Abstract
Macroscopic assays that are traditionally used to investigate the adhesion behaviour of microbial cells provide averaged information obtained on large populations of cells and do not measure the fundamental forces driving single-cell adhesion. Here, we use single-cell force spectroscopy (SCFS) to quantify the specific and non-specific forces engaged in the adhesion of the human fungal pathogen Candida albicans. Saccharomyces cerevisiae cells expressing the C. albicans adhesion protein Als5p were attached on atomic force microscopy tipless cantilevers using a bioinspired polydopamine wet polymer, and force-distance curves were recorded between the obtained cell probes and various solid surfaces. Force signatures obtained on hydrophobic substrates exhibited large adhesion forces (1.25 ± 0.2 nN) with extended rupture lengths (up to 400 nm), attributed to the binding and stretching of the hydrophobic tandem repeats of Als5p. Data collected on fibronectin (Fn) -coated substrates featured strong adhesion forces (2.8 ± 0.6 nN), reflecting specific binding between Fn and the N-terminal immunoglobulin-like regions of Als5p, followed by weakly adhesive macromolecular bonds. Both hydrophobic and Fn adhesion forces increased with contact time, emphasizing the important role that time plays in strengthening adhesion. Our SCFS methodology provides a versatile platform in biomedicine for understanding the fundamental forces driving adhesion and biofilm formation in fungal pathogens.
Keywords: adhesion, Als proteins, atomic force microscopy, Candida albicans, force spectroscopy, fungal pathogens, single-cells
Introduction
Single-cell force spectroscopy (SCFS) is a powerful tool for measuring the forces that drive cell-cell and cell-substrate interactions.1–4 The general principle is to immobilize a single living cell on an atomic force microscopy (AFM) cantilever and to record force-distance curves between the cell probe and a substrate or another cell. Protocols involving specific receptor-ligand interactions are available to attach animal cells on cantilevers.3,4 Because these approaches are not suited for yeast cells, alternate procedures have been developed, including fixation with glue,5 immobilization via hydrophobic interactions,6 and use of hollow cantilevers (“fluidFM”7–9). The first approach is not recommended because it leads to cell death and denaturation of the cell surface. The second method uses interactions that are too weak to immobilize most yeast cells. FluidFM has a strong potential for the SCFS analysis of yeasts,7–9 but it is not yet compatible with most commercial microscopes. Hence, there is a need for versatile and non-destructive methods for the reliable SCFS analysis of yeast cells.
Adhesion of the human pathogen Candida albicans to surfaces - prosthetics, catheters, host cells and tissues - is a major factor leading to fungal infections.10 Key players in this process are cell-surface proteins known as Als (Agglutinin-like sequence) proteins.11,12 Als proteins possess four functional regions, i.e. an N-terminal immunoglobulin (Ig) -like region, which initiates cell adhesion, followed by a threonine-rich region (T), an hydrophobic tandem repeat (TR) region that participates in cell-cell aggregation, and a stalk region projecting the molecule away from the cell surface (for a schematic view of Als proteins please refer to a recent review10). Because multiple Als adhesins are expressed on C. albicans, most of our current knowledge of the functional roles of individual Als proteins derives from Saccharomyces cerevisiae surface display models.13,14 Microscopic assays using these models have revealed that Als-mediated adherence involves two steps, i.e. initial adhesion via the specific binding of the Ig-region to peptide ligands, followed by cell-cell aggregation mediated by the TR regions.13,14 Recent single-molecule AFM studies have unravelled the remarkable biophysical properties of Als proteins, both on isolated proteins15,16 and on live cells.17,18 These properties include strong recognition events between the Ig-like regions,15 strong hydrophobic interactions associated with unfolding of the TR regions,17 and amyloid-mediated clustering and interaction of the T regions.16,17 Yet, two crucial questions remain unanswered: how do Als properties contribute to the adhesion of whole cells, and how do they mediate attachment to host cells? Addressing these issues requires the development of suitable SCFS methodologies. In this article, we report a non-invasive method for the SCFS analysis of Als-mediated cell adhesion, using a bioinspired polydopamine adhesive. We show that the method is suitable for quantifying the specific and non-specific forces engaged in fungal adhesion.
