Abstract
Ribonucleotides are frequently incorporated into DNA during replication. They are recognized and processed by several cellular enzymes, and their continued presence in the yeast nuclear genome results in replicative stress and genome instability. Thus, it is important to understand the effects of isolated ribonucleotide incorporation on DNA structure. Towards this goal, here we describe the NMR structure of the self-complementary Dickson dodecamer sequence [d(CGC)rGd(AATTCGCG)]2 containing two symmetrically positioned riboguanosines. The absence of an observable H1-H2 scalar coupling interaction indicates a C3'-endo conformation for the ribose. Longer-range structural perturbations resulting from the presence of the ribonucleotide are limited to the adjacent and trans-helical nucleotides, while the global B-form DNA structure is maintained. Since crystallographic studies have indicated that isolated ribonucleotides promote global B→A transitions, we also performed molecular modeling analyses to evaluate the structural consequences of higher ribonucleotide substitution levels. Increasing the ribonucleotide content increased the minor groove width toward values more similar to A-DNA, but even 50% ribonucleotide substitution did not fully convert the B-DNA to A-DNA. Comparing the present structure with the structure of an RNase H2-bound DNA supports the conclusion that, as with other DNA-protein complexes, the DNA conformation is strongly influenced by the interaction with the protein.
Introduction
The integrity of DNA-based genomes rests on the ability of replicative DNA polymerases to select not only the correct bases, but also the 2'-deoxy form of the incoming nucleotide. This selection must be accomplished against a background of a significant ribo/deoxyribo-nucleotide excess.1–3 The extent to which DNA polymerases prevent rNTP insertion depends on the polymerase and the base examined, with selectivity for insertion of dNTPs over rNTPs varying from 10-fold to greater than 106-fold.4,5 That sugar discrimination is imperfect is illustrated by the fact that S. cerevisiae replicative DNA polymerases 〈, ™ and Σ stably incorporate rNMPs into DNA both in vitro3 and in vivo.3,5 The presence of ribonucleotides in newly synthesized DNA has several important implications, including reduced genome stability and potentially significant perturbations of interactions with proteins and enzymes that recognize DNA. Examples include DNA synthesis by DNA polymerase Σ , which is impeded by a single rNMP in a DNA template,3 DNA backbone incision by topoisomerase 1, which preferentially cleaves duplex DNA containing a single ribonucleotide,6,7 and assembly of nucleosomes onto duplex DNA, which occurs with reduced efficiency if a ribonucleotide is present in duplex DNA.8 It therefore becomes important to understand the structural and biochemical perturbations that result from the presence of ribonucleotides that have been incorporated into DNA.
Both NMR and X-ray crystallography have been used previously to characterize the structure of dsDNA containing short sequences of ribonucleotides that substitute for the corresponding deoxyribonucleotides.9–17 Such sequences are related to Okazaki fragments and to retroviral tRNA primer-DNA junctions,12,13,18 but they do not correspond to the isolated ribonucleotides expected to be incorporated into newly synthesized DNA by DNA polymerases. Several studies also have been directed at characterizing dsDNA containing isolated ribonucleotide substitutions.17,19–22 Interestingly, all of the X-ray studies indicate that these isolated substitutions alter the global conformation of the DNA from B-form to A-form, with most of the deoxyribose sugars adopting C3'-endo or closely related conformations, while the single solution state study indicates only a localized perturbation in the region of the ribonucleotide. Hence, these studies represent one of the clearest examples of structural results that strongly correlate with the method of determination.
Since the NMR study by Jaishree et al.17 utilized a shorter, less frequently analyzed sequence and did not provide detailed structural parameters (no pdb file is available for this study), we have investigated the conformational effect of an isolated ribonucleotide by introduction of a guanosine residue at position 4 in the extensively characterized self-complementary Dickerson dodecamer sequence.23 An analogous model structure has previously been used to evaluate the conformational effects of an isolated arabinonucleotide in dsDNA.24 In order to facilitate the presentation, in the present study we have referred to the ribonucleotide-containing dodecamer structure and to other dsDNA sequences contaminated with isolated ribonucleotides as “rcDNA”. Molecular modeling studies were also performed in order to provide further insights into the extent to which a local perturbation of this nature influences global conformational properties.
Materials and Methods
Sample Preparation and NMR Experiments
The [d(CGC)r(G)d(AATTCGCG)]2 oligomer was purchased from Integrated DNA Technologies (Coralville, IA). The oligomer was dissolved in either 99.9% D2O or 90% H2O/10% D2O at ~2 mM duplex concentration. Each sample also contained 50 mM NaCl, 50 mM sodium phosphate at pH 7.0, and 0.5 mM EDTA. Residual dipolar coupling (RDC) measurements were obtained by comparing spectra acquired with the isotropic D2O sample with spectra obtained with 20 mg/ml Pf1 phage (Asla Biotech, Latvia) added to the sample. The D2O and H2O spectra were acquired in Shigemi NMR tubes (Shigemi Inc., Allison Park, PA) containing 200 µl of sample. A third D2O sample in a standard 5 mm tube was used to make the RDC measurements.
