Abstract
Proliferating cell nuclear antigen (PCNA) monomers assemble to form a ring-shaped clamp complex that encircles duplex DNA. PCNA binding to other proteins tethers them to the DNA providing contacts and interactions for many other enzymes essential for DNA metabolic processes. Most eukarya and euryarchaea have only one PCNA homolog but Thermococcus kodakarensis uniquely has two, designated PCNA1 and PCNA2, encoded by TK0535 and TK0582, respectively. Here, we establish that both PCNA1 and PCNA2 form homotrimers that stimulate DNA synthesis by archaeal DNA polymerases B and D and ATP hydrolysis by the replication factor C complex. In exponentially growing cells, PCNA1 is abundant and present at an ~100-fold higher concentration than PCNA2 monomers. Deletion of TK0582 (PCNA2) had no detectable effects on viability or growth whereas repeated attempts to construct a T. kodakarensis strain with TK0535 (PCNA1) deleted were unsuccessful. The implications of these observations for PCNA1 function and the origin of the two PCNA-encoding genes in T. kodakarensis are discussed.
Keywords: Archaea, DNA replication, Genetics, PCNA, Structure, Thermococcus kodakarensis
Introduction
In archaea and eukarya monomers of proliferating cell nuclear antigen (PCNA) assemble to form a ring-shaped trimeric clamp complex that encircles duplex DNA and is essential for many DNA metabolic processes (Jeruzalmi et al. 2002; Indiani and O’Donnell 2006; Kelch et al. 2012). The PCNA clamp does not assemble autonomously but is loaded onto DNA by the replication factor C (RFC) clamp loader complex. RFC recognizes the 3′ end of a single-strand/duplex DNA (primer–template) junction and uses the energy of ATP hydrolysis to assemble the PCNA ring around the primer (Jeruzalmi et al. 2002; Kelch et al. 2012; Yao and O’Donnell 2012). PCNA was first reported as a processivity factor for replicative DNA polymerases but subsequent studies have established that both archaeal and eukaryal PCNAs also associate with, and modulate the activities of many other proteins involved in nucleic acid metabolic transactions (Vivona and Kelman 2003).
There are three major archaeal lineages, the Euryarchaeota, Crenarchaeota and Thaumarchaeota (Brochier-Armanet et al. 2008) and based on genome and metagenome sequencing members of the euryarchaeal and thaumarchaeal lineages have only one PCNA homolog whereas Crenarchaea have three PCNA homologs that co-assemble to form active heterotrimeric complexes in vitro (Spang et al. 2010; Pan et al. 2011a). Most archaeal genomes also encode two RFC homologs, designated the RFC large (RFC-L) and small (RFC-S) subunits, that assemble to form a pentameric complex that contains one RFC-L and four RFC-S subunits (Chia et al. 2010; Ishino and Ishino 2012).
Thermococcus kodakarensis is a genetically tractable hyperthermophilic heterotroph (Atomi et al. 2004) that has become a model species for studies of archaeal biology (Hileman and Santangelo 2012) but is unique, as an euryarchaeon, in having two PCNA homologs, designated PCNA1 and PCNA2, encoded by TK0535 and TK0582, respectively. Previous studies established that both genes are expressed in vivo (Li et al. 2010; Kuba et al. 2012) and that the encoded proteins assemble in vitro to form homotrimeric rings (Ladner et al. 2011; Kuba et al. 2012). Here, we report that both PCNA homologs are functional, with similar biochemical properties in vitro. However, whereas PCNA1 is abundant in vivo and is apparently essential for viability, PCNA2 is present at an ~100-fold lower concentration in growing cells and deletion of TK0582 had no discernible effects on growth or viability.
Materials and methods
Protein expression and purification
PCNA1 and PCNA2, the RFC complex and DNA polymerase B (PolB) were purified as previously described (Ladner et al. 2011; Chemnitz Galal et al. 2012). The gene (TK1902) encoding the small subunit of DNA polymerase D (DP1) was PCR amplified from genomic DNA and cloned into pET-21a (Novagen) to generate pET-TK1902. The gene (TK1903) encoding the large subunit of PolD (DP2) without the intein was cloned by GeneArt into pET-21a (Novagen) to generate pET-TK1903. The PolD sub-units encoded by these plasmids have C-terminal His6-tags and so were purified, after expression in Escherichia coli BL21 DE3 Rosetta cells, by Ni2+-affinity chromatography column as previously described for the purification of PolB (Ladner et al. 2011). Following elution from the Ni2+-affinity column, the proteins were dialyzed against buffer containing 50 mM Tris–HCl (pH 8.0), 500 mM NaCl, 0.5 mM EDTA, 2 mM DTT and 10 % glycerol (v/v). The PolD complex was formed by incubation of the two proteins mixed in a 1:1 molar ratio at 25 °C for 1 h.
Gel-filtration analysis
Aliquots (200 μg) of PCNA1 or PCNA2 and the RFC complex in 200 μl buffer containing 25 mM Tris–HCl (pH 7.5), 500 mM NaCl and 10 % glycerol (v/v) were fractionated through a Superdex-200 gel-filtration column (HR10/30; GE Healthcare) pre-equilibrated with 25 mM Tris–HCl (pH 7.5), 500 mM NaCl and 10 % glycerol (v/v) at 22 °C. The proteins present in fractions (15 μl) were separated by electrophoresis through a 10 % SDS-PAGE and visualized by staining with Coomassie brilliant blue (R250).