Materials and methods
Microorganisms and cultures
S. cerevisiae W3031B harboring plasmids pJL1 and pJL1-EV were grown on SC-trp plates.18 Two or three colonies from the SC-trp plate used as inoculum were transferred into SC-trp medium (1,7 g/L yeast extract w/o amino acids and w/o ammonium sulfate, 1.92 g/L yeast synthetic drop-out medium supplements w/o Trp, 5 g/L ammonium sulfate, and 20 g/L galactose). Cells were agitated at 30°C, grown overnight, and harvested by centrifugation. They were washed three times with deionized water (for hydrophobic force measurements) or PBS (for fibronectin force measurements) and resuspended to a concentration of ~106 cells per mL. Note that deionized water was chosen for hydrophobic force measurements despite its known osmotic influence on yeast cells because buffer solutions were found to alter the force signatures. As a matter of fact, we found that in buffer solution, ionic species can adsorb on the surfaces and lower the measured adhesion force.
Atomic force microscopy
AFM measurements were performed at room temperature (20°C) in deionized water or PBS, using a Bioscope Catalyst AFM (Bruker AXS Corporation, Santa Barbara, CA) and tipless microfabricated Si3N4 cantilevers with a nominal spring constant of ~0.01 N/m (MSCT levers, Bruker AXS Corporation). The spring constants of the cantilevers were measured using the thermal noise method. Force measurements were recorded in the force-volume mode by recording arrays of 16 × 16 or 32 × 32 force curves, using a maximum applied force of 1 nN, and tip approach and pulling velocities of 1000 nm/s. During contact time, the system is maintained at constant height. For each condition, at least 3 independent probe-substrate combinations were tested.
Cell probes
A tipless cantilever was immersed for 1 h in a 10 mM Tris buffer solution (pH 8.5) containing 4 mg/mL dopamine hydrochloride (99%, Sigma-Aldrich). The cantilever was then washed and dried under N2, brought into contact with an isolated cell for 1 min, and the obtained cell probe was then transferred, in the same glass petri dish, without dewetting over a solid substrate for further force measurements.
Substrates
Hydrophobic and hydrophilic substrates were prepared by immersing overnight gold-coated substrates in solutions of 1 mM 1-Dodecanethiol (Sigma-Aldrich, 98%) or 1 mM 11-Mercapto-1-undecanol (Sigma-Aldrich, 97%), rinsing them with ethanol and drying them under N2. Fibronectin (Fn) substrates were prepared by depositing 200 μL drops of a PBS solution containing 100 μg/mL Fn (Sigma-Aldrich) on gold-coated substrates, rinsing them gently with PBS after 12 hr contact time. All substrates were transparent, enabling us to observe cell probes by optical microscopy during the course of the experiments.
Viability tests
The viability of attached cells was tested using a LIVE/DEAD yeast viability kit (Molecular Probes). Prior to attachment, a cell suspension (106 cell/mL in 2% glucose Hepes solution) was mixed with FUN1 cell stain (5 μM), mixed thoroughly and incubated for 30 min in the dark at 30°C. Labelled cells were then attached to polydopamine probes or substrates, and their viability checked using a Zeiss Axio Observer Z1 equipped with a Hamamatsu camera C10600.