Standard 2D NMR experiments were used to calculate the solution structure of the rG4-substituted oligomer25–27. In order to compare the structure to the unsubstituted Dickerson dodecamer, a calculation was performed on the unsubstituted molecule using similar experimental restraints to those provided with the XPLOR-NIH release. In the remainder of this manuscript, the NMR-calculated rG4-substituted structure will be referred to as rG4-DNA, and the newly calculated unsubstituted Drew-Dickerson dodecamer structure as dd-DNA. See supporting information for details of the rG4-DNA and dd-DNA 2D NMR experiments and structure calculations.
Molecular Dynamics Simulations
Using the ten best NMR structures as the starting configurations, ten different molecular dynamics (MD) trajectories were calculated for rG4-DNA. MD simulations were also performed on dd-DNA (details provide as supplementary information). In order to evaluate the effect of a higher level of ribonucleotide incorporation, we also performed simulations on the sequences CGC(rG)AA(rU)TCG(rC)G (25% ribonucleotide) and C(rG)C(rG)A(rA)T(rU)C(rG)C(rG) (50% ribonucleotide). These sequences, containing the same base sequence used in the rG4-DNA model, were selected to minimize both the linear and cross-strand proximity of the ribonucleotides. The initial configurations were derived from the idealized template of RNA in its canonical A-form. Appropriate O2'H groups were replaced from this template to obtain the dodecamers with the desired compositions.
Results
Chemical Shifts and H-bonding
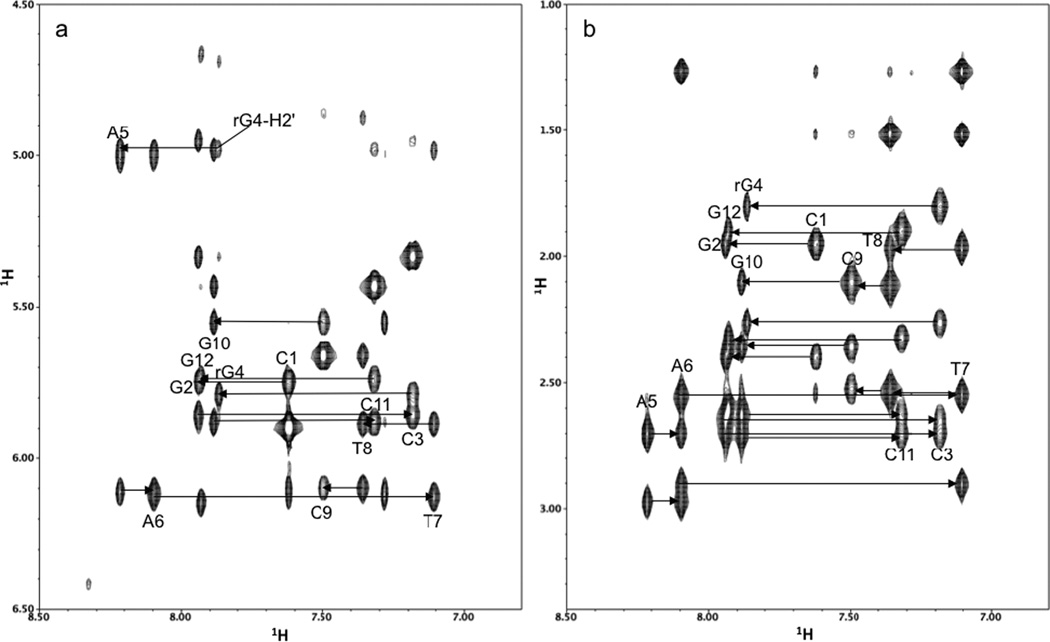
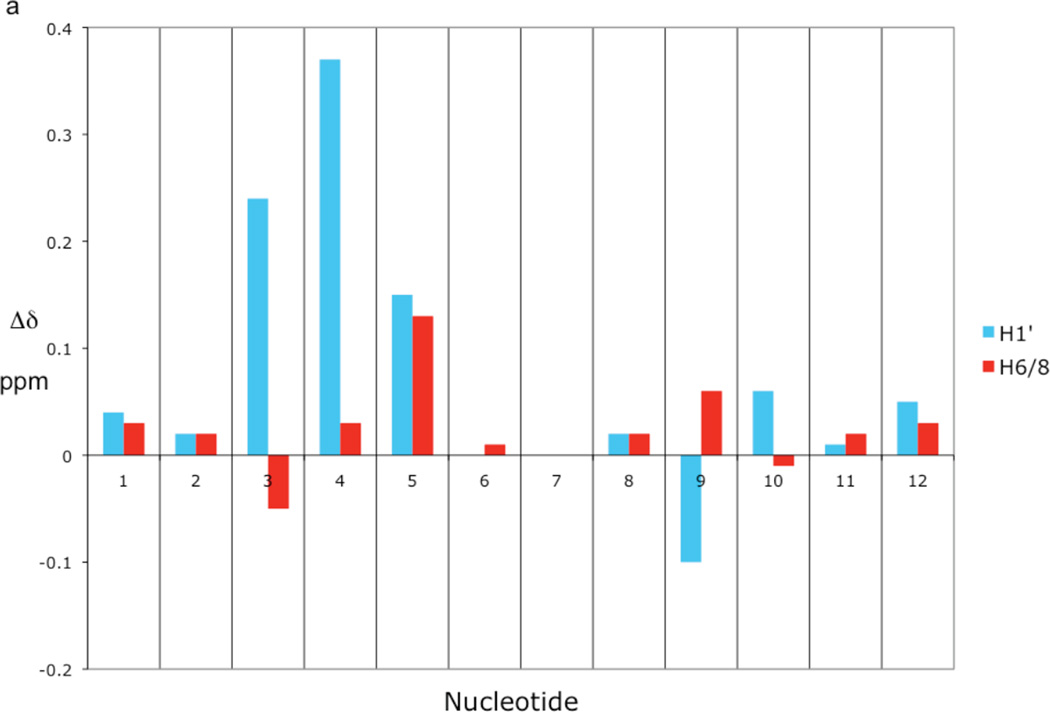
The sequential chemical shift assignments between the base aromatic protons and the sugar H1', H2' and H2" protons were made from the 2D NOESY spectra as shown in Figure 1. Once these protons were assigned, the remaining sugar protons were assigned from a combined analysis of 2D NOESY, TOCSY and 1H/31P COSY spectra. The 1H/31P COSY spectrum was particularly useful for assigning the H5' and H5", as well as for assigning the 31P chemical shifts. A comparison of the base H6/H8 and sugar H1', H2', H2", H3', and H4' chemical shifts with those reported in the literature for the Dickerson dodecamer30 show only minor shift changes localized in the vicinity of the ribonucleotide substitution (Table 1a, Figure 2a). Besides the rG4 anomeric proton, only the anomeric protons of the adjacent nucleotides, C3 and A5, and the complementary nucleotide to rG4, C9, change by more than 0.1 ppm compared to the unmodified DNA shift. The only base aromatic proton exhibiting a shift change greater than 0.1 ppm is the A5-H8 proton. This increase in chemical shift may be caused by greater overlap of the rG4 and A5 bases (see supporting information, Figure S1). The other base H6/H8 protons, including the rG4-H8 proton, show surprisingly small shift changes. The base imino and most of the amino protons were assigned from the 200 ms NOESY spectrum in H2O. Only the rG4 imino proton chemical shift in rG4-DNA shows any appreciable change compared to the ddDNA imino proton shifts reported by Moe and Russu31 (Table 1b). These chemical shift results imply that the DNA solution structure is only locally perturbed by the ribonucleotide substitution, and the imino proton shifts indicate that the base pair hydrogen bonding is only affected at the site of the ribose substitution. As mentioned above, the imino proton region of the 200 ms NOESY spectrum in H2O is little changed from that reported for the unsubstituted dodecamer,32 indicating that the base pair hydrogen bonding is not appreciably affected by the ribonucleotide substitution.
Figure 1. NOESY connectivities of base H6/H8 and H1'/H2',H2" protons in rG4-substituted Dickerson dodecamer.
(a) Finger print region of the 100 ms mixing time NOESY spectrum of rG4-DNA, showing the aromatic to H1' intra-residue and sequential connectivities. (b) Finger print region of the 100 ms mixing time NOESY spectrum of rG4-DNA, showing the aromatic to H2'/2" intra-residue and sequential connectivities.
Table 1.
| a. Comparison of rG4-DNA and DNA 1H chemical shifts | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Base | H1'a | H6/8 | H2' | H2" | H3' | H4' | ||||||
| C1 | 5.75 | 5.71 | 7.62 | 7.59 | 1.96 | 1.89 | 2.31 | 2.35 | 4.69 | 4.65 | 4.06 | 4.15 |
| G2 | 5.86 | 5.84 | 7.94 | 7.92 | 2.65 | 2.59 | 2.70 | 2.68 | 4.95 | 4.91 | 4.33 | 4.31 |
| C3 | 5.79* | 5.55 | 7.19 | 7.24 | 1.80 | 1.81 | 2.15 | 2.22 | 4.84 | 4.78 | 4.16 | 4.17 |
| rG4/G4 | 5.79* | 5.42 | 7.86 | 7.83 | 4.74* | 2.63 | 2.74 | 4.86* | 4.98 | 4.30 | 4.29 | |
| A5 | 6.11* | 5.96 | 8.22* | 8.09 | 2.70 | 2.65 | 2.98 | 2.90 | 5.02 | 5.05 | 4.45 | 4.44 |
| A6 | 6.13 | 6.13 | 8.10 | 8.09 | 2.56 | 2.53 | 2.91 | 2.92 | 4.99 | 5.00 | 4.44 | 4.44 |
| T7 | 5.89 | 5.89 | 7.11 | 7.11 | 1.97 | 1.98 | 2.55 | 2.54 | 4.80 | 4.82 | 4.19 | 4.24 |
| T8 | 6.10 | 6.08 | 7.37 | 7.35 | 2.13 | 2.17 | 2.53 | 2.54 | 4.88 | 4.89 | 4.19 | 4.24 |
| C9 | 5.57* | 5.67 | 7.50 | 7.44 | 2.10 | 2.02 | 2.36 | 2.40 | 4.73 | 4.87 | 4.11 | 4.20 |
| G10 | 5.88 | 5.82 | 7.88 | 7.89 | 2.63 | 2.58 | 2.70 | 2.68 | 4.98 | 4.97 | 4.37 | 4.35 |
| C11 | 5.74 | 5.73 | 7.32 | 7.30 | 1.89 | 1.85 | 2.33 | 2.28 | 4.81 | 4.79 | 4.13 | 4.20 |
| G12 | 6.15 | 6.10 | 7.93 | 7.90 | 2.36 | 2.34 | 2.60 | 2.57 | 4.67 | 4.66 | 4.17 | 4.20 |
| b. Comparison of rG4-DNA and DNA 1H imino chemical shifts | ||
|---|---|---|
| Base pair | rG4-DNA imino shift | DNA imino shift |
| C1-G12 | N/Oa | N/Oa |
| G2-C11 | 13.03 | 13.02 |
| C3-G10 | 12.84 | 12.85 |
| rG4-C9/G4-C9 | 12.77* | 12.65 |
| A5-T8 | 13.71 | 13.72 |
| A6-T7 | 13.61 | 13.60 |
The first column under each proton heading contains the rG4-DNA proton chemical shifts; the second column contains the DNA shifts from Hare et al.30
rG4-DNA chemical shifts that deviate by ≥ 0.1 ppm from the corresponding DNA values.
Not observed at 25°C. The DNA imino chemical shifts are from Moe and Russu.31
rG4-DNA chemical shifts that deviate by > 0.01 ppm from the corresponding DNA values.
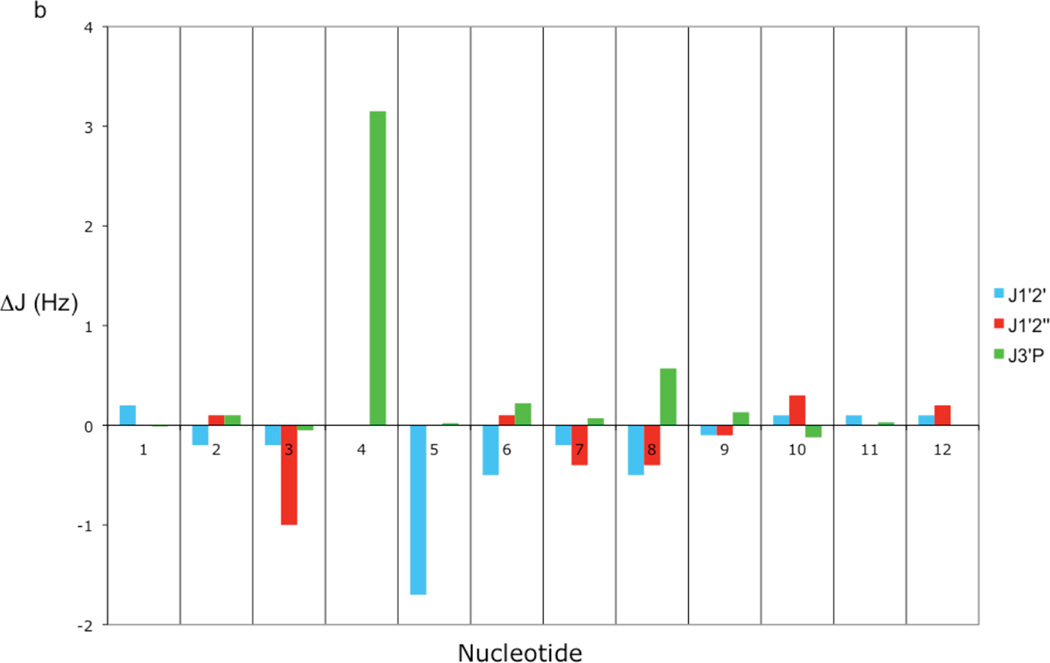
Figure 2. Comparison of chemical shift and scalar coupling of rG4-substituted and unsubstituted Dickerson dodecamer.