Light scattering of RFC
The molecular mass of the RFC complex was determined using 100 μg of protein dissolved in 20 μl of 25 mM Tris–HCl (pH 7.5), 50 mM NaCl and 10 % glycerol as previously reported for the PCNA proteins (Ladner et al. 2011). A 1200 series HPLC system (Agilent Technologies) with a Shodex KW-802.5 or a Shodex KW-804 column (Showa Denko K.K.) was used. The flow rate was 0.5 ml/min, in a solution containing 25 mM Tris–HCl (pH 7.5), 500 mM NaCl and 10 % glycerol (v/v). Light scattering was measured with a miniDawn Treos (Wyatt Technology) and the protein concentration was measured with an Optilab rEX differential refractometer (Wyatt Technology).
ATPase assays
The ATPase activity of RFC was assayed in reaction mixtures (15 μl) that contained 25 mM Tris–HCl (pH 8.0), 5 mM MgCl2, 1 mM DTT, 100 μg/ml BSA, 1.5 nmol of [γ-32P]ATP (3,000 Ci/mmol), 0.5 pmol of RFC and 0.01, 0.05, 0.1, 0.25 or 0.5 pmol of PCNA1 or PCNA2 (as trimers) as indicated in the figure legends, with or without 50 pmol of primed substrate formed by annealing oligo-nucleotides with the sequences 5′-GCGGCGAGTCCA GCTCAGGAGCTCGCGCCG and 5′-TTTGTTTGTTTGT TT GTTTGTTTGTTTGTTTGTTTGCGGCGCGAGCTC CTGAGCTGGACTCGCCGC. After incubation at 70 °C for 1 h, an aliquot (1 μl) of the reaction mixture was spotted onto a polyethyleneimine cellulose thin layer plate. ATP and Pi were separated by chromatography in 1 M formic acid containing 0.5 M LiCl, and the amount of ATP hydrolysis was calculated based on phosphorimaging quantification. The ATPase assays were repeated three times, and the averages of the results obtained with standard deviations are reported.
To establish the rate of ATP hydrolysis by RFC, reaction mixtures (45 μl) that contained 25 mM Tris–HCl (pH 8.0), 5 mM MgCl2, 1 mM DTT, 100 μg/ml BSA, 4.5 nmol [γ-32P]ATP (3,000 Ci/mmol), 0.5 pmol RFC and 0.05 pmol PCNA were incubated at 70 °C, with or without 50 pmol of the primed substrate described above. Aliquots (3 μl) of the reaction mixture were removed after 0, 5, 10, 15, 30 and 45 min, mixed with 1 μl of 0.5 M EDTA, and the extent of ATP hydrolysis determined using polyethyleneimine cellulose thin layer chromatography as described above. The assays were repeated three times, and the averages of the results obtained with standard deviations are reported.
DNA replication assay
Elongation assays of singly primed M13 DNA templates with PolB were performed as reported previously (Ladner et al. 2011) in reaction mixtures (20 μl) containing 40 mM Tris–HCl (pH 8.0), 250 mM NaCl, 1.5 mM DTT, 100 μg/ml BSA, 10 mM Mg-acetate, 2 mM ATP, 100 μM each of dTTP, dCTP and dGTP and 20 μM [α-32P]dATP and 10 fmol singly primed M13mp18. Elongation assays of singly primed M13 DNA templates with PolD were performed in reaction mixtures (20 μl) containing 30 mM Tris–HCl (pH 8.0), 100 mM NaCl, 2.5 mM DTT, 100 μg/ml BSA, 7.5 mM Mg-acetate, 1.5 mM ATP, 150 μM each of dTTP, dCTP and dGTP and 20 μM [α-32P]dATP and 20 fmol singly primed M13mp18.
The proteins used in the reaction are indicated in the figure legends. Following incubation at 70 °C for 20 min the reactions were stopped by the addition of EDTA to final concentration of 10 mM. The products were separated by electrophoresis through a 1.1 % alkaline agarose gel. Total nucleotide incorporation was quantified by liquid scintillation from 4 μl aliquots of each reaction mixture.
Construction of T. kodakarensis intermediate and deletion strains
The procedures and selections used to generate intermediate and markerless deletion strains from T. kodakarensis TS517 have been described in detail (Pan et al. 2011b). Briefly, plasmid pZLE032 (Fig. S2) and pZLE033 DNAs (Fig. S1) were used as donor DNAs to transform T. kodakarensis TS517 (ΔpyrF; ΔtrpE::pyrF; ΔTK0664) with transformants selected by growth on plates lacking tryptophan. Colonies containing cells in which a double crossover had integrated pZLE033 DNA into the T. kodakarensis genome at the desired locus were identified by diagnostic PCR. Suspensions of these cells were spread on plates containing 6-methyl purine (6MP) and the genome structure in a spontaneous 6MP-resistant isolate (6MPR), designated T. kodakarensis TS-ZLE033 (ΔTK0582) was confirmed by diagnostic PCR and Southern blotting. Tryptophan-independent transformants of T. kodakarensis TS517 were readily obtained by transformation with ZLE033 DNA (Fig. S1) but no such transformants were obtained with pZLE032 (Fig. S2) as the donor DNA.
Southern blot confirmation of genome structures
Southern blots were performed as described (Santangelo et al. 2007), using 3 μg of BglII-digested T. kodakarensis TS517 and T. kodakarensis TS-ZLE033 genomic DNAs per lane. Probes complementary to TK0582 and TK0535 were generated by PCR amplification using primer pairs with the sequences GTACTTCAGAAGCAGTGGTATGTG and GC TTGCAAGGAGCAGGGATACGC (see Fig. S1), and GAC CAGGAAGGGCCTCTTCTATCAG and CTCGCGGCTT GGGAGGTTGCGATAG (see Fig. S2).