Results and discussion
Living cell probes
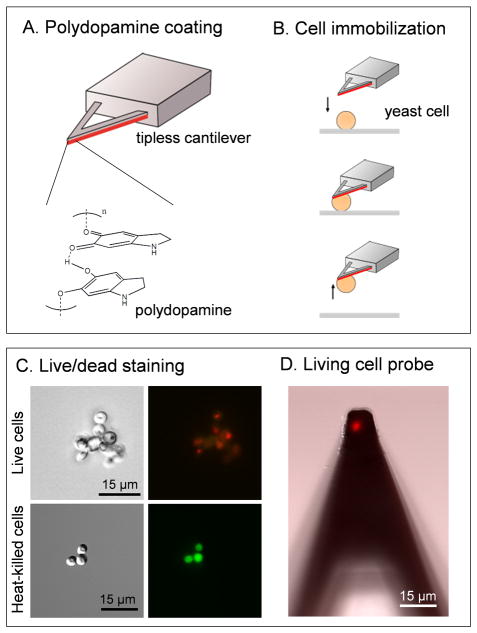
Cell probes were prepared using an integrated platform combining an AFM with an inverted light microscope. Single surface display S. cerevisiae cells were attached on AFM cantilevers using polydopamine.19 Tipless cantilevers were coated with a thin film of polydopamine by immersing them into a solution of 4 mg/mL dopamine for 60 min (Fig. 1A). Polydopamine-coated cantilevers were then slowly approached toward single cells deposited on a glass petri dish in buffer, kept in contact for 1 min, and then withdrawed while checking for proper cell attachment using the optical microscope (Fig. 1B). The viability of the cells was checked by staining them using a LIVE/DEAD yeast viability kit and observed by fluorescence microscopy. Live and dead cells exhibit red fluorescence and green fluorescence, respectively (Fig. 1C). Fig. 1D shows that single cells attached on polydopamine-coated cantilevers were alive, thus confirming that the cell probe preparation method is minimally-invasive.
Fig. 1.
Non-destructive method for the single-cell force spectroscopy of fungal adhesion. (A, B) Preparation of the cell probes involves coating tipless cantilevers with polydopamine (adapted from Dreyer et al.30) (A), followed by the controlled immobilization of a single S. cerevisiae cell (B). (C, D) Cell attachment with polydopamine does not alter cell viability: (C) phase contrast (left) and fluorescence (right) images of live (top) and heat-killed (60°C, 30 min) (bottom) yeast cells labelled with LIVE/DEAD yeast viability kit; (D) fluorescence image (overlayed with DIC) of a single yeast cell attached on a polydopamine-coated cantilever.
Als5p mediates hydrophobic interactions
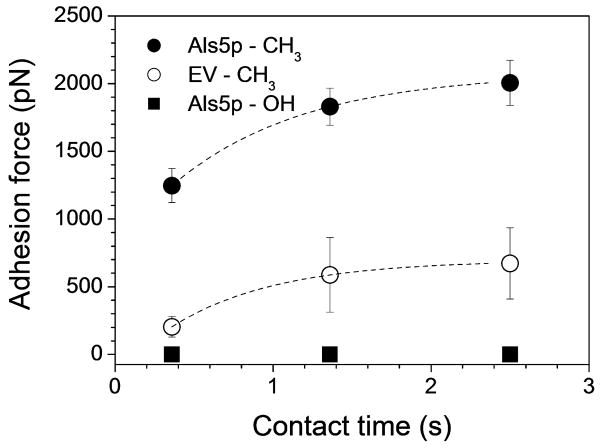
Hydrophobic interactions are thought to play a role in C. albicans adhesion,20,21 but their molecular origin is still a matter of debate. Als proteins contribute to cell surface hydrophobicity via their hydrophobic TR domains that mediate adhesion to hydrophobic surfaces.14,22 However, the extent to which the hydrophobic properties of Als proteins mediate the adhesion of whole cells is not clear. To address this issue, multiple force-distance curves (n = 1024) were recorded between S. cerevisiae yeast cells expressing the widely investigated Als5p adhesin and hydrophobic substrates. Figs. 2A–2C show the adhesion force histogram and representative force curves recorded between a single cell and a methyl-terminated self-assembled monolayer (SAM). All curves showed strong adhesion forces, of 1.25 ± 0.2 nN (mean ± s.d.; n = 1024) magnitude, with sequential rupture events and rupture distances up to 400 nm. This profile indicates that multiple bonds were formed and did not necessarily rupture simultaneously during detachment. The general features of the curves, including their maximum adhesion strength, did not substantially changed when recording consecutive force curves, when probing different regions of the substrates, or when comparing different cells. In many curves, we also observed a second small peak (<250 pN) following rupture of the first large peak at ~500 nm. By contrast, a complete lack of adhesion was observed between Als5p yeast cells and hydrophilic hydroxyl-terminated SAMs (Figs 2D–2F); also, only low adhesion force peaks, of 204 ± 76 pN (mean ± s.d.; n = 1024) magnitude, were measured between cells from an empty vector (EV) strain lacking Als5p and hydrophobic substrates (Figs. 2G–2I). These observations lead us to conclude that the large adhesion forces measured above reflect primarily hydrophobic interactions associated with multiple Als5p proteins, the small extended peaks reflecting the full unfolding of single Als5p molecules.15 The weak adhesion forces measured for the EV control are likely to be due to the contribution of other proteins from the S. cerevisiae cell wall. Interestingly, the magnitude of hydrophobic forces increased with contact time (from 1.25 ± 0.2 nN at 350 ms, to 2.0 ± 0.2 nN at 2.5 s; Fig. 3), indicating that Als- mediated hydrophobic interactions increase with time.