(a) Bar graph showing the differences between rG4-DNA and DNA H1' and H6/8 chemical shifts for each nucleotide. The differences were computed by subtracting the DNA value from the rG4-DNA value. The DNA chemical shifts are the values reported by Hare et al.30 (b) Bar graph showing the differences between rG4-DNA and DNA scalar coupling constants 3J1'2'3J1'2", and 3J3'(i)P(i+1). The differences were computed by subtracting the DNA value from the rG4-DNA value. The 3J1'2' and 3J1'2" scalar coupling values are from Bax and Lerner33 and the 3J3'(i)P(i+1) scalar coupling values are from Wu et al.34
Coupling Constants
A comparison of the 3J1'2' and 3J1'2" coupling constants with those determined for the unsubstituted Dickerson dodecamer33 show only minor changes localized in the vicinity of the ribonucleotide substitution (Table 2, Figure 2b). In fact, other than the 3J1'2' coupling of the ribose sugar of rG4, which is not observed in the COSY and TOCSY spectra, only the 3J1'2' coupling of the ribose sugar of A5 and the 3J1'2" coupling of the ribose sugar of C3 are appreciably perturbed by the ribose substitution. These changes may be reflected by the increase in pseudorotation angles of the C3 and A5 sugars in the rG4-DNA structures relative to the dd-DNA structures (see Table 4). In the case of the heteronuclear H3'-31P couplings, 3J3'P, only the ribose rG4-H3' to A5-31P coupling is significantly perturbed (Table 2 and Figure 2b). The value of 3J3'P increases from 4.01 Hz in the Dickerson dodecamer34 to 7.16 Hz in the ribonucleotide substituted dodecamer. The remaining 3J3'P couplings agree well with those measured for the Dickerson dodecamer.
Table 2.
Comparison of rG4-DNA and DNA scalar coupling constantsa.
| rG4-DNA | DNA | rG4-DNA | DNA | rG4-DNA | DNA | |
|---|---|---|---|---|---|---|
| Base | 3J1'2' | 3J1'2" | 3JH3'(i)-P(i+1) | |||
| C1 | 8.4 | 8.2 | 6.1 | 6.1 | 5.95 | 5.96 |
| G2 | 9.9 | 10.1 | 5.8 | 5.7 | 3.86 | 3.76 |
| C3 | 8.6 | 8.8 | 5.2* | 6.2 | 5.5 | 5.30 |
| G4 | N/Ob | 10.2 | N/Ac | 5.1 | 7.16* | 4.01 |
| A5 | 8.0* | 9.7 | 5.7 | 5.7 | 3.03 | 3.01 |
| A6 | 8.8 | 9.3 | 6.1 | 6.0 | 2.65 | 2.43 |
| T7 | 8.3 | 8.5 | 5.8 | 6.2 | 2.81 | 2.74 |
| T8 | 9.0 | 9.5 | 5.6 | 6.0 | 3.86 | 3.29 |
| C9 | 8.6 | 8.7 | 5.9 | 6.0 | 5.29 | 5.16 |
| G10 | 9.8 | 9.7 | 5.8 | 5.5 | 3.84 | 3.96 |
| C11 | 8.5 | 8.4 | 6.2 | 6.2 | 5.35 | 5.32 |
| G12 | 8.2 | 8.1 | 6.5 | 6.3 | N/Ad | N/Ad |
The DNA 3J1'2' and 3J1'2" scalar coupling values are from Bax and Lerner,33; the DNA, 3JH3'(i)-P(i+1) scalar couplings are from Wu et. al.34
The H1'-H2' cross-peak is not observed in the 2D COSY spectrum.
The H2" proton is replaced by an hydroxyl group in the ribonucleotide.
G12 is the final 3' nucleotide.
rG4-DNA coupling constants that differ by ≥ 1 Hz from the corresponding DNA coupling constants.
Table 4.
| Nucleotide | rG4-DNA | dd-DNA | ||
|---|---|---|---|---|
| P (°) | Pucker | P (°) | Pucker | |
| C1 | 153 ± 39c | C2'-endo | 89 ± 4 | O4'-endo |
| G2 | 157 ± 2 | C2'-endo | 161 ± 1 | C2'-endo |
| C3 | 143 ± 6 | C1'-exo | 100 ± 4 | O4'-endo |
| rG4/G4 | 33 ± 2 | C3'-endo | 153 ± 1 | C2'-endo |
| A5 | 165 ± 2 | C2'-endo | 154 ± 1 | C2'-endo |
| A6 | 155 ± 1 | C2'-endo | 150 ± 2 | C2'-endo |
| T7 | 133 ± 1 | C1'-exo | 132 ± 1 | C1'-exo |
| T8 | 152 ± 1 | C2'-endo | 146 ± 2 | C2'-endo |
| C9 | 160 ± 1 | C2'-endo | 152 ± 1 | C2'-endo |
| G10 | 161 ± 1 | C2'-endo | 152 ± 1 | C2'-endo |
| C11 | 150 ± 2 | C2'-endo | 114 ± 3 | C1'-exo |
| G12 | 149 ± 2 | C2'-endo | 146 ± 1 | C2'-endo |
Average sugar pseudorotation angles and sugar puckers of the five lowest energy structures obtained from 3DNA version 2.0.35 The standard deviations calculated for the five lowest energy structures underestimate the true uncertainty in the pseudorotation angle.
The C1 pseudorotation angle of one of the structures was 84° which accounts for the large standard deviation and may be a reflection of the greater conformational averaging of terminal base pair.
Solution Structure and Comparison with the Dickerson Dodecamer
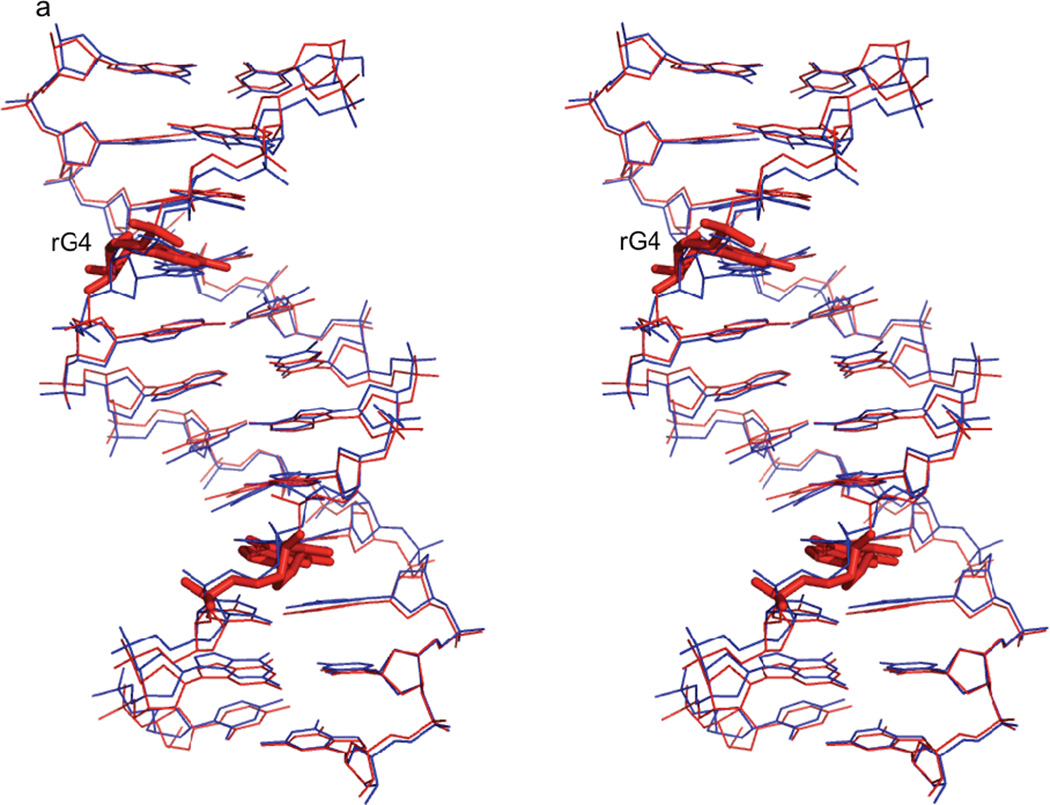
The structural statistics for the calculation of the rG4-DNA molecule for the five best structures is given in Table 3. These structures have the lowest energies and restraint violations from the ensemble of ten structures, and superimpose on their average structure with an RMSD of 0.211 ± 0.075 Å for all the atoms. The five best dd-DNA structures, calculated with similar restraints, exhibit an even lower average RMSD of 0.164 ± 0.028 Å, so the low RMSD does not appear to be caused by overly tight restraints. In fact, the low RMSD for the structures was not significantly affected by loosening the NOE distance restraints by binning the restraints as strong, medium, weak, and very weak, corresponding to distances of 1.8 – 2.8, 1.8 – 3.4, 1.8 – 5.0, and 1.8 – 6.0 Å, respectively. The five best rG4-DNA structures have been deposited in the RCSB PDB database (PDB ID code 2L7D). A 3DNA v2.0 analysis35 classifies all but the 4th and 8th base-pair steps, which contain the ribonucleotide substitutions, as B-like for all five structures. The lowest energy solution structure of the rG4-DNA molecule superimposes on the lowest energy dd-DNA structure, calculated with similar restraints as described above, with an RMSD of 0.750 Å for all atoms (Figure 3a). An expanded view of the overlay of the lowest energy rG4-DNA and dd-DNA model structures showing the rG4/G4 and flanking nucleotides (Figure 3b) illustrates the local perturbations to the structure caused by the ribose substitution. The primary perturbation is a change in the sugar pucker conformation, which goes from C2'-endo in the dd-DNA model to C3'-endo in the rG4-DNA model, resulting in a decrease in the average δ torsion angle from 138° to 81°. This change in the sugar pucker also causes a slight change in the position of the guanosine base in the rG4-DNA structure relative to the dd-DNA structure (Figure 3b).
