Christensen H, Minnikin DE, Adegbola RA, Barer MR. Intracellular lipophilic inclusions of mycobacteria in vitro and in sputum. Microbiology. 2002;148:2951–2958. doi: 10.1099/00221287-148-10-2951. [DOI] [PubMed] [Google Scholar]
- Gee CL, Papavinasasundaram KG, Blair SR, Baer CE, Falick AM, King DS, Griffin JE, Venghatakrishnan H, Zukauskas A, Wei JR, et al. A phosphorylated pseudokinase complex controls cell wall synthesis in mycobacteria. Sci Signal. 2012;5:ra7. doi: 10.1126/scisignal.2002525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gengenbacher M, Rao SP, Pethe K, Dick T. Nutrient-starved, non-replicating Mycobacterium tuberculosis requires respiration, ATP synthase and isocitrate lyase for maintenance of ATP homeostasis and viability. Microbiology. 2010;156:81–87. doi: 10.1099/mic.0.033084-0. [DOI] [PubMed] [Google Scholar]
- Georgellis D, Barlow T, Arvidson S, von Gabain A. Retarded RNA turnover in Escherichia coli: a means of maintaining gene expression during anaerobiosis. Mol Microbiol. 1993;9:375–381. doi: 10.1111/j.1365-2958.1993.tb01698.x. [DOI] [PubMed] [Google Scholar]
- Gill WP, Harik NS, Whiddon MR, Liao RP, Mittler JE, Sherman DR. A replication clock for Mycobacterium tuberculosis. Nat Med. 2009;15:211–214. doi: 10.1038/nm.1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong C, Bongiorno P, Martins A, Stephanou NC, Zhu H, Shuman S, Glickman MS. Mechanism of nonhomologous end-joining in mycobacteria: a low-fidelity repair system driven by Ku, ligase D and ligase C. Nat Struct Mol Biol. 2005;12:304–312. doi: 10.1038/nsmb915. [DOI] [PubMed] [Google Scholar]
- Gray JV, Petsko GA, Johnston GC, Ringe D, Singer RA, Werner-Washburne M. “Sleeping beauty”: quiescence in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 2004;68:187–206. doi: 10.1128/MMBR.68.2.187-206.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R, Lavollay M, Mainardi JL, Arthur M, Bishai WR, Lamichhane G. The Mycobacterium tuberculosis protein LdtMt2 is a nonclassical transpeptidase required for virulence and resistance to amoxicillin. Nat Med. 2010;16:466–469. doi: 10.1038/nm.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honaker RW, Dhiman RK, Narayanasamy P, Crick DC, Voskuil MI. DosS responds to a reduced electron transport system to induce the Mycobacterium tuberculosis DosR regulon. J Bacteriol. 2010;192:6447–6455. doi: 10.1128/JB.00978-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugonnet JE, Tremblay LW, Boshoff HI, Barry CE, 3rd, Blanchard JS. Meropenem-clavulanate is effective against extensively drug-resistant Mycobacterium tuberculosis. Science. 2009;323:1215–1218. doi: 10.1126/science.1167498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain S, Malik M, Shi L, Gennaro ML, Drlica K. In vitro model of mycobacterial growth arrest using nitric oxide with limited air. Antimicrob Agents Chemother. 2009;53:157–161. doi: 10.1128/AAC.00442-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreidl KO, Green GR, Wren SM. Intravascular hemolysis from a Clostridium perfringens liver abscess. J Am Coll Surg. 2002;194:387. doi: 10.1016/s1072-7515(01)01169-3. [DOI] [PubMed] [Google Scholar]
- Jona G, Choder M, Gileadi O. Glucose starvation induces a drastic reduction in the rates of both transcription and degradation of mRNA in yeast. Biochim Biophys Acta. 2000;1491:37–48. doi: 10.1016/s0167-4781(00)00016-6. [DOI] [PubMed] [Google Scholar]