Fig. 2.
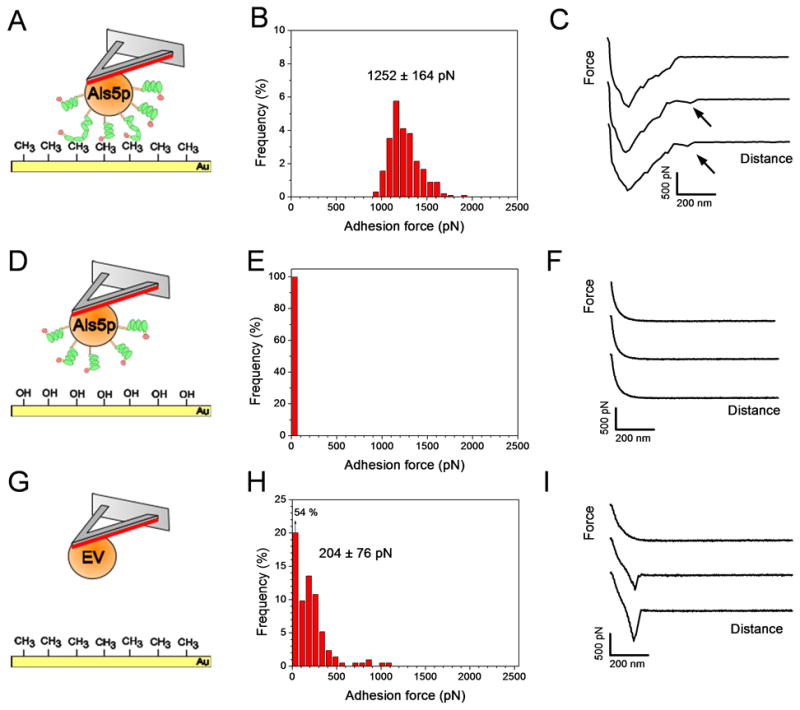
Measuring Als5p-mediated hydrophobic forces. (A, D, G) Schemes of the experimental set-ups in which the TR and Ig regions of Als5p are shown in green and red, respectively. (B, E, H) Adhesion force histograms (n = 1024 force curves) and (C, F, I) representative retraction force curves recorded in deionized water between single S. cerevisiae cells expressing Als5p proteins and hydrophobic (CH3) (A–C) or hydrophilic (OH) (D–F) substrates, and between single S. cerevisiae cells lacking Als5p (EV) and hydrophobic substrates (G–I). Adhesion force values used to build the histograms correspond to the largest adhesion events seen in the curves. Arrows in Fig. 2c indicate that large adhesion signatures were followed by weaker events rupturing at ~500 nm. The contact time was 350 ms. For each condition, similar data were obtained using at least 3 different cells from independent cultures and 3 different substrates.