Western-blot quantification of PCNA1 and PCNA2 concentrations in vivo
Cultures (50 ml) of T. kodakarensis KW128 were grown, as previously reported (Santangelo and Reeve 2010), to an OD600 of 0.5–0.6 that contained 2–5 × 108 cells/ml. The cells were harvested by centrifugation at 3,000 rpm for 20 min at 23 °C, the cell pellet resuspended in 500 μl 25 mM Tris–HCl (pH 8.0), 500 mM NaCl and 10 % glycerol, and the cells ruptured by sonication. The lysates were clarified by centrifugation at 13,000 rpm for 10 min at 4 °C and known amounts of recombinant T. kodakarensis PCNA1 and PCNA2 were subjected to electrophoresis through 12 % SDS-PAGE in lanes adjacent to aliquots of the clarified cell extracts. The gels were electroblotted onto a nitrocellulose membrane (Protran BA83 0.2 μm Nitrocellulose membrane, Whatman Inc.).
Western analysis was performed using a combination of guinea pig polyclonal antibodies (Cocalico Biologicals Inc) generated against the recombinant PCNA proteins and rabbit anti-guinea pig antibodies coupled to horse-radish peroxidase (Sigma-Aldrich); blots were developed using enhanced chemiluminescence (ECL, GE Healthcare). The intensity of each band was quantified using ImageQuant TL (GE Healthcare). The experiment was repeated three times. The average results of three experiments were used to calculate the in vivo levels of the PCNA proteins.
Results
PCNA1, PCNA2 and RFC oligomerization
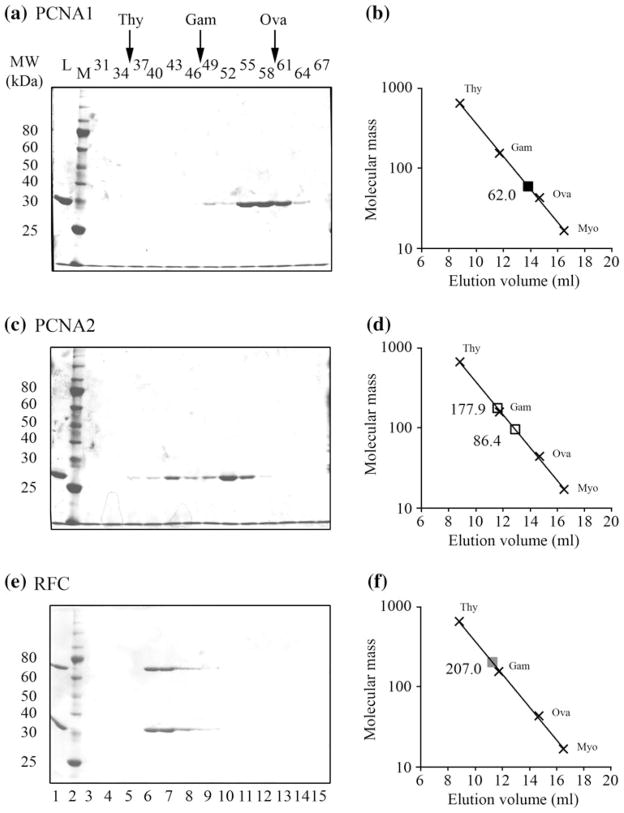
Homotrimers of T. kodakarensis PCNA1 and PCNA2 assemble to form very similar ring-shaped structures but differences at the subunit-interaction sites (Ladner et al. 2011). Based on light scattering assays, the PCNA1 and PCNA2 homotrimers have different in vitro stabilities, with only PCNA2 forming stable complexes at low concentrations (Ladner et al. 2011). Size exclusion chromatography has confirmed the light scattering results. PCNA1 (Mr = 29.5 kDa) eluted from the column as a broad peak correlating with a protein with a molecular mass of ~62 kDa (Fig. 1a, b), most consistent with PCNA1 solutions containing an equilibrium mixture of dimers (58.1 kDa) and trimers (87.2 kDa). The elution profile of PCNA2, in contrast, contained two discrete peaks revealing assembly into stable complexes with molecular masses of ~86 kDa and ~178 kDa (Fig. 1c, d), consistent with trimeric (87.8 kDa) and hexameric (175.6 kDa) complexes, respectively. There is evidence that human PCNA may also form hexameric ring structures (Naryzhny et al. 2005).
Fig. 1.
Size exclusion chromatography of PCNA1, PCNA2 and RFC complexes. Solutions of a PCNA1, c PCNA2 and e RFC were subjected to Superdex-200 gel filtration. The proteins present in an aliquot (15 μl) of each fraction obtained were separated by electrophoresis through 10 % acrylamide-SDS gels and visualized by Coomassie Blue staining. The elution positions of thyroglobulin (Thy 669 kDa), gamma globulin (Gam 158 kDa) and ovalbumin (Ova 44 kDa) are indicated at the top of the figure. Control lanes contained a sample of the material loaded onto the gel-filtration column (L) and molecular mass standards (M). The position(s) at which the experimental samples eluted from the column are indicated (open squares; filled squares) relative to the mass standards in b PCNA1, d PCNA2 and f RFC
Based on gel-filtration analyses, T. kodakarensis RFC (Fig. 1e, f) monomers assembled to form a single complex with molecular mass of ~208 kDa. As with most other archaeal RFCs investigated (Chia et al. 2010; Ishino and Ishino 2012), the T. kodakarensis RFC complex therefore most likely contains one RFC-L (58.1 kDa) and four RFC-S (37.2 kDa) subunits, a pentameric complex with a molecular mass of 206.9 kDa. Light scattering assays were also consistent with this result, indicating that the T. kodakarensis RFC complex had a molecular mass of ~203 kDa and therefore the predicted pentameric structure.