- Jones SE, Lennon JT. Dormancy contributes to the maintenance of microbial diversity. Proc Natl Acad Sci USA. 2010;107:5881–5886. doi: 10.1073/pnas.0912765107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadouri D, Jurkevitch E, Okon Y, Castro-Sowinski S. Ecological and agricultural significance of bacterial polyhydroxyalkanoates. Crit Rev Microbiol. 2005;31:55–67. doi: 10.1080/10408410590899228. [DOI] [PubMed] [Google Scholar]
- Kalscheuer R, Stoveken T, Malkus U, Reichelt R, Golyshin PN, Sabirova JS, Ferrer M, Timmis KN, Steinbuchel A. Analysis of storage lipid accumulation in Alcanivorax borkumensis: Evidence for alternative triacylglycerol biosynthesis routes in bacteria. J Bacteriol. 2007;189:918–928. doi: 10.1128/JB.01292-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohanski MA, Dwyer DJ, Hayete B, Lawrence CA, Collins JJ. A common mechanism of cellular death induced by bactericidal antibiotics. Cell. 2007;130:797–810. doi: 10.1016/j.cell.2007.06.049. [DOI] [PubMed] [Google Scholar]
- Kumar P, Arora K, Lloyd JR, Lee IY, Nair V, Fischer E, Boshoff HI, Barry CE., 3rd Meropenem inhibits D,D-carboxypeptidase activity in Mycobacterium tuberculosis. Mol Microbiol. 2012;86:367–381. doi: 10.1111/j.1365-2958.2012.08199.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel TA. DNA replication fidelity. J Biol Chem. 2004;279:16895–16898. doi: 10.1074/jbc.R400006200. [DOI] [PubMed] [Google Scholar]
- Lam H, Oh DC, Cava F, Takacs CN, Clardy J, de Pedro MA, Waldor MK. D-amino acids govern stationary phase cell wall remodeling in bacteria. Science. 2009;325:1552–1555. doi: 10.1126/science.1178123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavollay M, Arthur M, Fourgeaud M, Dubost L, Marie A, Veziris N, Blanot D, Gutmann L, Mainardi JL. The peptidoglycan of stationary-phase Mycobacterium tuberculosis predominantly contains cross-links generated by L,D-transpeptidation. J Bacteriol. 2008;190:4360–4366. doi: 10.1128/JB.00239-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenaerts AJ, Gruppo V, Brooks JV, Orme IM. Rapid in vivo screening of experimental drugs for tuberculosis using gamma interferon gene-disrupted mice. Antimicrob Agents Chemother. 2003;47:783–785. doi: 10.1128/AAC.47.2.783-785.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis DL, Gattie DK. Ecology of quiescent microbes, ASM News. 1991;57:27–32. [Google Scholar]
- Lewis K. Persister cells, dormancy and infectious disease. Nat Rev Microbiol. 2007;5:48–56. doi: 10.1038/nrmicro1557. [DOI] [PubMed] [Google Scholar]
- Martinez A, Kolter R. Protection of DNA during oxidative stress by the nonspecific DNA-binding protein Dps. J Bacteriol. 1997;179:5188–5194. doi: 10.1128/jb.179.16.5188-5194.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marton LJ, Pegg AE. Polyamines as targets for therapeutic intervention. Annu Rev Pharmacol Toxicol. 1995;35:55–91. doi: 10.1146/annurev.pa.35.040195.000415. [DOI] [PubMed] [Google Scholar]
- McDermott W. Microbial persistence. Yale J Biol Med. 1958;30:257–291. [PMC free article] [PubMed] [Google Scholar]
- Mir M, Asong J, Li X, Cardot J, Boons GJ, Husson RN. The extracytoplasmic domain of the Mycobacterium tuberculosis Ser/Thr kinase PknB binds specific muropeptides and is required for PknB localization. PLoS Pathog. 2011;7:e1002182. doi: 10.1371/journal.ppat.1002182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison DA, Coates AR. Predictive in vitro models of the sterilizing activity of anti-tuberculosis drugs. Curr Pharm Des. 2004;10:3285–3295. doi: 10.2174/1381612043383269. [DOI] [PubMed] [Google Scholar]