Table 3.
Structural Statistics
| Experimental Restraints | RMSD | Violations | Number |
|---|---|---|---|
| NOE distance (Å) | 0.071 ± 0.002 | 0 (> 0.5 Å) | 286a |
| δ (°) | 0.186 ± 0.415 | 0 (> 5°) | 24 |
| DHH (Hz) | 1.117 ± 0.055 | 158 | |
| D3'P (Hz) | 0.189 ± 0.010 | 22 | |
| 3J3'P (Hz) | 0.186 ± 0.009 | 22 |
Total distance and H-bond restraints.
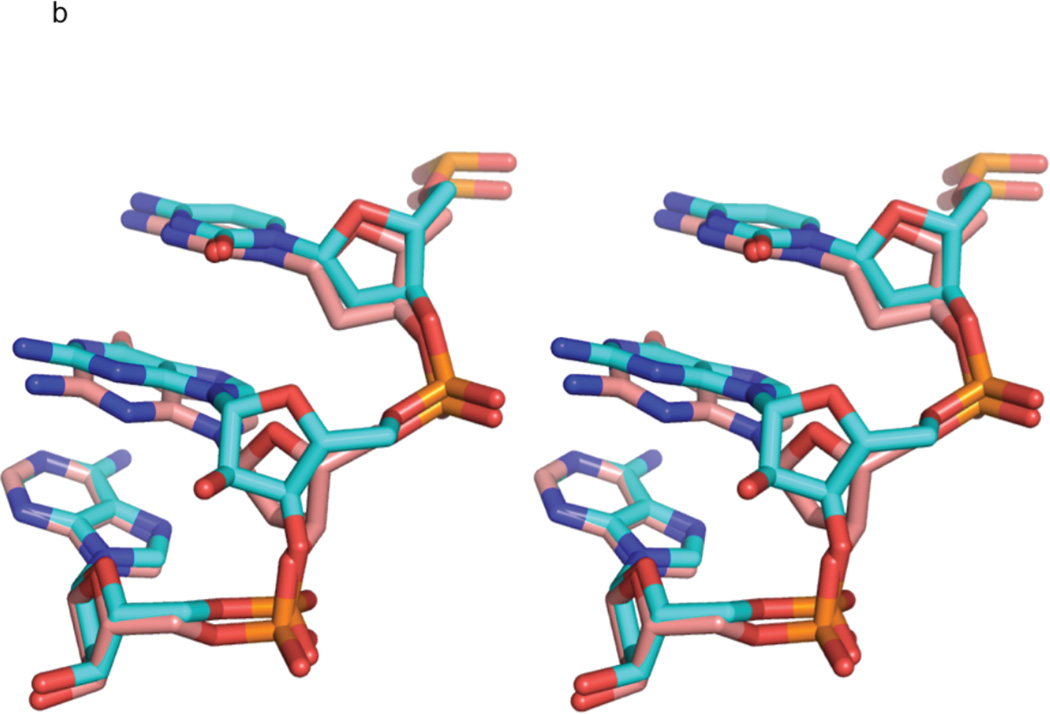
Figure 3. Comparison of rG4-substituted and unsubstituted Dickerson dodecamer NMR solution structures.
a) Stereo view of the superposition of the lowest energy rG4-DNA (red) and dd-DNA (blue) structures. The structures superimpose with an RMSD of 0.750 Å for all atoms. The rG4 ribonucleotide and its complement are highlighted as a stick rendering. The dd-DNA structures were computed using the same XPLOR-NIH simulated annealing calculation as the rG4-DNA with a similar set of experimental restraints as described in the text. The view is into the major groove along the major helical axis. 3b) A stereo view of nucleotides C3, rG4/G4, and A5 of the lowest energy rG4-DNA (cyan) and dd-DNA (coral) structures, showing the slight change in guanosine base position and sugar pucker from C2'-endo in the dd-DNA structure to C3'-endo in the rG4-DNA structure. The view is looking into the minor groove.
The average differences between the five best rG4-DNA and dd-DNA structures for all the backbone torsion angles are plotted in Figure 4. Changes in the rG4-DNA backbone due to the ribose substitution are located primarily at the site of the substitution and the flanking nucleotides. The decrease in δ in the rG4-DNA molecule appears to be largely compensated by an increase in ζ, and to a lesser extent increases in ε and γ. The average ζ, torsion angle of nucleotide 4 increases from −113° in the dd-DNA structures to −73° in the rG4-DNA structures. Other changes in the torsion angles of the ribonucleotide and flanking deoxynucleotides help to accommodate the change in sugar pucker conformation and to rotate the A5-C5' methylene group away from the bulky C2' hydroxyl group of the ribonucleotide as shown in Figure 3b.
Figure 4. Comparison of backbone torsion angles in rG4-substituted and unsubstituted Dickerson dodecamer NMR solution structures.
Plot of the average differences in backbone torsion angles of the five best rG4-DNA and dd-DNA solution structures for each nucleotide. The differences were computed by subtracting the dd-DNA values from the rG4-DNA values.
The average pseudorotation phase angles and sugar puckers of the five lowest energy structures are given in Table 4. Other than the first deoxyribose sugar at C1 and the site of the ribose substitution at rG4, the sugar pucker conformations are similar and fall primarily in the southern region of the pseudorotation wheel.36 As mentioned above, the ribose sugar pucker at rG4 has an N-type C3'-endo sugar pucker (P = 33), while the corresponding deoxyribose sugar pucker in the dd-DNA structure is an S-type C2'-endo pucker (P = 153). This is not surprising since these are the predominant sugar puckers for ribose sugars in RNA and deoxyribose sugars in DNA.37 The drop in pseudo rotation angle gives rise to an increase in the average glycosidic torsion angle from -103 in the dd-DNA structures to -152 in the rG4-DNA structures. The average C1 deoxyribose sugar pucker of the rG4-DNA structures is also C2'-endo, but the average C1 deoxyribose sugar pucker of the dd-DNA structures is O4'-endo, E-type configuration. The reason for the difference in these two sugar puckers is probably fraying of the terminal base pairs, e.g., the imino protons of the C1-G12 base pair are not observed at 25 °C, indicating greater disorder at the ends of the duplex. Additionally, the NOE distance restraints and DHH restraints used for the dd-DNA structure calculation were obtained from Tjandra et al.26 and Wu et al.27, and are somewhat different than the restraints obtained in this study used to compute the rG4-DNA structures.
Base-pair Parameters Shear, Stretch, Opening and Buckle
The base-pair parameters were obtained using 3DNA version 2.0.35 As mentioned above, a 3DNA analysis of the best five rG4-DNA structures classifies all but the fourth and eighth nucleotide pairs, i.e., rG4-C9 to A5-T8 and T8-A5 to C9-rG4, respectively, as B-like for all five structures. The base pair parameters shear, stretch, and opening are important for characterizing the hydrogen bonding between the base pairs.35 Shear and stretch define the relative offset of the bases in the base pair plane, and opening is the angle between two bases in the base pair plane. The local base pair parameters shear, stretch, and opening for the average of the NMR-calculated rG4-DNA (blue curves) and dd-DNA (orange curves) structures are plotted in Figure 5a-c. Other than slight changes near the ribonucleotide substitutions, these parameters are not appreciably altered by the substitutions.