Both PCNA1 and PCNA2 stimulate DNA synthesis by T. kodakarensis PolB and PolD
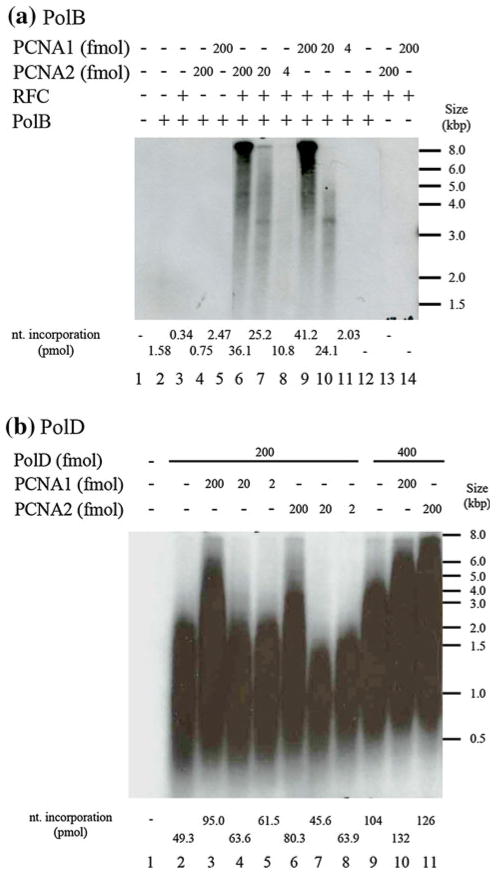
PCNAs increase the processivity of DNA polymerases (Jeruzalmi et al. 2002). The ability of PCNA1 and PCNA2 to stimulate DNA synthesis by T. kodakarensis PolB (encoded by TK0001) and PolD (encoded by TK1902 and TK1903) was therefore measured using a primer extension assay on a M13 DNA template. Primer extension by PolB was completely dependent on the presence of RFC plus either PCNA1 or PCNA2, and the rate of DNA synthesis increased when the PCNA1 or PCNA2 concentration was increased (Fig. 2a, lanes 9–11 and 6–8). At the same low PCNA concentrations, the extension products generated with PCNA1 were shorter than those generated with PCNA2 (Fig. 2a, compare lanes 10 and 11 to lanes 7 and 8), most likely reflecting the lower stability of PCNA1 versus PCNA2 trimers at low solution concentrations [Fig. 1a, b, (Ladner et al. 2011)]. In contrast to PolB, PolD had polymerase activity in the absence of a PCNA (Fig. 2b lanes 2, and 9) but, with RFC and either PCNA1 or PCNA2 present, the primer extension activity of PolD was stimulated and more full-length products were synthesized (Fig. 2b compare lanes 3 and 6 to lane 2; lanes 10 and 11 to lane 9). These results differ from those reported by Kuba et al. (2012), in that both PCNA1 and PCNA2 stimulated PolD activity (Fig. 2b) whereas only PCNA1 was reported to do so by Kuba et al. (2012). This discrepancy presumably reflects differences in the experimental conditions and/or the proteins purified by Kuba et al. (2012) without a tag, and here with His6-tags.
Fig. 2.
PCNA1 and PCNA2 stimulate primer extension by T. kodakarensis DNA polymerases B and D. a DNA elongation assay with PolB. Reaction mixtures (20 μl) containing 440 fmol of PolB, 430 fmol of RFC (as pentamers), 10 fmol of M13 primed template DNA, [α-32P]dNTP and PCNA1 or PCNA2, as indicated, were incubated for 20 min at 70 °C. An aliquot (4 μl) was removed and 32P incorporation measured to quantify the DNA synthesis. 32P-labeled DNA molecules in the remaining solution were separated by electrophoresis through alkaline 1.1 % (w/v) agarose gels and visualized, after gel drying, by autoradiography. b DNA elongation assay with PolD. Reaction mixtures (20 μl) containing 500 fmol of RFC (as pentamers), 20 fmol of M13 primed template DNA, [α-32P]dNTP, 200 or 400 fmol of PolD, as indicated, and 2, 20 or 200 fmol of PCNA1 or PCNA2, as indicated, were incubated for 20 min at 70 °C. An aliquot (4 μl) was removed and 32P incorporation measured to quantify the DNA synthesis. 32P-labeled DNA molecules in the remaining solution were separated by electrophoresis through alkaline 1.1 % (w/v) agarose gels and visualized, after gel drying, by autoradiography
Both PCNA1 and PCNA2 stimulate the ATPase activity of RFC
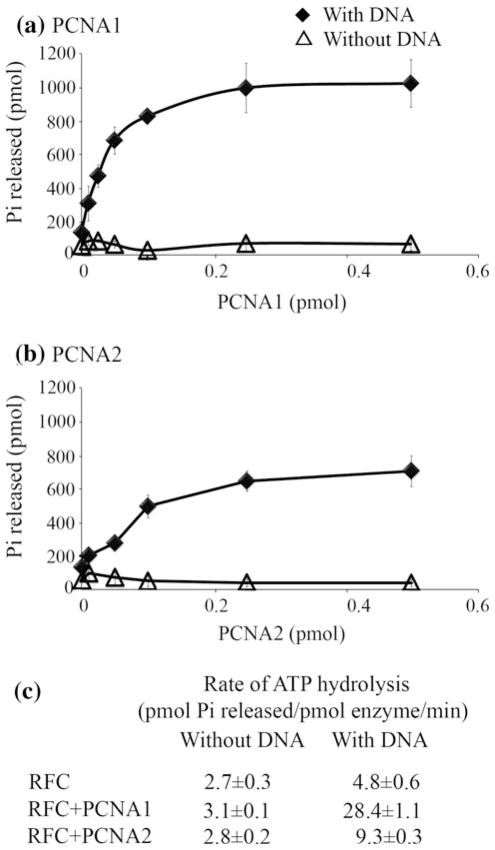
The ATPase activity of other archaeal and eukaryotic RFCs is stimulated by the presence of PCNA plus a primed DNA template (Kelman and Hurwitz 2000; Cann et al. 2001). Consistent with these results, both PCNA1 (Fig. 3a) and PCNA2 (Fig. 3b) stimulated ATP hydrolysis by T. kodakarensis RFC when incubated in the presence of a primed DNA substrate. In both cases, the extent of ATP hydrolysis was dependent on the PCNA concentration, but the rate of ATP hydrolysis was ~3-fold higher with PCNA1 than with PCNA2 (Fig. 3c).