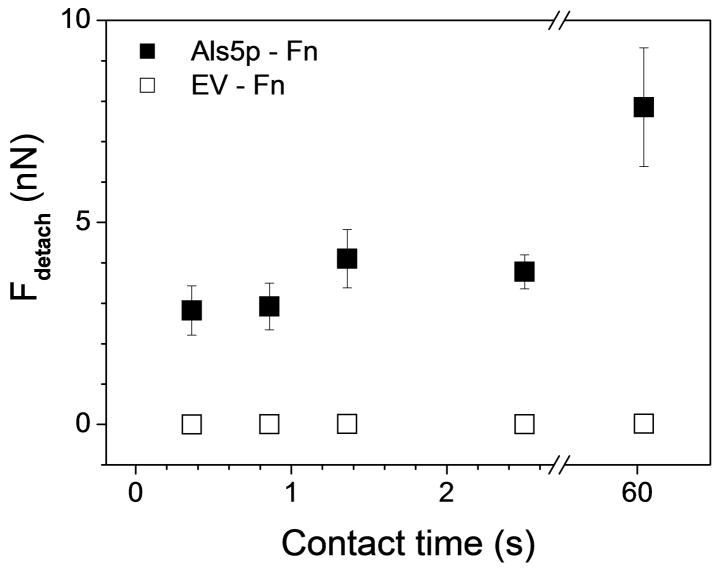
Fig. 3.
Als5p-mediated hydrophobic forces strengthen with time. Variation of the Als5p-CH3, EV- CH3 and Als5p-OH adhesion forces with contact time, using the same conditions as in Fig. 2. The data represent the mean ± standard deviation (S.D.; n = 256) and are fitted with an exponential decay function. Similar data were obtained using at least 3 different cells from independent cultures and 3 different substrates.
Our findings are consistent with a mechanism in which C. albicans hydrophobicity originates from the hydrophobic sequences in Als proteins. We expect that, in nature, mechanical contact between a cell and a surface will trigger the force-induced unfolding of Als TR domains (see cartoon in Fig. 2A, green color),15,17,23 thereby leading to extended conformations in which hydrophobic groups are freshly exposed and promote hydrophobic interactions. Increasing the interaction time before pulling would increase the probability that more TR regions bind to the hydrophobic substrate. Such a model resembles cryptic sequences in animal cell proteins like fibronectin and cadherins, which are known to be unraveled by force and to lead to alterations in molecular binding sites.24
Als5p specifically binds to fibronectin
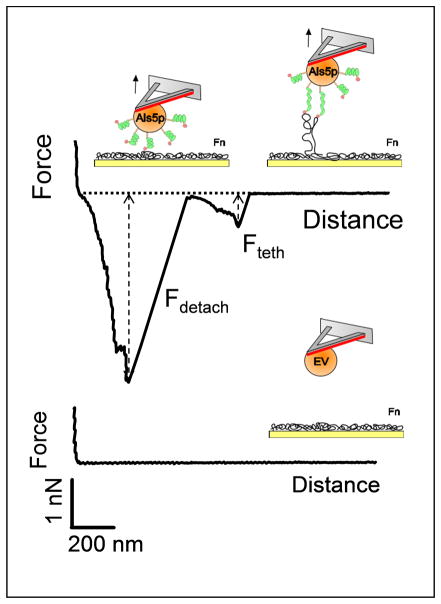
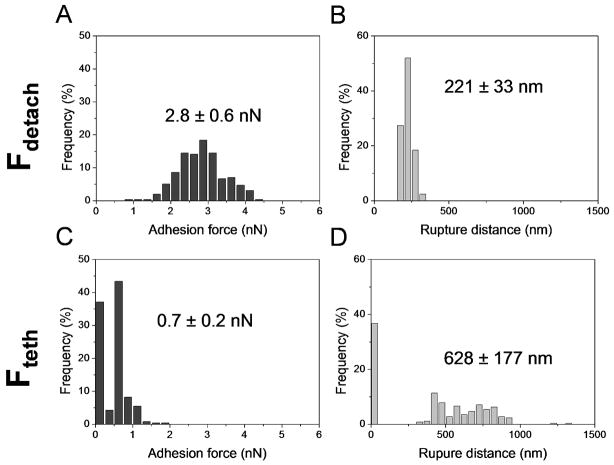
The N-terminal Ig-like domains of Als proteins mediate specific adhesion to host constituents, including epithelial cells, endothelial cells, and extracellular matrix proteins such as fibronectin (Fn).11,12 To quantify this interaction at the single-cell level, the forces between individual S. cerevisiae cells expressing Als5p and Fn-coated substrates were measured. Before SCFS measurements, Fn-coated substrates were first imaged in contact mode to confirm homogeneous protein coating. As shown in Figs. 4 and 5, adhesive signatures were observed in all curves and were quite different from those observed on hydrophobic substrates. Two consecutive force peaks were always observed, a first peak with strong adhesion values (2.8 ± 0.6 nN; mean ± s.d.; n = 1024) and single sharp ruptures (221 ± 33 nm), and a second peak of weaker adhesion (0.7 ± 0.2 nN; mean ± s.d.; n = 1024) at extended rupture lengths (628 ± 177 nm), observed in 63 % of the curves. Note that discrete rupture steps were seen before rupture of the first and second peaks. The general features of the curves did not substantially changed when recording consecutive force curves or when comparing different cells. The large adhesion force, also named maximum detachment force, represents the maximum cell-substrate binding strength at close contact.3 Detachment of a cell from a solid substrate is a complex process depending on multiple properties like cell geometry, cell elasticity, as well as receptor binding strength, cooperativity and clustering.3 In the present study, detachment forces were never seen with EV cells lacking Als5p (Fig. 4), demonstrating they were due to the multi-point interaction between Als5p Ig regions and Fn molecules (see upper left cartoon in Fig. 4). It is worth noting that, unlike hydrophobic bonds, detachment forces seemed to rupture simultaneously rather than sequentially since rupture events were sharp and linear.
Fig. 4.
Measuring specific Als5p-fibronectin (Fn) forces. Representative retraction force curves recorded in PBS buffer between single S. cerevisiae cells expressing (top) or not (bottom) Als5p proteins and Fn substrates (see text for details).