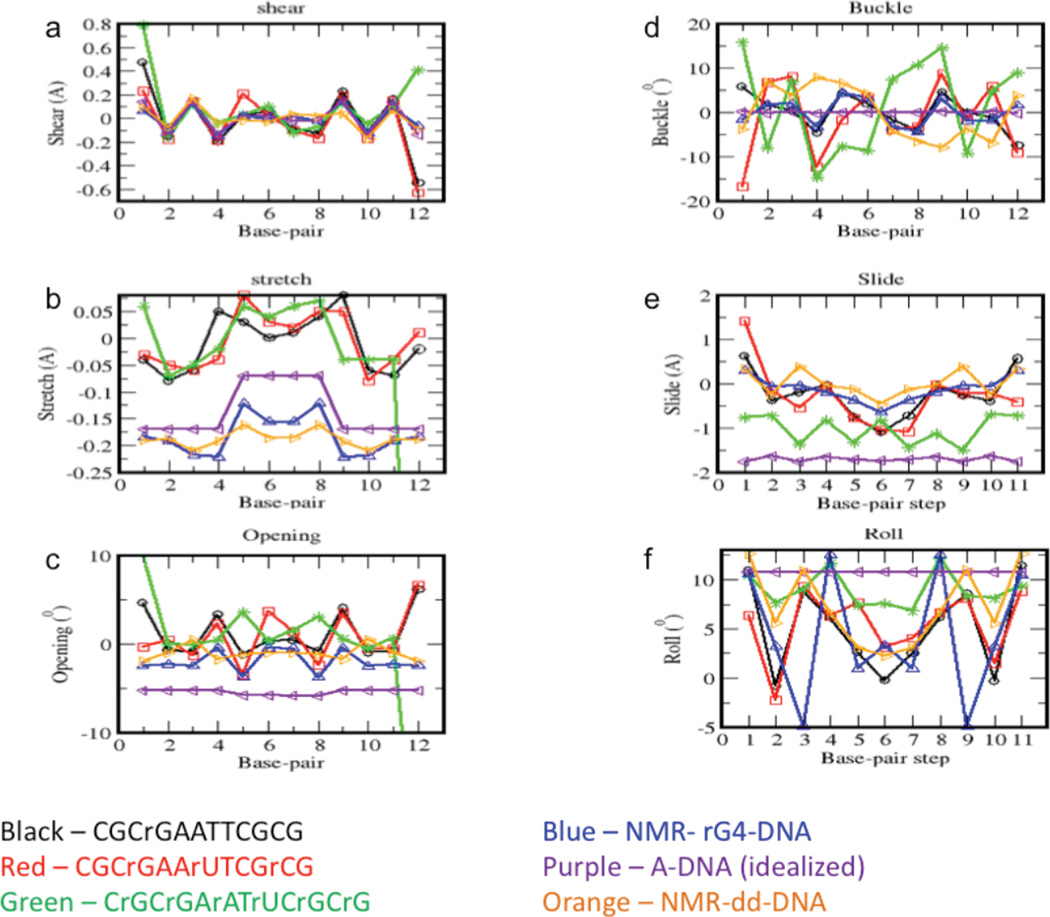
Figure 5. Structural parameters for NMR experimental and modeled rcDNA.
Plots of average calculated values of the shear (a), stretch (b), opening (c), buckle (d), roll (e), and twist (f) for the two NMR-based structures, the dd-DNA (orange) and rG4-DNA (blue), and for four model rcDNA structures. The modeled structures correspond to: CGCrGAATTCGCG = rG4-DNA (black); CGCrGAArUTCGrCG (red); CrGCrGArAUrTCrGCrG (green); and Dickerson dodecamer sequence in an idealized A-DNA geometry (purple).
Buckle defines the angle between the planes of the base pairs, and it has been observed that buckle changes sign from positive to negative at DNA-RNA junctions.11,13 This change in sign is also observed in the present set of structures. The average rG4-DNA buckle of base pair 3, C3-G10, is 1.98° and drops to −3.26° for base pair 4, rG4-C9, containing the ribonucleotide (Figure 5d, blue curve). The largest difference in average rG4-DNA buckle compared to the dd-DNA buckle (Figure 5d, orange curve) also occurs at base pair 4. The average buckle of base pair 4 drops from 7.93° in the dd-DNA structures to −3.26° in the rG4-DNA structures. The average buckle of the remaining base pairs is similar between the two sets of structures, but the average buckle of base pair 2, G2-C11, drops slightly from 6.91° in the dd-DNA structures to 1.83° in the rG4-DNA structures (Figure 7d). Due to the symmetry of the duplex, the buckle values reverse sign in the second half of the duplex.
Figure 7. Effect of structural regularity on crystallization of ribonucleotide-containing DNA.
Repetitive conformational regularity may result in better lattice contacts, providing a partial explanation for the crystallographic selectivity for minor but more regular solution conformations. The ribonucleotide-containing base pair is indicated as having an A' or A-like conformation.
Base-pair Step Parameters Slide and Roll
Slide specifies the lateral displacement between successive base pairs, and roll specifies the angle between successive base pairs. Canonical B-DNA is characterized by no slide or roll, and canonical A-form DNA is characterized by −1.5 Å of slide and +12° of roll.38,39 Figure 5e shows that the average base-pair slide values for the rG4-DNA (blue curves) and dd-DNA (orange curves) structures are similar, with the exception of base-pair step 3, C3-G10 to rG4-C9, containing the ribose substitution in the second base pair. The average slide of base-pair step 3 drops from 0.39 Å in the dd-DNA structures to −0.04 Å in the rG4-DNA structures. None of the values approach −1.5 Å characteristic of A-DNA.40 MD simulations described below show that incorporation of 50 % ribonucleotides did lower the slide value to ~ −1.0. Due to symmetry, the values of slide for the second half of the duplex are mirror images of the values for the first half of the duplex.
The average roll values of the rG4-DNA (blue curves) and dd-DNA (orange curves) structures are shown in Figure 5f. Only base pair steps 3, 4, 8, and 9 containing the ribonucleotide substitutions have appreciably altered roll values in the rG4-DNA structures compared to the dd-DNA structures. The roll of base-pair step 3, C3-G10 to rG4-C9, actually decreases from the more A-like value of 11.00° in the dd-DNA structures to −4.93° in the rG4-DNA structures. This decrease in roll is partially offset by an increase in the roll of base-pair step 4, rG4-C9 to A5-T8, which increases from 6.65° in the dd-DNA structures to the A-like value of 12.53° in the rG4-DNA structures. Again, due to the symmetry of the duplex, the average base-pair step roll values in second half of the duplex are essentially mirror images of the values in the first half of the duplex (Figure 5f). Other than the roll of base-pair steps containing the ribonucleotides, only the first and last base-pair steps have elevated roll characteristic of A-DNA. All the other base-pair steps are characteristic of B-DNA. The 3DNA analyses of the DNA structures computed by Kuszewski et al.25 and Wu et al.27 also reveals elevated roll for the first and last base-pair steps, which may be the result of fraying and greater disorder of the these terminal base-pair steps. In general, the Amber simulations described below indicate that the roll parameter is highly sensitive to the specific nature of the ribonucleotide substitution pattern. However, as with the slide parameter, the roll values of the 50% substituted DNA show a clear trend toward the A-form value (Figure 5f, green curve).
Comparison of Molecular Dynamics Simulations with NMR-calculated Structures
Molecular dynamic simulations based on the Amber force field were performed on the same rcDNA dodecamer sequence containing a guanidine residue at position 4. Results from ten different MD trajectories were averaged. Selected results from the 3DNA analysis are shown in Figure 5 (black curves) along with the results from the dodecamers with higher percentages of ribonucleotides (red, green, and purple curves) and the NMR-calculated structures rG4-DNA (blue curve) and dd-DNA (orange curve). In general, the modeling studies (black curves) showed a reasonable agreement with the NMR-based structural analysis (blue curves). Typically, only the results for the terminal nucleotides differ significantly, and this results largely from fraying and greater disorder of the terminal nucleotides in dsDNA. However, the roll values of the simulated rG4-DNA structures (Figure 5f, black curve) are closer to the NMR-calculated dd-DNA structures (Figure 5f, orange curve) than the NMR-calculated rG4-DNA structures (Figure 5f, blue curve). In particular, the large drop in roll value towards the B-DNA value is not observed for base-pair step 3 in the simulated structures. In addition, the opening value for the rG4 base pair increases from −0.4° in NMR-calculated rG4-DNA (Figure 5c, blue curve) structures to +3.7° in the simulated structure (Figure 5c, black curve). These differences may be due to the fact that no experimental restraints were used in the Amber simulations. Most of the other local fluctuations in the parameters are reproduced in the theoretical analysis. A systematic discrepancy of ~ 0.2 Å is apparent for the base-pair stretch of simulated versus NMR-calculated structures (Figure 5b), which may reflect a difference between the Amber99 and XPLOR-NIH force fields. Overall, the MD simulation results also indicate the localized nature of the structural perturbations resulting from ribonucleotide substitution within DNA.