- Mukamolova GV, Kaprelyants AS, Young DI, Young M, Kell DB. A bacterial cytokine. Proc Natl Acad Sci USA. 1998;95:8916–8921. doi: 10.1073/pnas.95.15.8916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Elias EJ, McKinney JD. Mycobacterium tuberculosis isocitrate lyases 1 and 2 are jointly required for in vivo growth and virulence. Nat Med. 2005;11:638–644. doi: 10.1038/nm1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Elias EJ, Timm J, Botha T, Chan WT, Gomez JE, McKinney JD. Replication dynamics of Mycobacterium tuberculosis in chronically infected mice. Infect Immun. 2005;73:546–551. doi: 10.1128/IAI.73.1.546-551.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musk DJ, Jr, Hergenrother PJ. Chemical countermeasures for the control of bacterial biofilms: effective compounds and promising targets. Curr Med Chem. 2006;13:2163–2177. doi: 10.2174/092986706777935212. [DOI] [PubMed] [Google Scholar]
- Neidhardt FC. Bacterial growth: constant obsession with dN/dt. J Bacteriol. 1999;181:7405–7408. doi: 10.1128/jb.181.24.7405-7408.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen D, Joshi-Datar A, Lepine F, Bauerle E, Olakanmi O, Beer K, McKay G, Siehnel R, Schafhauser J, Wang Y, et al. Active starvation responses mediate antibiotic tolerance in biofilms and nutrient-limited bacteria. Science. 2011;334:982–986. doi: 10.1126/science.1211037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen S. The chain growth rate for protein synthesis varies in Escherichia coli. In: Schaechter M, Neidhardt FC, Ingraham JL, Kjeldgaard NO, editors. The Molecular Biology of Bacterial Growth. Boston, Mass; Jones and Bartlett, Inc.: 1986. pp. 13–20. [Google Scholar]
- Pernthaler A, Amann R. Simultaneous fluorescence in situ hybridization of mRNA and rRNA in environmental bacteria. Appl Environ Microbiol. 2004;70:5426–5433. doi: 10.1128/AEM.70.9.5426-5433.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potera C. Antibiotic Resistance: Biofilm dispersing agent rejuvenates older antibiotics. Environ Heath Perspect. 2010;118:A288. [Google Scholar]
- Primm TP, Andersen SJ, Mizrahi V, Avarbock D, Rubin H, Barry CE., 3rd The stringent response of Mycobacterium tuberculosis is required for long-term survival. J Bacteriol. 2000;182:4889–4898. doi: 10.1128/jb.182.17.4889-4898.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radunz A, Schmid GH. Wax esters and triglycerides as storage substances in seeds of Buxus sempervirens. Eur J Lipid Sci Tech. 2000;102:734–738. [Google Scholar]
- Rao PK, Li Q. Protein turnover in mycobacterial proteomics. Molecules. 2009;14:3237–3258. doi: 10.3390/molecules14093237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SP, Alonso S, Rand L, Dick T, Pethe K. The protonmotive force is required for maintaining ATP homeostasis and viability of hypoxic, nonreplicating Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 2008;105:11945–11950. doi: 10.1073/pnas.0711697105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeve CA, Amy PS, Matin A. Role of protein synthesis in the survival of carbon-starved Escherichia coli K-12. J Bacteriol. 1984;160:1041–1046. doi: 10.1128/jb.160.3.1041-1046.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rustad TR, Minch KJ, Brabant W, Winkler JK, Reiss DJ, Baliga NS, Sherman DR. Global analysis of mRNA stability in Mycobacterium tuberculosis. Nucleic Acids Res. 2013;41:509–517. doi: 10.1093/nar/gks1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sala C, Dhar N, Hartkoorn RC, Zhang M, Ha YH, Schneider P, Cole ST. Simple model for testing drugs against nonreplicating Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2010;54:4150–4158. doi: 10.1128/AAC.00821-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarathy J, Dartois V, Dick T, Gengenbacher M. Reduced Drug Uptake in Phenotypically Resistant Nutrient-Starved Nonreplicating Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2013;57:1648–1653. doi: 10.1128/AAC.02202-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer U, Lasko DR, Fiaux J, Hochuli M, Glaser R, Szyperski T, Wuthrich K, Bailey JE. Metabolic flux ratio analysis of genetic and environmental modulations of Escherichia coli central carbon metabolism. J Bacteriol. 