Fig. 5.
The Als5p-fibronectin (Fn) interaction is characterized by a detachment force peak followed by a tether force peak. (A, C) Adhesion force histograms (n = 1024) and (B, D) histograms of rupture distances of the maximum detachment force peaks (A, B) and of the tether peaks (C, D) measured in PBS buffer between Als5p cells and Fn substrates. The contact time was 350 ms. Similar data were obtained using at least 3 different cells from independent cultures and 3 different substrates.
After the cell body detached from the substrate, smaller force peaks with non-linear force loading behaviour were observed during which the cell was still in contact with the substrate through extended macromolecular bonds, referred to “tethers” (Figs. 4 and 5).3,25 Because these features were not observed with the EV strain (Fig. 4), we suggest they reflect the stretching of Fn molecules, possibly with some stretched Als5p proteins as well (see upper right cartoon in Fig. 4). As the lifetime of the cell-substrate bond is given by the bond rupture length divided by the pulling speed, we note that these extended macromolecular bonds substantially increase the lifetime of the adhesive bond, a phenomenon which in nature may contribute to strengthen the overall cell adhesion26.
As the adhesion strength of animal cells is known to increase with contact time,3 we next asked whether this also applies to the Als5p-Fn interaction. Increasing the contact time from 350 ms to 60 s strongly increased the magnitude of the maximum detachment force (Fig. 6), with tether forces and rupture distances remaining essentially unchanged. These results are consistent with the behaviour of animal cells, where the adhesion force increases with time due to increase of receptor-ligand pairs anchoring the cell to the substrate3. Yet, we cannot exclude that this increase in adhesion force with time could result from increased applied force due to thermal drift. This time-dependency therefore provides further evidence that most detachment forces measured here reflects the rupture of multiple Als5p-Fn bonds. As the specific binding force between Fn and microbial Fn-attachment proteins was previously measured to be ~50 pN,27 we estimate that the 2.8 nN and 7.8 nN maximum detachment forces measured at short and long contact times would correspond to ~50 and ~150 Als5p-Fn bonds. Note that this calculation assumes parallel loading of all bonds and linearly-additive adhesion forces, which may not be strictly correct for such single-live cell experiments. Nevertheless, the obtained values may be converted into protein surface densities, considering the cell-substrate contact area. As a rough approximation, the contact zone of a deformable sphere pressed on a rigid flat maybe estimated by the following equation:28,29 A = π R δ, in which in which A is the contact area, R the radius of the cell, and δ the cell deformation. Considering a cell radius of 2.5 μm and a deformation of 75 nm (estimated from indentation curves), we found a contact area of ~1 μm2, thus yielding a protein surface density ranging from 50 to 150 proteins/μm2. The 150 proteins/μm2 value is in the range of the average cell surface concentration expected for yeast adhesins,10 and in remarkable agreement with the 172 ± 16 proteins/μm2 minimum density that we measured earlier for the same strain using single-molecule AFM imaging.17
Fig. 6.