Fig. 3.
PCNA1 and PCNA2 stimulate ATP hydrolysis by T. kodakarensis RFC. ATPase assays were carried out in reaction mixtures that contained increasing amounts of a PCNA1 or b PCNA2 with (diamonds) or without (triangles) a primed DNA substrate. c The average values obtained for the rates of ATP hydrolysis by RFC determined in three separate experiments
PCNA1 is sufficient for T. kodakarensis viability
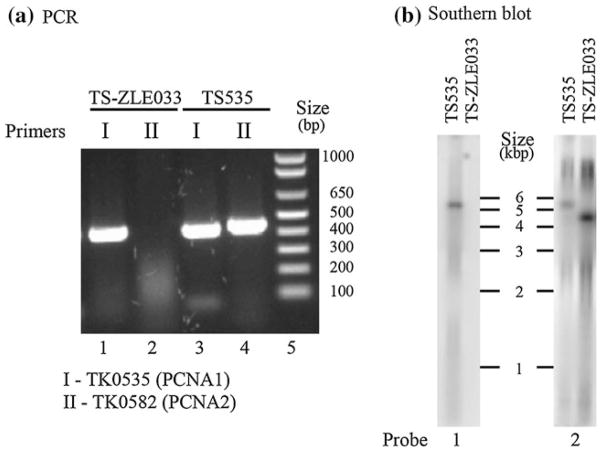
As both PCNA proteins stimulated PolB and PolD (Fig. 2) and RFC ATPase (Fig. 3), it seemed possible that either would be sufficient for T. kodakarensis viability. We therefore tried to construct stains with the PCNA1- or PCNA2-encoding genes, TK0535 and TK0582, respectively, individually deleted. T. kodakarensis TS–ZLE033 (ΔTK0582) was readily obtained with the genome structure confirmed by diagnostic PCR and Southern blots, as illustrated in Fig. 4a and b. The gene encoding PCNA2 is located in a region of the chromosome thought to be of a viral origin and the entire region can also be deleted from the chromosome (Tagashira et al. 2013).
Fig. 4.
Confirmation of the genome structure of T. kodakarensis TS-ZLE033 (ΔTK0582). a Electrophoretic separation of PCR amplicons generated from genomic DNA of T. kodakarensis TS517 and TS-ZLE033. The primers generated amplicons (450–500 bp) from within (I) TK0535 and (II) TK0582. The approximate locations of the primer pair I (TK0535-For and TK0535-Rev) used to generate the TK0535 amplicon indicated by arrows in Figure S2 and primer pair II (TK0582-For and TK0582-Rev) used to generate the TK0582 amplicon indicated by arrows in Figure S1. DNA size markers are shown in lane 5. b Southern blot analysis of genomic DNA from T. kodakarensis TS517 and TS-ZLE033. DNA molecules in aliquots (3 μg) of BglII-digested genomic DNA were separated by agarose gel electrophoresis, transferred to a membrane, denatured and allowed to hybridize to [32P]-labeled probe #1 (complementary to TK0582; see Fig. S1) or probe #2 (complementary to TK0579; see Fig. S1). As shown, probe #1 did not hybridize to a defined restriction fragment in BglII digest of T. kodakarensis TS-ZLE033 genomic DNA consistent with the deletion of TK0582. The size of the restriction fragment that hybridized with probe #2 confirmed the markerless deletion of TK0582 and the T. kodakarensis TS-ZLE033 genome structure diagrammed in Figure S1. The locations of BglII sites in the T. kodakarensis TS517 and TS-ZLE033 genomic DNA are indicated in Figure S1
The strain in which TK0582 is deleted (ΔTK0582) had no detectable viability or growth defects consistent with the presence of PCNA1 alone being fully sufficient to support T. kodakarensis genome replication as was previously reported (Kuba et al. 2012). In contrast, however, despite repeated attempts using the same transformation procedure and selection, but with pZLE032 as the donor DNA (Fig. S2), we were unable to generate a strain with TK0535 deleted, as was previously reported (Kuba et al. 2012), suggesting that PCNA1 is essential for T. kodakarensis viability. The structure of plasmid pZLE032 and the gene organization surrounding TK0535 in the T. kodakarensis genome are illustrated in Figure S1.