1999;181:6679–6688. doi: 10.1128/jb.181.21.6679-6688.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnorpfeil M, Janausch IG, Biel S, Kroger A, Unden G. Generation of a proton potential by succinate dehydrogenase of Bacillus subtilis functioning as a fumarate reductase. Eur J Biochem. 2001;268:3069–3074. doi: 10.1046/j.1432-1327.2001.02202.x. [DOI] [PubMed] [Google Scholar]
- Shah IM, Laaberki MH, Popham DL, Dworkin J. A eukaryotic-like Ser/Thr kinase signals bacteria to exit dormancy in response to peptidoglycan fragments. Cell. 2008;135:486–496. doi: 10.1016/j.cell.2008.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaikh AS, Tang YJ, Mukhopadhyay A, Martin HG, Gin J, Benke PI, Keasling JD. Study of stationary phase metabolism via isotopomer analysis of amino acids from an isolated protein. Biotechnol Prog. 2010;26:52–56. doi: 10.1002/btpr.325. [DOI] [PubMed] [Google Scholar]
- Sherrid AM, Rustad TR, Cangelosi GA, Sherman DR. Characterization of a Clp protease gene regulator and the reaeration response in Mycobacterium tuberculosis. PLoS One. 2010;5:e11622. doi: 10.1371/journal.pone.0011622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L, Sutter BM, Ye X, Tu BP. Trehalose is a key determinant of the quiescent metabolic state that fuels cell cycle progression upon return to growth. Mol Biol Cell. 2010;21:1982–1990. doi: 10.1091/mbc.E10-01-0056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi W, Zhang X, Jiang X, Yuan H, Lee JS, Barry CE, 3rd, Wang H, Zhang W, Zhang Y. Pyrazinamide inhibits trans-translation in Mycobacterium tuberculosis. Science. 2011;16:1630–1632. doi: 10.1126/science.1208813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuman S, Glickman MS. Bacterial DNA repair by non-homologous end joining. Nat Rev Microbiol. 2007;5:852–861. doi: 10.1038/nrmicro1768. [DOI] [PubMed] [Google Scholar]
- Sillje HH, Paalman JW, ter Schure EG, Olsthoorn SQ, Verkleij AJ, Boonstra J, Verrips CT. Function of trehalose and glycogen in cell cycle progression and cell viability in Saccharomyces cerevisiae. J Bacteriol. 1999;181:396–400. doi: 10.1128/jb.181.2.396-400.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Manjunatha U, Boshoff HI, Ha YH, Niyomrattanakit P, Ledwidge R, Dowd CS, Lee IY, Kim P, Zhang L, et al. PA-824 kills nonreplicating Mycobacterium tuberculosis by intracellular NO release. Science. 2008;322:1392–1395. doi: 10.1126/science.1164571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirakova TD, Deb C, Daniel J, Singh HD, Maamar H, Dubey VS, Kolattukudy PE. Wax ester synthesis is required for Mycobacterium tuberculosis to enter in vitro dormancy. PLoS One. 2012;7:e51641. doi: 10.1371/journal.pone.0051641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RM, Williams SB. Circadian rhythms in gene transcription imparted by chromosome compaction in the cyanobacterium Synechococcus elongatus. Proc Natl Acad Sci USA. 2006;103:8564–8569. doi: 10.1073/pnas.0508696103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stallings CL, Stephanou NC, Chu L, Hochschild A, Nickels BE, Glickman MS. CarD is an essential regulator of rRNA transcription required for Mycobacterium tuberculosis persistence. Cell. 2009;138:146–159. doi: 10.1016/j.cell.2009.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinhaus EA, Birkeland JM. Studies on the Life and Death of Bacteria: I. The Senescent Phase in Aging Cultures and the Probable Mechanisms Involved. J Bacteriol. 1939;138:249–261. doi: 10.1128/jb.38.3.249-261.1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stragier P, Losick R. Molecular genetics of sporulation in Bacillus subtilis. Annu Rev Genet. 1996;30:297–241. doi: 10.1146/annurev.genet.30.1.297. [DOI] [PubMed] [Google Scholar]
