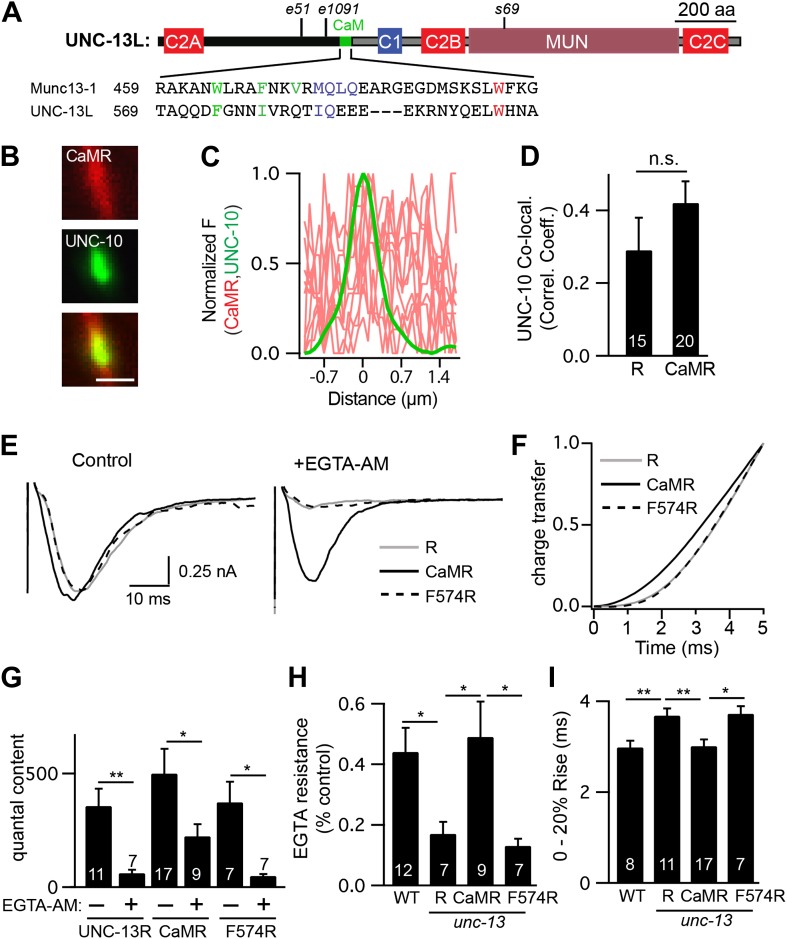
Figure 9. A predicted calmodulin-binding site in UNC-13L accelerates release.
The predicted calmodulin (CaM)-binding site of UNC-13L (residues 556–610) was fused to the R domain. CaMR had a diffuse axonal distribution, was poorly colocalized with UNC-10/RIM, but exhibited faster and more EGTA-resistant evoked ACh release. The effects of the CaM-binding site on release were eliminated by the F574R mutation, which is predicted to disrupt calmodulin binding (Junge et al., 2004). (A) Position of the putative CaM-binding site in UNC-13L and its alignment with the rat Munc13-1 CaM-binding site sequence are shown. Hydrophobic residues anchoring the C-terminal (green) and N-terminal (red) lobes of CaM, and predicted apo-calmodulin binding sites (blue) are indicated (Rodriguez-Castaneda et al., 2010; Lipstein et al., 2012). (B–D) The distribution of mCherry-tagged CaMR at active zones (labeled with GFP-tagged UNC-10/RIM) are shown. Tagged proteins were expressed in DA and DB motor neurons of wild-type animals (using the unc-129 promoter). Representative images (B), line scans (C), and correlation coefficients (D) for UNC-10/RIM and CaMR fluorescence at synaptic puncta are shown. Scale bar indicates 1 μm. (E and H) Stimulus-evoked EPSCs were recorded from body wall muscles in control saline and after the addition of EGTA-AM. Averaged traces (E) and summary data for time course of initial evoked charge transfer (0–5 ms, F), quantal content (G), EGTA resistance (H), and the latency of release (0–20% rise time, I) are shown. Values that differ significantly are indicated (**p<0.01; *p<0.05; n.s., not significant). The number of animals analyzed is indicated for each genotype. Error bars indicate SEM.