PCNA1 is abundant in vivo
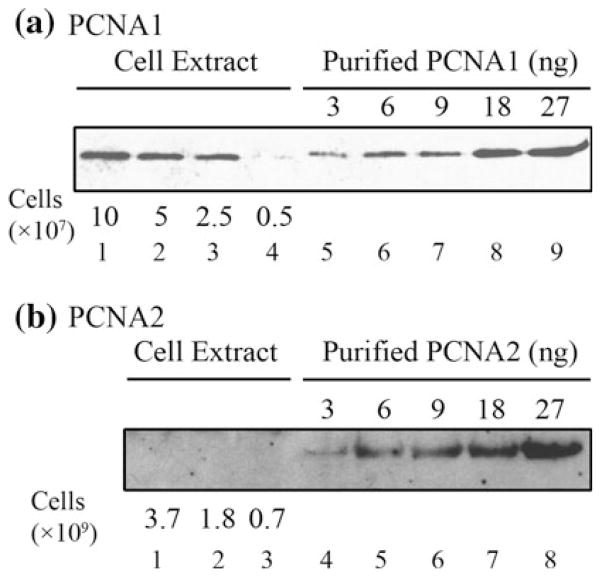
The concentrations of PCNA1 and PCNA2 in vivo were determined by quantitative Western-blot measurements (Fig. 5). PCNA1 was found to be abundant, with 5,000–10,000 molecules present per cell, a concentration likely sufficient for stable trimer formation in vivo [Fig. 1a, (Ladner et al. 2011)]. The concentration of PCNA2, in contrast, was ~100-fold lower, with <60 molecules present per cell, below the minimum detectable amount. The low sensitivity of the antibodies and the low expression of PCNA2 prevent us from detecting the protein in the Western-blot analysis. Previous studies, however, with more sensitive antibodies detected the PCNA proteins in cell extract and estimated the presence of 10–30 molecules of PCNA2 per cell (Kuba et al. 2012).
Fig. 5.
Western-blot quantification of PCNA1 and PCNA2 abundances in vivo. The proteins in aliquots of clarified cell extracts, derived from the numbers of T. kodakarensis cells indicated, were separated by electrophoresis in lanes adjacent to samples of purified recombinant (a) PCNA1 and (b) PCNA2. Western blots were generated and quantified as described in “Materials and Methods”
Possibly, our inability to delete TK0535 (PCNA1) reflected this low concentration of PCNA2 being insufficient for PCNA function in vivo and, to address this, we tried to increase PCNA2 synthesis in vivo by ectopic over-expression of TK0582 cloned into a replicating plasmid downstream from a strong constitutive promoter and by replacing the promoter of TK0582 with the promoter of the more highly expressed TK0535 (PCNA1). Neither procedure was successful. We were unable to isolate a strain in which TK0582 was over-expressed from a plasmid, and although many clones were obtained with the desired promoter substitution, in every strain investigated, sequencing revealed that the substituted TK0535 promoter had acquired inactivating mutations. It appears therefore that T. kodakarensis cells do not tolerate a higher intracellular concentration of PCNA2. The reason for this observation is not clear. It may be that PCNA2 can interact with the same proteins as PCNA1 but cannot perform all the activities of PCNA required for cell viability. Under this scenario, the extra PCNA2 may bind those essential factors, titrating them out, and thus prevent their interaction with PCNA1. When expressed normally this is not observed because PCNA2 expression is very low.
Discussion
T. kodakarensis is the only Euryarchaeon currently known to have more than one PCNA homolog and deletion of TK0582 (PCNA2) has confirmed that T. kodakarensis, like all other Euryarchaea, can grow normally with only one PCNA. PCNA1 and PCNA2 have very similar biochemical activities, and could potentially have redundant roles in vivo, but at the low concentration present, PCNA2 alone was unable to support viability. Intriguingly, despite repeated attempts to address this, we were unable to generate a T. kodakarensis strain with a much higher intracellular abundance of PCNA2. Our inability to delete TK0535 does not provide definitive proof that PCNA1 is essential for viability but, in addition to its predominant abundance, additional details support the argument that PCNA1 is normally the replicative PCNA in T. kodakarensis. In many archaeal genomes, the single PCNA-encoding gene is part of an operon that also contains a gene encoding a subunit of the GINS replication factor (Berthon et al. 2008; MacNeill 2010). TK0535 encodes PCNA1 and TK0536 that encodes the GINS15 subunit are present in the same operon (Fig. S2). TK0582, in contrast, is not adjacent to a GINS-encoding gene but is within a region of the T. kodakarensis genome thought to be of a vestige of a viral infection (Fukui et al. 2005; Tagashira et al. 2013). As previously concluded for two of the three minichromosome maintenance (MCM) homologs in T. kodakarensis (Pan et al. 2011b; Ishino et al. 2011), PCNA2 was most likely therefore acquired as a component of a viral replisome. Furthermore, although the atomic structures of the PCNA1 and PCNA2 complexes are overall very similar, the details of the monomer–monomer interfaces are significantly different with PCNA1 interfaces more similar to those in other archaeal PCNA complexes (Ladner et al. 2011).