- Summers EL, Meindl K, Uson I, Mitra AK, Radjainia M, Colangeli R, Alland D, Arcus VL. The structure of the oligomerization domain of Lsr2 from Mycobacterium tuberculosis reveals a mechanism for chromosome organization and protection. PLoS One. 2012;7:e38542. doi: 10.1371/journal.pone.0038542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talukder AA, Yanai S, Nitta T, Kato A, Yamada M. RpoS-dependent regulation of genes expressed at late stationary phase in Escherichia coli. FEBS Lett. 1996;386:177–180. doi: 10.1016/0014-5793(96)00426-7. [DOI] [PubMed] [Google Scholar]
- Valcourt JR, Lemons JM, Haley EM, Kojima M, Demuren OO, Coller HA. Staying alive: metabolic adaptations to quiescence. Cell Cycle. 2012;11:1680–1696. doi: 10.4161/cc.19879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayan V, Zuzow R, O'Shea EK. Oscillations in supercoiling drive circadian gene expression in cyanobacteria. Proc Natl Acad Sci USA. 2009;106:22564–22568. doi: 10.1073/pnas.0912673106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S, Zimmermann M, Goodwin MB, Sauer U, Barry CE, 3rd, Boshoff HI. Fumarate reductase activity maintains an energized membrane in anaerobic Mycobacterium tuberculosis. PLoS Pathog. 2011;7:e1002287. doi: 10.1371/journal.ppat.1002287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayne LG. Dynamics of submerged growth of Mycobacterium tuberculosis under aerobic and microaerophilic conditions. Am Rev Respir Dis. 1976;114:807–811. doi: 10.1164/arrd.1976.114.4.807. [DOI] [PubMed] [Google Scholar]
- Wayne LG. Synchronized replication of Mycobacterium tuberculosis. Infect Immun. 1977;17:528–530. doi: 10.1128/iai.17.3.528-530.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner-Washburne M, Braun EL, Crawford ME, Peck VM. Stationary phase in Saccharomyces cerevisiae. Mol Microbiol. 1996;19:1159–1166. doi: 10.1111/j.1365-2958.1996.tb02461.x. [DOI] [PubMed] [Google Scholar]
- Wolfe AJ. The acetate switch. Microbiol Mol Biol Rev. 2005;69:12–50. doi: 10.1128/MMBR.69.1.12-50.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Siddiqi N, Rubin EJ. Differential antibiotic susceptibilities of starved Mycobacterium tuberculosis isolates. Antimicrob Agents Chemother. 2005;49:4778–4780. doi: 10.1128/AAC.49.11.4778-4780.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Wu J, Oliver SG. Gis1 is required for transcriptional reprogramming of carbon metabolism and the stress response during transition into stationary phase in yeast. Microbiology. 2009;155:1690–1698. doi: 10.1099/mic.0.026377-0. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Wade NM, Scorpio A, Zhang H, Sun Z. Mode of action of pyrazinamide: disruption of Mycobacterium tuberculosis membrane transport and energetics by pyrazinoic acid. J Antimicrob Chemother. 2003;52:790–795. doi: 10.1093/jac/dkg446. [DOI] [PubMed] [Google Scholar]
- Zhou X, Cegelski L. Nutrient-dependent structural changes in S. aureus peptidoglycan revealed by solid-state NMR spectroscopy. Biochemistry. 2012;51:8143–8153. doi: 10.1021/bi3012115. [DOI] [PMC free article] [PubMed] [Google Scholar]