Als5p-fibronectin (Fn) interactions strengthen with time. Variation of the Als5p-Fn maximum detachment force with contact time, using the same conditions as in Fig. 5 (closed symbols). As a control, data obtained for the EV strain lacking Als5p are also shown (open symbols). The data represent the mean ± standard deviation (S.D.; n = 256). Similar data were obtained using at least 3 different cells from independent cultures and 3 different substrates.
Conclusions
We have shown that the non-destructive attachment of single yeast cells to AFM cantilevers using a polydopamine wet adhesive is a reliable approach for the SCFS analysis of fungal adhesion. Application of the method to Als5p-mediated adhesion reveals that the protein promotes strong hydrophobic interactions, as well as specific binding to fibronectin, that involve the Als5p TR and Ig-like regions, respectively. Both types of interactions strengthen with time. Our SCFS platform allows us, for the first time, to compare binding characteristics on whole cells to that generated in single-molecule studies. Such data will be critical for our understanding of the forces involved in pathogen-to-host adhesion and in biofilm formation.
Acknowledgments
Work at the Université catholique de Louvain was supported by the National Foundation for Scientific Research (FNRS), the Université catholique de Louvain (Fonds Spéciaux de Recherche), the Région Wallonne, the Federal Office for Scientific, Technical and Cultural Affairs (Interuniversity Poles of Attraction Programme), and the Research Department of the Communauté française de Belgique (Concerted Research Action). Work at Brooklyn College was supported by NIH grant R01 GM 098616. Y.F.D. and D.A. are Senior Research Associate and Postdoctoral Researcher of the FRS-FNRS. We thank Caleen Ramsook for technical support.
References
- 1.Benoit M, Gabriel D, Gerisch G, Gaub HE. Nat Cell Biol. 2000;2(6):313–317. doi: 10.1038/35014000. [DOI] [PubMed] [Google Scholar]
- 2.Krieg M, Arboleda-Estudillo Y, Puech PH, Kafer J, Graner F, Muller DJ, Heisenberg CP. Nat Cell Biol. 2008;10(4):429–436. doi: 10.1038/ncb1705. [DOI] [PubMed] [Google Scholar]
- 3.Helenius J, Heisenberg CP, Gaub HE, Muller DJ. J Cell Sci. 2008;121(11):1785–1791. doi: 10.1242/jcs.030999. [DOI] [PubMed] [Google Scholar]
- 4.Friedrichs J, Helenius J, Müller DJ. Nat Protoc. 2010;5(7):1353–1361. doi: 10.1038/nprot.2010.89. [DOI] [PubMed] [Google Scholar]
- 5.Bowen WR, Lovitt RW, Wright CJ. J colloid and interface Sci. 2001;237:54–61. doi: 10.1006/jcis.2001.7437. [DOI] [PubMed] [Google Scholar]
- 6.Emerson RJIV, Camesano TA. Appl Environ Microbiol. 2004;70(10):6012–6022. doi: 10.1128/AEM.70.10.6012-6022.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meister A, Gabi M, Behr P, Studer P, Vörös J, Niedermann P, Bitterli J, Polesel-Maris J, Liley M, Heinzelmann H, Zambelli T. Nano Lett. 2009;9(6):2501–2507. doi: 10.1021/nl901384x. [DOI] [PubMed] [Google Scholar]
- 8.Dörig P, Stiefel P, Behr P, Sarajlic E, Bijl D, Gabi M, Vörös J, Vorholt JA, Zambelli T. Appl Phys Lett. 2010;97(2):023701. [Google Scholar]