This report provides the quantification of PCNA in vivo in an Archaeon. Given the similar sizes of bacterial and archaeal genomes, their common circular structures and bidirectional replication from a single origin of replication, it seems unlikely that archaeal genome replication would require a much higher number of clamp complexes than bacterial genome replication. In Bacteria, the β-subunit of DNA polymerase is a structural and functional homolog of PCNA (Kelman and O’Donnell 1995; Jeruzalmi et al. 2002) although the β-subunit is a polypeptide dimer whereas PCNA complexes are trimers (Kelman and O’Donnell 1995). E. coli has ~600 β-subunit monomers per cell allowing the assembly of ~300 clamp complexes (Turner 1998). T. kodakarensis cells would therefore need to have ~900 PCNA monomers for the assembly of a similar number of clamp complexes but, in fact, has substantially more, 5,000–10,000 per cell, given the Western-blot quantifications (Fig. 5). It is possible therefore that PCNA1 plays additional roles in T. kodakarensis although this monomer abundance may, in fact, simply be needed to direct and maintain stable trimerization in vivo.
Supplementary Material
Acknowledgments
This work was supported by grant MCB-0815646 from National Science Foundation to ZK, and grants GM073336 to TJS, GM53185 to JNR and TJS and GH034559 to JH from the National Institutes of Health. Certain commercial materials, instruments, and equipment are identified in this paper to specify the experimental procedure as completely as possible. In no case does such identification imply a recommendation or endorsement by the National Institute of Standards and Technology nor does it imply that the materials, instruments, or equipment identified are necessarily the best available for the purpose.
Abbreviations
- PolB
DNA polymerase B
- PolD
DNA polymerase D
- PCNA
Proliferating cell nuclear antigen
- RFC
Replication factor C
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s00792-013-0526-8) contains supplementary material, which is available to authorized users.
Contributor Information
Miao Pan, Institute for Bioscience and Biotechnology Research, 9600 Gudelsky Drive, Rockville, MD 20850, USA.
Thomas J. Santangelo, Department of Microbiology, Ohio State University, Columbus, OH 43210, USA
Ľbomíra Čuboňová, Department of Microbiology, Ohio State University, Columbus, OH 43210, USA.
Zhuo Li, Institute for Bioscience and Biotechnology Research, 9600 Gudelsky Drive, Rockville, MD 20850, USA.
Harlette Metangmo, Institute for Bioscience and Biotechnology Research, 9600 Gudelsky Drive, Rockville, MD 20850, USA.
Jane Ladner, Institute for Bioscience and Biotechnology Research, 9600 Gudelsky Drive, Rockville, MD 20850, USA. National Institute of Standards and Technology, 9600 Gudelsky Drive, Rockville, MD 20850, USA.
Jerard Hurwitz, Program of Molecular Biology, Memorial Sloan Kettering Cancer Center, 1275 York Avenue, New York, NY 10065, USA.
John N. Reeve, Department of Microbiology, Ohio State University, Columbus, OH 43210, USA
Zvi Kelman, Email: zkelman@umd.edu, Institute for Bioscience and Biotechnology Research, 9600 Gudelsky Drive, Rockville, MD 20850, USA. National Institute of Standards and Technology, 9600 Gudelsky Drive, Rockville, MD 20850, USA.
References
- Atomi H, Fukui T, Kanai T, Morikawa M, Imanaka T. Description of Thermococcus kodakaraensis sp nov, a well studied hyperthermophilic archaeon previously reported as Pyrococcus sp KOD1. Archaea. 2004;1:263–267. doi: 10.1155/2004/204953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthon J, Cortez D, Forterre P. Genomic context analysis in Archaea suggests previously unrecognized links between DNA replication and translation. Genome Biol. 2008;9:R71. doi: 10.1186/gb-2008-9-4-r71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brochier-Armanet C, Boussau B, Gribaldo S, Forterre P. Mesophilic Crenarchaeota: proposal for a third archaeal phylum, the Thaumarchaeota. Nat Rev Microbiol. 2008;6:245–252. doi: 10.1038/nrmicro1852. [DOI] [PubMed] [Google Scholar]
- Cann IK, Ishino S, Yuasa M, Daiyasu H, Toh H, Ishino Y. Biochemical analysis of replication factor C from the hyperthermophilic archaeon Pyrococcus furiosus. J Bacteriol. 2001;183:2614–2623. doi: 10.1128/JB.183.8.2614-2623.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chemnitz Galal W, Pan M, Kelman Z, Hurwitz J. Characterization of the DNA primase complex isolated from the archaeon, Thermococcus kodakaraensis. J Biol Chem. 2012;287:16209–16219. doi: 10.1074/jbc.M111.338145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia N, Cann I, Olsen GJ. Evolution of DNA replication protein complexes in eukaryotes and archaea. PLoS ONE. 2010;5:e10866. doi: 10.1371/journal.pone.0010866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukui T, Atomi H, Kanai T, Matsumi R, Fujiwara S, Imanaka T. Complete genome sequence of the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1 and comparison with Pyrococcus genomes. Genome Res. 2005;15:352–363. doi: 10.1101/gr.3003105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hileman TH, Santangelo TJ. Genetics techniques for Thermococcus kodakarensis. Front Microbiol. 2012;3:195. doi: 10.3389/fmicb.2012.00195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indiani C, O’Donnell M. The replication clamp-loading machine at work in the three domains of life. Nat Rev Mol Cell Biol. 2006;7:751–761. doi: 10.1038/nrm2022. [DOI] [PubMed] [Google Scholar]