Simulations were also performed to evaluate the effects of additional, non-sequential ribonucleotides on the conformational behavior of the dodecamer. These studies were motivated by an effort to bridge the gap between the crystallographic and solution state results by providing a further indication of how the fractional incorporation of ribonucleotides might influence the global conformational preferences of rcDNA. In these calculations, ribonucleotides were inserted at positions 4, 7, and 11 (25 % ribonucleotides, Figure 5, red curves), and at each even position (50 % ribonucleotides, Figure 5, green curves) of each self-complementary 12-mer. In all cases, there were no adjacent ribonucleotides or pairs of hydrogen bonded ribonucleotides. For comparison, curves corresponding to B-form DNA and to idealized A-form DNA are shown in orange and purple, respectively. Buckle, slide and roll show a significant dependence on the ribonucleotide substitution pattern, while shear, stretch, and opening are less sensitive to the substitution pattern and provide a better readout of the global conformational preference. For the slide parameter, the 50 % level of ribonucleotide substitution does result in a decrease which makes the behavior more A-like, but the results still differ from those obtained using an idealized A-form geometry. The influence of the fractional ribonucleotide incorporation level is more apparent in plots of the minor groove width, described below.
Minor Groove Width
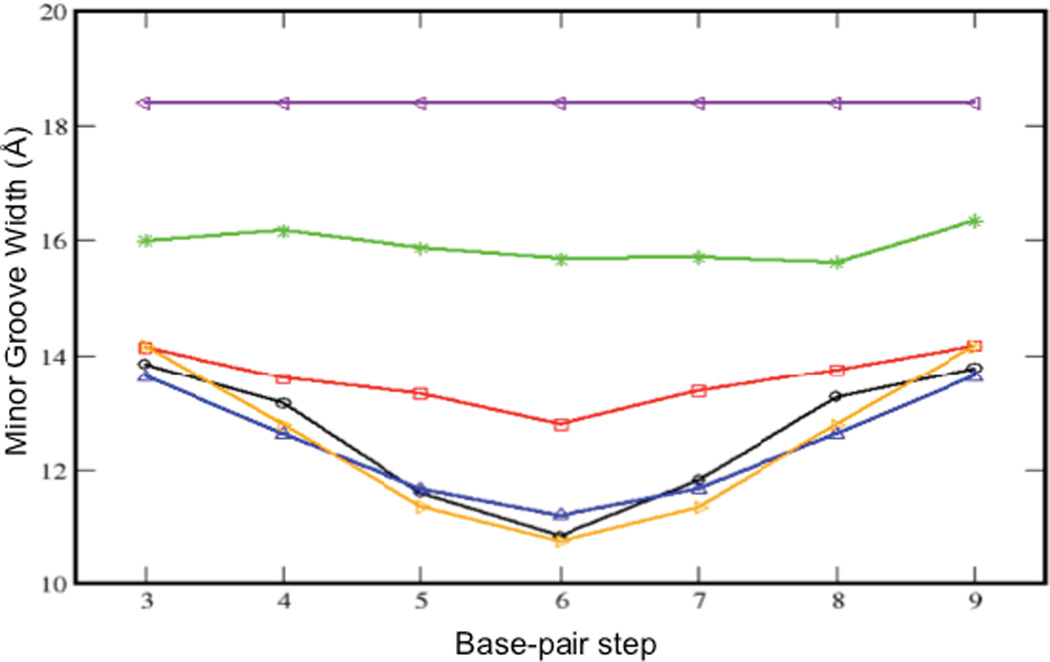
The minor groove width, calculated using 3DNA 2.0 analysis of the NMR-based rG4-DNA structure (Figure 6, blue curve) is in close agreement with the Amber-based calculation for the same sequence (black curve). The results are also in close agreement with the calculation for NMR-determined dd-DNA structure (orange curve). The rG4-DNA structure is apparently able to accommodate the bulky C2' hydroxyl group of the ribose sugar pointing out of the minor groove (Figure 3b), without increasing the minor groove width. As discussed below, the Amber MD simulations demonstrate that progressive increase in the ribonucleotide substitution level results in a more uniform minor groove width, with values that are closer to A-DNA.
Figure 6. Dependence of the minor groove width on ribonucleotide substitution.
Plot of the average minor groove widths for each base-pair step. Results are presented for two NMR-derived conformations and four modeled structures, color-coded as in Figure 7. The blue and black curves correspond to the same rG4-DNA sequence. The progressive effect of ribonucleotide substitution level on the minor groove width is apparent.
The simulations for the unsubstituted DNA (Figure 6, black curve) and the NMR-calculated rG4-DNA (Figure 6, blue curve) and dd-DNA (Figure 6, orange curve) exhibit a significant bowing in the center of the sequence, which appears to be a consequence of the greater tendency of the AT-rich sequence to adopt a more canonical B-form structure characterized by a larger minor groove width.41 In the sequence containing a 25 % substitution level, one of the T residues in the center is replaced by a uridine, and this sequence (Figure 6, red curve) shows significantly less bowing than the unsubstituted DNA and rG4-DNA. Further ribonucleotide incorporation results in an even more significant decrease in minor groove width, although even at the 50 % ribonucleotide substitution level (Figure 6, green curve), the mean minor groove width is significantly below that of the pure A-form (Figure 6, purple curve). This is surprising, given that the crystallographic studies showed that even a single ribonucleotide substitution is sufficient to cause a global transition in short stretches of DNA.19–22
Discussion
Recent studies indicate that replicative DNA polymerases frequently incorporate ribonucleotides into DNA during replication of the nuclear genome in S. cerevisiae3 and in S. pombe.42 The major roles of DNA polymerases epsilon and delta at the eukaryotic replication fork are evolutionarily conserved.42 Some consequences of ribonucleotide incorporation become unmasked in yeast strains lacking RNase H2, which initiates efficient repair of newly incorporated ribonucleotides. Such strains exhibit the characteristics of replicative stress, including slow progression through S phase and genome instability.5 The present investigation was motivated by the need to understand the structural implications of the presence of such isolated ribonucleotides in DNA. There have been several reported studies of the structural perturbations resulting from the introduction of isolated ribonucleotides into double stranded DNA.17,19–22 In each case, the crystallographic result was a global conversion of the entire DNA strand from B-form to A-form, even in the cases in which the ribonucleotide was present at the terminal position. Alternatively, in the one previously reported NMR study of a short sequence containing an isolated cytidine nucleoside, the structural perturbation was much more localized.17 In the present study, we have investigated the structure of the extensively characterized Dickerson dodecamer, modified here to contain a single guanosine ribonucleotide at (equivalent) positions 4 and 21.
Even prior to any structural analysis, the highly localized nature of the perturbation introduced by the ribonucleotide is apparent from a comparison of the 1H shift data and 3JHH data with the results reported for the unsubstituted dodecamer (Tables 1, 2). Excluding the modified position and its two nearest neighbors, all of the proton shifts in the sugar moieties are within 0.1 ppm, and in most cases, much smaller. The ribonucleotide substitution increases the value of the A5 3J1'2' by 1.7 Hz, and reduces rG4-A5 3J3‘p scalar coupling by −3.15 Hz, but introduces no other variations greater than 1 Hz.
The results of the NMR study are supported by a series of AMBER calculations, which also indicate the localized nature of the conformational perturbation introduced by the 2'-OH group. The overall conclusions are similar to those reported for studies of DNA containing an isolated arabinonucleoside,24 2'-fluoro-2'-deoxyribonucleosides,43 as well as theoretical analyses,44 all of which show only localized structural perturbations that do not significantly alter the global B-form conformation. The localized nature of the structural perturbation is also consistent with recent structural data for an RNase H2-rcDNA complex.45 The rcDNA present in the complex, with sequence: d(GACAC)r(C)d(TGATTC)•d(GAATCAGGTGTC) is characterized by a stretch of approximately 7 nucleotides at the protein interface, centered around the ribonucleotide, that adopt an A-form conformation. Nevertheless, although the DNA-enzyme interactions are sufficient to induce A-form geometry over a localized 7-nucleotide segment, the DNA residues beyond this region exhibit a minor groove width more characteristic of B-DNA. Hence, even with the more extensive enzyme-induced A-form geometry, there is no global transition to A-form DNA. Of course, it is not unusual for macromolecular interactions to strongly perturb DNA conformation.
The factors that bias crystal structures toward A-form DNA have been well discussed in the literature and include dehydration and cationic salts that reduce electrostatic repulsion of the phosphate groups in the major groove.46–51 However, such factors would also be operative for the unsubstituted Dickerson dodecamer, which was also crystallized in the presence of spermine and MPD; yet this sequence has been observed to adopt a B-form geometry in multiple crystallographic studies.23,52 The present results for the well studied dd-DNA sequence also demonstrate that the inconsistent method-dependent results are not a consequence of differences in nucleotide sequence, as has also been suggested.53 The introduction of the isolated ribonucleotide may increase the probability of global A-form rcDNA conformers, however the NMR parameters for nucleotides further than ± 1 position from the substitution do not reveal any significantly increased A-form bias. One constraint that has received less explicit discussion but appears to be of particular relevance to the present study involves the symmetry preferences at lattice contacts. The repetitive structural regularity of DNA characterized by a consistent global geometry allows for multiple lattice contacts along the length of the molecule. From this perspective, either A-form or B-form rcDNA is preferable to B-form rcDNA with a localized A-form perturbation. If the less symmetric form identified in solution does not crystallize well, then the crystal selection will be between the more regular A- and B-forms (Figure 7). Due to the significant energy penalty of placing the ribonucleotide in a B-form geometry, any B-form rcDNA that forms is expected to be very short lived. In summary, it is likely that lattice packing constraints that favor conformational regularity, rather than more general effects that bias DNA structures toward A-form geometry, represent the primary basis for the systematic difference between crystallographic and solution-based determinations for the structure of dsDNA containing isolated ribonucleotides.