- 9.Potthoff E, Guillaume-Gentil O, Ossola D, Polesel-Maris J, LeibundGut-Landmann S, Zambelli T, Vorholt JA. PLoS ONE. 2012;7(12):e52712. doi: 10.1371/journal.pone.0052712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dranginis AM, Rauceo JM, Coronado JE, Lipke PN. Microbiol Mol Biol Rev. 2007;71(2):282–294. doi: 10.1128/MMBR.00037-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoyer LL. Trends Microbiol. 2001;9(4):176–180. doi: 10.1016/s0966-842x(01)01984-9. [DOI] [PubMed] [Google Scholar]
- 12.Lipke PN, Garcia MC, Alsteens D, Ramsook CB, Klotz SA, Dufrene YF. Trends Microbiol. 2012;20(2):59–65. doi: 10.1016/j.tim.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klotz SA, Gaur NK, Lake DF, Chan V, Rauceo J, Lipke PN. Infect Immun. 2004;72(4):2029–2034. doi: 10.1128/IAI.72.4.2029-2034.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rauceo JM, De Armond R, Otoo H, Kahn PC, Klotz SA, Gaur NK, Lipke PN. Eukaryot Cell. 2006;5(10):1664–1673. doi: 10.1128/EC.00120-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alsteens D, Dupres V, Klotz SA, Gaur NK, Lipke PN, Dufrêne YF. ACS Nano. 2009;3:1677–1682. doi: 10.1021/nn900078p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alsteens D, Ramsook CB, Lipke PN, Dufrêne YF. ACS Nano. 2012;6(9):7703–7711. doi: 10.1021/nn3025699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alsteens D, Garcia MC, Lipke PN, Dufrene YF. Proc Natl Acad Sci U S A. 2010;107(48):20744–20749. doi: 10.1073/pnas.1013893107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garcia MC, Lee JT, Ramsook CB, Alsteens D, Dufrene YF, Lipke PN. PLoS ONE. 2011;6(3):e17632. doi: 10.1371/journal.pone.0017632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee H, Dellatore SM, Miller WM, Messersmith PB. Science. 2007;318(5849):426–430. doi: 10.1126/science.1147241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yoshijima Y, Murakami K, Kayama S, Liu D, Hirota K, Ichikawa T, Miyake Y. Mycoses. 2010;53(3):221–226. doi: 10.1111/j.1439-0507.2009.01694.x. [DOI] [PubMed] [Google Scholar]
- 21.Masuoka J, Hazen KC. Microbiology. 1997;143(9):3015–3021. doi: 10.1099/00221287-143-9-3015. [DOI] [PubMed] [Google Scholar]
- 22.Frank AT, Ramsook CB, Otoo HN, Tan C, Soybelman G, Rauceo JM, Gaur NK, Klotz SA, Lipke PN. Eukaryot Cell. 2010;9(3):405–414. doi: 10.1128/EC.00235-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beaussart A, Alsteens D, El-Kirat-Chatel S, Lipke PN, Kucharíková S, Van Dijck P, Dufrêne YF. ACS Nano. 2012;6:10950–10964. doi: 10.1021/nn304505s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vogel V, Sheetz M. Nat Rev Mol Cell Biol. 2006;7(4):265–275. doi: 10.1038/nrm1890. [DOI] [PubMed] [Google Scholar]
- 25.Sun M, Graham JS, Hegedüs B, Marga F, Zhang Y, Forgacs G, Grandbois M. Biophys J. 2005;89(6):4320–4329. doi: 10.1529/biophysj.104.058180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krieg M, Helenius J, Heisenberg CP, Muller DJ. Angewandte Chemie (English Edition) 2008;47(50):9775–9777. doi: 10.1002/anie.200803552. [DOI] [PubMed] [Google Scholar]
- 27.Verbelen C, Dufrêne YF. Integr Biol. 2009;1(4):296–300. doi: 10.1039/b901396b. [DOI] [PubMed] [Google Scholar]
- 28.Chatterjee B, Sahoo P. Adv Tribology. 2012:472794. [Google Scholar]
- 29.Kogut L, Etsion I. J Appl Mech-T ASME. 2002;69(5):657–662. [Google Scholar]
- 30.Dreyer DR, Miller DJ, Freeman BD, Paul DR, Bielawski CW. Langmuir. 2012;28(15):6428–6435. doi: 10.1021/la204831b. [DOI] [PubMed] [Google Scholar]