- Ishino Y, Ishino S. Rapid progress of DNA replication studies in Archaea, the third domain of life. Sci China Life Sci. 2012;55:386–403. doi: 10.1007/s11427-012-4324-9. [DOI] [PubMed] [Google Scholar]
- Ishino S, Fujino S, Tomita H, Ogino H, Takao K, Daiyasu H, Kanai T, Atomi H, Ishino Y. Biochemical and genetical analyses of the three mcm genes from the hyperthermophilic archaeon, Thermococcus kodakarensis. Genes Cells. 2011;16:1176–1189. doi: 10.1111/j.1365-2443.2011.01562.x. [DOI] [PubMed] [Google Scholar]
- Jeruzalmi D, O’Donnell M, Kuriyan J. Clamp loaders and sliding clamps. Curr Opin Struct Biol. 2002;12:217–224. doi: 10.1016/s0959-440x(02)00313-5. [DOI] [PubMed] [Google Scholar]
- Kelch BA, Makino DL, O’Donnell M, Kuriyan J. Clamp loader ATPases and the evolution of DNA replication machinery. BMC Biol. 2012;10:34. doi: 10.1186/1741-7007-10-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelman Z, Hurwitz J. A unique organization of the protein subunits of the DNA polymerase clamp loader in the archaeon Methanobacterium thermoautotrophicum ΔH. J Biol Chem. 2000;275:7327–7336. doi: 10.1074/jbc.275.10.7327. [DOI] [PubMed] [Google Scholar]
- Kelman Z, O’Donnell M. Structural and functional similarities of prokaryotic and eukaryotic DNA polymerase sliding clamps. Nucleic Acids Res. 1995;23:3613–3620. doi: 10.1093/nar/23.18.3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuba Y, Ishino S, Yamagami T, Tokuhara M, Kanai T, Fujikane R, Daiyasu H, Atomi H, Ishino Y. Comparative analyses of the two proliferating cell nuclear antigens from the hyperthermophilic archaeon, Thermococcus kodakarensis. Genes Cells. 2012;17:923–937. doi: 10.1111/gtc.12007. [DOI] [PubMed] [Google Scholar]
- Ladner JE, Pan M, Hurwitz J, Kelman Z. Crystal structures of two active proliferating cell nuclear antigens (PCNAs) encoded by Thermococcus kodakaraensis. Proc Natl Acad Sci USA. 2011;108:2711–2716. doi: 10.1073/pnas.1019179108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Santangelo TJ, Čuboňová L, Reeve JN, Kelman Z. Affinity purification of an archaeal DNA replication protein network. MBio. 2010;1:e00221–00210. doi: 10.1128/mBio.00221-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacNeill SA. Structure and function of the GINS complex, a key component of the eukaryotic replisome. Biochem J. 2010;425:489–500. doi: 10.1042/BJ20091531. [DOI] [PubMed] [Google Scholar]
- Naryzhny SN, Zhao H, Lee H. Proliferating cell nuclear antigen (PCNA) may function as a double homotrimer complex in the mammalian cell. J Biol Chem. 2005;280:13888–13894. doi: 10.1074/jbc.M500304200. [DOI] [PubMed] [Google Scholar]
- Pan M, Kelman LM, Kelman Z. The archaeal PCNA proteins. Biochem Soc Trans. 2011a;39:20–24. doi: 10.1042/BST0390020. [DOI] [PubMed] [Google Scholar]
- Pan M, Santangelo TJ, Li Z, Reeve JN, Kelman Z. Thermococcus kodakarensis encodes three MCM homologs but only one is essential. Nucleic Acids Res. 2011b;39:9671–9680. doi: 10.1093/nar/gkr624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santangelo TJ, Reeve JN. Genetic tools and manipulations of the hyperthermophilic heterotrophic archaeon Thermococcus kodakarensis. In: Horikoshi K, editor. Extremophiles handbook. Springer; Japan: 2010. [Google Scholar]
- Santangelo TJ, Cubonova L, James CL, Reeve JN. TFB1 or TFB2 is sufficient for Thermococcus kodakaraensis viability and for basal transcription in vitro. J Mol Biol. 2007;367:344–357. doi: 10.1016/j.jmb.2006.12.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spang A, Hatzenpichler R, Brochier-Armanet C, Rattei T, Tischler P, Spieck E, Streit W, Stahl DA, Wagner M, Schleper C. Distinct gene set in two different lineages of ammonia-oxidizing archaea supports the phylum Thaumarchaeota. Trends Microbiol. 2010;18:331–340. doi: 10.1016/j.tim.2010.06.003. [DOI] [PubMed] [Google Scholar]
- Tagashira K, Fukuda W, Matsubara M, Kanai T, Atomi H, Imanaka T. Genetic studies on the virus-like regions in the genome of hyperthermophilic archaeon, Thermococcus kodakarensis. Extremophiles. 2013;17:153–160. doi: 10.1007/s00792-012-0504-6. [DOI] [PubMed] [Google Scholar]
- Turner JL. The use of the DNA sliding clamp in E. coli replication—how the clamp loader put clamps onto DNA and how it takes them off. Cornell University Medical College; New York: 1998. [Google Scholar]
- Vivona JB, Kelman Z. The diverse spectrum of sliding clamp interacting proteins. FEBS Lett. 2003;546:167–172. doi: 10.1016/s0014-5793(03)00622-7. [DOI] [PubMed] [Google Scholar]
- Yao NY, O’Donnell M. The RFC clamp loader: structure and function. Subcell Biochem. 2012;62:259–279. doi: 10.1007/978-94-007-4572-8_14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.