Supplementary Material
Acknowledgements
The authors thank Dr. Joseph M. Krahn for many helpful discussions regarding the conformational selectivity of crystallization, and Drs. Rajendrakumar A. Gosavi and Nisha A. Cavanaugh for thoughtful comments on the manuscript.
Funding: This research was supported by Research Project Number Z01-ES050111 to REL and by Project Z01 ES065070 to TAK, both in the Intramural Research Program of the National Institute of Environmental Health Sciences, National Institutes of Health. E.F.D. is supported by National Institutes of Health, NIEHS, under Delivery Order HHSN273200700046U.
Abbreviations
- COSY
2D correlation spectroscopy
- DIPSI
Decoupling In the Presence of Scalar Interactions
- DSS
4.4-dimethyl-4-silapentane-1-sulfonic acid
- HSQC
2D single quantum coherence
- NOESY
2D nuclear Overhauser effect spectroscopy
- ROESY
2D rotational nuclear Overhauser effect spectroscopy
- T-ROESY
2D transverse rotational nuclear Overhauser effect spectroscopy
- TOCSY
2D total correlation spectroscopy
- rcDNA
ribonucleotide-containing double stranded DNA that includes at least one ribonucleotide
- rG4-DNA
Dickerson dodecamer sequence in which deoxyguanidine at position 4 has been replaced by guanosine
- dd-DNA
Drew-Dickerson dodecamer dsDNA
- RDC
residual dipolar coupling
- WATERGATE
WATER suppression by GrAdient Tailored Excitation;
Footnotes
Supporting Information
A detailed description of the 2D NMR methods used to determine the solution structure of the rG4-DNA and dd-DNA dodecamers, as well as the MD simulations, is given in the supporting information. The supporting information also contains a discussion of the comparison of the base stacking in rG-DNA and dd-DNA. These supporting materials may be accessed free of charge online at http://pubs.acs.org.
References
- 1.Kornberg A, Baker T. DNA replication. 2nd. New York: Freeman; 1992. [Google Scholar]
- 2.Traut TW. Physiological concentrations of purines and pyrimidines. Mol. Cell. Biochem. 1994;140:1–22. doi: 10.1007/BF00928361. [DOI] [PubMed] [Google Scholar]
- 3.Nick-McElhinny SA, Watts BE, Kumar D, Watt DL, Lundstrom E-B, Burgers PMJ, Johansson E, Chabes A, Kunkel TA. Abundant ribonucleotide incorporation into DNA by yeast replicative polymerases. Proc. Natl.Acad. Sci. U.S.A. 2010;107:4949–4954. doi: 10.1073/pnas.0914857107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cavanaugh N, Beard WA, Wilson SH. DNA Polymerase beta Ribonucleotide Discrimination: insertion, misinsertion, extension, and coding. J. Biol. Chem. 2010;67:3633–3647. doi: 10.1074/jbc.M110.132407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nick-McElhinny SA, Kumar D, Clark AB, Watt DL, Watts BE, Lundstrom E-B, Johansson E, Chabes A, Kunkel TA. Genome instability due to ribonucleotide incorporation into DNA. Nature Chemical Biol. 2010;6:774–781. doi: 10.1038/nchembio.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sekiguchi J, Shuman S. Site-specific ribonuclease activity of eukaryotic DNA topoisomerase I. Mol. Cell. 1997;1:89–97. doi: 10.1016/s1097-2765(00)80010-6. [DOI] [PubMed] [Google Scholar]
- 7.Kim N, Huang SYN, Williams JS, Li YC, Clark AB, Cho JE, Kunkel TA, Pommier Y, Jinks-Robertson S. Mutagenic Processing of Ribonucleotides in DNA by Yeast Topoisomerase I. Science. 2011;332:1561–1564. doi: 10.1126/science.1205016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hovatter KR, Martinson HG. Ribonucleotide-induced helical alteration in DNA prevents nucleosome formation. Proc. Natl. Acad. Sci. U.S.A. 1987;84:1162–1166. doi: 10.1073/pnas.84.5.1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang AH-J, Fujii S, van Boom JH, van der Marel GA, van Boeckel SAA, Rich A. Molecular structure of r(GCG)d(TATACGC): a DNA-RNA hybrid helix joined to double helical DNA. Nature. 1982;299:601–604. doi: 10.1038/299601a0. [DOI] [PubMed] [Google Scholar]
- 10.Egli M, Usman N, Zhang S, Rich A. Crystal structure of an Okazaki fragment at 2-Å resolution. Proc. Natl. Acad. Sci. U.S.A. 1992;89:534–538. doi: 10.1073/pnas.89.2.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Salazar M, Fedoroff OY, Zhu L, Reid BR. The solution structure of the r(gcg)d(TATACCC)•d(GGGTATACGC) Okazaki fragment contains two distinct duplex morphologies connected by a junction. J. Mol. Biol. 1994;241:440–455. doi: 10.1006/jmbi.1994.1519. [DOI] [PubMed] [Google Scholar]
- 12.Salazar M, Fedoroff OY, Reid BR. Structure of Chimeric Duplex Junctions: Solution Conformation of the Retroviral Okazaki-like Fragment r(ccca)d(AATGA)•d(TCATTTGGG) from Moloney Murine Leukemia Virus. Biochemistry. 1996;35:8126–8135. doi: 10.1021/bi9528917. [DOI] [PubMed] [Google Scholar]
- 13.Federoff OY, Salazar M, Reid BR. Structural variation among retroviral primer-DNA junctions: Solution structure of the HIV-1 strand Okazaki fragement r(gcca)d(CTGC)•d(GCAGTGGC) Biochemistry. 1996;35:11070–11080. doi: 10.1021/bi9607822. [DOI] [PubMed] [Google Scholar]
- 14.Nishizaki T, Iwai S, Ohkubo T, Kojima C, Nakamura H, Kyogoku Y, Eiko Ohtsuka E. Solution Structures of DNA Duplexes Containing a DNA,RNA Hybrid Region, d(GG)r(AGAU)d(GAC),d(GTCATCTCC) and d(GGAGA)r(UGAC),d(GTCATCTCC) Biochemistry. 1996;35:4016–4025. doi: 10.1021/bi9519821. [DOI] [PubMed] [Google Scholar]
- 15.Hsu S-T, Chou M-T, Chou S-H, Huang W-C, Cheng J-W. Hydration of [d(GCG)r(aaa)d(TTTGCG)]2 . J. Mol. Biol. 2000;295:1129–1137. doi: 10.1006/jmbi.1999.3388. [DOI] [PubMed] [Google Scholar]
- 16.Tsao Y-P, Wang L-Y, Hsua S-T, Jaina ML, Chou S-H, Huang W-C, Cheng J-W. The Solution Structure of [d(CGC)r(amamam)d(TTTGCG)]2 . J. Biomol. NMR. 2001;21:209–220. doi: 10.1023/a:1012924932513. [DOI] [PubMed] [Google Scholar]
- 17.Jaishree TN, va der Marel GA, van Boom JH, Wang H-J. Structural Influence of RNA Incorporation in DNA: Quantitative Nuclear Magnetic Resonance Refinement of d(CG)r(CG)d(CG) and d(CG)r(C)d(TAGCG) Biochemistry. 1993;32:4903–4911. doi: 10.1021/bi00069a027. [DOI] [PubMed] [Google Scholar]
- 18.Szyperski T, Gotte M, Billeter M, Perola E, Cellai L, Heumann H, Wuthrich K. NMR structure of the chimeric hybrid duplex r(gcaguggc)•r(gcca)d(CTGC) comprising the tRNA•DNA junction formed during initiation of HIV-1 reverse transcription. J. Biomol. NMR. 1999;13:343–355. doi: 10.1023/a:1008350604637. [DOI] [PubMed] [Google Scholar]
- 19.Egli M, Usman N, Rich A. Conformational Influence of the Ribose 2' - Hydroxyl Group: Crystal Structures of DNA-RNA Chimeric Duplexes. Biochemistry. 1993;32:3221–3237. [PubMed] [Google Scholar]
- 20.Ban C, Ramakrishnan B, Sundaralingam M. A single 2'-hydroxyl group converts B-DNA to A-DNA. J. Mol. Biol. 1994;236:275–285. doi: 10.1006/jmbi.1994.1134. [DOI] [PubMed] [Google Scholar]
- 21.Ban C, Ramakrishnan B, Sundaralingam M. Crystal structure of the highly distorted chimeric decamer r(C)d(CGGCGCCG)r(G)*spermine complex spermine binding to phosphate only and minor groove tertiary base-pairing. Nucleic Acids Res. 1994;22:5466–5476. doi: 10.1093/nar/22.24.5466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wahl MC, Sundaralingam M. B-form to A-form conversion by a 3'- terminal ribose: crystal structure of the chimera d(CCACTAGTG)r(G) Nucleic Acids Res. 2000;28:4356–4363. doi: 10.1093/nar/28.21.4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Drew HR, Wing RM, Takano T, Broka T, Tanaka S, Itakura K, Dickerson RE. Structure of a B-DNA dodecamer: Conformation and dynamics. Proc. Natl. Acad. Sci. U.S.A. 1981;78:2179–2183. doi: 10.1073/pnas.78.4.2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schweitzer BI, Mikita T, Kellogg GW, Gardner KH, Beardsley GP. Solution structure of a DNA dodecamer containing the anti-neoplastic agent arabinosylcytosine: combined use of NMR, restrained molecular dynamics, and full relaxation matrix refinement. Biochemistry. 1994;33:11460–11475. [PubMed] [Google Scholar]
- 25.Kuszewski J, Schweiters C, Clore GM. Improving the accuracy of NMR structures of DNA by means of a database potential of mean force describing base-base positional interactions. J. Am. Chem. Soc. 2001;123:3903–3918. doi: 10.1021/ja010033u. [DOI] [PubMed] [Google Scholar]
- 26.Tjandra N, Tate S, Ono A, Kainosho M, Bax A. The NMR structure of a DNA dodecamer in a n aqueous dilute liquid crystalline phase. J. Am. Chem. Soc. 2000;122:6190–6200. [Google Scholar]
- 27.Wu Z, Delaglio F, Tjandra N, Zhurkin VB, Bax A. Overall structure and sugar dynamics of a DNA dodecamer from homo- and heteronuclear dipolar coupling and 31P chemical shift anisotropy. J. Biomol. NMR. 2003;26:297–315. doi: 10.1023/a:1024047103398. [DOI] [PubMed] [Google Scholar]
- 28.Case DA, Darden TA, Cheatham TE, III, Simmerling CL, Wang J, Duke RE, Luo R, Walker RC, Zhang W, Merz KM, Roberts B, Wang B, Hayik S, Roitberg A, Seabra G, Kolossvai I, Wong KF, Paesani F, Vanicek J, Liu J, Wu X, Brozell SR, Steinbrecher T, Gohlke H, Cai Q, Ye X, Wang J, Hsieh M-J, Cui G, Roe DR, Mathews DH, Seetin MG, Sagui C, Babin V, Luchko T, Gusarov S, Kovalenko A, Kollman PA. AMBER 11. San Francisco: University of California; 2010. [Google Scholar]
- 29.Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG. A Smooth Particle Mesh Ewald Method. J. Chem. Phys. 1995;103:8577–8593. [Google Scholar]
- 30.Hare DR, Wemmer DE, Chou S-H, Drobny G. Assignment of the non-exchangeable proton resonances of d(CGCGAATTCGCG) using two-dimensional nuclear magnetic resonance methods. J. Mol. Biol. 1983;171:319–336. doi: 10.1016/0022-2836(83)90096-7. [DOI] [PubMed] [Google Scholar]
- 31.Moe JG, Russu IM. Proton exchange and base-pair opening kinetics in 5'-d(CGCGAATTCGCG)-3' and related dodecamers. Nucleic Acids Res. 1990;18:821–827. doi: 10.1093/nar/18.4.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rajagopal P, Gilbert DE, van der Marel GA, Jacques H, van Boom JH, Feigon J. Observation of Exchangeable Proton Resonances of DNA in Two-Dimensional NOE Spectra Using a Presaturation Pulse; Application to d(CGCGAATTCGCG) and d(CGCGAm6ATTCGCG) J. Magn. Reson. 1988;78:526–537. [Google Scholar]
- 33.Bax A, Lerner L. Measurement of 1H-1H coupling constants in DNA fragments by 2D NMR. J. Magn. Reson. 1988;79:429–438. [Google Scholar]
- 34.Wu Z, Tjandra N, Bax A. Measurement of 1H3'-31P dipolar couplings in a DNA oligonucleotide by constant-time NOESY difference spectroscopy. J. Biomol. NMR. 2001;19:367–370. doi: 10.1023/a:1011292803363. [DOI] [PubMed] [Google Scholar]
- 35.Lu X-J, Olson WK. 3DNA a software package for the analysis, rebuilding and visualization three-dimensional nucleic acid structures. Nucleic Acids Res. 2003;31:5108–5121. doi: 10.1093/nar/gkg680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Altona C, Sundaralingam M. Conformational analysis of sugar ring in nucleosides and nucleotides – a new description using the concept of pseudorotation. J. Am. Chem. Soc. 1972;94:8205–8212. doi: 10.1021/ja00778a043. [DOI] [PubMed] [Google Scholar]
- 37.Saenger W. Principles of Nucleic Acid Structures. New York: Springer Verlag; 1984. [Google Scholar]
- 38.Dickerson RE, Ng H-L. DNA structure from A to B Proc. Natl. Acad. Sci. U.S.A. 2001;98:6986–6988. doi: 10.1073/pnas.141238898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Calladine CR, Drew HR. A base-centered explanation of the B-to-A transition in DNA. J. Mol. Biol. 1984;178:773–781. doi: 10.1016/0022-2836(84)90251-1. [DOI] [PubMed] [Google Scholar]
- 40.Olson WK, Bansal M, Burley SK, Dickerson RE, Gerstein M, Harvey SC, Heinemann U, Lu X-J, Neidle S, Shakked Z, Sklenar H, Suzuki M, Tung C-S, Westhor E, Wolberger C, Berman HMA. Standard Reference Frame for the Description of Nucleic Acid Base-pair Geometry. J. Mol. Biol. 2001;313:229–237. doi: 10.1006/jmbi.2001.4987. [DOI] [PubMed] [Google Scholar]
- 41.Folloppe N, MacKerrell AD., Jr Intrinsic Conformational Properties of Deoxyribonucleosides: Implicated Role for Cytosine in the Equilibrium Among the A, B, and Z Forms of DNA. Biophys. J. 1999;78:3208–3218. doi: 10.1016/S0006-3495(99)77472-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miyabe I, Kunkel TA, Carr AM. The major roles of DNA polymerases epsilon and delta at the eukaryotic replication fork are evolutionarily conserved. PLoS Genetics. 2011;7:e1002407. doi: 10.1371/journal.pgen.1002407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ikeda H, Fernandez R, Wilk A, Barchi JJ, Jr, Huang X, Marquez VE. The effect of two antipodal fluorine-induced sugar puckers on the conformation and stability of the Dickerson-Drew dodecamer duplex [d(CGCGAATTCGCCG)]2 . Nucleic Acids Res. 1998;26:2237–2244. doi: 10.1093/nar/26.9.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lane AN. Influence of conformational averaging on 1H-1H NOEs and structure determination in DNA. Magn. Reson. Chem. 1996;34:S3–S10. [Google Scholar]
- 45.Rychlik MP, Chon H, Cerritelli SM, Klimek P, Crouch RJ, Nowotny M. Crystal structures of RNase H2 in complex with nucleic acid reveal the mechanism of RNA-DNA junction recognition and cleavage. Mol. Cell. 2010;40:658–670. doi: 10.1016/j.molcel.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aboul-Ela F, Varani G, Walker GT, Tinoco I., Jr The TFIIIA recognition fragment d(GGATGGGAG)•d(CCCCATCC) is B-form in solution. Nucleic Acids Res. 1988;16:3559–3572. doi: 10.1093/nar/16.8.3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Clark GR, Brown DG, Sanderson MR, Chwalinski T, Neidle S, Veal JM, Jones RL, Wilson WD, Zon G, Garman E, Stuart DI. Crystal and solution structures of the oligonucleotide d(ATGCGCAT)2: a combined X-ray and NMR study. Nucl. Acids. Res. 1990;18:5521–5528. doi: 10.1093/nar/18.18.5521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vorlickova M, Subirana JA, Chladkova J, Tejralova I, Huynh-Dinh T, Arnold L, Kypr J. Comparison of the solution and crystal conformations of (G + C)-Rich fragments of DNA. Biophys. J. 1996;71:1530–1538. doi: 10.1016/S0006-3495(96)79355-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kypr J, Chladkova, Zimulova M, Vorlickova M. Aqueous trifluorethanol solutions simulate the environment of DNA in the crystalline state. Nucleic Acids Res. 1999;27:3466–3473. doi: 10.1093/nar/27.17.3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Robinson H, Wang AH-J. Neomycin, spermine and hexaaminecobalt(III) share common structural motifs in converting B- to A-DNA. Nucleic Acids Res. 1996;24:676–682. doi: 10.1093/nar/24.4.676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lane AN, Jenkins TC. Thermodynamics of nucleic acids and their interactions with ligands. Quart. Rev. Biophys. 2000;33:255–306. doi: 10.1017/s0033583500003632. [DOI] [PubMed] [Google Scholar]
- 52.Ghosh A, Bansal M. Structural features of B-DNA dodecamer crystal structures; influence of crystal packing vs. base sequence. Indian J. Biochem. Biophys. 2001;38:7–15. [PubMed] [Google Scholar]
- 53.Verbeure B, Lescrinier E, Wang J, Herdewijn P. RNase H mediated cleavage of RNA by cyclohexene nucleic acid (CeNA) Nucleic Acids. Res. 2001;29:4941–4947. doi: 10.1093/nar/29.24.4941. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.